Abstract
Marine sponges are ancient metazoans that are populated by distinct and highly diverse microbial communities. In order to obtain deeper insights into the functional gene repertoire of the Mediterranean sponge Aplysina aerophoba, we combined Illumina short-read and PacBio long-read sequencing followed by un-targeted metagenomic binning. We identified a total of 37 high-quality bins representing 11 bacterial phyla and two candidate phyla. Statistical comparison of symbiont genomes with selected reference genomes revealed a significant enrichment of genes related to bacterial defense (restriction-modification systems, toxin-antitoxin systems) as well as genes involved in host colonization and extracellular matrix utilization in sponge symbionts. A within-symbionts genome comparison revealed a nutritional specialization of at least two symbiont guilds, where one appears to metabolize carnitine and the other sulfated polysaccharides, both of which are abundant molecules in the sponge extracellular matrix. A third guild of symbionts may be viewed as nutritional generalists that perform largely the same metabolic pathways but lack such extraordinary numbers of the relevant genes. This study characterizes the genomic repertoire of sponge symbionts at an unprecedented resolution and it provides greater insights into the molecular mechanisms underlying microbial-sponge symbiosis.
Introduction
Marine sponges (Porifera) are evolutionary ancient metazoans dating back to Precambrian times (Li et al., 1998; Love et al., 2009). By filtering extensive volumes of seawater—up to thousands of liters per kg sponge daily (Reiswig, 1974)—they take in food bacteria, but also potential pathogens, toxins and physical stress factors (De Goeij et al., 2009). Rapid cell turnover rates accompanied by extensive detritus production are likely a means of avoiding permanent, stress-induced damage to the sponge (De Goeij et al., 2009; Alexander et al., 2014). In line with the holobiont concept (Bordenstein and Theis, 2015), the highly diverse and distinct symbiotic microbial communities of marine sponges are thought to play a crucial role in their evolutionary success (Easson and Thacker, 2014; Tian et al., 2014; Webster and Thomas, 2016). 16S rRNA gene amplicon studies discovered an unusually high phylum-level diversity and stability of microbial associations in marine sponges comprising phototrophic as well as heterotrophic symbionts (Schmitt et al., 2012a; Easson and Thacker, 2014; Thomas et al., 2016; Webster and Thomas, 2016). The sponge microbiome includes as many as 52 microbial phyla and candidate phyla with the diversity and abundance varying between sponge species (Webster and Thomas, 2016). The most dominant symbiont groups belong to the phyla Proteobacteria (mainly gamma- and alphaproteobacteria), Actinobacteria, Chloroflexi, Nitrospirae, Cyanobacteria, candidatus phylum Poribacteria, and Thaumarchaea (Webster and Thomas, 2016).
Comparisons of sponge-associated and seawater microbial consortia have identified a number of genomic features that seem to facilitate bacterial adaptation to a symbiotic existence within sponges, for example, transposable elements, defense mechanisms, and eukaryote-like proteins (Thomas et al., 2010; Fan et al., 2012; Hentschel et al., 2012; Horn et al., 2016). Studies on individual clades of the microbial consortium have revealed specific features of sponge symbionts, such as adaptations of the lipopolysaccharide by the cyanobacterium ‘Candidatus Synechococcus spongiarum’ presumably to avoid host phagocytosis (Gao et al., 2014; Burgsdorf et al., 2015) and specific patterns for carbon degradation by Poribacteria (Kamke et al., 2013).
PacBio long-read sequencing is widely used in isolate genomics as a stand-alone tool or in combination with Illumina short-read sequencing (for example, Beims et al., 2015; Koren and Phillippy, 2015; Ricker et al., 2016). The error-prone PacBio reads need to be corrected either with themselves—if sufficient sequencing depth is provided—or with Illumina reads of far lower error-rate (Koren et al., 2012; Ono et al., 2013). The combination of PacBio and Illumina sequencing data in hybrid assemblies enables the closure of assembly gaps, for example, by spanning over long repeats, to merge contigs and thereby to reconstruct the genome architecture (Koren et al., 2012). In metagenomics, the Illumina-PacBio hybrid assembly approach has recently been shown to improve the quality of assemblies (Frank et al., 2016; Tsai et al., 2016). Although the improvements in targeted binning of the dominant members of the microbiomes in these studies have been demonstrated (Frank et al., 2016; Tsai et al., 2016), un-targeted binning and performance for less abundant members of the microbial communities have not been evaluated.
Even though considerable metagenomic information from sponge microbiomes has been accrued, only few symbiont genomes have been reconstructed by single-cell genomics or binning (Siegl et al., 2011; Kamke et al., 2013; Gao et al., 2014, Burgsdorf et al., 2015). Therefore, correlations between phylogeny and function have only rarely been possible. In the present study, we aimed at obtaining a larger number and greater diversity of sponge symbiont genomes. We present the first metagenomic hybrid assembly derived from Illumina short-read and the PacBio long-read data with subsequent un-targeted differential coverage binning. The highly complex microbiome of the Mediterranean sponge Aplysina aerophoba was used towards this goal. By applying an un-targeted binning technique, we aimed to include also the less abundant members of the microbial community. We provide statistical evidence for gene networks that are enriched in the symbiont genomes over selected reference genomes and we discuss the role of these genomic adaptations in context of a symbiotic existence in the sponge matrix. Furthermore, a comparison between symbiont genomes revealed a specialization into three distinct, yet phylogenetically diverse groups within the consortium, of which two appear to metabolize distinct components of the sponge extracellular matrix.
Materials and methods
Sample collection
Aplysina aerophoba specimens were collected from the Mediterranean Sea near Piran, Slovenia (45.517680, 13.567991). One specimen was collected in May 2013 for Illumina sequencing and one specimen was collected in May 2014 for PacBio sequencing. Both were collected from ca. 5 m depth and transported to the laboratory in natural seawater at ambient temperature. Sponge pinacoderm (outer layer) and mesohyl (inner core), visually distinguishable by the reddish-greenish color of the cyanobacteria-containing pinacoderm, were separated with a sterile scalpel blade and microbial cell enrichment was performed by differential centrifugation (Fieseler et al., 2006). These sponge-associated prokaryotes (SAPs) were frozen with 15% glycerin at −80 °C.
DNA extraction and sequencing
DNA of sponge-associated prokaryotes (SAPs) obtained from either pinacoderm or mesohyl tissue (three technical replicates each) was extracted with the FastDNA SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA, USA). Different cell lysis protocols were applied for each triplicate to obtain differential sequencing coverage for downstream binning as previously described (Albertsen et al., 2013; Alneberg et al., 2014): (i) bead beating, following the manufacturer’s protocol, (ii) freeze-thaw cycling (3 cycles of 20 min at −80 °C and 20 min at 42 °C), (iii) proteinase K digestion for 1 h at 37 °C (TE buffer with 0.5% SDS and proteinase K at 100 ng ml−1 final concentration). Metagenomic DNA was sequenced on an Illumina HiSeq2000 platform (150-bp paired-end reads) and quality filtered at the DOE Joint Genome Institute (Walnut Creek, CA, USA) following the JGI sequencing and the data processing pipeline (Markowitz et al., 2012). Additionally, V4 iTag sequences were obtained by Illumina MiSeq sequencing and analyzed in the respective iTagger pipeline at JGI (for more information, see https://bitbucket.org/berkeleylab/jgi_itagger and http://jgi.doe.gov/wp-content/uploads/2013/05/iTagger-methods.pdf). For the PacBio data set, DNA was extracted with the above-mentioned kit following the manufacturer’s protocol (cell lysis by bead beating) and sequenced on a PacBio RS II platform using 8 SMRT cells by GATC Biotech (Konstanz, Germany).
Assembly, binning, and annotation
Illumina reads were coverage-normalized with bbnorm of BBMap v. 34 (https://sourceforge.net/projects/bbmap/) at default settings. PacBio reads were corrected with all (non-normalized) Illumina reads using proovread (Hackl et al., 2014) optimized for handling the metagenomic data (Hackl, 2016). Only corrected PacBio reads longer than 1000 bp were used for further analyses. To assess the improvement of the assembly by adding PacBio long-reads compared to only Illumina short-reads, we assembled two sets of data as follows: (i) only the Illumina reads (Illumina-only assembly) and (ii) Illumina and PacBio reads together (hybrid assembly). The two independent assemblies were calculated with SPAdes v. 3.5.0 (Bankevich et al., 2012) for kmers 21, 33, 55, 77, 99 and 127, and with the single-cell and only-assembler options enabled. Illumina-only contigs and corrected PacBio reads were both mapped to the hybrid assembly with blasr v. 1.3.1 (Chaisson and Tesler, 2012) to assess whether all available information was incorporated into the hybrid assembly. Only contigs of at least 1000 bp length were used for further analyses.
Binning was performed with CONCOCT v. 0.4.0 (Alneberg et al., 2014). For this, the data were prepared as follows. Contigs longer than 20 000 bp were split into sub-contigs of at least 10 000 bp length with the provided script (Alneberg et al., 2014). The non-normalized Illumina reads of the six Illumina data sets were mapped to the sub-contigs with bowtie2 v. 2.2.2 at default settings (Langmead and Salzberg, 2012). The resulting SAM files were converted to BAM, sorted and indexed with samtools v. 0.1.18 (Li et al., 2009), and duplicates were marked according to the script map-bowtie2-markduplicates.sh provided with the CONCOCT package (Alneberg et al., 2014). Samtools v. 0.1.18 was also used for depth calculation (Li et al., 2009). The in-house python script avgcov_from_samtoolsout.py (https://github.com/bslaby/scripts/) was used to calculate the average coverage of each sub-contig. The coverage tables for each mapping were merged into one for binning with CONCOCT v. 0.4.0 (Alneberg et al., 2014) at default settings. A fasta file for each bin was created with the in-house python script mkBinFasta.py (https://github.com/bslaby/scripts/). Sub-contigs were merged into the original contigs again. If sub-contigs of one contig were assigned to different bins, the contig was placed in the bin by majority-vote. Assembly statistics were obtained from QUAST v. 3.1 (Gurevich et al., 2013). To assess similarity of Illumina-only and hybrid assembly as well as assembly improvements by adding of PacBio long-reads on the genome level, the contigs of an Illumina-only bin were mapped to the contigs of the corresponding hybrid assembly bin with nucmer of MUMmer 3.0 (Kurtz et al., 2004) and visualized with AliTV (Ankenbrand et al., 2017).
Open reading frames (ORFs) were called with prodigal v. 2.6.1 (Hyatt et al., 2010) with -m and -p meta options enabled, and the completeness of genomic bins was estimated by hmmsearch (HMMER 3.1b1) against a database of 111 essential genes with –cut_tc and –notextw options (Finn et al., 2011; Albertsen et al., 2013). Contamination levels were assessed by a blastp search (BLAST 2.2.28+) of the essential genes against the refseq_protein database (release number 81) at an e-value cutoff of 1e-5 followed by determination of the last common ancestor for each gene by MEGAN version 6.4.3 (Pruitt et al., 2007; Camacho et al., 2009; Huson et al., 2016). Only reference genomes>90% and bins>70% completeness were used in further analyses.
The Illumina-only and the PacBio-Illumina hybrid assemblies were deposited on MG-RAST (Meyer et al., 2008) (Table 1). Additionally, the raw Illumina sequencing data were deposited under GOLD Study ID Gs0099546 (Reddy et al., 2014). Uncorrected and corrected PacBio reads were deposited on MG-RAST (Meyer et al., 2008) with the IDs mgm4670967.3 and mgm4670966.3, respectively. The accession numbers for all bins >70% completeness are listed in Table 2. The Illumina-only assembly is also deposited on GenBank with the accession MKWU00000000.
Table 1. Comparison of Illumina-only and Illumina-PacBio hybrid assemblies.
| Illumina-only | Illumina-PacBio hybrid | |
|---|---|---|
| MG-RAST ID | mgm4671062.3 | mgm4671058.3 |
| Contig number (⩾1000 bp) | 110 609 | 31 187 |
| Size (Mb) | 490 | 301 |
| N50 | 8958 | 33 831 |
| N75 | 2873 | 12 184 |
| L50 | 8886 | 1980 |
| L75 | 34 979 | 5726 |
| CDSs | 509 054 | 289 685 |
| Bin number | 217 | 137 |
| >90% completeness (with 16S rRNA gene) | 25 (12) | 26 (22) |
| 85–90% completeness (with 16S rRNA gene) | 12 (4) | 6 (6) |
| 70–85% completeness (with 16S rRNA gene) | 5 (0) | 5 (4) |
Table 2. Binned genomes of Illumina-PacBio hybrid assembly.
| Accession | Bin | Phylogeny | Symbiont guild | % est. com. | Contig no. | Times cov. | Est. size (Mb) | % GC | N50 | CDS no. | % in COGs | Dupl. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MPNP00000000 | bin131 | Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceaea | III | 93.69 | 416 | 612 | 4 | 41.99 | 19 278 | 3392 | 59.58 | 1 |
| MPMP00000000 | bin36 | Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Albidovuluma | II | 89.19 | 201 | 189 | 6.3 | 58.04 | 44 410 | 5122 | 64.02 | 0 |
| MPMX00000000 | bin65 | Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae; unculturedb | I | 91.89 | 94 | 396 | 4.7 | 66.16 | 72 338 | 4036 | 76.39 | 0 |
| MPNO00000000 | bin129 | Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae; Defluviicoccusb | I | 82.88 | 122 | 237 | 5.8 | 69.54 | 56 772 | 4742 | 72.75 | 0 |
| MPMV00000000 | bin56 | Proteobacteria; Alphaproteobacteria; Rickettsiales; EF100-94H03b | I | 93.69 | 102 | 54 | 4.8 | 63.69 | 78 682 | 4292 | 70.36 | 0 |
| MPMT00000000 | bin52 | Proteobacteria; Alphaproteobacteriac | I | 92.79 | 120 | 234 | 4.5 | 66.54 | 52 938 | 3989 | 72.75 | 1 |
| MPNG00000000 | bin98 | Proteobacteria; Alphaproteobacteriaa | III | 85.59 | 105 | 96 | 1.9 | 40.65 | 46 493 | 1455 | 33.68 | 0 |
| MPNF00000000 | bin95 | Proteobacteria; Alphaproteobacteriac | I | 75.68 | 582 | 152 | 4.5 | 66.27 | 6974 | 3890 | 64.16 | 0 |
| MPMI00000000 | bin18 | Proteobacteria; Deltaproteobacteria; Desulfurellales; Desulfurellaceae; unculturedb | I | 87.39 | 83 | 38 | 6 | 57.83 | 103 191 | 5238 | 65.65 | 0 |
| MPMQ00000000 | bin40 | Proteobacteria;Gammaproteobacteria;Oceanospirillales;Hahellaceae;Kistimonas;a | III | 93.69 | 215 | 76 | 4 | 57.27 | 38 525 | 2848 | 46.91 | 0 |
| MPMU00000000 | bin55 | Proteobacteria; Gammaproteobacteriaa | III | 84.68 | 183 | 47 | 3.5 | 47.27 | 24 711 | 2562 | 67.49 | 1 |
| MPNI00000000 | bin106 | Proteobacteriaa | III | 90.09 | 148 | 53 | 2.9 | 39.61 | 55 882 | 2088 | 41.62 | 0 |
| MPNJ00000000 | bin107 | Nitrospinae/tectomicrobia group; Nitrospinaec | I | 90.09 | 60 | 440 | 4.9 | 59.46 | 165 774 | 4046 | 69.7 | 1 |
| MPMZ00000000 | bin75 | Nitrospirae; Nitrospira; Nitrospirales; Nitrospiraceae; Nitrospira;b | III | 91.89 | 115 | 65 | 3.3 | 56.24 | 44 884 | 3093 | 56.58 | 2 |
| MPMH00000000 | bin63 | SBR1093;EC214;d | III | 88.29 | 150 | 479 | 2.6 | 50.41 | 30 980 | 2180 | 68.21 | 4 |
| MPNK00000000 | bin110 | Acidobacteria; Holophagae; Subgroup 10; TK85b | I | 79.28 | 758 | 549 | 7.9 | 67.45 | 12 332 | 5726 | 55.43 | 4 |
| MPMW00000000 | bin61 | Acidobacteria; Acidobacteriaa | III | 70.27 | 207 | 117 | 4.1 | 67.65 | 19 828 | 2561 | 55.92 | 0 |
| MPMY00000000 | bin70 | Candidatus Poribacteria;c | II | 91.89 | 106 | 351 | 5.5 | 40.34 | 70 347 | 4254 | 59.07 | 6 |
| MPMS00000000 | bin44 | Candidatus Poribacteria; Poribacteria genera incertae sedisa | II | 91.89 | 465 | 265 | 7.7 | 47.18 | 23 989 | 6288 | 54.28 | 10 |
| MPNB00000000 | bin80 | Bacteroidetes; Cytophagia; Order II; Rhodothermaceae; unculturedb | III | 92.79 | 192 | 453 | 4.4 | 50.96 | 34 696 | 3555 | 50.44 | 0 |
| MPMN00000000 | bin25 | Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceaea | III | 90.09 | 124 | 589 | 3.3 | 36.18 | 40 599 | 2420 | 51.03 | 0 |
| MPMR00000000 | bin43 | Gemmatimonadetes; Gemmatimonadetes; BD2-11 terrestrial groupb | III | 92.79 | 65 | 633 | 4.8 | 67.96 | 132 700 | 3664 | 60.84 | 1 |
| MPNE00000000 | bin94 | Gemmatimonadetesc | III | 91.89 | 83 | 190 | 4.8 | 66.9 | 89 414 | 3702 | 59.62 | 1 |
| MPNH00000000 | bin103 | Spirochaetae; Spirochaetes; Spirochaetales; Spirochaetaceaea | II | 87.39 | 96 | 66 | 5.5 | 67.36 | 71 825 | 4338 | 67.75 | 0 |
| MPNA00000000 | bin76 | Actinobacteria; Acidimicrobiia; Acidimicrobiales; OM1 cladeb | I | 90.09 | 82 | 102 | 3.9 | 61.59 | 108 948 | 3269 | 65.65 | 0 |
| MPNQ00000000 | bin134 | Actinobacteria; Acidimicrobiia; Acidimicrobiales; Sva0996 marine groupb | I | 90.09 | 77 | 224 | 4.1 | 64.29 | 91 761 | 3487 | 68.71 | 1 |
| MPNL00000000 | bin119 | Deinococcus-Thermus; Deinococci;Deinococcales; Trueperaceae; Trueperaa | II/III | 91.89 | 91 | 62 | 3.5 | 62.23 | 62 429 | 2876 | 69.47 | 1 |
| MPMK00000000 | bin9 | Cyanobacteria; ;SubsectionI; FamilyI; unculturedb | III | 89.19 | 391 | 157 | 3 | 58.71 | 12 771 | 2808 | 50.68 | 2 |
| MPML00000000 | bin5 | Chloroflexi; Caldilineae; Caldilineales; Caldilineaceae; unculturedb | II | 92.79 | 120 | 81 | 6 | 58.5 | 64 429 | 4593 | 68.26 | 1 |
| MPMO00000000 | bin34 | Chloroflexi; Caldilineae; Caldilineales; Caldilineaceae; unculturedb | II | 90.99 | 111 | 46 | 5.1 | 63.15 | 63 615 | 3982 | 63.01 | 0 |
| MPMM00000000 | bin22 | Chloroflexi; SAR202 cladec | I | 90.99 | 58 | 94 | 4.8 | 59.2 | 163 655 | 4049 | 57.08 | 4 |
| MPNN00000000 | bin127 | Chloroflexi; SAR202 cladec | II/III | 90.09 | 21 | 74 | 3.3 | 56.35 | 309 970 | 2976 | 59.98 | 0 |
| MPND00000000 | bin90 | Chloroflexi; SAR202 cladeb | I | 89.19 | 213 | 61 | 5.2 | 57.14 | 50 603 | 4453 | 54.91 | 5 |
| MPMJ00000000 | bin16 | Chloroflexi; SAR202 cladeb | I | 88.29 | 101 | 70 | 3.7 | 65.63 | 62 928 | 3253 | 60.62 | 0 |
| MPNC00000000 | bin87 | Chloroflexi; SAR202 cladec | I | 90.99 | 67 | 331 | 5.4 | 62.79 | 269 076 | 4711 | 55.47 | 1 |
| MPNM00000000 | bin125 | Chloroflexia | I | 92.79 | 66 | 757 | 4 | 62.27 | 125 355 | 3410 | 61.73 | 1 |
| MPMG00000000 | bin20 | Chloroflexia | II | 91.89 | 22 | 245 | 4 | 59.31 | 250 998 | 3218 | 72.5 | 2 |
Abbreviations: dupl, duplicates; com, completeness; cov, coverage; est, estimated; Phylogenetic information.
Only duplicate genes other than PF00750, PF01795, and TIGR00436 were counted, as these genes are known to occur in multiple copies (Albertsen et al., 2013).
RDPclassifier; d concatenated gene tree.
LCA SILVA (SINA).
concatenated gene tree + 16S rRNA gene tree.
LCA greengenes (SINA).
Comparative analysis
A total of 27 reference genomes were chosen based on phylogeny and environment (Supplementary Table 1). Close taxonomic relatedness to the symbiont genomes, closed genomes, as well as marine (or at least aquatic) environments were preferably selected. In order to be able to validate the binning process, we included the sponge symbiont genomes ‘Ca. S. spongiarum’ 15L (Burgsdorf et al., 2015) and ‘Ca. Poribacterium’ (Kamke et al., 2013) in the analyses. We retrieved nucleic acid fasta files for all selected references from GenBank and MG-RAST (Benson et al., 2007; Meyer et al., 2008), which were then processed like the symbiont bins with respect to ORF prediction and annotation. Five additional references were added for 16S rRNA gene tree calculation for better phylogenetic resolution (see Supplementary Figure 3A). The annotation of rRNA genes was performed with rRNA prediction at default settings (Wu et al., 2011). The 16S rRNA genes were taxonomically assigned using the RDPclassifier at a 80% confidence cutoff (Wang et al., 2007) and the classification tool of SINA 1.2.11 (Pruesse et al., 2012) using the SILVA and Greengenes databases (DeSantis et al., 2006; Quast et al., 2013). Gap-only sites were removed from the SINA alignment of both, bins and references, in SeaView 4.5.2 (Gouy et al., 2010). A Neighbor Joining tree (GTR+G+I), which was determined to be the most suitable DNA/protein model for the data, was calculated in MEGA7 with 100 bootstrap replications (Kumar et al., 2016). Additionally, a concatenated gene tree of 29 essential genes was created (see Supplementary Table 2 for a list of genes). Alignments for every gene individually using the muscle algorithm in MEGA7 (Edgar, 2004; Kumar et al., 2016) were merged with a sequence of 20 Ns between the genes. After identifying the most suitable DNA/protein model for the data, a maximum likelihood tree (LG+G+I) was calculated in MEGA7 with 100 bootstrap replications (Kumar et al., 2016). Bins lacking 16S rRNA genes or with an ambiguous classification of this gene were phylogenetically classified according to their placement in the concatenated tree.
To assess the distribution of the binned sponge symbionts among different sponge species, a BLAST search was conducted for the available 16S rRNA genes against a database of the representative amplicon OTUs by Thomas et al. (2016). For each bin, the best three hits were obtained, the sequences were aligned with SINA, and a Neighbor Joining tree (K2P) was calculated in SeaView with 1000 bootstrap replications (Gouy et al., 2010; Pruesse et al., 2012). The closest OTU for each bin was determined based on BLAST results and the phylogenetic tree. Information on the distribution of the selected OTUs was obtained from Thomas et al. (2016).
ORFs were annotated with rpsblast+ of BLAST 2.2.28+ against a local version of the COG database (ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd/, download on 28 May 2015) (Tatusov et al., 2003; Camacho et al., 2009). Only annotations with an e-value⩽1e-6 were used for further analyses, and only one annotation per ORF was kept ranked by e-value, length and bitscore. Because many sponge–symbiont lineages, in some cases whole phyla, are not abundant in seawater, we have opted for an approach different from previous publications, where only seawater metagenomes were used for comparison (for example, Thomas et al., 2010). We selected reference genomes based on phylogenetic similarity and on genome completeness. Marine sources were preferred over other sources.
To discover statistically significant differences between the sponge symbiont genomes and reference genomes, Welch’s t-test was performed in STAMP 2.0.9 (Parks et al., 2014) with Storey FDR and a q-value cutoff of 0.01. This was performed on the COG class level, double-counting COGs that belong to multiple classes, as well as on the COG level. Interactions between the significantly sponge-enriched COGs were explored using STRING v10 networks (Szklarczyk et al., 2014) and a heatmap was created in R version 3.2.3 (https://www.r-project.org). The phylo.heatmap function of phytools package version 0.5.30 (Revell, 2012) was used to complement the heatmap with phylogeny. The phylogenetic tree accompanying the heatmap is a simplified version (bins only) of the concatenated gene phylogeny.
The symbiont genomes were compared by applying a principle component analysis (PCA) in R with FactoMineR package version 1.33 (Lê et al., 2008), factoextra package version 1.0.3 (https://cran.r-project.org/web/packages/factoextra/index.html), and ggplot2 version 2.2.0 (http://ggplot2.org).
Results and discussion
Two metagenome assemblies were obtained, one only from Illumina HiSeq short-reads (Illumina-only assembly), and one from the same Illumina short-reads set, but combined with pre-corrected PacBio long-reads (hybrid assembly). The two assemblies differed notably in number of contigs and total size (Table 1). The Illumina-only assembly comprised >100 000 contigs with a total length of 490 Mbp, the hybrid assembly consisted of >30 000 contigs with a total length of 301 Mbp. Mappings of all contigs of the Illumina-only assembly and the corrected PacBio reads to the hybrid assembly showed that 100% of each data set mapped to the hybrid assembly. This demonstrates that all information had been transferred to the hybrid assembly. The hybrid assembly is smaller because it is better resolved, thus leading to a lower number of ambiguities. Only contigs >=1000 bp were considered. The addition of the PacBio reads to the assembly increased the N50 value 3.8-fold, from about 9 kbp to 34 kbp. While the number of highly complete genome bins (>70% completeness) decreased (42 Illumina-only bins vs 37 hybrid bins), the portion of full-length 16S rRNA gene containing bins doubled from 16 in the Illumina-only assembly to 32 in the hybrid assembly. To assess if contigs from the Illumina-only assembly were reappearing in the hybrid assembly and if the PacBio reads merged them into larger contigs, an Illumina-only bin was mapped to the corresponding hybrid bin. This allowed a visual comparison of the assemblies (Supplementary Figure 1). This mapping shows that the two assemblies corresponded well because contigs that had been constructed out of the Illumina data reappeared upon addition of the PacBio reads. Moreover, they were merged into even larger contigs, thus resulting in a higher-quality bin.
To obtain the short-read data optimized for differential coverage binning, six DNA samples from the same sponge specimen were extracted with varied lysis protocols, and deeply sequenced on an Illumina HiSeq2000 instrument (see Supplementary Figure 2 of JGI Project ID 1024999 for the additional ribosomal 16S rRNA V4 iTag data of this sequencing project). Although we already obtained a large number of high-completeness bins from the Illumina-only assembly, only 38% of the binned genomes contained a 16S rRNA gene. Contrasting, in the PacBio-Illumina hybrid assembly 86% of the bins contained a 16S rRNA gene (Table 1). Furthermore, with a 3.8-fold higher N50 hybrid assembly was more contiguous. For these reasons, all downstream analyses were carried out with the genomes binned from the PacBio-Illumina hybrid assembly.
Thirty-seven high-quality sponge symbiont genomes representing 13 bacterial phyla
The 37 binned genomes belonged to 11 bacterial phyla and 2 candidate phyla, which are representative of the sponge symbiont consortium: Proteobacteria (Alpha, Gamma, and Delta), Chloroflexi, Acidobacteria, Actinobacteria, Bacteroidetes, Gemmatimonadetes, Deinococcus-Thermus, Nitrospirae, Nitrospinae, Cyanobacteria, Spirochaetes and the candidate phyla Poribacteria and SBR1093 (Table 2). This composition is congruent with the known microbial diversity of A. aerophoba (Hentschel et al., 2002; Schmitt et al., 2012b). The bins varied in total number of contigs from 21 to 758. Large numbers of contigs did not correlate with low sequence coverage: the bin with lowest coverage (bin18 with 38-times coverage), for example, was composed of as few as 83 contigs and was 87% complete. Estimated genome sizes, based on total length and estimated genome completeness, ranged from 1.9 Mbp (Alphaproteobacterium bin98) to 7.9 Mbp (Acidobacterium bin110). With respect to GC content, the genomes ranged from 36% (Bacteroidetes bin25) to nearly 70% (Alphaproteobacterium bin129). Overall, the sponge symbionts had genomes of high GC-content, which are as follows: 13 were between 50 and 60%, 17 of symbiont genomes comprised >60% of GC-bases. Comparably high average GC contents are a known feature of sponge metagenomes (Horn et al., 2016). The N50 values also showed variability, with the smallest being 6974 bp for Alphaproteobacterium bin95 and the largest being 309 970 bp for Chloroflexi bin127. The number of coding sequences (CDSs) in the symbiont genomes ranged from 1455 (Alphaproteobacterium bin98) to 6288 (Ca. Poribacterium bin44). The number of COGs annotated for each genome ranged between 490 (bin98) and 3450 (Alphaproteobacterium bin129), which translates to 34% (bin98) and 76% (Alphaproteobacterium bin65) CDSs in COGs.
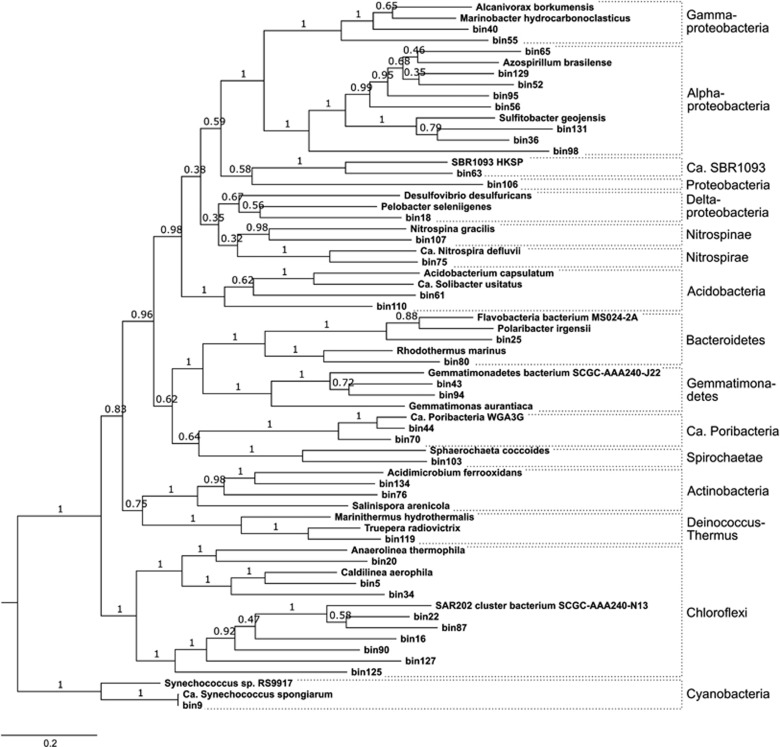
In order to resolve the phylogenies of the recovered bins, a concatenated tree (Figure 1) of 29 essential single-copy genes (Supplementary Table 2) as well as a 16S rRNA gene tree were constructed (Supplementary Figure 3A). Overall, the phylogeny of the binned bacterial genomes reflected the major phylogenetic lineages known to inhabit sponges (Supplementary Figure 3B; Thomas et al., 2016). This finding suggests that the sequenced lineages are prevalent in A. aerophoba, as more abundant taxa were more likely sequenced than rare lineages from this diverse metagenome. Our hypothesis that the binned genomes derive from symbionts and not from environmental bacteria was further supported by the 16S rRNA gene data. The best BLAST hits for all 34 bin-derived 16S rRNA genes were from sponge-associated or sponge/coral-associated bacteria (Supplementary Table 3A). Because the remaining three bins did not contain a 16S rRNA gene, their identity could not be confirmed by BLAST alone.
Figure 1.
Maximum likelihood (LG+G+I) phylogenetic tree based on the amino acid sequences of 29 essential genes, calculated in MEGA7 with 100 bootstrap replications. Cyanobacteria were used as outgroup, because they were closest to the archaeal outgroup in the 16S rRNA gene phylogeny (Supplementary Figure 3A).
The concatenated tree shows the phylogenetic placement of all 37 bins and their references, which had been selected based on genome completeness, phylogenetic similarity, and habitat (marine preferred over other habitats) (Supplementary Tables 1 and 3B). It was in overall agreement with the 16S rRNA gene tree regarding the phylogenetic placement of the bins containing this gene and furthermore provides placement for the three bins missing the 16S rRNA gene.
Sponge symbiont genomes are enriched in defense and in matrix interactions
In order to identify the gene functions that are enriched in the genomes of sponge symbionts, we compared the pool of symbiont genomes against the pool of selected reference genomes. Significant differences were identified between the symbiont genomes and reference genomes on the level of COG classes. While COG classes R (‘General function prediction only’), E (‘amino acid transport and metabolism’), L (‘replication, recombination and repair’), and Q (‘secondary metabolites biosynthesis, transport and catabolism’) are enriched in the symbionts, the classes T (‘signal transduction mechanisms’), K (‘transcription’), M (‘cell wall/membrane/envelope biogenesis’) and N (‘cell motility’) were depleted in comparison to the reference genomes (Supplementary Figure 4A).
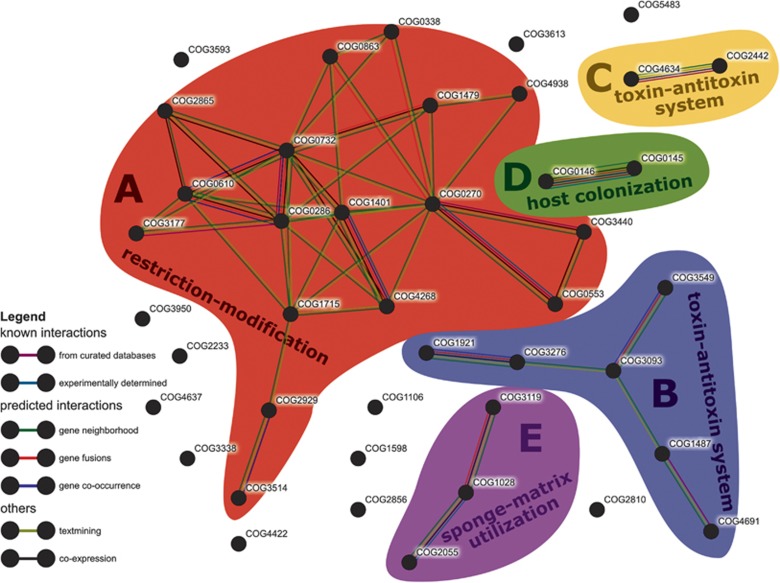
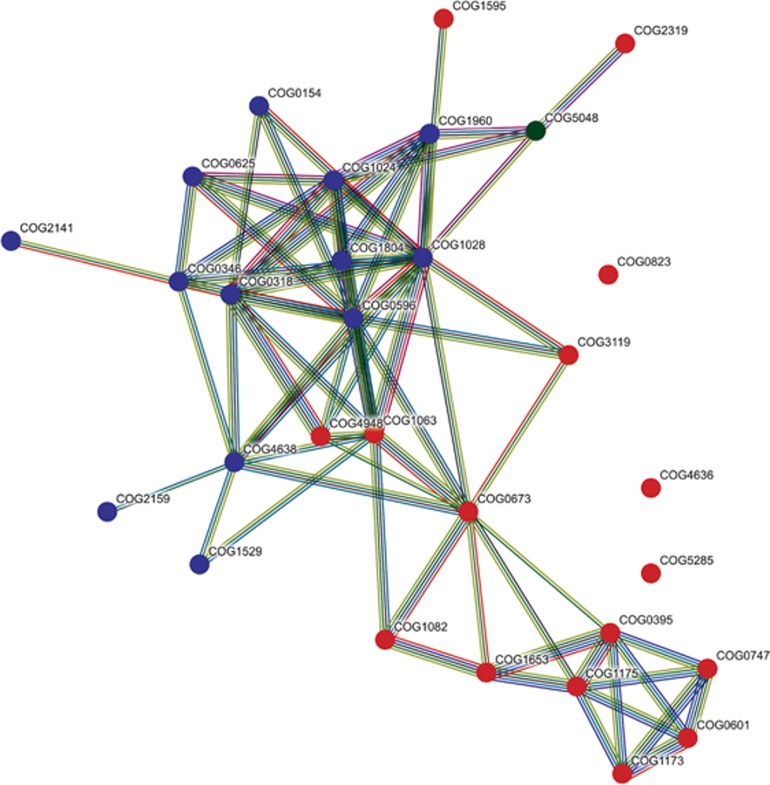
When comparing on the level of individual COGs, 42 symbiont-enriched genes were identified (Supplementary Figure 4B). Most of them (43%) belonged to COG classes R and S (‘general function prediction only’ and ‘function unknown’), a large fraction (19%) belonged to class V (‘defense mechanisms’), and 5 (12%) to class L (‘replication, recombination and repair’). According to the STRING database, many of these significantly symbiont-enriched COGs were likely interacting (Figure 2). At a high confidence cutoff (0.700 minimum required interaction score), five networks (A–E) comprising 17, 6, 3, 2 and 2 COGs were obtained. The remaining 12 symbiont-enriched COGs did not interact with any other COGs in the list. The set includes a restriction endonuclease (COG2810) and a bacteriophage protein gp37 (COG4422).
Figure 2.
STRING network of significantly sponge symbiont-enriched COGs. Colored areas mark COGs that belong to the same network (A–E). Colors of the connectors indicate the type of evidence of the predicted interaction between the two connected COGs. Only connections of ‘high confidence’ (minimum required interaction score: 0.700) are shown.
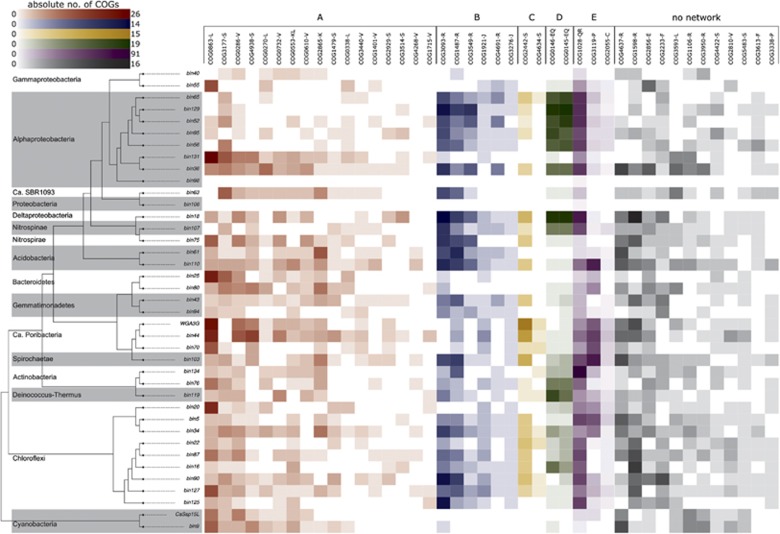
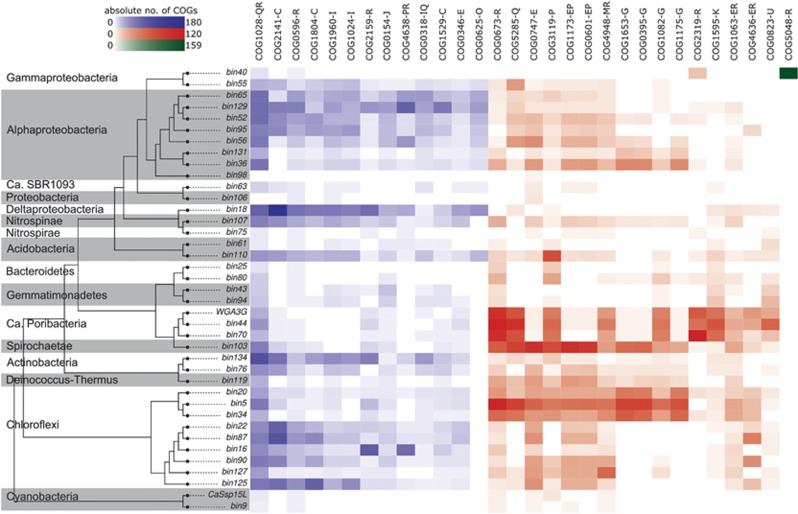
The largest STRING network was built of sponge-enriched COGs related to restriction-modification (RM) with endonucleases, helicases and methylases (cluster A in Figure 2, see Supplementary Table 4 for COG annotation). It was present in all sponge symbiont phyla in this study (Figure 3). RM systems represent one major line of defense against incoming, foreign DNA, a feature frequently referred to as bacterial immunity (Vasu and Nagaraja, 2013). RM systems are also known to play a role in symbioses (for example, Zheng et al., 2016) and have recently also been described in sponge symbionts (Tian et al., 2015; Gauthier et al., 2016; Horn et al., 2016). Many of the COGs of network A were previously described as sponge-enriched (Thomas et al., 2010; Fan et al., 2012; Gao et al., 2014; Burgsdorf et al., 2015). This recurring finding of RM in symbionts of a variety of sponges from different geographic locations, and the abundance of RM in all 13 bacterial phyla in our data set underscore the apparent significance for sponge symbioses.
Figure 3.
Heatmap of absolute counts of significantly sponge symbiont-enriched COGs in the genomes binned from the PacBio-Illumina hybrid assembly. Phylogenetic relationships of the genomes are indicated by a simplified version of the tree in Figure 1 (only sponge symbionts are shown here). Possibly interacting COGs as shown in Figure 2 are grouped and colored accordingly and marked by the letters A–E. The letters next to each COG indicate the according COG class.
Most COGs of STRING network B were related to toxin–antitoxin (TA) systems that supposedly play a role in phage defense, stress response, and programmed cell death (for example, Sberro et al., 2013). COG3549 and COG3093 form the HigAB TA plasmid maintenance system, and COG1487 encodes for the toxin in a TA system of the VapBC family (Makarova et al., 2009; Sberro et al., 2013). COG4691 is a plasmid stability protein and encodes for a proposed antitoxin of a VapBC TA system (Chen, 2007). COG1921 (SelA) and COG3276 (SelB), involved in selenocysteine production (Stolz et al., 2006), co-occurred in the majority of symbiont bins of various phyla but were missing in the majority of their closely related references (Supplementary Table 4). STRING network C consists of COG4634 and COG2442, two uncharacterized conserved proteins according to the NCBI annotation. COG4634 is hypothesized to be a fine-tuning modulator in conjugative plasmid transfer (López-Fuentes et al., 2015), and COG2442 is a PIN-associated antitoxin in a widespread TA system most abundant in Cyanobacteria and Chloroflexi (Makarova et al., 2009). Furthermore, COG2929 and COG3514, which are part of network A, were predicted to form a TA system as well (Makarova et al., 2009). Both COGs co-localize on a plasmid of the cyanobacterium Synechococcus elongatus PCC7942 where this TA system plays a crucial role in plasmid maintenance (Chen, 2007). In our data set, both COGs co-occurred in 16 sponge symbiont bins of various bacterial phyla, but only once in the reference group, in the acidobacterium Solibacter usitatus. The abundance and distribution of multiple RM and TA systems in their genomes suggests that defense against foreign DNA is an important feature of sponge symbionts being consistent with the previously stated concept of their convergent evolution (Thomas et al., 2010; Fan et al., 2012; Horn et al., 2016). These defense mechanisms are possibly a necessary countermeasure against the exposure to free DNA resulting from the sponge’s extensive filtration and phagocytosis activity (Reiswig, 1974).
Symbiont-enriched STRING networks D and E are related to colonization of the host and possibly utilization of the host matrix. COG0145 (hyuA) and COG0146 (hyuB) of network D have been hypothesized to play an important role for Helicobacter pylori in the colonization of mice (Zhang et al., 2009). These genes are known to be involved in the metabolism of hydantoin (Kim et al., 2000). The abundance and distribution of network D across various phyla of sponge-associated bacteria in our study suggests that it may also be of importance for the colonization of sponge hosts. COG1028 (FabG) and COG3119 (arylsulfatase A) of network E displayed the highest counts within the sponge-enriched COGs. Arylsulfatase A might allow the symbionts to metabolize sulfated polysaccharides from the sponge extracellular matrix, where their abundance has been documented in a number of sponge species, including the related species Aplysina fulva (Zierer and Mourão, 2000; Vilanova et al., 2009).
Sponge symbionts display metabolic specialization
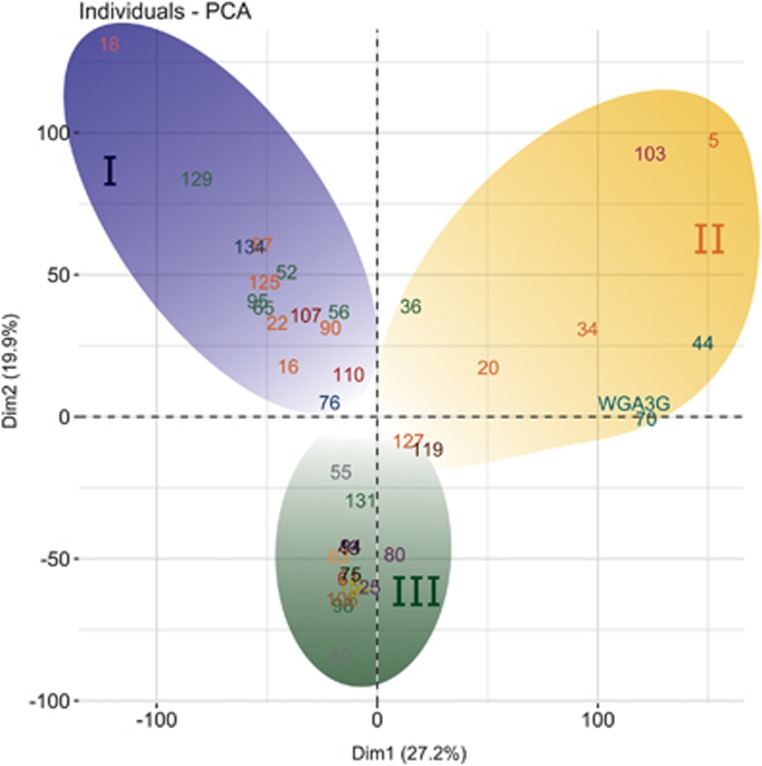
In order to compare the symbiont genomes among each other and to identify functional groups, a principle component analysis (PCA) was performed, clustering the sponge symbiont clades into three functional groups (Figure 4). The 30 COGs with the greatest influence on the functional grouping are shown in Supplementary Figure 5. Most COGs of symbiont groups I, II and III are strongly connected according to a STRING network with the COGs enriched in groups I and II clustering on different sides of the network (Figure 5). The correlation between the 30 COGs and their phylogenetic context is shown in Figure 6. The functional grouping is only partly coherent with phylogeny. While for example, Gemmatimonadetes cluster closely together, Chloroflexi are split up in two groups, which are as follows: (i) SAR202 clustering with a group of Alphaproteobacteria, Deltaproteobacteria, Nitrospinae and Actinobacteria, and (ii) Caldilineae and Anaerolineae that built a group with Poribacteria and Spirochaetae.
Figure 4.
PCA plot comparing the genomes of the sponge-symbionts to each other based on their COG annotation. Phylogenetic affiliation is indicated by font colors (see Table 2 for details). The symbionts build three groups I–III marked by background color (blue, yellow and green, respectively).
Figure 5.
STRING network of the 30 COGs contributing most to the grouping of the sponge-symbionts in Figure 4. Circles representing the COGs’ position in the network are colored according to the symbiont group where they are overrepresented (group I–III: blue, red and green, respectively). Colors of the connectors indicate the type of evicence of the predicted interactions between the two connected COGs as shown in Figure 2. Only connections of ‘high confidence’ (minimum required interaction score: 0.700) are shown.
Figure 6.
Heatmap of absolute counts of the 30 COGs contributing most to the grouping of the sponge-symbionts as shown in Figure 4. Phylogenetic relationships of the genomes are indicated by a simplified version of the tree in Figure 1 (only sponge symbionts are shown here). Colors represent the symbiont group, where the regarding COGs is overrepresented (group I–III: blue, red and green, respectively). The letters next to each COG indicate the according COG class.
According to our analysis, the COGs enriched in symbiont group I are mainly involved in metabolism and energy production. Most enriched in this group are COGs related to carnitine metabolism. Carnitine is an organic compatible solute that some bacteria can use as a source for carbon, nitrogen, and energy (Meadows and Wargo, 2015). It is produced by most eukaryotes, including sponges (Fraenkel, 1954) and we posit that it may be taken up by symbiotic bacteria from the readily available sponge-derived detritus consisting largely of shed sponge cells (De Goeij et al., 2009; Alexander et al., 2014). Uptake of carnitine by bacteria can also serve as protection against environmental stress like variation in water content, salinity, or temperature (Meadows and Wargo, 2015).
Symbiont group II is characterized by high numbers of arylsulfatase A genes (COG3119), various ABC transporters and dehydrogenases. This phylogenetically heterogeneous guild of microorganisms seems to be specialized on the utilization of sulfated polysaccharides, as described above for symbiont-enriched COG network E. Inspection of the genomic context on the bin-level shows that the arylsulfatase repeatedly clusters with the ABC transporters and the dehydrogenase that are likewise enriched in symbiont group II (Supplementary Figure 6 and Supplementary Table 5). This further supports our hypothesis that this gene cluster is of importance for sponge symbionts, and especially for the members of symbiont group II.
The genomes of symbiont group III did not show an enrichment of any particular COGs. They also contained the COGs of symbiont groups I and II, but not in as high numbers. We therefore posit that symbiont group III is not metabolically specialized and may represent a group of metabolic generalists. Within the 30 COGs most responsible for the grouping, only COG5048 (FOG: Zinc-finger) was enriched in bin40 of this group with a total of 159 copies. Zn-fingers are small structural protein motifs that have general cellular roles in binding nucleic acids and proteins. They are commonly found in eukaryotes, but also present in prokaryotes, where they are likely involved in virulence or symbiosis (Malgieri et al., 2015).
Conclusions
The complementation of Illumina short-read with PacBio long-read sequencing for metagenomic binning of highly complex environmental samples greatly improves the overall assembly statistics. It also improves the quality of binned genomes and eases, often newly enables phylogenetic classification of the binned genomes. The statistical comparison revealed an enrichment of genes related to RM and TA systems in most symbiont genomes over the reference genomes. This implies that the defense against incoming foreign DNA is of high importance for a symbiotic existence within the sponge mesohyl. This finding is particularly relevant in the context of the extensive animal’s filtration and phagocytosis activities, resulting in an ample exposure of the symbionts to free DNA. Secondly, host colonization and host matrix utilization were identified as significantly enriched features in sponge symbionts. The within-symbionts genome comparison revealed a nutritional specialization, where one guild of symbionts appears to metabolize carnitine, while the other appears to metabolize sulfated polysaccharides, both of which are abundant molecules of the sponge extracellular matrix. We hypothesize that the sponge symbionts feed on the sponge cells that are shed as part of the cell turnover, and on components of the sponge extracellular matrix. A third guild of symbionts may be viewed as nutritional generalists, whose precise function within this consortium remains to be identified. The presence of guilds specializing on different metabolic traits, such as butyrate or lactate production or mucin degradation, is also known from the human gut microbiome (Shetty et al., 2017). Complex microbial communities may structure themselves around nutritional niches provided by the specific host-related environment. The unprecedented resolution of the genomic repertoire was enabled by binning of a metagenomic hybrid assembly of hitherto unprecedented depth for sponge symbioses. The hypotheses on niche specializations by the symbionts could be tested in feeding studies combined with metatranscriptomics and/or imaging techniques.
Acknowledgments
This project was funded by Grant Number CSP1291 of the DOE Joint Genome Institute, the European Commission within its FP7 Program, under the thematic area KBBE.2012.3.2.-01 with Grant Number 311932 (‘SeaBioTech’), and by the European Union’s Horizon 2020 research and innovation program under Grant Agreement No. 679849 (‘SponGES’). BMS was supported by a grant of the German Excellence Initiative to the Graduate School of Life Sciences, University of Würzburg. We thank Monika Bright (University of Vienna) and the Marine Biology Station Piran for support during sampling, Tanja Woyke (DOE Walnut Creek, CA, USA) for helpful discussions on differential coverage binning, Martin Jahn (GEOMAR) for suggestions on the interrogations of the amplicon data, as well as Frank Förster and Thomas Dandekar (Department of Bioinformatics, University of Würzburg, Würzburg, Germany) for useful advice and access to their servers.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. (2013). Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31: 533–538. [DOI] [PubMed] [Google Scholar]
- Alexander BE, Liebrand K, Osinga R, Van Der Geest HG, Admiraal W, Cleutjens JPM et al (2014). Cell turnover and detritus production in marine sponges from tropical and temperate benthic ecosystems. PLoS One 9: e109486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ et al (2014). Binning metagenomic contigs by coverage and composition. Nat Methods 11: 1144–1150. [DOI] [PubMed] [Google Scholar]
- Ankenbrand MJ, Hohlfeld S, Hackl T, Förster F. (2017). AliTV-interactive visualization of whole genome comparisons. PeerJ Comput Sci 3: e116. [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS et al (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beims H, Wittmann J, Bunk B, Spröer C, Rohde C, Günther G et al (2015). Paenibacillus larvae-directed bacteriophage HB10c2 and its application in American foulbrood-affected honey bee larvae. Appl Environ Microbiol 81: AEM.00804–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. (2007). GenBank. Nucleic Acids Res 35: D21–D25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein SR, Theis KR. (2015). Host biology in light of the microbiome: Ten principles of holobionts and hologenomes. PLOS Biol 13: e1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgsdorf I, Slaby BM, Handley KM, Haber M, Blom J, Marshall CW et al (2015). Lifestyle evolution in cyanobacterial symbionts of sponges. MBio 6: e00391–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K et al (2009). BLAST+: Architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJ, Tesler G. (2012). Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. (2007).Functional genomics of the unicellular cyanobacterium Synechococcus elongatus PCC 7942. PhD thesis. Texas A&M University, Kingsville, TX, USA.
- De Goeij JM, De Kluijver A, Van Duyl FC, Vacelet J, Wijffels RH, De Goeij AFPM et al (2009). Cell kinetics of the marine sponge Halisarca caerulea reveal rapid cell turnover and shedding. J Exp Biol 212: 3892–3900. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easson CG, Thacker RW. (2014). Phylogenetic signal in the community structure of host-specific microbiomes of tropical marine sponges. Front Microbiol 5: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Reynolds D, Liu M, Stark M, Kjelleberg S, Webster NS et al (2012). Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc Natl Acad Sci USA 109: E1878–E1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fieseler L, Quaiser A, Schleper C, Hentschel U. (2006). Analysis of the first genome fragment from the marine sponge-associated, novel candidate phylum Poribacteria by environmental genomics. Environ Microbiol 8: 612–624. [DOI] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39: W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraenkel G. (1954). The distribution of vitamin BT (carnitine) throughout the animal kingdom. Arch Biochem Biophys 50: 486–495. [DOI] [PubMed] [Google Scholar]
- Frank JA, Pan Y, Tooming-Klunderud A, Eijsink VGH, McHardy AC, Nederbragt AJ et al (2016). Improved metagenome assemblies and taxonomic binning using long-read circular consensus sequence data. Sci Rep 6: 25373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z-M, Wang Y, Tian R-M, Wong YH, Batang ZB, Al-Suwailem AM et al (2014). Symbiotic adaptation drives genome streamlining of the cyanobacterial sponge symbiont 'Candidatus Synechococcus spongiarum'. mBio 5: e00079–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier M-EA, Watson JR, Degnan SM. (2016). Draft genomes shed light on the dual bacterial symbiosis that dominates the microbiome of the coral reef sponge Amphimedon queenslandica. Front Mar Sci 3: 196. [Google Scholar]
- Gouy M, Guindon S, Gascuel O. (2010). SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224. [DOI] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29: 1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackl T. (2016).A draft genome for the Venus flytrap, Dionaea muscipula. PhD thesis. Julius-Maximilians-Universität Würzburg, Germany.
- Hackl T, Hedrich R, Schultz J, Förster F. (2014). proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 30: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel U, Hopke J, Horn M, Friedrich AB, Wagner M, Hacker J et al (2002). Molecular evidence for a uniform microbial community in sponges from different oceans. Appl Environ Microbiol 68: 4431–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel U, Piel J, Degnan SM, Taylor MW. (2012). Genomic insights into the marine sponge microbiome. Nat Rev Microbiol 10: 641–654. [DOI] [PubMed] [Google Scholar]
- Horn H, Slaby BM, Jahn MT, Bayer K, Moitinho-Silva L, Förster F et al (2016). An enrichment of CRISPR and other defense-related features in marine sponge-associated microbial metagenomes. Front Microbiol 7: 1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Beier S, Flade I, Górska A, El-Hadidi M, Mitra S et al (2016). MEGAN Community Edition—Interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput Biol 12: e1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamke J, Sczyrba A, Ivanova N, Schwientek P, Rinke C, Mavromatis K et al (2013). Single-cell genomics reveals complex carbohydrate degradation patterns in poribacterial symbionts of marine sponges. ISME J 7: 2287–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GJ, Lee DE, Kim HS. (2000). Functional expression and characterization of the two cyclic amidohydrolase enzymes, allantoinase and a novel phenylhydantoinase, from Escherichia coli. J Bacteriol 182: 7021–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Phillippy AM. (2015). One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly. Curr Opin Microbiol 23: 110–120. [DOI] [PubMed] [Google Scholar]
- Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, Ganapathy G et al (2012). Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol 30: 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 7: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C et al (2004). Versatile and open software for comparing large genomes. Genome Biol 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê S, Josse J, Husson F. (2008). FactoMineR : An R package for multivariate analysis. J Stat Softw 25: 1–18. [Google Scholar]
- Li C-W, Chen J-Y, Hua T-E. (1998). Precambrian sponges with cellular structures. Science 279: 879–882. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Fuentes E, Torres-Tejerizo G, Cervantes L, Brom S. (2015). Genes encoding conserved hypothetical proteins localized in the conjugative transfer region of plasmid pRet42a from Rhizobium etli CFN42 participate in modulating transfer and affect conjugation from different donors. Front Microbiol 5: 793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love GD, Grosjean E, Stalvies C, Fike DA, Grotzinger JP, Bradley AS et al (2009). Fossil steroids record the appearance of Demospongiae during the Cryogenian period. Nature 457: 718–721. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Koonin EV. (2009). Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct 4: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malgieri G, Palmieri M, Russo L, Fattorusso R, Pedone PV, Isernia C. (2015). The prokaryotic zinc-finger: Structure, function and comparison with the eukaryotic counterpart. FEBS J 282: 4480–4496. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, Chen I-MA, Chu K, Szeto E, Palaniappan K, Grechkin Y et al (2012). IMG/M: the integrated metagenome data management and comparative analysis system. Nucleic Acids Res 40: D123–D129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows JA, Wargo MJ. (2015). Carnitine in bacterial physiology and metabolism. Microbiology 161: 1161–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M et al (2008). The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y, Asai K, Hamada M. (2013). PBSIM: PacBio reads simulator—toward accurate genome assembly. Bioinformatics 29: 119–121. [DOI] [PubMed] [Google Scholar]
- Parks DH, Tyson GW, Hugenholtz P, Beiko RG. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30: 3123–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glöckner FO. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. (2007). NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35: D61–D65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res 41: 590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy TBK, Thomas AD, Stamatis D, Bertsch J, Isbandi M, Jansson J et al (2014). The Genomes OnLine Database (GOLD) v.5: a metadata management system based on a four level (meta)genome project classification. Nucleic Acids Res 43: D1099–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiswig HM. (1974). Water transport, respiration and energetics of three tropical marine sponges. J Exp Mar Bio Ecol 14: 231–249. [Google Scholar]
- Revell LJ. (2012). phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol Evol 3: 217–223. [Google Scholar]
- Ricker N, Shen S, Goordial J, Jin S, Fulthorpe RR. (2016). PacBio SMRT assembly of a complex multi-replicon genome reveals chlorocatechol degradative operon in a region of genome plasticity. Gene 586: 239–247. [DOI] [PubMed] [Google Scholar]
- Schmitt S, Tsai P, Bell J, Fromont J, Ilan M, Lindquist N et al (2012. a). Assessing the complex sponge microbiota: Core, variable and species-specific bacterial communities in marine sponges. ISME J 6: 564–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S, Hentschel U, Taylor MW. (2012. b). Deep sequencing reveals diversity and community structure of complex microbiota in five Mediterranean sponges. Hydrobiologia 687: 341–351. [Google Scholar]
- Sberro H, Leavitt A, Kiro R, Koh E, Peleg Y, Qimron U et al (2013). Discovery of functional toxin/antitoxin systems in bacteria by shotgun cloning. Mol Cell 50: 136–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty SA, Hugenholtz F, Lahti L, Smidt H, de Vos WM. (2017). Intestinal microbiome landscaping: Insight in community assemblage and implications for microbial modulation strategies. FEMS Microbiol Rev 41: 182–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegl A, Kamke J, Hochmuth T, Piel J, Richter M, Liang C et al (2011). Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J 5: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz JF, Basu P, Santini JM, Oremland RS. (2006). Arsenic and selenium in microbial metabolism. Annu Rev Microbiol 60: 107–130. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J et al (2014). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43: D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV et al (2003). The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Moitinho-Silva L, Lurgi M, Björk JR, Easson C, Astudillo-García C et al (2016). Diversity, structure and convergent evolution of the global sponge microbiome. Nat Commun 7: 11870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Rusch D, DeMaere MZ, Yung PY, Lewis M, Halpern A et al (2010). Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J 4: 1557–1567. [DOI] [PubMed] [Google Scholar]
- Tian R-M, Sun J, Cai L, Zhang W-P, Zhou G-W, Qiu J-W et al (2015). The deep-sea glass sponge Lophophysema eversa harbors potential symbionts responsible for the nutrient conversions of carbon, nitrogen and sulfur. Environ Microbiol 18: 2481–2494. [DOI] [PubMed] [Google Scholar]
- Tian R-M, Wang Y, Bougouffa S, Gao Z-M, Cai L, Bajic V et al (2014). Genomic analysis reveals versatile heterotrophic capacity of a potentially symbiotic sulfur-oxidizing bacterium in sponge. Environ Microbiol 16: 3548–3561. [DOI] [PubMed] [Google Scholar]
- Tsai Y-C, Conlan S, Deming C NISC Comparative Sequencing ProgramSegre JA, Kong HH et al (2016). Resolving the complexity of human skin metagenomes using single-molecule sequencing. MBio 7: e01948–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu K, Nagaraja V. (2013). Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol Biol Rev 77: 53–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilanova E, Coutinho CC, Mourão PAS. (2009). Sulfated polysaccharides from marine sponges (Porifera): An ancestor cell-cell adhesion event based on the carbohydrate-carbohydrate interaction. Glycobiology 19: 860–867. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster NS, Thomas T. (2016). The sponge hologenome. MBio 7: e00135–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Zhu Z, Fu L, Niu B, Li W. (2011). WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics 12: 444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M-J, Zhao F, Xiao D, Gu Y-X, Meng F-L, He L-H et al (2009). Comparative proteomic analysis of passaged Helicobacter pylori. J Basic Microbiol 49: 482–490. [DOI] [PubMed] [Google Scholar]
- Zheng H, Dietrich C, Hongoh Y, Brune A. (2016). Restriction-modification mystems as mobile genetic elements in the evolution of an intracellular symbiont. Mol Biol Evol 33: 721–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierer MS, Mourão PAS. (2000). A wide diversity of sulfated polysaccharides are synthesized by different species of marine sponges. Carbohydr Res 328: 209–216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.