Abstract
The growing demand for renewable, carbon-neutral materials and energy is leading to intensified forest land-use. The long-term ecological challenges associated with maintaining soil fertility in managed forests are not yet known, in part due to the complexity of soil microbial communities and the heterogeneity of forest soils. This study determined the long-term effects of timber harvesting, accompanied by varied organic matter (OM) removal, on bacterial and fungal soil populations in 11- to 17-year-old reforested coniferous plantations at 18 sites across North America. Analysis of highly replicated 16 S rRNA gene and ITS region pyrotag libraries and shotgun metagenomes demonstrated consistent changes in microbial communities in harvested plots that included the expansion of desiccation- and heat-tolerant organisms and decline in diversity of ectomycorrhizal fungi. However, the majority of taxa, including the most abundant and cosmopolitan groups, were unaffected by harvesting. Shifts in microbial populations that corresponded to increased temperature and soil dryness were moderated by OM retention, which also selected for sub-populations of fungal decomposers. Biogeographical differences in the distribution of taxa as well as local edaphic and environmental conditions produced substantial variation in the effects of harvesting. This extensive molecular-based investigation of forest soil advances our understanding of forest disturbance and lays the foundation for monitoring long-term impacts of timber harvesting.
Introduction
The growing renewable resource sector is driving demand for forest biomass and the intensification of forest land-use, resulting in shorter crop cycles, more densely replanted forests and increased harvesting of residual woody biomass (Fox, 2000; Allmér et al., 2009; Achat et al., 2015). Sustaining the productivity of future plantations depends upon prudent soil management, which includes monitoring and assessing the influence of microbially mediated processes. Management practices can be improved with a better understanding of the composition and function of soil microbial communities, their differences according to biogeography and forest type, and how populations are affected by harvesting over the long term. Timber harvesting is known to affect the ecology of belowground communities in the short- and long-term (Supplementary Table 1), yet the inherent variability of soils and diversity of microbial inhabitants necessitates a more comprehensive investigation than has been conducted to date.
Following harvesting and before canopy closure, soils experience significant environmental changes that include higher average temperatures and lower average moisture content as well as more frequent fluctuation and higher extrema (Childs and Flint, 1987; Adams et al., 1991; Kranabetter and Chapman, 1999; Redding et al., 2003; Kulmala et al., 2014). These changes are known to affect microbial processes, such the uptake of atmospheric methane which is reduced over the short- (Castro et al., 2000; Zerva and Mencuccini, 2005; Takakai et al., 2008; Kulmala et al., 2014) and long-term (Wu et al., 2011) owing to reduced populations of high-affinity methanotrophs (Nazaries et al., 2011). Other examples include substantive and persistent changes in populations of ectomycorrhizal fungi due to the loss of tree hosts (Hartmann et al., 2012), broad changes in the ratio of Basidiomycota to Ascomycota (Bader et al., 1995; Hartmann et al., 2012; McGuire et al., 2015) and the rise of stress-tolerant cellulolytic populations and reduction in cellulolytic activity (Wilhelm et al., 2017b). However, recent next-generation sequencing-based surveys of microbial communities find that edaphic and geographic factors outweigh the effects of harvesting, accounting for between 4- and 14-fold more variation over the long-term (Hartmann et al., 2012; Cardenas et al., 2015; Leung et al., 2016). Given the extent of variation, it is necessary to conduct a comprehensive comparative study across various forest and soil types to test the robustness of previous findings and identify potentially novel, generalizable long-term responses to harvesting.
The long-term soil productivity study (LTSP) was initiated in 1989 to assess changes in forest productivity following harvesting and emulates varying degrees of soil compaction and harvesting of residual organic matter (OM), such as woody debris and organic soil (Powers et al., 2005). The LTSP is a field experiment replicated at sites in some of the most productively managed forested regions, or ecozones, in North America, which include British Columbia, California, Ontario and Texas. The extensive LTSP network provided a sufficiently broad scope to assess and contrast characteristics of long-term changes in soil bacterial and fungal communities. The existence of generalizable effects across ecozones may reveal common abiotic or biotic factors shaping post-harvest soil communities. Conversely, the extent of localized effects may reveal the influence of biogeography, succession and climate on the effects of harvesting. We set out to weigh the effects of harvesting at local and global levels and contrast the effect size among ecozones.
The retention of coarse woody debris following forest harvesting is a management practice that has multiple benefits (Gustafsson et al., 2012), potentially including the mitigation of long-term changes in soil communities. Not only does the retention of woody debris change the quality and quantity of OM input to soils, it also mitigates chemical and physical changes in pH, moisture and temperature (Entry et al., 1986; Bååth et al., 1995; Kranabetter and Chapman, 1999). Negligible differences in microbial biomass, respiration and methanotrophic activity were found between partially logged and unharvested forests, in contrast to significant impacts from clearcutting (Wu et al., 2011; Holden and Treseder, 2013a). Even at clear-cut sites, the retention of coarse woody debris tempered long-term shifts in microbial community composition and selected for unique sub-populations (Hartmann et al., 2012) and increased the diversity of wood-rot fungi relative to unharvested plots (Brazee et al., 2014). The LTSP experiment was designed to test the effects of retention (OM1) or removal (OM2) of coarse woody debris along with an extreme degree of organic matter removal, where the organic soil layer is removed (OM3) as an experimental endpoint for maximal OM loss. We hypothesized that the retention of coarse woody debris would differentiate soil communities in harvested plots by selecting for populations involved in early-stage wood-decay and decomposition, resulting in differences in soil carbon and nitrogen content.
We set out to determine long-term effects on forest soil communities in harvested plots with varying amounts of OM retention at sites across North America using bacterial (16 S rRNA gene) and fungal (ITS) phylogenetic gene marker libraries, accompanied by whole community shotgun metagenomes. The main objectives of this study were (1) to determine the extent to which previous findings from the LTSP and other studies could be generalized (that is, their ecological validity), (2) to determine how slash-retention and extreme OM removal modulate effects of forest harvesting, (3) to identify indicator taxa relevant to monitoring forest regeneration in accordance with the goals of the LTSP (Powers, 2006), and (4) to compare long-term effects of harvesting to those of natural disturbance reported in the literature. This study presents the most comprehensive long-term perspective on the effects of timber harvesting to date, shedding light on the extent and nature of ecological change and offering a new perspective on the potential impacts of intensified forest land-use.
Materials and methods
Overview of sites and sample collection
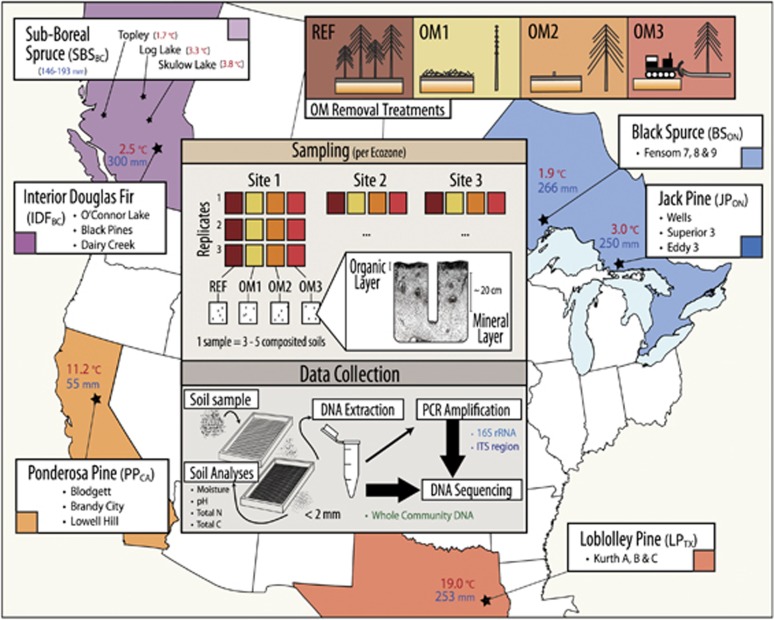
Soils were collected from a total of eighteen reforested LTSP sites, between 11 and 17 years old, located in six conifer-dominated North American ecozones with three sites per ecozone (Figure 1; Detailed Site Information in Supplementary Table 2). Each site contained plots corresponding to four experimental treatments: an unharvested reference, REF, and three harvested treatments with varying degrees of OM removal: OM1, where tree boles (stems) were debranched and removed, leaving woody debris in place; OM2, where whole trees including branches were removed and, OM3, where whole trees were removed and the upper organic layer of forest floor scraped away. Plots were replanted with the native tree species commonly used in commercial forestry in each ecozone (Supplementary Table 2) and no additional manipulations were performed after the initial installation of harvested treatments. The composition of plant communities differed among ecozones, but, for the most part, did not differ among harvested treatment plots (details in Supplementary Materials 1). Plant community composition was slightly different between OM3 versus OM1 and OM2 in SBSBC and BSON (see Supplementary Materials I). Soil compaction was controlled at three levels during harvesting and samples were collected from the lowest intensity compaction treatment in order to focus on the effects of OM removal. Sampling from SBSBC and IDFBC included libraries from moderate (C1) and severe (C2) compaction treatments previously shown to have minor influence on community composition (Hartmann et al., 2012). Three replicates were collected from each treatment at each of the eighteen sites. In Ontario (BSON and JPON), three replicate plots were available at each site for each treatment, while in the other four ecozones triplicate samples were collected from one single, larger treatment plot per site. During sampling, the litter layer was first removed and organic layer samples (O-horizon) were collected with a trowel. Then, the top 20 cm of mineral soil (including the A and occasionally upper B-horizon) was collected using a Stoney auger (5 cm diameter). Sampling was performed to reflect consistent soil characteristics among treatments and sites. To account for heterogeneity at the plot level and ensure sufficient soil material, sub-samples from three to five points (consistent per ecozone) were composited to produce the previously described samples. Samples were stored at 4 °C during transport, sieved through 2-mm mesh and stored at −80 °C until DNA was extracted within three months of sampling date.
Figure 1.
Overview of experimental design superimposed on a map of sampling locations in each of the six ecozones (three replicate sites per ecozone). Ecozones were named after the predominant tree species in the region. Mean annual temperature and precipitation during the warmest quarter are provided. Samples from SBSBC and IDFBC were originally collected by Hartmann et al. (2012). The methods of sample processing and sequencing were identical. Where available, photographs of treatment plots at the time of harvesting and at time of sampling can be found in the Supplementary Materials III.
Preparation of 16 S rRNA gene and ITS region pyrotag libraries
DNA was extracted from soil (0.5 g) using the FastDNA™ Spin Kit for Soil (MPBio, Santa Ana, CA) according to the manufacturer’s protocol. PCR amplification was performed on bacterial 16 S rRNA gene (V1–V3) using primers 27 F/519 R (Lane, 1991; Amann et al., 1995) and fungal internal transcribed spacer region (ITS2) using primers ITS3/ITS4 (White et al., 1990) according to methods described in Hartmann et al. (2012). PCR reactions were performed in triplicate and pooled before purification and quantification. All DNA quantitation was performed using Pico-Green fluorescent dye (ThermoFisher, Waltham, MA, USA). Samples were sequenced using the Roche 454 Titanium platform (GS FLX+) at the McGill University and Genome Québec Innovation Centre, yielding an average of 7800 bacterial and 8000 fungal quality filtered reads per sample. The ITS and 16 S amplicon libraries from SBSBC and IDFBC used in this study were obtained from Hartmann et al. (2012). Pyrotag libraries were quality filtered and processed using mothur according to the Schloss ‘454 SOP’ (accessed November 2015; Schloss et al., 2009). 16 S rRNA gene libraries were clustered into operational taxonomic units (OTUs) at 1% dissimilarity to produce a count matrix. Fungal sequences were clustered at 5.5% dissimilarity, due to the hypervariability of the ITS region, using CrunchClust (Hartmann et al., 2012). Taxonomic classification was performed using the RDP Classifier (Wang et al., 2007) with the Greengenes database for 16 S rRNA genes (database gg_13_8_99; August 2013) and the mothur-formatted release of UNITE database for ITS (sh_mothur_release_08.12.2013; August 2013). All OTU counts were normalized to total counts per thousand reads. Supplementary Table 3 provides an account of all sequenced samples used in analysis according to ecozone, soil layer and OM treatment. Raw sequencing data can be retrieved from the Short Read Archive under the study accessions: PRJEB12501 (ITS), PRJEB8599 (16 S rRNA) and PRJEB8420 (shotgun metagenomes), and additional sequencing data from related LTSP projects can be found in Wilhelm et al. (2017a). All metadata used in analyses can be found embedded in phyloseq objects available as Supplementary Data.
Preparation of shotgun metagenome libraries
Whole shotgun metagenomes were prepared from samples from a single site in each of four ecozones, BSON (A8), JPON (JW), PPCA (BL) and LPTX (TXA), using freshly prepared DNA extracts distinct from those for pyrotag libraries (n=21 per ecozone). Illumina paired-end, 150-bp libraries were generated at the Joint Genome Institute (Walnut Creek, CA, USA). Previously published paired-end, 75-bp metagenomic libraries for IDFBC (Cardenas et al., 2015) were also included in analyses. Raw sequence data for the IDF ecozone is available at European Nucleotide Archive (Study accession PRJEB8420, sample accessions ERS656878 to ERS65689) and, for the other four ecozones, at the JGI Genome portal (JGI proposal ID 543). Raw sequences were quality-processed as previously reported (Cardenas et al., 2015).
Bioinformatic analysis
Statistics were performed using R v. 3.1.0 (R Core Team, 2008) with general dependency on the following packages: reshape2, ggplot2, plyr (Wickham, 2007, 2009), combinat (Chasalow, 2012), limma (Ritchie et al., 2015), Hmisc (Harrell and Dupont, 2015) and phyloseq (McMurdie and Holmes, 2014). Where necessary, P-values were adjusted according to the Benjamini and Hochberg (1995) false discovery rate (FDR) correction using the qval function of the qvalue R-package (v2.2.2; Storey et al., 2015). Chao1 richness and Shannon diversity estimates were calculated on rarefied data (nmin= 1300 reads) using the ‘plot_richness’ function (phyloseq) and estimates represent the average of 500 random samplings. The ‘vegan’ R-package (Oksanen et al., 2015) provided tools to calculate rarefaction curves (‘rarecurve’), non-parametric multidimensional scaling (‘metaMDS’), analysis of similarities (‘anosim’ nperm=999) and permutational analysis of variance (‘adonis’ nperm=999). The latter three analyses were performed using Bray-Curtis dissimilarities calculated using the function ‘vegdist.’ Correlations between OTUs were made using SparCC (Friedman and Alm, 2012) and co-association networks were constructed from significant (P<0.01) and relatively strong (r>| 0.3 |) negative and positive correlations using Gephi (v.0.9.3; Bastian et al., 2009). Indicator species analysis was performed using the R-package ‘indicspecies’ (De Cáceres and Legendre, 2009) on normalized count data for OTUs as well as binned taxonomic classification, in which case the analysis was repeated for all taxonomic ranks. Indicator analysis was also performed on data subsetted by ecozone and soil layer as well as aggregated by all ecozones and soil layers. Results can be found in the Supplementary Data package along with all R code to reproduce the analyses. Phylogenetic trees were prepared using MEGA6 (Tamura et al., 2013), including sequence alignment, trimming and building of maximum-likelihood trees (bootstrap n=500; Tamura-Nei substitution model). Analysis of fungal functional guilds was performed using FUNGuild (v. 1.0; Nguyen et al., 2015), verification of FUNGuild analysis of EM designation was performed based on a selection of 55 genera known to contain EM species (listed in Supplementary Materials II). A genus was deemed to possess radiation, desiccation, and heat-tolerant taxa if it was reported as having notable abundance in lithic or desert environments or if cultured representatives had documented exceptional tolerances.
The abundance of KEGG orthologous genes in the unassembled genomes were obtained from IMG/M pipeline annotation (Markowitz et al., 2008) and normalized to counts per million. Differences in abundance among treatments was calculated for each ecozone-soil layer combination, as well as a combination of all samples for each soil layer using ANOVA tests, and corrected for multiple testing (P-values and q-values<0.05).
Results
Overview of community composition
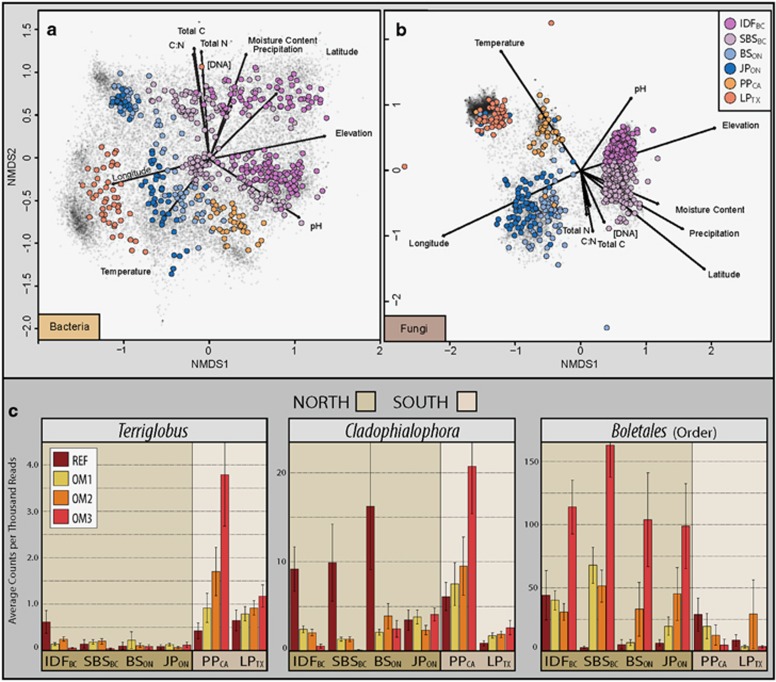
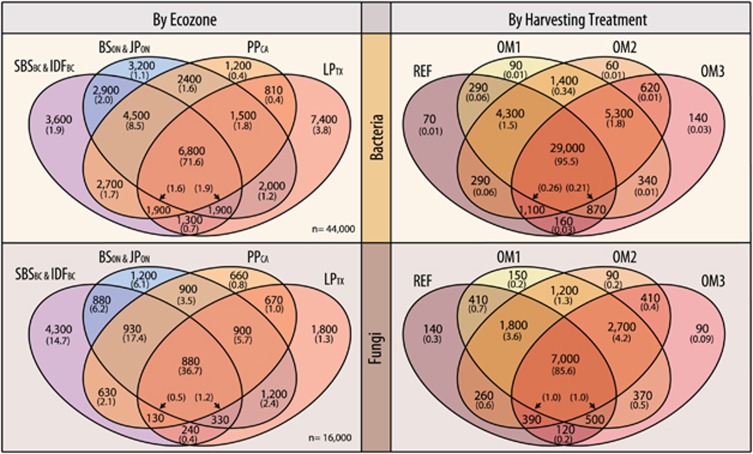
The majority of variation in beta-diversity of bacterial and fungal communities was attributed to ecozone and soil layer. Of the total variation explained by experimental factors in PERMANOVA (Supplementary Figure 1), ecozone accounted for 64 and 67% of variation in bacterial and fungal pyrotags, respectively, followed by site (15 and 21%), soil layer (18 and 7%) and OM treatment (3 and 5%). The dominant effect of ecozone is evident in ordinations, as is the stronger effect of soil layer on bacteria versus fungal communities (Figure 2). The core set of bacterial OTUs present at all sites and ecozones (15% of OTUs @ 99% identity) accounted for 72% of total reads, despite the large variation in overall community composition among ecozones. Core fungal OTUs were less cosmopolitan (5.5% of OTUs @ 94.5%) and less abundant (37% of total reads) than bacteria (Figure 3). Shannon diversity (alpha-diversity) significantly differed among ecozones (Tukey HSD, padj<0.001), though proximal ecozones shared similar diversity (Supplementary Figure 2) and the greatest proportion of OTUs (Supplementary Table 4). A sampling depth of ~8000 reads per sample did not saturate OTU richness at sites according to rarefaction curves (Supplementary Figure 2).
Figure 2.
Geographic differences in community structure illustrated by non-parametric multidimensional scaling of bacterial (a) and fungal (b) pyrotag libraries and by trends in relative abundance of specific taxa whose response to harvested treatments differed between northern and southern sites (c). NMS was based on Bray-Curtis dissimilarities and individual OTUs were mapped as black crosses (~44 000 bacterial and 16 000 fungal OTUs) and samples were mapped as colored circles. Unlike ITS libraries, bacterial 16 S rRNA gene libraries exhibited a clear split between organic (top) and mineral (bottom) layers, though samples from the organic layer in PPCA and LPTX were not mapped due to incomplete environmental data. Experimental factors were fitted to ordination with arrow length proportional to the correlation between variable and ordination axes.
Figure 3.
A Venn diagram displaying the extent of OTUs shared among ecozones (left) and OM removal treatments (right) for bacterial and fungal pyrotag libraries. The total number of OTUs is given followed by their total relative abundance (%) in parentheses. Proximal ecozones in Ontario and British Columbia are combined due to their similarities. Supplementary Table 4 provides a complete breakdown of all individual ecozones.
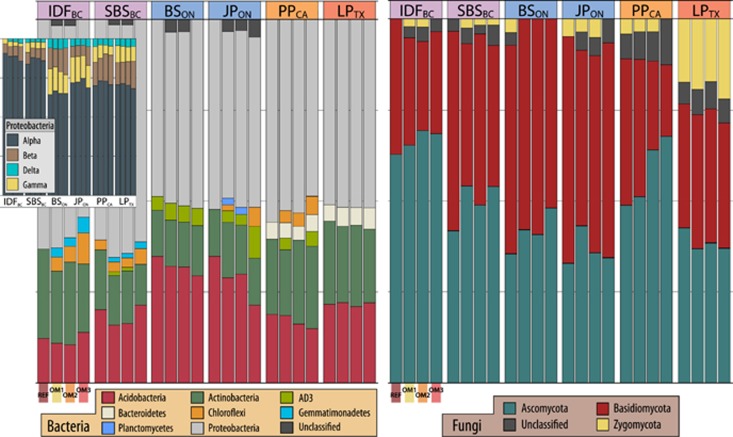
Cosmopolitan and abundant bacterial OTUs were classified as Rhodoplanes (5–7% of total libraries), Mycobacterium (0.4–4%), Burkholderia (0.5–3%), Reyranella (0.4–2%) and two candidate acidobacterial genera: Koribacter (0.2–1.5%) and Solibacter (0.2–1%). Members of Bradyrhizobiaceae were by far the most abundant taxonomic group, ranging from 25% (in IDFBC and SBSBC) to 8% (in LPTX). Although fungal OTUs were not cosmopolitan among ecozones, a few fungal families did predominate, these included: Atheliaceae (3–24%), Russulaceae (3.5–18%) and Suillaceae (3–9%). The exceptions were PPCA, dominated by Trichocomaceae (8–14%), and LPTX, dominated by Mortierellaceae (~15%). Differences in the abundance of certain phyla were observed between soil layers. Alphaproteobacteria, Gammaproteobacteria, Bacteroidetes and Actinobacteria were most abundant in organic layers, while candidate phylum AD3, Chloroflexi, Betaproteobacteria, Deltaproteobacteria, Firmicutes and Acidobacteria were most abundant in mineral layers. No fungal phyla were differentially abundant between soil layers.
Impacts of OM removal on soil conditions
Powers (2006) reported that 40–70% of aboveground biomass was removed in OM1, 70–90% in OM2 and nearly 100% in OM3 plots. At the time of our sampling, we observed gradients in total carbon and nitrogen in organic layer soils that corresponded with the original intensity of OM removal. (Supplementary Figure 3; full details Supplementary Table 5). OM gradients did not exist in any mineral layer samples or the organic layer of JPON at time of sampling. Organic layer soil depth was reduced according to the OM removal gradient. For instance, average organic layer depth at the BSON site was 0.4 cm (OM3), 3.2 cm (OM2), 4.0 cm (OM1) and 4.8 (REF). Organic layers were absent in OM3 in JPON and both BC ecozones. Soil pH was slightly higher in harvested plots versus REF in northern, but not southern, ecozones (Supplementary Table 5).
Mean daily soil temperature during periods before our soil sampling (recorded in PPCA, BSON and JPON) was significantly higher in harvested plots and increased with OM removal (Supplementary Figure 4). Five years after harvesting at Californian sites, summer soil temperatures were consistently 1.5 °C (OM1) and 6 °C (OM3) warmer than REF plots(Paz 2001). At Ontario sites, where long-term soil temperature data was available, OM3 was on average ~4 °C warmer during summer than OM2 over the first 5 years after harvesting (5 cm depth), though the difference diminished over time. In JPON, variation in daily temperature was on average ~60% greater in OM3 than in OM2 (t=−3.8; P<0.001), amounting to a variation of ~1.8 °C during the month of July. At the time of sampling, soil moisture was inversely related to the level of OM removal (Supplementary Figure 3).
Impacts of OM removal on soil communities
OM removal treatments had a relatively small, though significant (P<0.01), effect on overall bacterial and fungal beta-diversity (Supplementary Figure 1). These treatments accounted for ~5% of variation in both fungal and bacterial community composition, with British Columbian ecozones showing the lowest variation for bacteria (SBSBC: 2% and IDFBC: 3.6%) and LPTX (3%) for fungi. Conversely, PPCA showed the highest variation due to treatments in both bacterial (9%) and fungal (8%) populations. In agreement with the relatively small effects of OM removal, the vast majority of bacterial and fungal OTUs were common to all OM removal treatments (Figure 3). Pairwise comparisons between treatments using ANOSIM revealed that communities in OM1 and OM2 were the least distinct, while communities in REF and OM3 were the most differentiated (summary: Supplementary Figure 1; complete: Supplementary Table 6). These trends were mirrored in the greater number of OTUs shared between OM1 and OM2 versus between REF and OM3 (Figure 3). The long-term impacts of harvesting on Shannon diversity varied among ecozones (Supplementary Figure 5). Fungal populations had consistently higher Shannon diversity estimates in mineral layers of harvested plots relative to REF in all ecozones, though statistically significant in only SBSBC and JPON (Supplementary Figure 5). The fungal alpha-diversity in mineral soils was greatest in OM1 in all ecozones except LPTX, but significantly greater in only SBSBC (Tukey HSD; padj=0.03).
Differences in community composition between REF and harvested plots were apparent at the phylum level for both fungi and bacteria (Figure 4). The relative abundance of Chloroflexi, candidate phylum AD3 and Gemmatimonadetes (all mineral layer associated) and Cyanobacteria (organic layer associated) increased with increasing OM removal in at least five of six ecozones. Conversely, populations of Acidobacteria, Actinobacteria and Gammaproteobacteria declined with increasing OM removal, though not across all ecozones. Fungal communities exhibited a decline in the ratio of Basidiomycota to Ascomycota with increasing OM removal everywhere except for LPTX (Supplementary Figure 6). The proportion of unclassifiable sequences also increased with OM removal (Supplementary Figure 7), though the most pronounced and statistically significant effects were observed in OM3 (~2-fold greater than to REF).
Figure 4.
Relative abundance of bacteria and fungi among treatments at the phylum level faceted by ecozone. Inset: relative abundance of divisions within the phylum Proteobacteria. Phyla with low abundances (<0.075% of total reads) are not shown.
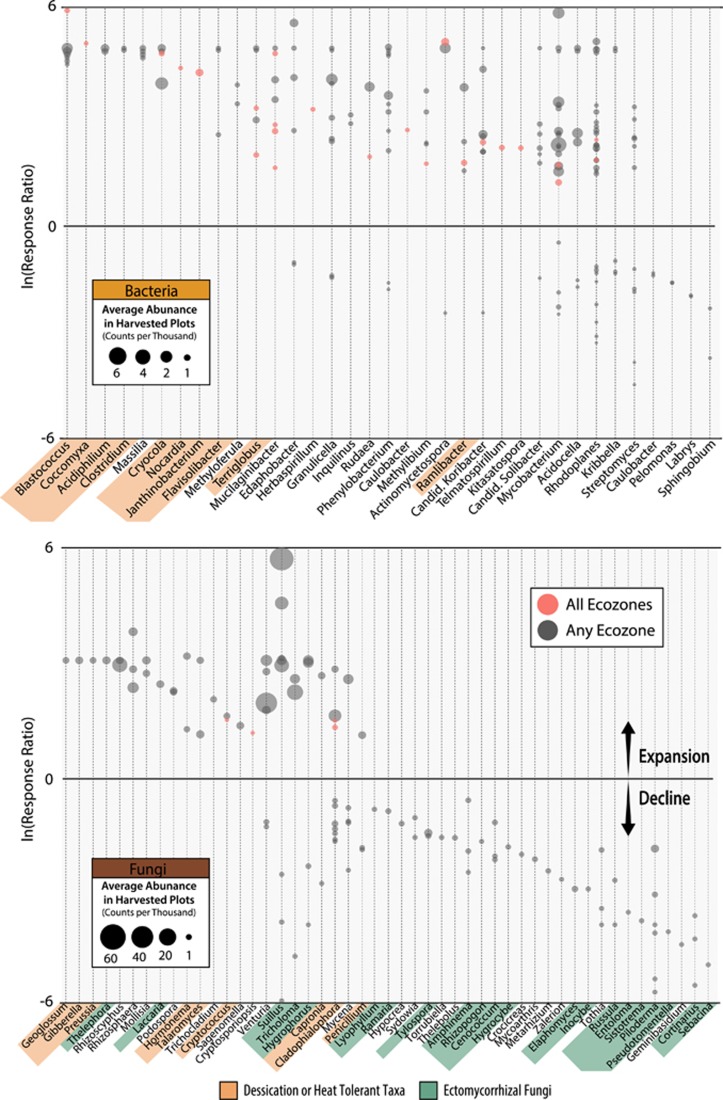
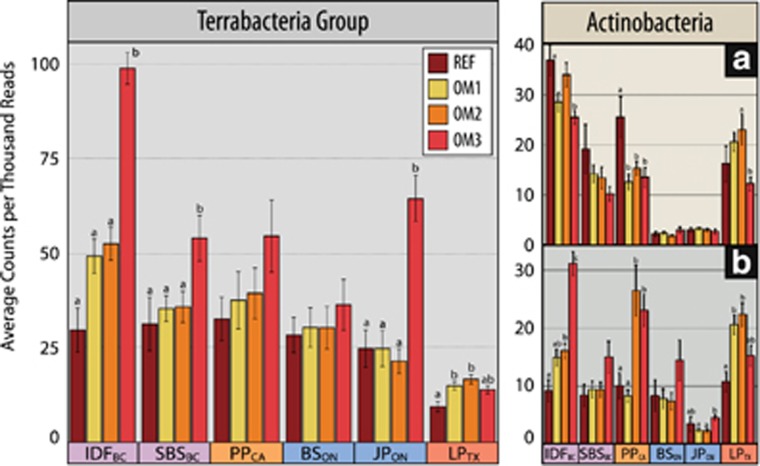
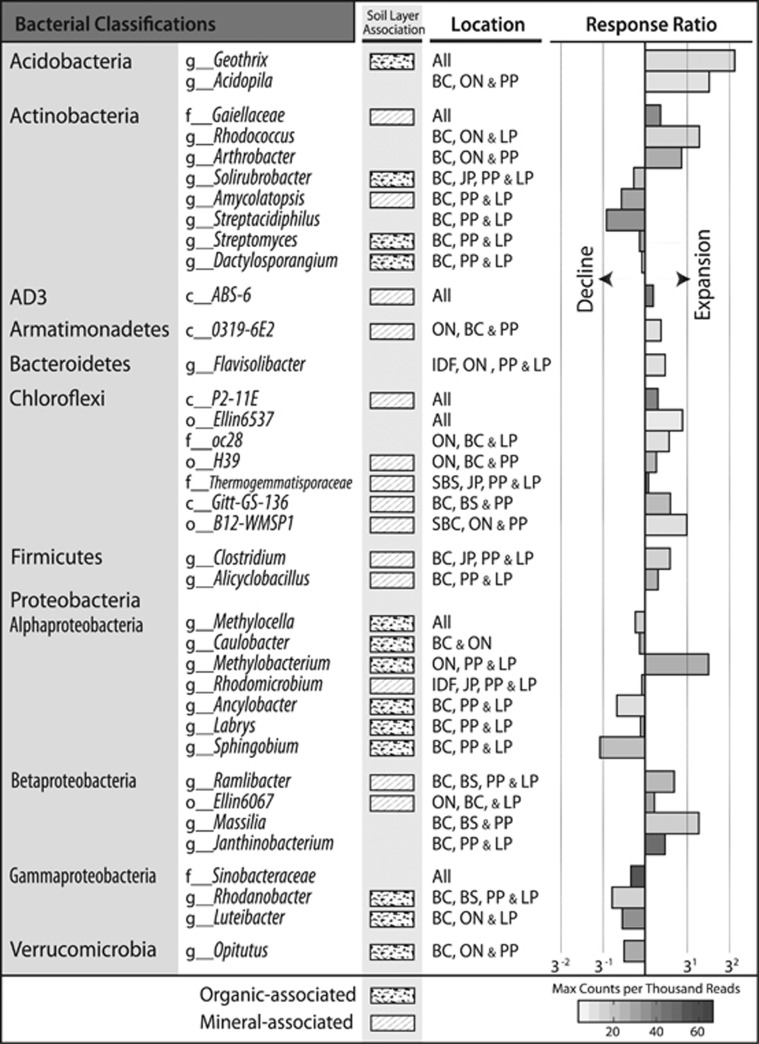
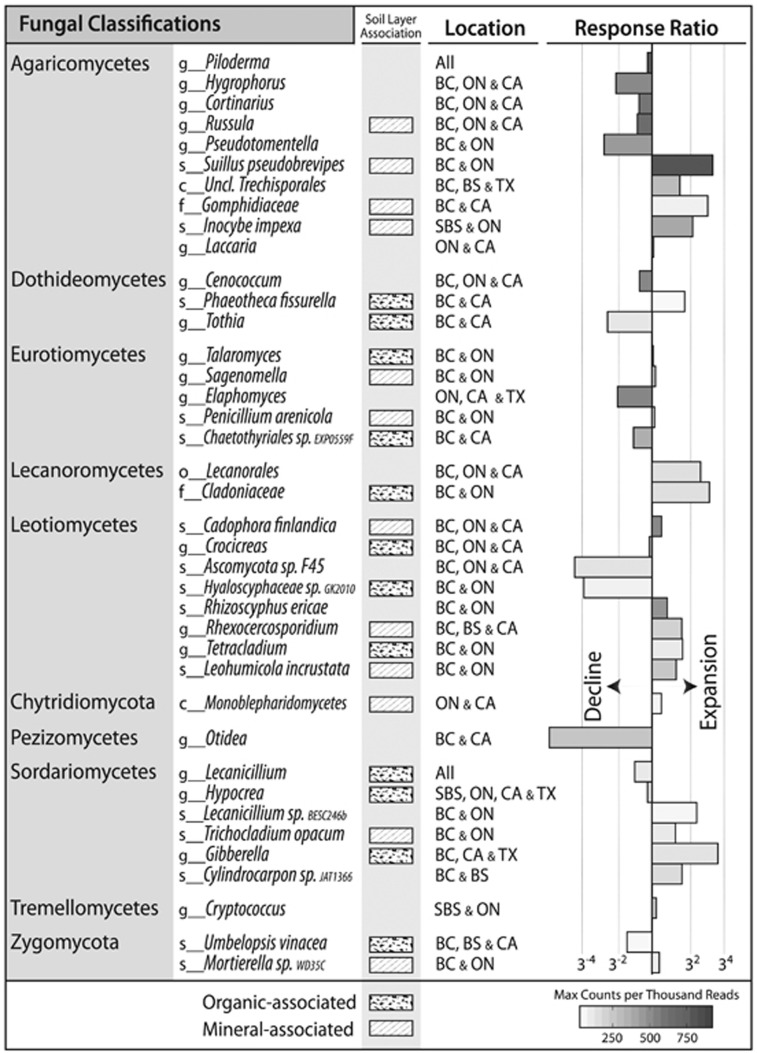
Three abundance patterns were apparent for both bacterial and fungal OTUs: (i) an increased relative abundance with increasing OM removal; (ii) a decline with increasing OM removal, and (iii) an increase at intermediate intensities. These three trends occurred statistically more often than expected by random chance (3σ or P<0.01), while all other permutations of abundance patterns occurred at less than or equal to random frequency. Similar abundance patterns were observed in clusters (or ‘modules’) of fungal OTUs in the co-association network (Supplementary Figure 8; modules 11, 12, 13 and 14). Bacterial networks had no apparent modularity. Indicator analysis provided the most detailed account of OTUs affected by harvesting (Figure 5). Among indicator OTUs showing the greatest increase in relative abundance following harvesting were members of a number of radiation, desiccation and heat-tolerant taxa, including the superphylum ‘Terrabacteria group’ (Battistuzzi and Hedges, 2009). Terrabacterial phyla (Actinobacteria, Armatimonadetes, Chloroflexi, Cyanobacteria and Firmicutes) were all more abundant in harvested treatments, particularly in OM3. However, some families within the Actinobacteria declined with increasing OM removal (Figure 6). There was also an expansion in harvested treatments of several stress-tolerant fungal taxa, including known pyrophilous fungi (Supplementary Figure 9), lichenized (Lecanorales) and lichenolous fungi, and melanized rock-inhabiting fungi, such as members of the genus Phaeotheca (Sterflinger, 2000), and desert-adapted taxa such as Talaromyces (Stolk, 1965), Hormonema (Burford et al., 2003) and Preussia (Rao et al., 2016). Populations of Glomeromycota, a phylum of arbuscular mycorrhiza, were substantially expanded only in harvested sites in BSON (undetected in REF and ~1% of total ITS reads in OM3). Indicators of harvested treatments that were common to four or more ecozones are summarized in Figure 7, while a complete list of all indicator taxa can be found in Supplementary Table 7.
Figure 5.
Bacterial (top) and fungal (bottom) genera containing indicator OTUs that expanded or declined in harvested plots relative to reference plots. The response ratio corresponds to the natural log of the average abundance in all three harvested treatments divided by the average abundance in REF. Genera (x-axis) are ordered from left to right by average abundance in harvested plots. Dots correspond to individual indicator OTUs with an indicator value >0.5 and padj <0.01. Dot area is scaled to total average counts. Red dots show which OTU were indicators across all ecozones. Orange background labels indicate genera with reported tolerance of radiation, desiccation and/or heat. Green background labels indicate genera containing ectomycorrhizal fungi.
Figure 6.
Total abundance of phyla from Terrabacteria (Actinobacteria, Armatimonadetes, Chloroflexi, Cyanobacteria and Firmicutes) according to ecozones and OM treatment. All terrabacterial phyla, except for Actinobacteria, increased with increasing OM removal and were therefore aggregated. Actinobacteria were plotted separately. The following actinobacterial families exhibited decreased with increasing OM removal (summed in a): Actinospicaceae, Micromonosporaceae, Solirubrobacteraceae, Streptosporangiaceae, Thermomonosporaceae, and Streptomycetaceae. While, the following actinobacterial families had the opposite trend (summed in b): Gaiellaceae, Geodermatophilaceae and Micrococcaceae. Significant (Tukey HSD; padj<0.05) pairwise differences are grouped by lettering.
Figure 7.
A table of bacterial and fungal taxa that expanded or declined in response to harvesting in four or more ecozones. The lowest depth of classification supported by bootstrapping (>80) is provided and is prefaced by rank (c, class; o, order; f, family; g, genus; s, species). Mineral layer and organic layer-associations are noted by shaded squares. Response ratio barplots show the average abundance in all three harvested treatments divided by the average abundance in REF. Bars are shaded according to the maximum observed relative abundance of each taxon indicated in scale bar. Changes observed in both Ontario ecozones (BSON and JPON) were denoted by ‘ON’ and, similarly, effects observed in both British Columbian ecozones (IDFBC and SBSBC) were denoted by ‘BC’.
Fungal indicator OTUs were more likely to decline following harvesting than bacterial ones (odds ratio 4.5, P<0.001). Ectomycorrhizal (EM) fungi were highly abundant overall (~10–40% of total libraries) and EM indicator OTUs were more likely to decline following harvesting compared with indicators from saprotrophic groups (o.r. 7.1, P=0.002). The diversity of EM fungi decreased with increasing OM removal and was most pronounced in the organic layer (Supplementary Figure 10). A majority of EM genera declined in harvested plots, such as Russula, Cenoccocum, Cortinarius, Otidea, Piloderma, Hygrophorus and Pseudotomentella, while a minority consistently and substantially expanded in harvested plots, such as Suillus (3- to 6-fold increases), Thelephora, Tomentella and Wilcoxina. The most abundant bacterial taxa that declined following harvesting were Verrucomicrobia (Opitutus sp.), Gammaproteobacteria (unclassified Sinobacteraceae, Rhodanobacter and Luteibacter sp.), and Alphaproteobacteria (Rhodomicrobium and Ancylobacter) and the commonly methanotrophic genus Methylocapsa (Supplementary Figure 11), which was negatively correlated with pH (r=−0.39, P<0.001).
Twenty-four fungal taxa and three bacterial genera exhibited combined increases in both OM1 and OM2 (Supplementary Table 8). Their abundance patterns were less consistent across ecozones than taxa exhibiting expansion or decline with increasing OM removal. IDFBC and PPCA were the only two ecozones in which community composition in OM1 and OM2 significantly differed (Supplementary Table 6). Several genera designated as ‘wood saprotrophs’ by FUNGuild had higher relative abundance in OM1, and to some extent OM2, including Coniophora, Gymnopilus, Serpula, Perenniporia and Trechispora (Supplementary Figure 12). Other poorly classified members of saprotrophic groups such as several unclassified Agaricales OTUs (rC:N=0.30, P<0.001), Dermataceae (rC:N=0.36, P<0.001) and Agaricomycotina (n.s.) were also more abundant in OM1 (Supplementary Figure 12). One EM species, classified as Thelephora sp. ECM1, had consistently higher abundances in OM1 (except in LPTX; Supplementary Figure 12) and was positively correlated with the C:N ratio (rC:N=0.25, P<0.001).
Shotgun metagenomic analyses revealed a total of 128 KEGG orthologous genes were significantly affected by harvesting. Genes related to photosynthesis (n=15), porphyrin and chlorophyll metabolism (n=11), carotenoid biosynthesis (n=8) and bacterial anoxygenic photosynthesis (n=4), all increased in harvested treatments. The increase of xerophilic, endolithic Geodermatophilaceae in harvested plots was confirmed by read mapping to representative genomes (Supplementary Figure 13). Support for the decline in Methylocapsa populations in shotgun metagenomic libraries was equivocal. The estimated abundance of Methylocapsa acidophila, based on read mapping to a representative genome (NCBI accession: NZ_ATYA00000000.1), declined only in IDFBC and did not significantly differ between treatments (Supplementary Figure 11). The abundance of methane oxidases, pmoA and mmoX were also lower in IDFBC (at least in OM3); however, these patterns did not occur in other ecozones, and the abundances of pmoA and mmoX increased in harvested plots in several instances (Supplementary Figure 11).
Ecozone-specific impacts of OM removal
Variation in the effects of OM removal among ecozones was expected given the generally large geographic differences in community composition. LPTX shared the fewest common OTUs with other ecozones and the smallest number of common taxa indicative of harvesting. Common responses to harvesting were typical of proximal ecozones, such as the expansion of Nostoc in both British Columbian ecozones (Supplementary Figure 14), reflecting the similarity of environmental conditions (Figures 2a and b). Several taxa exhibited contrasting responses along a North-South axis, such as Terriglobus (bacterium) and Cladophialophora (fungus), which had declining populations in northern sites and the reverse pattern at southern sites (Figure 2c). Conversely, fungi from the order Boletales increased with greater OM removal in northern sites, but declined in southern sites. Northern and southern sites exhibited marked differences in soil temperature, precipitation and soil moisture. Similarly, several indicator taxa were exclusive to eastern (Cupriavidus and candidate phylum GAL15) or western ecozones (Limnohabitans, Nostoc and AKIW781, an order of Chloroflexi), while other taxa exhibited contrasting responses between eastern and western ecozones, such as Rudaea, Kitasatospora and members of Atheliales (Supplementary Figure 14).
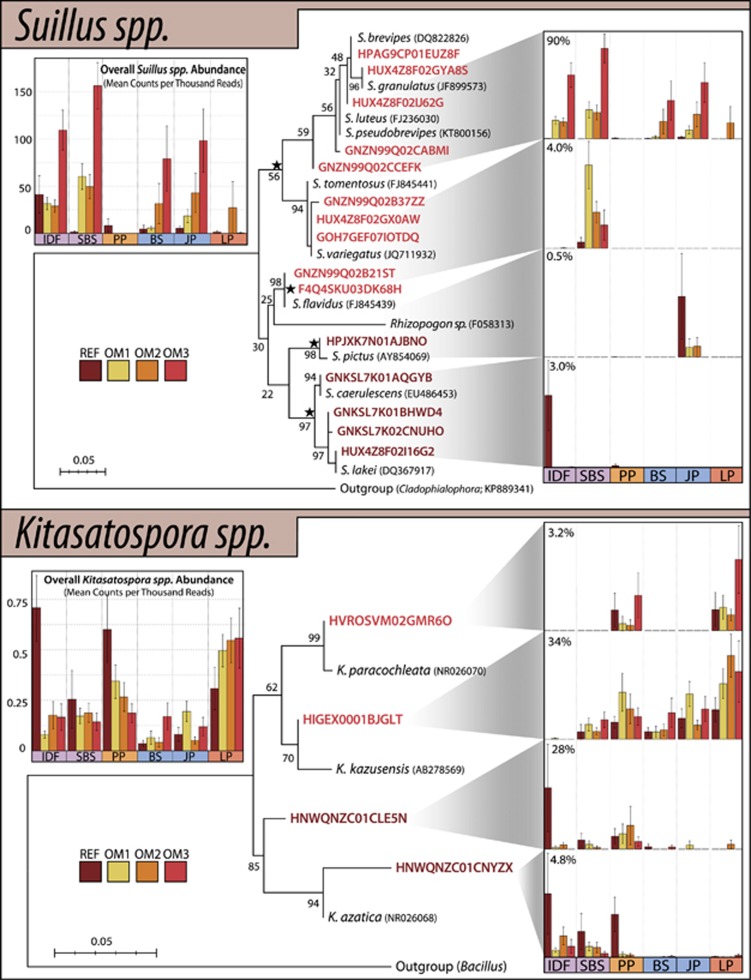
There were several cases where OTUs within the same genus exhibited opposite responses to harvesting and a subset of these could be attributed to phylogenetic differences in closely related species. In the EM genus Suillus, OTUs that showed substantially increased relative abundance belonged to separate clades from those which declined in abundance (Figure 8). Rhizopogon species also exhibited similar species-level differences in response to harvesting (Supplementary Figure 15) along with the bacterial genus, Kitasatospora (Figure 8). EM genera within the same family, Tomentella and Pseudotomentella, exhibited divergent responses to harvesting (Supplementary Figure 16). However, the majority of these OTU-level contrasting responses did not correspond to phylogeny.
Figure 8.
Maximum-likelihood phylogenetic trees for predominant OTUs from the fungal genus, Suillus, and bacterial genus, Kitasatospora, accompanied by their abundances in harvested treatments. OTUs names are colored according to whether they exhibited expanding (red) or declining (brown) relative abundances in harvested treatments. Barplots on the left show the sum of counts for each genus, while barplots on the right show the abundances of individual clades (marked with stars). For simplification, the Y-axis on the right-side plots corresponds to percent abundance of each clade in its respective genus. Overall, Suillus spp. accounted for an average of 5% of ITS reads per library, while Kitasatospora spp. accounted for an average of 0.05% of 16 S reads per library. Aligned sequences were trimmed to 355 bp (Suillus) and 250 bp (Kitasatospora) before tree building.
Discussion
Over a decade after LTSP sites were harvested, we observed gradients in soil conditions that corresponded to initial levels of OM removal. Yet, OM removal had a relatively small effect on beta-diversity, accounting for far less variability than occurred among ecozones and, within most ecozones, did not account for much more variability than occurred among sites. The OM removal treatments had no clear effect on populations of the most abundant and cosmopolitan taxa, like members of the highly abundant Bradyrhizobiaceae, reported here and elsewhere (VanInsberghe et al., 2015). Despite the minimal effects on overall community composition, OM removal treatments did have significant effects on a variety of populations of which EM fungi and stress-tolerant taxa were most clearly impacted. For most of those taxa, the effects were observed in OM1 and/or OM2, indicating the effects were the result of harvesting and not solely due to the removal of the forest floor (OM3). The magnitude of changes in community composition due to harvesting was comparable among ecozones, though PPCA and LPTX, exhibited the most and least pronounced changes, respectively. The extent of ecozone-specific responses to harvesting demonstrates the need for microbial assessments at regional scales, though the many cross-ecozone effects amount to potential indicators for monitoring and evaluating microbial succession in long-term managed forests.
The consistent increase in desiccation- and heat-tolerant taxa in both soil layers reflected the warmer, drier conditions in harvested plots. While harvesting was sufficient to produce this effect, the removal of the forest floor had a strong impact, demonstrating the importance of the forest floor in shading and insulating underlying soil. The changes we observed in stress-tolerant taxa resembled reported long-term changes following forest fire, indicative of comparable post-disturbance environmental regimes. The expansion of terrabacterial phyla has been observed following forest fire (Tas et al., 2014; Xiang et al., 2014) and in other exposed soil environments such as glacial forefelds (Rime et al., 2015). The decline in relative abundance of Basidiomycota, predominantly EM fungi, in favor of Ascomycota was also consistent with long-term changes in fire-affected soils (Holden et al., 2013b; Buscardo et al., 2015). This pattern likely reflects the fact that most thermo-tolerant fungi characterized to date belong to Ascomycota (Morgenstern et al., 2012), including several pyrophilous taxa. The absence of significant trends in alpha- and beta-diversity is also in agreement with studies of wildfire-affected soils, suggesting both disturbances produce relatively minor restructuring of communities (Weber et al., 2014; Oliver et al., 2015). Overall, these results suggest that the forestry strategy of emulating natural disturbance, particularly wildfire (Long, 2009; Stockdale et al., 2016), extends to belowground communities, though a more direct comparison is needed.
EM fungi were the most prominent trophic guild of fungi, constituting between 10 and 40% of total ITS libraries, and their overall decline was a major inter-ecozone effect of harvesting. In contrast, arbuscular mycorrhiza populations increased in harvested plots likely due to their common symbioses with successional plant cover, as observed post-wildfire (Xiang et al., 2015). The decline of EM communities in SBSBC and IDFBC was previously reported by Hartmann et al., (2012), and, here, we have shown it to be a general response across distinct ecozones. The subset of EM genera, including Rhizopogon, Suillus and Thelephora, that expanded in most ecozones following harvesting have all been reported to thrive in the early-stages of forest succession at local scales (Simard, 2009; Buscardo et al., 2015; Glassman et al., 2015; Oliver et al., 2015). Interestingly, the predominance of Suillus may partly result from their recruitment by young trees to fulfill nutritional needs, given the nitrogen fixing activity present in S. tomentosus tubers (Kranabetter, 2004; Paul et al., 2012). Our results provide new evidence that the expansion of these taxa is a common feature of early-stage plantations in diverse North American forests and that their dominance lasts, at least, 17 years post harvest.
Harvesting produced changes in the relative abundance of several bacterial groups involved in soil processes important to forest ecosystems. These included declining populations of Verrucomicrobia (Spartobacteria and Opitutus) and Streptomycetaceae, previously identified as major cellulolytic taxa affected by harvesting (Wilhelm et al., 2017b) and by forest fire (Tas et al., 2014; Weber et al., 2014). Forest floor removal produced significant expansion in Syntrophobacteraceae, a group of thermophilic sulfate reducers (Kuever, 2014), while harvesting, in general, produced a near universal expansion of metal-reducing bacteria from genus Geothrix. These trends suggest soil conditions may favor anaerobic respiration, perhaps due to soil compaction from even light machinery. Populations of Methylocapsa also declined following harvesting in most ecozones, which was previously linked to a reduction in atmospheric methane oxidation in soils (Nazaries et al., 2011). However, we could not establish a link between changes in community structure and function, since the changes in Methylocapsa abundance did not correspond to a decrease in relative abundance of pmoA genes. The decline in Methylocapsa may be due to altered environmental conditions in the organic layer where their populations were most abundant and negatively correlated with pH.
The retention of coarse woody debris (OM1 vs OM2) did not produce any major restructuring of the microbial community over the long-term. Across ecozones, OM1 and OM2 shared the greatest proportion of overlapping OTUs and were undifferentiated in most analyses, extending the findings of Hartmann et al. (2012). Yet, there were noteworthy exceptions that suggest retention may yield marginal benefits. For example, the only ecozones where overall communities significantly differed between OM1 and OM2 (IDFBC and PPCA) were the same ecozones where soil carbon content was significantly higher in OM1 relative to OM2. Furthermore, fungal communities were most diverse in OM1 compared with any treatment, and, as hypothesized, OM1 selected for certain decomposer populations, like wood-rot fungi. Many differentially abundant taxa between OM1 and OM2 were identified, but these differences tended to occur in only one or a few ecozones. Inconsistency among ecozones may partly reflect the local communities, such as the adaptation of endemic taxa to local sources of OM (Ayres et al., 2009; Freschet et al., 2012). Yet, most of the differentially abundant taxa are unclassified and poorly characterized, so it is premature to draw conclusions about them. Overall, these observations indicate that woody debris retention has a minimal impact on the overall soil community in diverse ecozones. In contrast, OM retention had a clear effect in terms of buffering abiotic changes, moderating the expansion and decline of certain populations.
Removal of forest floor (OM3) had the greatest impact on the soil community by nearly all measures. This extreme treatment was not intended to test a forest management practice, but rather to determine the consequences of maximum OM removal. The inclusion of OM3 made for a starker increase in the expansion of stress-tolerant populations in all harvested sites, a phenomenon which has yet been described in the forest disturbance literature. Despite the severity of this treatment, and its major effects on individual populations, even OM3 did not have a great impact on overall community composition. This suggests that the community is largely resilient to perturbation, though future work is required to determine whether greater changes manifest as forest plantations mature.
The ecozone-specific impacts of harvesting on microbial communities is consistent with the variability in forest productivity reported in other LTSP publications (Fleming et al., 2006; Thiffault et al., 2011; Ponder et al., 2012; Holub et al., 2013). Ecozone-specific impacts were mainly attributable to the irregular distribution of taxa, which was not unexpected given the large differences among ecozones in climate, edaphic factors, and plant communities. Notably, some ecozone-specific effects could be attributed to divergent responses of closely related species, such as those within Kitasatospora, Rhizopogon and Suillus. Substantial differences were observed even between proximal ecozones that shared similar climates and plant communities, such as the expansion in populations of candidate phyla AD3 and GAL in JPON, but not in BSON. The trends in certain microbial populations along north-south and east-west axes demonstrated the effects of biogeography and differences in local conditions brought about by harvesting. The expansion of Boletales in harvested plots in northern ecozones exemplifies how soil warming and drying in these cooler climates may foster growth, while post-harvesting conditions in already hot and dry climates exceeded tolerance thresholds leading to their decline in southern soils. Integrating microbial ecology into forest management practices must, therefore, be evaluated at regional scales, despite the occurrence of general trends. This will be particularly necessary for management considerations pertaining to fungal communities which exhibit greater endemicity at the continental scale than bacteria (Talbot et al., 2014).
Populations that were consistently affected by harvesting across all other ecozones were unchanged between harvested and references plots in Texas (LPTX). The uniquely rapid rate of forest regeneration and land-use legacy at LPTX sites provide interesting context for a possible explanation. The rate of regeneration at LPTX was nearly twice the rate of other ecozones (~25-year reforestation cycles) and this rapid return to environmental conditions resembling a pre-harvest state may hasten the recovery of microbial communities. Additionally, the LPTX reference plots were previously harvested (~75 years before the LTSP installation), and, historically have been managed by humans with fire. This history of disturbance may contribute to the similarity of soil communities between unharvested and harvested plots. As such, LPTX sites likely represent a case study on the resilience of soil communities to repeated disturbances and long-term management practices, though more longitudinal evidence is clearly necessary.
This study offers insight into some of the belowground ecological changes that take place in the decades after timber harvesting, providing a long list of indicators in bacterial and fungal communities. Changes resulting from harvesting were relatively minor in comparison to the variability between soil layers and among geographic regions. However, the relative magnitude of ostensibly unaffected populations may be inflated due to substantial presence of relic DNA in soil (Carini et al., 2016). If these populations prove to be metabolically active and are truly unaffected by harvesting, their resilience would serve as a compelling area of future research. Overall, it remains to be seen whether the changes we observed in microbial populations persist as the forests mature, and whether changes compound over repeated cycles of harvest and regeneration. Our efforts to identify specific taxa affected by harvesting will enable researchers to track populations over time to answer long-standing questions about the impacts of forest disturbance and various management practices. Yet, at present, our understanding of the contributions of soil communities to forest productivity and succession is limited. This limited understanding was highlighted in our study by the increased proportion of unclassifiable OTUs in harvested plots. The full extent of the impact of harvesting on soil communities cannot be known until those knowledge gaps are filled.
Acknowledgments
This study was supported by grants from Genome Canada and Genome BC. We thank the US Forestry Service, Natural Resources Canada, the Canadian Forest Service, Ontario Ministry of Natural Resources and the British Columbia Ministry of Forests, Lands and Natural Resources for their access to the LTSP sites and logistical support. E Cardenas was supported by a postdoctoral fellowship from the Tula Foundation and R. Wilhelm by an NSERC graduate scholarship. Sequencing conducted by the US Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the US Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Achat D, Deleuze C, Landmann G, Pousse N, Ranger J, Augusto L. (2015). Quantifying consequences of removing harvesting residues on forest soils and tree growth – a meta-analysis. Forest Ecol Manag 348: 124–141. [Google Scholar]
- Adams PW, Flint AL, Fredriksen RL. (1991). Long-term patterns in soil moisture and revegetation after a clearcut of a Douglas-fir forest in Oregon. Forest Ecol Manag 41: 249–263. [Google Scholar]
- Allmér J, Stenlid J, Dahlberg A. (2009). Logging-residue extraction does not reduce the diversity of litter-layer saprotrophic fungi in three Swedish coniferous stands after 25 years. Can J Forest Res 39: 1737–1748. [Google Scholar]
- Amann RI, Ludwig W, Schleifer KH. (1995). Phylogenetic identification and in situ detection of individual microbial-cells without cultivation. Microbiol Rev 59: 143–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres E, Steltzer H, Berg S, Wall DH. (2009). Soil biota accelerate decomposition in high‐elevation forests by specializing in the breakdown of litter produced by the plant species above them. J Ecol 97: 901–912. [Google Scholar]
- Bååth E, Frostegård Å, Pennanen T, Fritze H. (1995). Microbial community structure and pH response in relation to soil organic matter quality in wood-ash fertilized, clear-cut or burned coniferous forest soils. Soil Biol Biochem 27: 229–240. [Google Scholar]
- Bader P, Jansson S, Jonsson B. (1995). Wood-inhabiting fungi and substratum decline in selectively logged boreal spruce forests. Biol Conserv 72: 355–362. [Google Scholar]
- Bastian M, Heymann S, Jacomy M. (2009). Gephi: an open source software for exploring and manipulating networks. ICWSM 8: 361–362. [Google Scholar]
- Battistuzzi FU, Hedges SB. (2009). A major clade of prokaryotes with ancient adaptations to life on land. Mol Biol Evol 26: 335–343. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Met 57: 289–300. [Google Scholar]
- Brazee NJ, Lindner DL, D’Amato AW, Fraver S, Forrester JA, Mladenoff DJ. (2014). Disturbance and diversity of wood-inhabiting fungi: effects of canopy gaps and downed woody debris. Biodivers Conserv 23: 2155–2172. [Google Scholar]
- Burford EP, Kierans M, Gadd GM. (2003). Geomycology: fungi in mineral substrata. Mycologist 17: 98–107. [Google Scholar]
- Buscardo E, Rodríguez-Echeverría S, Freitas H, De Angelis P, Pereira JS, Muller LA. (2015). Contrasting soil fungal communities in Mediterranean pine forests subjected to different wildfire frequencies. Fungal Divers 70: 85–99. [Google Scholar]
- Cardenas E, Kranabetter JM, Hope G, Maas KR, Hallam S, Mohn WW. (2015). Forest harvesting reduces the soil metagenomic potential for biomass decomposition. ISME J 9: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carini P, Marsden PJ, Leff JW, Morgan EE, Strickland MS, Fierer N. (2016). Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat Microbiol 2: 16242. [DOI] [PubMed] [Google Scholar]
- Castro MS, Gholz HL, Clark KL, Steudler PA. (2000). Effects of forest harvesting on soil methane fluxes in Florida slash pine plantations. Can J Forest Res 30: 1534–1542. [Google Scholar]
- Chasalow S. (2012), combinat: combinatorics utilities. R package version 0.0-8. http://CRAN.R-project.org/package=combinat.
- Childs S, Flint L. (1987). Effect of shadecards, shelterwoods, and clearcuts on temperature and moisture environments. Forest Ecol Manag 18: 205–217. [Google Scholar]
- De Cáceres M, Legendre P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90: 3566–3574. [DOI] [PubMed] [Google Scholar]
- Entry JA, Stark NM, Loewenstein H. (1986). Effect of timber harvesting on microbial biomass fluxes in a northern Rocky Mountain forest soil. Can J Forest Res 16: 1076–1081. [Google Scholar]
- Fleming RL, Powers RF, Foster NW, Kranabetter M, Scott AD, Ponder F et al. (2006). Effects of organic matter removal, soil compaction, and vegetation control on 5-year seedling performance: a regional comparison of long-term soil productivity sites. Can J Forest Res 36: 529–550. [Google Scholar]
- Fox TR. (2000). Sustained productivity in intensively managed forest plantations. Forest Ecol Manag 138: 187–202. [Google Scholar]
- Freschet GT, Aerts R, Cornelissen JH. (2012). Multiple mechanisms for trait effects on litter decomposition: moving beyond home‐field advantage with a new hypothesis. J Ecol 100: 619–630. [Google Scholar]
- Friedman J, Alm EJ. (2012). Inferring correlation networks from genomic survey data. PLoS Comput Biol 8.9: e1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassman SI, Levine CR, DiRocco AM, Battles JJ, Bruns TD. (2015). Ectomycorrhizal fungal spore bank recovery after a severe forest fire: some like it hot. ISME 10: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson L, Baker SC, Bauhus J, Beese WJ, Brodie A, Kouki J et al. (2012). Retention forestry to maintain multifunctional forests: a world perspective. BioScience 62: 633–645. [Google Scholar]
- Harrell FE, Dupont C. (2015). Hmisc: Harrell Miscellaneous. R package version 3.17-1. https://CRAN.R-project.org/package=Hmisc.
- Hartmann M, Howes CG, VanInsberghe D, Yu H, Bachar D, Christen R et al. (2012). Significant and persistent impact of timber harvesting on soil microbial communities in Northern coniferous forests. ISME J 6: 2199–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden SR, Treseder KK. (2013. a). A meta-analysis of soil microbial biomass responses to forest disturbances. Front Microbiol 4: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden SR, Gutierrez A, Treseder KK. (2013. b). Changes in soil fungal communities, extracellular enzyme activities, and litter decomposition across a fire chronosequence in Alaskan boreal forests. Ecosystems 16: 34–46. [Google Scholar]
- Holub SM, Terry TA, Harrington CA, Harrison RB, Meade R. (2013). Tree growth ten years after residual biomass removal, soil compaction, tillage, and competing vegetation control in a highly-productive Douglas-fir plantation. Forest Ecol Manag 305: 60–66. [Google Scholar]
- Kuever J. (2014) The family Syntrophobacteraceae. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (eds). The Prokaryotes. Springer: Berlin, pp 289–299. [Google Scholar]
- Kranabetter J, Chapman B. (1999). Effects of forest soil compaction and organic matter removal on leaf litter decomposition in central British Columbia. Can J Soil Sci 79: 543–550. [Google Scholar]
- Kranabetter JM. (2004). Ectomycorrhizal community effects on hybrid spruce seedling growth and nutrition in clearcuts. Can J Bot 82: 983–991. [Google Scholar]
- Kulmala L, Aaltonen H, Berninger F, Kieloaho AJ, Levula J, Bäck J et al. (2014). Changes in biogeochemistry and carbon fluxes in a boreal forest after the clear-cutting and partial burning of slash. Agr Forest Meteorol 188: 33–44. [Google Scholar]
- Lane DJ. (1991) 16 S/23 S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds). Nucleic Acid Techniques in Bacterial Systematics. John Wiley and Sons: Chichester, NY, pp 115–175. [Google Scholar]
- Leung HT, Maas KR, Wilhelm RC, Mohn WW. (2016). Long-term effects of timber harvesting on hemicellulolytic microbial populations in coniferous forest soils. ISME J 10: 363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J. (2009). Emulating natural disturbance regimes as a basis for forest management: a North American view. Forest Ecol Manag 257: 1868–1873. [Google Scholar]
- Markowitz VM, Ivanova NN, Szeto E, Szeto E, Palaniappan K, Chu K et al. (2008). IMG/M: a data management and analysis system for metagenomes. Nucl Acids Res 36: D534–D538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire KL, D’Angelo H, Brearley FQ, Gedallovich SM, Babar N, Yang N et al. (2015). Responses of soil fungi to logging and oil palm agriculture in Southeast Asian tropical forests. Microb Ecol 69: 733–747. [DOI] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. (2014). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8: e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern I, Powlowski J, Ishmael N, Darmond C, Marqueteau S, Moisan MC et al. (2012). A molecular phylogeny of thermophilic fungi. Fungal Biol 116: 489–502. [DOI] [PubMed] [Google Scholar]
- Nazaries L, Tate KR, Ross DJ, Singh J, Dando J, Saggar S et al. (2011). Response of methanotrophic communities to afforestation and reforestation in New Zealand. ISME J 5: 1832–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J et al. (2015). FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20: 241–248. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB et al. (2015), vegan: Community Ecology Package. R package version 2.3-1 http://CRAN.R-project.org/package=vegan.
- Oliver AK, Callaham MA, Jumpponen A. (2015). Soil fungal communities respond compositionally to recurring frequent prescribed burning in a managed southeastern US forest ecosystem. Forest Ecol Manag 345: 1–9. [Google Scholar]
- Paul LR, Chapman WK, Chanway CP. (2012). Diazotrophic bacteria reside inside Suillus tomentosus / Pinus contorta tuberculate ectomycorrhizae. Botany 91: 48–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz LW. (2001) Soil-water Characteristics and Hydrologic Implications Following Forest Soil Disturbance: The Relative Influence of Organic Residue and Soil Compaction on Permeability and Moisture Capacity: a Study on Cohasset Soil in the Sierra Nevada Mixed Conifer Zone. University of California: Berkeley. [Google Scholar]
- Ponder F, Fleming RL, Berch S, Busse M, Elioff JD, Hazlett PW et al. (2012). Effects of organic matter removal, soil compaction and vegetation control on 10th year biomass and foliar nutrition: LTSP continent-wide comparisons. Forest Ecol Manag 278: 35–54. [Google Scholar]
- Powers RF, Scott AD, Sanchez FG, Voldseth RA, Page-Dumroese D, Elioff JD et al. (2005). The North American long-term soil productivity experiment: findings from the first decade of research. Forest Ecol Manag 220: 31–50. [Google Scholar]
- Powers RF. (2006). Long-Term Soil Productivity: genesis of the concept and principles behind the program. Can J Forest Res 36: 519–528. [Google Scholar]
- R Core Team. (2008). R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria. Available from http://www.R-project.org.
- Rao S, Chan Y, Bugler-Lacap DC, Bhatnagar A, Bhatnagar M, Pointing SB. (2016). Microbial diversity in soil, sand dune and rock substrates of the Thar Monsoon Desert. India. Indian J Microbiol 56: 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding T, Hope G, Fortin MJ, Schmidt M, Bailey W. (2003). Spatial patterns of soil temperature and moisture across subalpine forest-clearcut edges in the southern interior of British Columbia. Can J Soil Sci 83: 121–130. [Google Scholar]
- Rime T, Hartmann M, Brunner I, Widmer F, Zeyer J, Frey B. (2015). Vertical distribution of the soil microbiota along a successional gradient in a glacier forefield. Mol Ecol 24: 1091–1108. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard SW. (2009) Response diversity of ectomycorrhizas in forest succession following disturbance. In: Azcón-Aguilar C, Barea JM, Gianinazzi S, Gianinazzi-Pearson V (eds). Mycorrhizas—Functional Processes and Ecological Impact. Springer: Heidelberg, Germany, pp 1–19. [Google Scholar]
- Sterflinger K. (2000). Fungi as Geologic Agents. Geomicrobiol J 17: 97–124. [Google Scholar]
- Stockdale C, Flannigan M, Macdonald E. (2016). Is the END (emulation of natural disturbance) a new beginning? A critical analysis of the use of fire regimes as the basis of forest ecosystem management with examples from the Canadian western Cordillera. Environ Rev 24: 233–243. [Google Scholar]
- Stolk AC. (1965). Thermophilic species of Talaromyces benjamin and Thermoascus miehe. A Van Leeuw J Microb 31: 262–276. [DOI] [PubMed] [Google Scholar]
- Storey JD, Bass AJ, Dabney A, Robinson D. (2015). qvalue: Q-value estimation for false discovery rate control. R package version 2.2.2.
- Takakai F, Desyatkin AR, Lopez CL, Fedorov AN, Desyatkin RV, Hatano R. (2008). Influence of forest disturbance on CO2, CH4 and N2O fluxes from larch forest soil in the permafrost taiga region of eastern Siberia. Soil Sci Plant Nutr 54: 938–949. [Google Scholar]
- Talbot JM, Bruns TD, Taylor JW, Smith DP, Branco S, Glassman SI et al. (2014). Endemism and functional convergence across the North American soil mycobiome. PNAS 111: 6341–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson P, Filipski A, Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tas N, Prestat E, McFarland JW, Wickland KP, Knight R, Berhe AA et al. (2014). Impact of fire on active layer and permafrost microbial communities and metagenomes in an upland Alaskan boreal forest. ISME J 8: 1904–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiffault E, Hannam KD, Paré D, Titus BD, Hazlett PW, Maynard DG et al. (2011). Effects of forest biomass harvesting on soil productivity in boreal and temperate forests—A review. Environ Rev 19: 278–309. [Google Scholar]
- VanInsberghe D, Maas KR, Cardenas E, Strachan CR, Hallam S, Mohn WW. (2015). Non-symbiotic Bradyrhizobium ecotypes dominate North American forest soils. ISME J 9: 2435–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CF, Lockhart JS, Charaska E, Aho K, Lohse KA. (2014). Bacterial composition of soils in ponderosa pine and mixed conifer forests exposed to different wildfire burn severity. Soil Biol Biochem 69: 242–250. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA for phylogenetics. In: Innis MA, Gelfand DH, Sninsko JJ, White TJ (eds). PCR Protocols: a Guide to Methods and Applications. Academic Press: San Diego, pp 315–322. [Google Scholar]
- Wickham H. (2007). Reshaping data with the reshape package. J Stat Soft 21: 1–20. [Google Scholar]
- Wickham H. (2011). The split-apply-combine strategy for data analysis. J Stat Soft 40: 1–29. [Google Scholar]
- Wickham H. (2009) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag: New York, 2009. [Google Scholar]
- Wilhelm RC, Cardenas E, Leung H, Maas K, Hartmann M, Hahn A et al. (2017. a). A metagenomic survey of forest soil microbial communities more than a decade after timber harvesting. Sci Data (in press; doi:10.1038/sdata.2017.92. [DOI] [PMC free article] [PubMed]
- Wilhelm RC, Cardenas E, Leung H, Szeitz A, Jensen LD, Mohn WW. (2017. b). Long-Term Enrichment of Stress-Tolerant Cellulolytic Soil Populations following Timber Harvesting Evidenced by Multi-Omic Stable Isotope Probing. Front Microbiol 8: 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Brüggemann N, Gasche R, Papen H, Willibald G, Butterbach-Bahl K. (2011). Long-term effects of clear-cutting and selective cutting on soil methane fluxes in a temperate spruce forest in southern Germany. Environ Pollut 159: 2467–2475. [DOI] [PubMed] [Google Scholar]
- Xiang X, Gibbons SM, Yang J, Kong J, Sun R, Chu H. (2015). Arbuscular mycorrhizal fungal communities show low resistance and high resilience to wildfire disturbance. Plant Soil 397: 347–356. [Google Scholar]
- Xiang X, Shi Y, Yang J, Kong J, Lin X, Zhang H et al. (2014). Rapid recovery of soil bacterial communities after wildfire in a Chinese boreal forest. Sci Rep 4: 3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerva A, Mencuccini M. (2005). Short-term effects of clearfelling on soil CO2, CH4, and N2O fluxes in a Sitka spruce plantation. Soil Biol Biochem 37: 2025–2036. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.