Abstract
The release of 700 million liters of oil into the Gulf of Mexico over a few months in 2010 produced dramatic changes in the microbial ecology of the water and sediment. Here, we reconstructed the genomes of 57 widespread uncultivated bacteria from post-spill deep-sea sediments, and recovered their gene expression pattern across the seafloor. These genomes comprised a common collection of bacteria that were enriched in heavily affected sediments around the wellhead. Although rare in distal sediments, some members were still detectable at sites up to 60 km away. Many of these genomes exhibited phylogenetic clustering indicative of common trait selection by the environment, and within half we identified 264 genes associated with hydrocarbon degradation. Alkane degradation ability was near ubiquitous among candidate hydrocarbon degraders, whereas just three harbored elaborate gene inventories for the degradation of alkanes and aromatic and polycyclic aromatic hydrocarbons (PAHs). Differential gene expression profiles revealed a spill-promoted microbial sulfur cycle alongside gene upregulation associated with PAH degradation. Gene expression associated with alkane degradation was widespread, although active alkane degrader identities changed along the pollution gradient. Analyses suggest that a broad metabolic capacity to respond to oil inputs exists across a large array of usually rare indigenous deep-sea bacteria.
Introduction
Marine oil spills are frequent occurrences that can have a severe impact on environmental health and dependent economies (Barron, 2012). In US marine environments alone, hundreds of oil spills occur annually, releasing millions of liters of oil per year on average (USCG, 2012). Although spills have typically occurred at shallow water depths, an expansion of drilling into the deep-sea led to the 2010 Deepwater Horizon (DWH) accident in the Gulf of Mexico (Peterson et al., 2012). DWH released over 700 million liters of oil from the Macondo MC252 wellhead at 1500 m depth (Atlas and Hazen, 2011). This yielded a vast sea surface oil slick (MacDonald et al., 2015), and an expansive plume of hydrocarbons at a water column depth of ~1100 m (Camilli et al., 2010). The leak also polluted deep-sea sediments up to tens of kilometers distance, owing to direct contamination and flocculent fallout from plumes (Valentine et al., 2014). Pollution of seafloor sediments persisted at least 3 months post spill (Zukunft, 2010), likely supported by ongoing inputs from sinking hydrocarbon-bearing particles (Yan et al., 2016). Post spill contamination of sediment was greater than in the water column and was greatest within 3 km of MC252, where total polyaromatic hydrocarbon (PAH) concentrations remained above the Environmental Protection Agency’s (EPA) Aquatic Life benchmark (Zukunft, 2010; Mason et al., 2014b).
Indigenous marine microorganisms are important ecosystem engineers capable of breaking down the complex mixtures of hydrocarbons in leaked oil (Pritchard et al., 1992; Brooijmans et al., 2009). Extensive degradation of hydrocarbons released during the DWH spill is largely attributed to microbial activity (Joye et al., 2014). Gammaproteobacteria—some closely related to known psychrotolerant or psychrophilic bacteria—dominated the deep-sea response (Hazen et al., 2010; Redmond and Valentine, 2012; Mason et al., 2014b), contrasting with higher alphaproteobacterial ratios in shallow environments (King et al., 2013). Genes or transcripts associated with hydrocarbon degradation (HCD) were found to be enriched or expressed in the plume (Mason et al., 2012; Rivers et al., 2013), polluted deep-sea sediments (Mason et al., 2014b) and beach sands (Lamendella et al., 2014; Rodriguez et al., 2015). In the deep-sea, the buoyant plume received far greater attention than the underlying seafloor sediments (Joye et al., 2014), although the sediments were observed to share some key taxa with the plume (a bacterium associated with the hydrocarbonoclastic genus Colwellia and an abundant undescribed gammaproteobacterium) (Mason et al., 2014b). In the plume, HCD during the spill (May–June) was linked to the single cell genomes of two alkane-degrading bacteria (Colwellia and a transcriptionally active Oceanospirillales) (Mason et al., 2012, 2014a), and indirectly to a small number of other transcriptionally active methane and alkane-degrading bacteria (Mason et al., 2012; Rivers et al., 2013). Plume-based gene expression levels for aromatic hydrocarbons and PAHs at this time were found to be low (Mason et al., 2012). However, the genetic capacity of several plume bacteria to degrade two important petroleum PAHs was demonstrated ex situ with the support of 13C substrate labeling (Dombrowski et al., 2016), and the PAH-degrading genus, Cycloclasticus, was enriched in the plume up until mid-August (Dubinsky et al., 2013). Seafloor sediments were collected at a later post spill date (September–October). They were enriched in genes associated with pathways for denitrification and the degradation of various aliphatic hydrocarbons and PAHs (that is, cyclohexane, benzene-toluene-ethylbenzene-xylene, biphenyl and undefined alkanes) (Kimes et al., 2013; Scott et al., 2014; Mason et al., 2014b), suggesting persistent seafloor pollution supported a well-developed community of hydrocarbon degraders. Nevertheless, the organism-specific metabolic response of seafloor communities to the spill has not been directly explored, including the hydrocarbon-degrading potential of numerous uncultivated oil-responsive bacteria, which were previously identified (Mason et al., 2014b).
Here we analyzed 13 metagenomes (Mason et al., 2014b) and 10 previously unreported metatranscriptomes to link community-wide HCD strategies with microbial taxonomy in the top one centimeter of deep-sea sediment, collected 3 months after the DWH wellhead was capped. We reconstructed bacterial genomes from the metagenomes, and mapped metatranscriptomes to these genomes to determine the activity of oil-responsive functional pathways across a hydrocarbon concentration gradient left by the spill. Studied communities included those from seven highly polluted ‘near-well’ sites around MC252 (0.3–2.7 km away), and six ‘distal’ sites that were distributed along a linear transect (10.1–59.5 km away) (Mason et al., 2014b), which followed the prevailing southwesterly deep plume path (Camilli et al., 2010) (Supplementary Figure S1). Distal-most sites were either un-impacted or minimally impacted, based on greatly diminishing plume hydrocarbon concentrations beyond ~30 km (Spier et al., 2013; Valentine et al., 2014) and oil-proxy sediment hopane concentrations beyond 40 km from the wellhead (Valentine et al., 2014). Results provide insights into the genomic potential and in situ transcriptional activity of dozens of spill-responsive bacteria, including >20 candidate hydrocarbon degraders.
Materials and methods
Sampling and nucleic acid sequencing
Thirteen seafloor sediment cores were collected between 28 September and 19 October 2010 at radially distributed locations around the capped MC252 wellhead (× 7 cores between 0.3 and 2.7 km from MC252), and along a distal southwesterly linear transect (× 6 cores between 10.1 and 59.5 km from MC252) (Supplementary Figure S1 and Supplementary Table S1) (Mason et al., 2014b). The outer surfaces of 0 to 1 cm deep cores were removed prior to DNA and RNA extractions (Mason et al., 2014b). DNA extraction and whole-genome shotgun sequencing are described by Mason et al. (2014b). In brief, DNA extracted from the cores was fragmented and prepared for sequencing using Illumina’s TruSeq DNA Sample Prep Kit. Each library was sequenced on a full HiSeq2000 lane at the Institute for Genomics and Systems Biology’s Next Generation Sequencing Core (Argonne National Laboratory). This yielded ~18 Gb of sequence per sample with 2 × 101 bp reads and insert sizes of ~135 bp. Low-quality reads were trimmed using Sickle v. 1.29 with a quality score threshold of Q=3 (based on Illumina 1.5 encoding whereby Q2 or ‘B’ quality scores denote poor quality bases ⩽Q15), and were removed if trimmed to <80 bp long (https://github.com/najoshi/sickle).
RNA was extracted in duplicate from cores using 0.5 g of sediment for each replicate. A modified hexadecyltrimethylammonium bromide (CTAB) extraction buffer was used as previously described (DeAngelis et al., 2010). Duplicate extracts were pooled, and purification was carried out using the Qiagen AllPrep DNA/RNA Mini Kit with on-column DNase digestion using the RNase-Free DNase Set (Qiagen, Valencia, CA, USA), yielding RNA integrity numbers from 8.2–9.2 for high yield samples (no RIN values were obtained for low yield samples). RNA from samples with low yields (<150 ng RNA: 0.3 km, 1.1 km, 1.3 km, 10.1 km, 15.1 km and 33.9 km samples) was amplified using the Ambion MessageAmp II aRNA Amplification Kit (Foster City, CA, USA). RNA was converted into double-stranded cDNA using the SuperScript Double-Stranded cDNA Synthesis Kit with random hexamers (Invitrogen, Carlsbad, CA, USA). Double-stranded cDNA was prepared for sequencing using the TruSeq Nano DNA kit (Illumina, San Diego, CA, USA). Prepared libraries (~440 bp long) were sequenced using an Illumina HiSeq 2500 at the Yale Center for Genomic Analysis, with three libraries per lane, and generating 150 bp paired-end reads. Adapter sequences were removed using Cutadapt (Martin, 2011), and reads were trimmed with Trimmomatic (Bolger et al., 2014) (sliding window quality score ⩾15) and removed if shorter than 60 bp.
Metagenome assembly
Metagenomic sequences from each sample were first assembled individually using the IDBA-UD v. 1.1.0 metagenome assembler (Peng et al., 2012). Consolidation and improved genome and HCD gene recovery was achieved via a co-assembly of three representative samples (0.5, 0.7, 0.9 km from MC252), owing to high compositional similarity among near-well communities. IDBA-UD was used for the co-assembly with an optimal kmer range of 45–75 and step size of 15. Improved recovery of the highly abundant GSC11 genome was achieved through selective re-assembly from the 0.5 km metagenome using Velvet (kmer size=63, expected kmer coverage=147, kmer coverage range=92–225) (Zerbino and Birney, 2008).
Genome binning, annotation, completion estimates and comparisons
To bin contigs ⩾2 kbp long, we used the multi-parameter approach previously described by Handley et al. (2013). To better separate closely related genomes using emergent self-organizing maps, contig tetranucleotide frequencies were augmented with the coverage of that contig in each spatially distinct sample (Supplementary Figure S2) based on the approach of Sharon et al. (2013). The differential coverage of contigs was determined by mapping reads to co-assembled contigs using bowtie2 (Langmead and Salzberg, 2012). Contig coverages per sample were scaled to 1 for emergent self-organizing maps. Genome bin abundance heat and line plots were created using gplots and ggplot2 packages in R, respectively.
Contigs were annotated using the Integrated Microbial Genomes pipeline (Markowitz et al., 2012), and the Rapid Annotations using Subsystems Technology (RAST) Server (Aziz et al., 2008). Predicted protein sequences of potential HCD genes were searched against the National Center for Biotechnology Information’s (NCBI’s) Conserved Domain Database (Marchler-Bauer et al., 2015).
Genome completion was estimated based on the presence of 107 single copy core genes (Dupont et al., 2012), excluding glyS, proS, pheT and rpoC, which were missing or poorly recovered in this study and according to Albertson et al. (2013). Core genes were detected using HMMER3 with the default cutoff (Eddy, 2011). Estimates were similar to those obtained using AMPHORA2 with a set of 31 universal bacterial protein-coding housekeeping genes (Wu and Scott, 2012).
Genomes were compared using average amino-acid identities (AAIs) including only BLASTp matches that shared ⩾30% identity over an alignable region of ⩾70% sequence length. 16S rRNA gene sequences and RecA and PAH dioxygenase predicted protein sequences were compared using MEGA6 (Tamura et al., 2013) ClustalW alignments and neighbor-joining or maximum-likelihood trees.
Transcriptome read mapping
Between 21 and 58 million trimmed paired-end reads were mapped to the co-assembly using Bowtie2 (Langmead and Salzberg, 2012) with end-to-end and sensitive settings. Alignments were sorted with Samtools (Li et al., 2009). Hits were enumerated and filtered using htseq-count in HTSeq v. 0.6.1p1 (Anders et al., 2015) with default settings. Counts were normalized to gene length and reads per sample by a modification of the approach described by Mortazavi et al. (2008), whereby normalization was to Reads Per Kilobase per Average library size (RPKA; average=44 million). To determine whether gene expression was up- or down-regulated spatially, RPKA values were also normalized to the genome coverage in each sample (RPKAC), and we required that at least one sample per gene had at least 10 mapped reads (un-normalized). To compare differential gene expression between proximal and distal sites we used edgeR (Robinson et al., 2010) on un-normalized data with RPKAC values supplied as an offset matrix of correction factors to the generalized linear model. Sample sizes were supplied as total library sizes.
Ribosomal RNA sequence assembly and clustering
To investigate beta diversity near full-length small subunit (SSU) rRNA (gene) sequences were reconstructed using the reference-guided Expectation Maximization Iterative Reconstruction of Genes from the Environment (EMIRGE) method (Miller et al., 2011). Whole-genome shotgun samples were rarefied to an equal depth of 64 million paired-end reads. Transcriptome samples were rarefied to between 22 and 32 million reads after first pre-selecting SSU rRNA-specific transcriptome kmers using bbduk (http://sourceforge.net/projects/bbmap) and the SILVA SSU rRNA database (Quast et al., 2013). Sequences were reconstructed over 80 iterations using EMIRGE with the SILVA 111 SSU rRNA database. Gapped and chimeric sequences were removed - the latter using 64-bit USEARCH v. 8.0 with the RDP Gold v. 9 database (http://drive5.com/). RNA and RNA gene sequences were co-clustered at ⩾97% similarity into operational taxonomic units (OTUs) using -cluster_otus (32-bit USEARCH v. 8.1) (Edgar, 2010) after sorting by size. OTU representative sequences were identified by 64-bit USEARCH global alignment to the full SILVA 111 SSU rRNA database, and by RDP classifier v. 2.6 (Wang et al., 2007) with a 0.8 bootstrap cutoff.
Sequence accession
Metagenomic and EMIRGE assemblies, and transcriptome reads are accessible via NCBI BioProject’s PRJNA258478 and PRJNA342256, respectively. Metagenome annotations are accessible via Integrated Microbial Genomes ID 3300003691.
Results and discussion
Genome recovery of diverse spill-responsive bacteria after wellhead capping
To establish a site-specific genomic database for metatranscriptome mapping, metagenomic sequences were co-assembled from three genomically representative (Figure 1) and comparatively well-assembling sediment samples (Supplementary Table S1) collected 0.5, 0.7 and 0.9 km from MC252. Contigs were binned into genomes aided by compositional information (Dick et al., 2009) and differential coverage (Sharon et al., 2013), which was obtained by mapping metagenomic reads from all 13 sites to the co-assembly. This also enabled us to determine the abundance of co-assembled genomes at each site. Most sequences (66% or 119 Mbp) were classified into 51 bins comprising 57 genomes (Supplementary Table S2, Supplementary Dataset S1) associated with the Gammaproteobacteria (Table 1), Alphaproteobacteria, Deltaproteobacteria and Bacteroidetes (Supplementary Table S2). A further 4.4% of contigs contained virus-related genes and were mainly associated with the Gammaproteobacteria. The relative abundance of the bacterial genome bins ranged from 0.6 to 13.1% (1.6% on average), and consisted of partial to near complete genomes estimated to be 7–97% complete (50% on average; Supplementary Table S2). Five bins contained two to three very closely related genomes with partial (insufficient) coverage separation (Supplementary Figure S3). Metatranscriptomic sequences, derived from 10 of the same samples as the metagenomes, were mapped to the co-assembly, generating genome-specific expression profiles up to 33.9 km from MC252 (Supplementary Dataset S1). This also enabled mRNA hits to be normalized to genome abundance per site to determine gene up/downregulation.
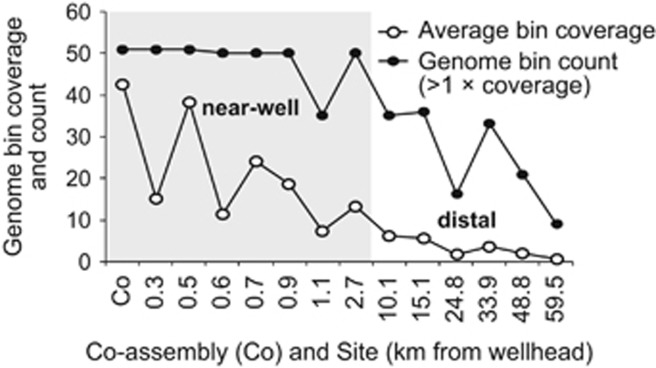
Figure 1.
Average genome bin coverage and count (for bins with >1 × coverage) in the co-assembly, and for the same collection of genome bins at each individual site; both decrease noticeably with increasing distance from the wellhead, although some of the genomes were still detectable 60 km away. Distance along the x axis is not shown to scale.
Table 1. Gammaproteobacterial genome bin designations.
| Bin | Taxonomic designation | Closest taxonomic match based on 16S rRNA OTUsa (% sequence identity) | Phylogenetic cluster |
|---|---|---|---|
| GSC1 | Ca. Thiotrichales | Ca. Halobeggiatoa sp. HMW-S2528 (99) | 1 |
| GSC2-3 | Ca. Thiotrichales | - | 1 |
| GSC4,7 | Ca. Colwellia | Colwellia psychrerythraea 34H (99) | 2 |
| GSC5-6 | Ca. Colwellia | Colwellia sp. MT41 (97) | 2 |
| GSC8 | Ca. Cycloclasticus | Cycloclasticus zancles 78-ME (98) | 3 |
| GSC9-10 | Ca. Cycloclasticus | Cycloclasticus zancles 78-ME (95) | 3 |
| GSC11-15 | Ca. Cellvibrionales | Porticoccus hydrocarbonoclasticus (93) | 4 |
| GSC16-24 | Gammaproteobacteria | - | - |
| GSC25-26 | Gammaproteobacteria | - | 5 |
See OTUs in Figure 2.
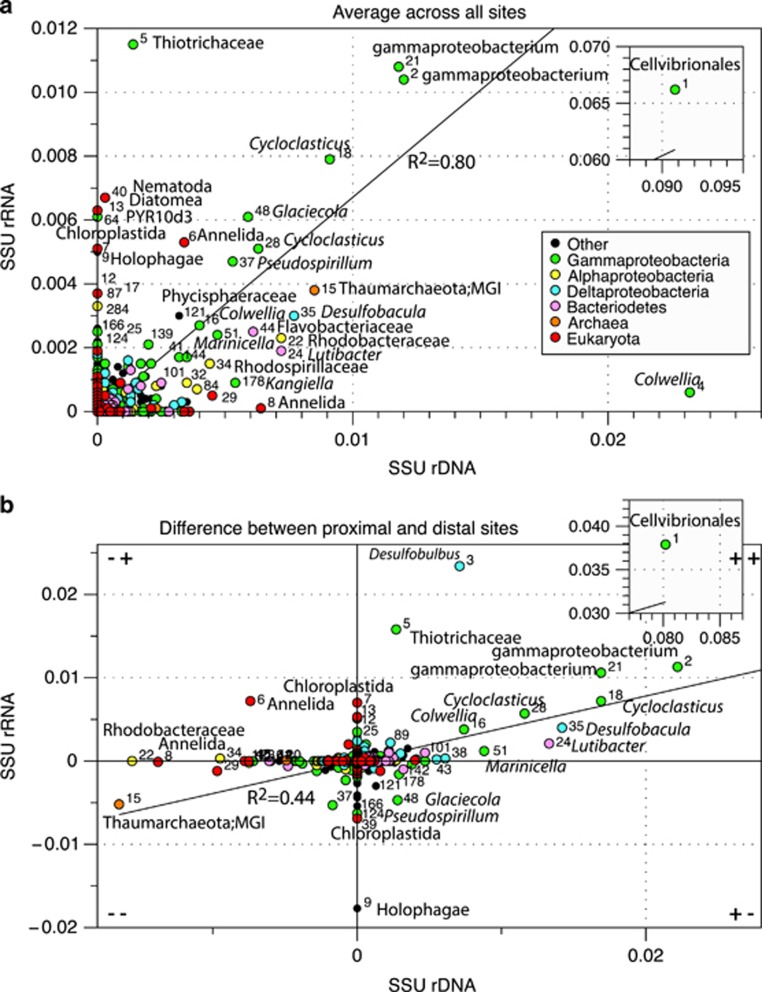
SSU rRNA genes and rRNA sequences from the metagenomes and metatranscriptomes were reconstructed using EMIRGE (Miller et al., 2011), and co-clustered into OTUs, including rRNA sequences from an additional unpaired metatranscriptome sample collected 1.3 km from MC252. Comparison of the relative abundances of bacterial, archaeal and eukaryotic SSU rRNA gene sequences demonstrated that bacteria near MC252 increased appreciably relative to archaea and eukaryotes (Supplementary Figure S4). Gammaproteobacteria predominated near MC252 (Mason et al., 2014b) and exhibited increased species richness (Supplementary Figure S4), analogous to the predominance of Gammaproteobacteria observed in the deep-sea plume (Redmond and Valentine, 2012). Although rRNA is not necessarily a good indicator of microbial growth (Blazewicz et al., 2013), we found rRNA gene and rRNA relative abundances were generally well correlated for prokaryotes (but not eukaryotes), particularly for bacteria enriched near the wellhead, and for a distally abundant OTU belonging to the Marine Group I Archaea (OTU-15; Figure 2). These data, along with mRNA expression profiles (Supplementary Dataset S1), imply spill-responsive bacterial communities were still viable and active ⩾75 days after well closure.
Figure 2.
(a) The correlation of EMIRGE 16 S rRNA gene and rRNA OTU abundances. (b) The difference between proximal and distal sites. Read clockwise: higher rRNA gene and rRNA in proximal locations (++); higher rRNA gene but lower rRNA proximally (+−); lower rRNA gene and rRNA proximally (– –); and lower rRNA gene but higher rRNA proximally (−+). (a, b) OTU numbers are given besides taxa points. Insets show the highly abundant OTU 1. Based on abundance and phylogenetic affiliation, OTU 1 corresponds to the Ca. Cellvibrionales GSC11-15 genomes (93% identity to P. hydrocarbonoclasticus). It also shares 100% identity (ID) with an iTag sequence (>97% similar to Greengenes OTU 248394) from a highly abundant uncultured gammaproteobacterium previously identified in the contaminated near-well sediments (Mason et al., 2014b). Thiotrichaceae OTU 5 corresponds to Ca. Thiotrichaceae GSC1 (99% ID to Ca. Halobeggiatoa sp. HMW-S2528); Cycloclasticus OTUs 18 and 28 respectively correspond to Ca. Cycloclasticus GSC8 and GSC9-10 (respectively 98 and 95% ID to Cycloclasticus zancles 78-ME); Colwellia OTU 4 corresponds to Ca. Colwellia GSC4 and GSC7 (99% ID to Colwellia psychrerythraea 34H); and Colwellia OTU 16 corresponds to Ca. Colwellia GSC5-6 (97% ID to Colwellia sp. MT41).
Well-conserved bacterial co-occurrences across the seafloor comprising both related groups and diverse bacteria
Bacterial genome bins were highly and exponentially enriched near MC252 (Supplementary Figure S5, Supplementary Table S3); the average genome read-coverage was only 0.7–6.2 × across widespread sites 10–60 km away, but was 7.3–42.6 × among sites within 2.7 km of MC252 (Figure 1,Supplementary Figure S6). The same genomes were found across all near-well sites, as well as at many less impacted or un-impacted distal locations, depicting a remarkably uniform community response across vast areas of the seafloor. This included six Gammaproteobacteria and two Bacteroidetes bins that were universally present at sites spanning the entire 60km (based on average genome mapped read coverages of >1 Figure 1, Supplementary Figure S7). When not considering an average coverage threshold, 54–100% of contigs from each of the 51 bins were detected across all 13 sites (Supplementary Table S4 and S5). The detection of spill-responsive bacteria up to 60 km from the wellhead, and beyond the extent of measurable plume fallout (Valentine et al., 2014), points to the enrichment of native benthic microorganisms capable of spill remediation. These bacteria are likely well adapted to hydrocarbons given the prevalence of natural seeps in the northern Gulf of Mexico (Atlas and Hazen, 2011; Joye et al., 2014; MacDonald et al., 2015). Although a few highly abundant and widely distributed OTUs were previously identified in the plume or along the seafloor (Hazen et al., 2010; Mason et al., 2012, 2014b), our genomic data indicate that the previously observed seafloor OTUs were part of a wider community-level response, whereby a large collection of the same bacteria co-occurred at each contaminated site. Smaller bacterial co-occurrence groups of ~15 organisms have been identified in the terrestrial subsurface (in sediments and groundwater), where taxa sharing similar spatial distribution and abundance patterns were termed ‘microbial species cohorts’ (Hug et al., 2015). The large DWH seafloor bacterial co-occurrence group, or cohort, may further be described as a collection of guilds engaged in the functionally equivalent processes of sulfate reduction, sulfur oxidation, denitrification, and aerobic n-alkane, aromatic hydrocarbon or PAH degradation (Figure 3), with some DWH bacteria occupying more than one guild (for example, denitrifiers such as GSC1, GSC3 and GSC16 that also oxidized hydrocarbons or sulfur). Hydrocarbon degrader guilds are defined here based only on the general class of hydrocarbons metabolized; the exact hydrocarbon substrates degraded are unknown, owing to the broad specificities of the enzymes involved (van Beilen et al., 1994; Bertoni et al., 1996; Jouanneau et al., 2006).
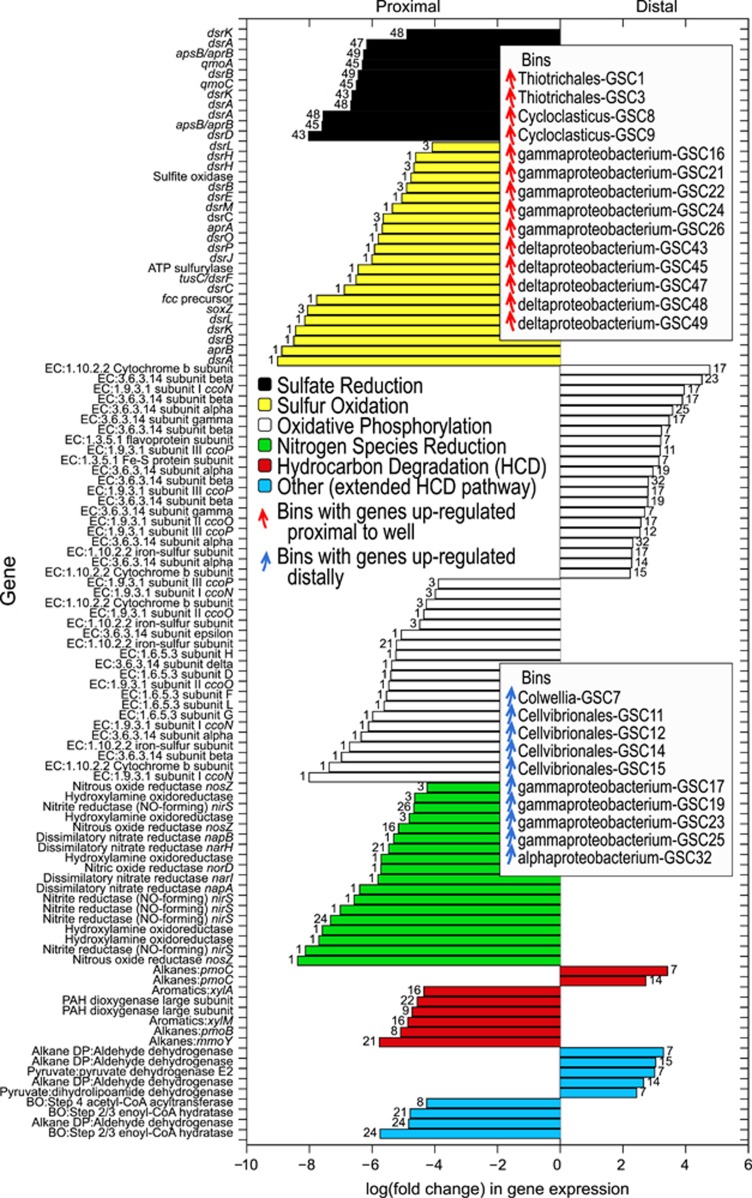
Figure 3.
Differentially expressed genes at proximal and distal sites associated with hydrocarbon degradation, oxidative phosphorylation, and S cycling and N reduction pathways. N species reduction was dominated by genes involved in the denitrification pathway. Genome bin numbers (GSC) are given beside each bar. Abbreviations: degradation pathway (DP); beta oxidation (BO).
Many of the DWH seafloor genomes were phylogenetically clustered (52%, co-binned populations excluded). Clusters in each of our sampled proteobacterial classes and the Bacteroidetes shared average amino-acid identities of 60–86%, which broadly equates to genus or family level relatedness (Konstantinidis and Tiedje, 2005). Of these, the Gammaproteobacteria exhibited five distinct inter-bin clusters (Table 1 and Supplementary Table S6). Prevalent phylogenetic clustering suggests strong habitat selection for traits shared among genetically similar organisms (Horner-Devine and Bohannan, 2006). Gammaproteobacterial clusters included genomes related to sulfur-oxidizing Candidatus Halobeggiatoa (Grünke et al., 2012), and to hydrocarbonoclastic Colwellia, Cycloclasticus and Porticoccus (Cellvibrionales) species (Geiselbrecht et al., 1998; Baelum et al., 2012; Gutierrez et al., 2012) (Table 1, Supplementary Table S7, Supplementary Figure S8). Of these, Colwellia and Cycloclasticus were typical plume genera (Redmond and Valentine, 2012). When compared with cultivated representatives, EMIRGE-reconstructed 16S rRNA gene sequences related to Cycloclasticus and Porticoccus formed distinct DWH seafloor clades, whereas extremely diverse sequences were associated with the Colwellia-Thalassomonas-Glaciecola group (Supplementary Figure S9).
Organisms within phylogenetic clusters shared common metabolic pathways (for example, sulfur oxidation or reduction, alkane degradation via AlkBTG and denitrification; Supplementary Table S8 and Figure 4). However, with the exception of sulfur metabolism, metabolic traits observed were not unique to clusters, and we found no evidence for niche differentiation amongst clusters based on unique functions. In addition, some metabolic pathways were shared by disparate organisms rather than among cluster members (for example, PAH degradation), although some of these mechanisms could be lacking from bins due to incomplete genome recovery. Regardless, the presence of multiple degradation mechanisms in some bins indicates that organisms within clusters were not strictly competitive, but were able to partition resources, as evidenced by the expression of distinct HCD genes by Ca. Cycloclasticus GSC8 and GSC9 (Figure 5). Despite this, some clusters exhibited markedly similar spatial metabolic response patterns, whereby all members upregulated key genes involved in S, N, O and hydrocarbon metabolism at either near-well or at distal sites (Figure 3; for example, phylogenetic clusters: 1, Thiotrichales; 3, Cycloclasticus; and 4, Cellvibrionales).
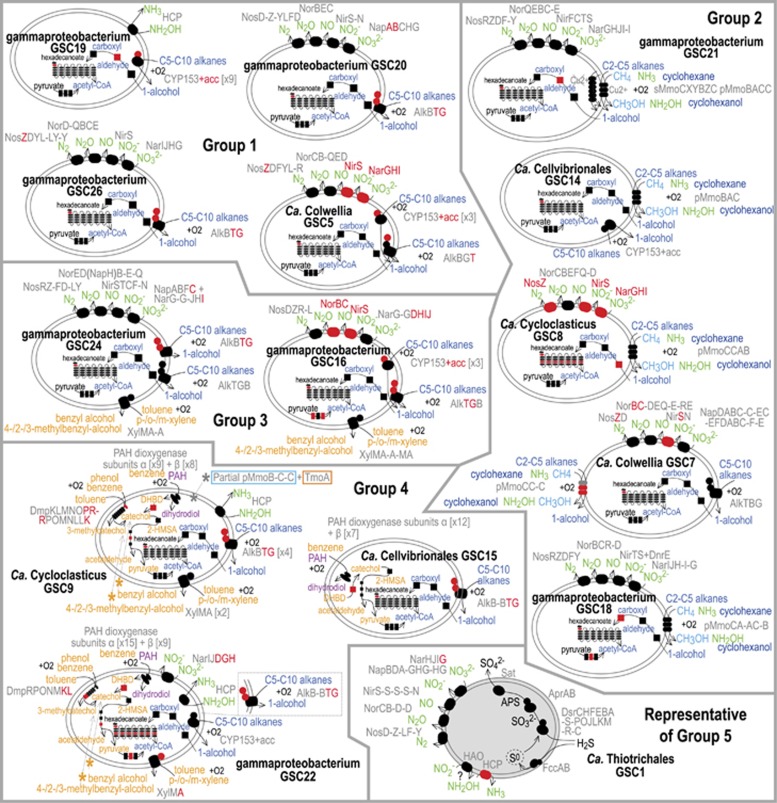
Figure 4.
Schematics of 14 gammaproteobacterial genome bins with the metabolic potential to degrade hydrocarbons (Groups 1–4), and a schematic showing the genes GSC1 likely uses to encode for denitrification and sulfide oxidation (representative bin of Group 5: GSC1–3). Genes present are represented by black symbols; those missing are red. Subunits are listed in order present in genomes, and separated by a dash if on different contigs. Substrates (and their products) are categorized by color: C2–C10 alkanes (dark blue, Groups 1–4), methane (light blue, Group 2), aromatic hydrocarbons (orange, panels bin groups 3–4), PAHs (purple, bin group 4), nitrogen species (green), and other (black). (Groups 1–4) All bins contain (near) complete beta-oxidation pathways that can be used to oxidize hexadecanoate or fatty acyl-CoA esters intermediates to acetyl-CoA. Accessory genes (acc); 2-Hydroxymuconate-semialdehyde (2-HMSA); cis-1,2-dihydrobenzene-1,2-diol (DHBD). (Group 4) A yellow * refers to pathways in Supplementary Figure S10a and b.
Figure 5.
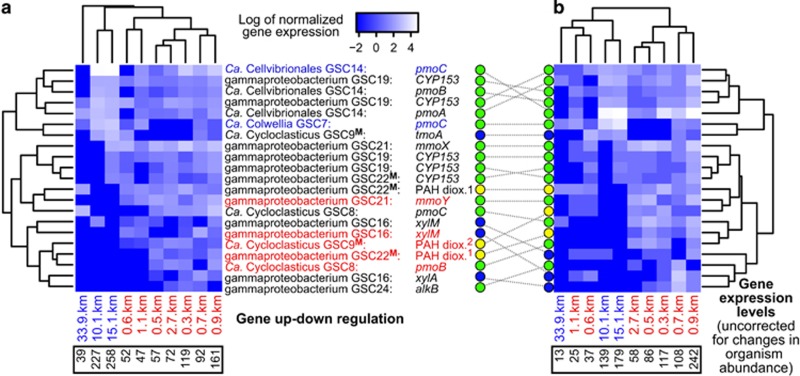
Spatial expression profiles of genes associated with hydrocarbon degradation: (a) genome bin coverage corrected values, and (b) coverage uncorrected values. Heatmap rows and columns are unscaled. Genome bin and gene identities are given for each row. Significantly differentially expressed genes at proximal locations are in red, and those at distal locations are in blue. Bins marked with a bolded superscript ‘M’ expressed genes encoding different proteins (that is, putative multi-tasking bacteria). PAH dioxygenases (dioxy.) are large subunits resembling 1naphthalene or 2anthranilate/(ortho-halo)benzoate dioxygenases. Circles indicate hydrocarbon classes targeted by genes: green=alkane; blue=aromatic; yellow=PAH. Lower black boxes contain sums of (a) RPKAC and (b) RPKA values per site.
Upregulation of sulfur and nitrogen cycling near the wellhead alongside HCD
The phylogenetic characteristics of the proximal sediment communities suggested a strong potential for sulfur and hydrocarbon metabolism; also supported by gene content and gene expression profiles. Among the most highly expressed genes were those exhibiting significant differential expression between proximal and distal sites for sulfur, hydrocarbon and nitrogen metabolism (Figure 3). Genes involved in sulfur oxidation by Thiotrichales GSC1 and GSC3 (closely related to Halobeggiatoa; Bin Group 5 Figure 4), and deltaproteobacterial sulfate reduction, were both significantly upregulated near MC252 (Figure 3), which implies an active spill-promoted sulfur cycle formed in the top 1 cm of seafloor sediment. In near-well sediments, we also observed the upregulation of oxidative phosphorylation genes by GSC1 and GSC3, concomitant with denitrification activity. This reflects the classic oscillating aerobic-anaerobic lifestyle of related Beggiatoa and Thiomargarita species, whereby anaerobically generated elemental sulfur (formed by nitrate-dependent sulfide oxidation) is further oxidized aerobically to sulfate (Schulz and Jorgensen, 2001).
An increase in genes involved in N metabolism was previously identified in DWH oil-polluted sediments (Scott et al., 2014; Mason et al., 2014b). Our data show these genes—involved in the denitrification pathway and nitrite/hydroxylamine oxidation/reduction—were significantly upregulated at proximal sites and this activity was associated with several diverse gammaproteobacterial bins. However, this activity was mostly linked to anaerobic sulfide oxidation by the two Thiotrichales genomes (GSC1 and GSC3; Figure 3). In addition to denitrification, GSC1 cells also expressed hydroxylamine oxidoreductase genes (EC 1.7.2.6), canonically used for the oxidation of hydroxylamine to nitrite. It seems likely that GSC1 and GSC3 used these hao-like genes as a mechanism for reducing nitrite to ammonia instead, although a hydroxylamine reductase required for the final step (hydroxylamine reduction to ammonia) was not detected in any Thiotrichales genome bin. Despite the incomplete pathway, dissimilatory nitrate reduction to ammonium is consistent with elevated pore water ammonium concentrations observed in locations near MC252 (Supplementary Figure S5). Moreover, ammonification via hao homologs has been predicted for related Beggiatoa and Ca. Thiopilula spp. (MacGregor et al., 2013; Jones et al., 2015) based on reverse operation in Nautilia profundicola (Hanson et al., 2013). It is thought duel denitrification and ammonification (via nirB) facilitates nitrate reduction under varied geochemical conditions in Thiomargarita nelsonii (Winkel et al., 2016), as demonstrated in Shewanella and marine intertidal sediments (Kraft et al., 2014; Yoon et al., 2015).
Overall, metabolic data indicate that the 1 cm deep sediment cores from contaminated near-well sites spanned a sharp redox gradient, encompassing aerobic sulfur and hydrocarbon oxidation, along with shallow anaerobic nitrate and sulfate reduction (Figure 3). Meanwhile, distal sediments appeared to be primarily dominated by aerobic processes (Figure 3), with a large number of bacteria engaged in aerobic oxidative phosphorylation (Figure 3,Supplementary Dataset S1). These data indicate that oil near MC252 accelerated oxygen consumption and shifted anaerobic processes towards the sediment air-water interface.
HCD gene associations and differential expression at contaminated and uncontaminated locations
Half of the bacterial genomes we reconstructed (n=25) contained genes associated with the degradation of hydrocarbons, which were largely concentrated in the Gammaproteobacteria (179 genes or 66% Table 2 and Supplementary Table S8). Most HCD genes belonged to aerobic pathways (Supplementary Table S8), and were observed almost exclusively within genomes in additive combinations targeting one or all three of the following hydrocarbon classes: (1) n-alkanes, (2) n-alkanes+aromatics or (3) n-alkanes+aromatics+PAHs (Figure 4 and Supplementary Figure S5). Genome bins endowed with aromatic hydrocarbon hydroxylases/monooxygenases and PAH dioxygenases appear evolutionarily well equipped with numerous mechanisms for hydrocarbon metabolism. However, alkane degradation potential was the common denominator, and genomes with genes for n-alkane degradation were cumulatively the most abundant near MC252 (Supplementary Figure S5). Correlations between genome bin relatedness and common and phylogenetically diverse alkB alkane degradation genes (Supplementary Figure S11) suggest DWH bacteria acquired these genes vertically rather than by recent lateral gene transfer, and hence also that the spill-enriched bacteria were indigenous deep-sea hydrocarbon degraders.
Table 2. Hydrocarbon degradation gene distributions.
| Group | HCD gene count | Relative abundance (%) | Normalized to CDS (%)a |
|---|---|---|---|
| Gammaproteobacteria | 180 | 66 | 55 |
| Unbinned | 67 | 25 | 22 |
| Deltaproteobacteria | 13 | 5 | 11 |
| Bacteroidetes | 9 | 3 | 7 |
| Alphaproteobacteria | 2 | 1 | 2 |
| Phage associated | 2 | 1 | 3 |
| Total | 273 | 100 | 100 |
Normalized to total coding DNA sequence (CDS) per group.
We observed the expression of 21 genes involved in the aerobic degradation of the three hydrocarbon classes across multiple seafloor sites, and some at all sites. At distal sites, HCD bacteria were likely metabolizing low levels of plume-derived MC252 oil, or possibly seep hydrocarbons emitted in the vicinity of MC252 (MacDonald et al., 2015). Isotope data reported by Kelly and Coffin (1998) suggest hydrocarbons could be a major source of organic carbon for bacteria in the northern Gulf of Mexico. Conditions at distal sites appeared to favor the expression of genes targeting n-alkanes over (poly)aromatic substrates, and HCD genes expressed there were only a subset of those expressed near MC252 (Figure 5). Although fewer genomes were active and genomes were at relatively lower abundances at distal locations, some HCD gene transcript abundances were comparable to those at proximal sites, and some HCD genes were more highly upregulated than any proximal HCD genes (Figure 5), suggesting a greater individual effort for alkane degradation at uncontaminated locations. Conversely, data indicate that bacterial competition for hydrocarbon substrate classes, and the diversity of substrates degraded, increased towards the wellhead. Overall a greater number and range of genes associated with HCD (of alkanes, aromatics and polyaromatics) were upregulated proximally (Figure 3), likely due to the higher average concentration of hydrocarbons near MC252, including higher aromatic and PAH levels (Supplementary Figure S5) (Mason et al., 2014b).
Bacteria that differentially expressed HCD genes, and associated oxidative phosphorylation genes, at proximal and distal sites partitioned into two largely distinct sets of Gammaproteobacteria (Figure 3). The distal upregulating set comprised Group 2 alkane-only degraders (Figure 4; Ca. Colwellia GSC7 and Ca. Cellvibrionales GSC14). In comparison, the proximal set comprised Group 2–4 bacteria (Figure 4) involved in the degradation of short to-mid-chain length n-alkanes, aromatics and PAHs (Ca. Cycloclasticus GSC8 and GSC9, Thiotrichales-like GSC21, and Gammaproteobacteria GSC16 and GSC22). Two of these genome bins (GSC9 and GSC22) also co-expressed genes targeting distinctly different substrates (Figure 5), suggesting that the bacterial populations represented by these genomes were multi-tasking. Although both distal versus near-well upregulating sets of bacteria were active across large distances (and all at proximal sites), the distal set appeared to be more active at 10 and 15 km distances (Figure 5) where total petroleum hydrocarbon and n-alkane concentrations were generally lower (Mason et al., 2014b; Supplementary Figure S5).
Abundant mechanisms for PAH and aromatic hydrocarbon oxidation among a small group of unrelated bacteria
Total PAH concentrations (particularly naphthalene>fluorene>phenanthrene>chrysenes) remained high in sediments surrounding MC252 in the months following the spill (Zukunft, 2010; Mason et al., 2014b), although they were no longer in excess of the EPA benchmark in the water column (Zukunft, 2010). We identified 3 candidate PAH degraders (Bin Group 4 Figure 4; Ca. Cycloclasticus GSC9, gammaproteobacterium GSC22, and Ca. Cellvibrionales GSC15) in seafloor sediments, of which GSC9 and GSC22 exhibited active expression of PAH dioxygenases, particularly near-well. Related Cycloclasticus species and Porticoccus hydrocarbonoclasticus (Cellvibrionales) are known for their ability to degrade various PAHs (Geiselbrecht et al., 1998; Gutierrez et al., 2012). All three candidate PAH degraders had broadly equivalent spatial abundances (Supplementary Figure S6), and each genome has 17–24 subunits (large and small) of diverse ring-hydroxylating dioxygenases that, except for 2, closely resemble dioxygenases used for the oxidation of PAHs and other aromatic hydrocarbons (Figure 4 and Supplementary Dataset S1) (Jouanneau et al., 2006). As previously observed in Cycloclasticus genomes (Lai et al., 2012; Cui et al., 2013), each of our three DWH genomes had a greater proportion of large versus small PAH dioxygenase subunits (58–63% were large in genomes and 64% in the unbinned fraction).
PAH dioxygenase sequences from the GSC9, GSC22 and GSC15 genomes chiefly resembled naphthalene dioxygenases (23 large and 17 small subunits)—the most abundant PAH present in the sediments (Zukunft, 2010)—whereas the remainder (20 large and 11 small subunits) were more closely related to biphenyl/benzene, anthranilate/(ortho-halo)benzoate or pyrene dioxygenases (Supplementary Figure S12 and Supplementary Table S8). In addition to the presence of diverse dioxygenases in these three bacterial genome bins, individual PAH dioxygenases, such as naphthalene dioxygenases, can act on a broad range of substrates (Jouanneau et al., 2006), meaning these bacteria likely had the capacity to degrade a range of PAHs. Of these, we observed the expression of dioxygenases resembling naphthalene dioxygenases (by Ca. Cycloclasticus GSC22) and anthranilate/(ortho-halo)benzoate dioxygenases (by GSC9) (Figure 5). PAH dioxygenases produce dihydrodiols (Jouanneau and Meyer, 2006). Canonical naphthalene, anthranilate and pyrene cis-dihydrodiol dehydrogenases (EC 1.3.1.29, EC 1.3.1.49) were not evident in our DWH genomes. However, all three genomes possessed cis-2,3-dihydrobiphenyl-2,3-diol dehydrogenase (bphB) and 2,3-dihydroxybiphenyl 1,2-dioxygenase (bphC) genes. BphB (EC 1.3.1.56) is a multi-substrate enzyme that acts on biphenyl-2,3-diol and a wide range of PAH dihydrodiols (Jouanneau and Meyer, 2006), including those relevant to MC252 oil, such as naphthalene 1,2-dihydrodiol, and phenanthrene and chrysene 3,4-dihydrodiols (Zukunft, 2010). BphC degrades the catechol product of BphB. These bacteria appear to use a universal pathway for the catabolism of early PAH degradation products, as opposed to distinct dihydrodiol dehydrogenases per PAH substrate identified in a collection of Gulf of Mexico seawater-associated Gammaproteobacteria (Dombrowski et al., 2016).
Genetic mechanisms for benzene-toluene-ethylbenzene-xylene degradation were previously found to be enriched at highly contaminated seafloor sites (Mason et al., 2014b). Through assembly and bin assignment (Bin Groups 3–4 Figure 4), we were able to link genes used for toluene, xylene and benzene degradation with at least four Gammaproteobacteria: GSC9, GSC22, GSC16 (related to n-alkane-degrading gammaproteobacterium HdN1 (Zedelius et al., 2011)) and GSC24 (related to Oceanospirillales Hahella chejuensis). Collectively, these four genomic bins represented 8% of the average genome abundance at the proximal sites (Supplementary Figure S5). All four bins had genes encoding xylene monooxygenase-like enzymes (Xyl), which oxidize toluene and xylenes (Harayama et al., 1989). xylAMM genes were expressed by GSC16 and were significantly higher at the proximal sites (Figures 3 and 5). In contrast, GSC9 demonstrated greater expression, albeit not significantly, of a gene (tmoA) encoding part of a largely unbinned aerobic toluene-4-monoxygenase system at distal sites, suggesting toluene/xylene metabolism very likely occurred across the seafloor, although the organisms and mechanisms varied. GSC9 and GSC22 also had genes encoding multicomponent phenol hydroxylase like enzymes (Dmp), which oxidize phenol, benzene and toluene (Cafaro et al., 2005). Further to these mechanisms, the three candidate PAH degraders (GSC9, GSC22 and GSC15) had putative benzene 1,2-dioxygenases (also similar to biphenyl dioxygenases) and catechol 2,3-dioxygenases (EC 1.13.11.2), suggesting these taxa could generate catechol by benzene oxidation, which could then be converted into 2-hydroxymuconate-semialdehyde, and sequentially transformed into pyruvate (Figure 4). Although HCD mechanisms identified in these surface sediment communities were overwhelmingly aerobic, there were a few exceptions in sequence data not binned to genomes. These include a single set of anaerobic ethylbenzene dehydrogenase genes (ebdACBA); and benzylsuccinate synthase genes (bssCAB and bssCA), which can be used for anaerobic toluene oxidation (Beller and Spormann, 1998), and were also observed in the lower anaerobic layer of seafloor sediments polluted with oil from MC252 (Kimes et al., 2013).
Phylogenetically widespread mechanisms for short- to mid-chain n-alkane oxidation
Widespread evidence for alkane oxidation is associated with three different mechanisms that target gaseous C2–C4 short-chain and liquid C5–C10 mid-chain length n-alkanes (Supplementary Table S8). Genes associated with the oxidation of both short to mid length n-alkane groups were expressed across at least 34 km of the Gulf of Mexico seafloor (Figure 5). Of the mechanisms present, transmembrane 1-alkane monooxygenase and membrane-bound cytochrome P450 CYP153 enzymes specifically act on mid-chain alkanes from pentane to decane (C5–C10) (van Beilen et al., 2006). Collectively, the transmembrane 1-alkane monooxygenase (alkB±alkGT rubredoxin/rubredoxin reductase) and CYP153 (± ferredoxin/ferredoxin reductase) hydroxylase genes are present in 11 out of 14 key gammaproteobacterial genome bins depicted in Figure 4 (Bin Groups 1–4). Although the alk genes were the most widespread, and were also found in other phyla (Supplementary Table S8), CYP153 genes were more commonly expressed (Figure 5). Pathway analysis of key gammaproteobacterial bins with these genes for alkane degradation suggests 1-alcohol generated by alkane oxidation could be converted sequentially to aldehydes by alcohol dehydrogenase (EC 1.1.1.1), carboxylates by aldehyde dehydrogenase (NAD, EC 1.2.1.3) and acetyl-CoA via beta oxidation (Figure 4 and Supplementary Figure S10).
Genes resembling particulate and soluble methane or ammonia monooxygenases were also common, and provide an alternative method for alkane metabolism by Group 2 populations in Figure 4. We recovered a diverse group of six genomes with particulate methane monooxygenase-like genes (Ca. Colwellia GSC7, Ca. Cycloclasticus GSC8-9, Ca. Cellvibrionales GSC14 and Thiotrichales-like GSC21, and gammaproteobacterium IMCC2047 relative GSC18). GSC21 also has genes encoding the components of a sMMO (soluble monooxygenase), which is used in place of the pMMO (particulate enzyme) under copper limiting conditions (Nielsen et al., 1997). Both types of methane monooxygenase-like genes were expressed across multiple seafloor locations; GSC7, GSC8 and GSC14 expressed particulate pmo genes, whereas GSC21 expressed soluble mmo genes (Figure 5).
These methane and ammonium monooxygenases constitute a group of related multi-substrate enzymes that in some other organisms preferentially target methane or ammonia (O'Neill and Wilkinson, 1977). They can also be used by methanotrophs, in the absence of methane, to oxidize short n-alkanes, namely C2–C4 gases (ethane, butane, propane) and C5 pentane (gas or liquid depending on incubation temperature) (Colby et al., 1977; Patel et al., 1979). Longer C6-C8 alkanes, can also be used, albeit at an appreciably slower rate (Colby et al., 1977). In comparison, Mycobacterium strains preferentially use sMMO to oxidize alkanes (Martin et al., 2014). In the current study, all 6 seafloor DWH genomes with pmo and mmo genes lack evidence for methanol or hydroxylamine oxidation, suggesting an inability to utilize the products of methane or ammonia oxidation (Supplementary Figure S10). However, all have the genetic capacity to convert 1-alcohols generated from n-alkane oxidation to acetyl-CoA (Figure 4 and Supplementary Figure S10), and some bins expressed pmo or mmo genes alongside genes in downstream alkane degradation pathways (Figure 3, Supplementary Dataset S1). For example, GSC21 co-expressed a short-chain alcohol dehydrogenase gene, resembling a NADH-dependent butanol dehydrogenase gene (bdhA), which it may have used to metabolize 1-alcohol produced by alkane degradation (Figure 4). In addition, we detected expression of parts of the downstream alkane degradation pathway by GSC7 and GSC8 (Supplementary Dataset S1), whereas all genes comprising the pathway from 1-alcohol to the first steps in the beta-oxidation pathway were expressed by GSC14, including multiple NAD-dependent alcohol dehydrogenase (EC 1.1.1.1) and aldehyde dehydrogenase genes. These data reflect the observation that a higher ratio of pmo to methanol dehydrogenase genes were expressed in the buoyant plume, suggesting not all pmo genes were used for methane oxidation by DWH bacteria (Rivers et al., 2013). We therefore predict that the seafloor bacteria primarily used pMMO±sMMO to oxidize n-alkanes, and specifically mid-chain length alkanes still trapped in the post-spill sediments (versus rapidly consumed C2–C4 gases). Accordingly, the lack of dedicated methanotrophs in the deep-sea sediments is consistent with the absence of methane in the late-stage plume (Kessler et al., 2011), and the expected absence of trapped methane in the post-spill sediments.
Conclusions
Gulf of Mexico microorganisms are naturally exposed to oil seeps (Joye et al., 2014; MacDonald et al., 2015) and frequent spills (USCG, 2012). Our genomic and metabolic reconstruction of seafloor communities indicates that diverse, indigenous bacteria from at least four phyla co-occurred across the seafloor, and responded to hydrocarbons associated with the DWH spill. Based on the widespread, and in some cases ubiquitous, presence of these spill-responsive bacteria it seems likely that they are indigenous members of the benthic community, although plume fallout did impact sediments at considerable distance from MC252. The DWH cohort consisted of guilds of functionally equivalent, phylogenetically clustered or un-clustered bacteria that actively competed for, or partitioned N, S or hydrocarbons in post-spill sediments. Strong environmental pressures were likely responsible for selecting clusters of genetically similar organisms, some of which exhibited spatially coordinated metabolic responses. Co-selection, implying the preservation or sharing of expressed traits, was important within phylogenetic clusters (such as opportunistic hydrocarbon oxidation and sulfur oxidation). However, competition was not uniquely defined by organism relatedness. The distribution of HCD mechanisms among gammaproteobacterial bins was not limited to phylogenetically similar organisms, and gene transcript data show that although some closely related groups of organisms actively competed (for example, for N and S), others were niche differentiated (for example, via use of distinct HCD strategies). Moreover, some binned populations appeared to have occupied more than one niche (or guild) by co-utilizing functionally distinct genes for HCD within the surface sediment layer. However, at the class level, many Gammaproteobacteria shared hydrocarbonoclastic traits, and several orthologous HCD genes were concomitantly expressed, indicating competition for certain types of hydrocarbons (for example, PAHs or mid-chain length n-alkanes). The large degree of diversity and apparent redundancy among HCD strategies used or possessed by seafloor bacteria suggests that the Gulf of Mexico harbors functionally robust communities that are poised to take advantage of petroleum hydrocarbon influxes.
Acknowledgments
This work was supported by Alfred P Sloan Foundation and Exxon-Mobile grants awarded to JAG, and a Royal Society of NZ Rutherford Discovery Fellowship awarded to KMH. Partial support was provided by the US Dept. of Energy under contracts DE-AC02-06CH11357 (ANL) and DE-AC05-76RL01830 (PNNL). We thank Christian Sieber (JGI) for transcriptome rRNA sequence assembly, and acknowledge resources provided by the University of Chicago Research Computing Center, NERSC, and the University of Auckland NeSI high-performance computing facilities and Centre for eResearch.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. (2013). Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31: 533–538. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlas RM, Hazen TC. (2011). Oil biodegradation and bioremediation: a tale of the two worst spills in US history. Environ Sci Technol 45: 6709–6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baelum J, Borglin S, Chakraborty R, Fortney JL, Lamendella R, Mason OU et al. (2012). Deep-sea bacteria enriched by oil and dispersant from the Deepwater Horizon spill. Environ Microbiol 14: 2405–2416. [DOI] [PubMed] [Google Scholar]
- Barron MG. (2012). Ecological impacts of the deepwater horizon oil spill: implications for immunotoxicity. Toxicol Pathol 40: 315–320. [DOI] [PubMed] [Google Scholar]
- Beller HR, Spormann AM. (1998). Analysis of the novel benzylsuccinate synthase reaction for anaerobic toluene activation based on structural studies of the product. J Bacteriol 180: 5454–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoni G, Bolognese F, Galli E, Barbieri P. (1996). Cloning of the genes for and characterization of the early stages of toluene and o-xylene catabolism in Pseudomonas stutzeri OX1. Appl Environ Microbiol 62: 3704–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazewicz SJ, Barnard RL, Daly RA, Firestone MK. (2013). Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J 7: 2061–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooijmans RJ, Pastink MI, Siezen RJ. (2009). Hydrocarbon-degrading bacteria: the oil-spill clean-up crew. Microb Biotechnol 2: 587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafaro V, Notomista E, Capasso P, Di Donato A. (2005). Regiospecificity of two multicomponent monooxygenases from Pseudomonas stutzeri OX1: molecular basis for catabolic adaptation of this microorganism to methylated aromatic compounds. Appl Environ Microbiol 71: 4736–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilli R, Reddy CM, Yoerger DR, Van Mooy BA, Jakuba MV, Kinsey JC et al. (2010). Tracking hydrocarbon plume transport and biodegradation at Deepwater Horizon. Science 330: 201–204. [DOI] [PubMed] [Google Scholar]
- Colby J, Stirling DI, Dalton H. (1977). The soluble methane mono-oxygenase of methylococcus capsulatus (Bath). Biochem J 165: 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z, Xu G, Li Q, Gao W, Zheng L. (2013). Genome sequence of the pyrene- and fluoranthene-degrading bacterium Cycloclasticus sp. Strain PY97M. Genome Announc 1: e00536-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis KM, Silver WL, Thompson AW, Firestone MK. (2010). Microbial communities acclimate to recurring changes in soil redox potential status. Environ Microbiol 12: 3137–3149. [DOI] [PubMed] [Google Scholar]
- Dick GJ, Andersson AF, Baker BJ, Simmons SL, Thomas BC, Yelton AP et al. (2009). Community-wide analysis of microbial genome sequence signatures. Genome Biol 10: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski N, Donaho JA, Gutierrez T, Seitz KW, Teske AP, Baker BJ. (2016). Reconstructing metabolic pathways of hydrocarbon-degrading bacteria from the Deepwater Horizon oil spill. Nat Microb 1: 16057. [DOI] [PubMed] [Google Scholar]
- Dubinsky EA, Conrad ME, Chakraborty R, Bill M, Borglin SE, Hollibaugh JT et al. (2013). Succession of hydrocarbon-degrading bacteria in the aftermath of the deepwater horizon oil spill in the gulf of Mexico. Environ Sci Technol/ 47: 10860–10867. [DOI] [PubMed] [Google Scholar]
- Dupont CL, Rusch DB, Yooseph S, Lombardo MJ, Richter RA, Valas R et al. (2012). Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J 6: 1186–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. (2011). Accelerated profile HMM searches. PLoS Comput Biol 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Geiselbrecht AD, Hedlund BP, Tichi MA, Staley JT. (1998). Isolation of marine polycyclic aromatic hydrocarbon (PAH)-degrading cycloclasticus strains from the gulf of mexico and comparison of their pah degradation ability with that of puget sound cycloclasticus strains. Appl Environ Microbiol 64: 4703–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünke S, Lichtschlag A, de Beer D, Felden J, Salman V, Ramette A et al. (2012). Mats of psychrophilic thiotrophic bacteria associated with cold seeps of the Barents Sea. Biogeosciences 9: 2947–2960. [Google Scholar]
- Gutierrez T, Nichols PD, Whitman WB, Aitken MD. (2012). Porticoccus hydrocarbonoclasticus sp. nov., an aromatic hydrocarbon-degrading bacterium identified in laboratory cultures of marine phytoplankton. Appl Environ Microbiol 78: 628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley KM, VerBerkmoes NC, Steefel CI, Williams KH, Sharon I, Miller CS et al. (2013). Biostimulation induces syntrophic interactions that impact C, S and N cycling in a sediment microbial community. ISME J 7: 800–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson TE, Campbell BJ, Kalis KM, Campbell MA, Klotz MG. (2013). Nitrate ammonification by Nautilia profundicola AmH: experimental evidence consistent with a free hydroxylamine intermediate. Front Microbiol 4: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S, Rekik M, Wubbolts M, Rose K, Leppik RA, Timmis KN. (1989). Characterization of five genes in the upper-pathway operon of TOL plasmid pWW0 from Pseudomonas putida and identification of the gene products. J Bacteriol 171: 5048–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N et al. (2010). Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330: 204–208. [DOI] [PubMed] [Google Scholar]
- Horner-Devine MC, Bohannan BJ. (2006). Phylogenetic clustering and overdispersion in bacterial communities. Ecology 87: S100–S108. [DOI] [PubMed] [Google Scholar]
- Hug LA, Thomas BC, Brown CT, Frischkorn KR, Williams KH, Tringe SG et al. (2015). Aquifer environment selects for microbial species cohorts in sediment and groundwater. ISME J 9: 1846–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DS, Flood BE, Bailey JV. (2015). Metatranscriptomic analysis of diminutive Thiomargarita-like bacteria (‘Candidatus Thiopilula’ spp.) from abyssal cold seeps of the Barbados Accretionary Prism. Appl Environ Microbiol 81: 3142–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouanneau Y, Meyer C. (2006). Purification and characterization of an arene cis-dihydrodiol dehydrogenase endowed with broad substrate specificity toward polycyclic aromatic hydrocarbon dihydrodiols. Appl Environ Microbiol 72: 4726–4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouanneau Y, Meyer C, Jakoncic J, Stojanoff V, Gaillard J. (2006). Characterization of a naphthalene dioxygenase endowed with an exceptionally broad substrate specificity toward polycyclic aromatic hydrocarbons. Biochemistry 45: 12380–12391. [DOI] [PubMed] [Google Scholar]
- Joye SB, Teske AP, Kostka JE. (2014). Microbial dynamics following the Macondo oil Well blowout across Gulf of Mexico environments. BioScience 64: 766–777. [Google Scholar]
- Kelley CA, Coffin RB. (1998). Stable isotope evidence for alternative bacterial carbon sources in the Gulf of Mexico. Limnol Oceanogr 43: 1962–1969. [Google Scholar]
- Kessler JD, Valentine DL, Redmond MC, Du M, Chan EW, Mendes SD et al. (2011). A persistent oxygen anomaly reveals the fate of spilled methane in the deep Gulf of Mexico. Science 331: 312–315. [DOI] [PubMed] [Google Scholar]
- Kimes NE, Callaghan AV, Aktas DF, Smith WL, Sunner J, Golding B et al. (2013). Metagenomic analysis and metabolite profiling of deep-sea sediments from the Gulf of Mexico following the Deepwater Horizon oil spill. Front Microbiol 4: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GM, Smith CB, Tolar B, Hollibaugh JT. (2013). Analysis of composition and structure of coastal to mesopelagic bacterioplankton communities in the northern gulf of Mexico. Front Microbiol 3: 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. (2005). Towards a genome-based taxonomy for prokaryotes. J Bacteriol 187: 6258–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft B, Tegetmeyer HE, Sharma R, Klotz MG, Ferdelman TG, Hettich RL et al. (2014). Nitrogen cycling. The environmental controls that govern the end product of bacterial nitrate respiration. Science 345: 676–679. [DOI] [PubMed] [Google Scholar]
- Lai Q, Li W, Wang B, Yu Z, Shao Z. (2012). Complete genome sequence of the pyrene-degrading bacterium Cycloclasticus sp. strain P1. J Bacteriol 194: 6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamendella R, Strutt S, Borglin S, Chakraborty R, Tas N, Mason OU et al. (2014). Assessment of the Deepwater Horizon oil spill impact on Gulf coast microbial communities. Front Microbiol 5: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald I, Garcia-Pineda O, Beet A, Daneshgar Asl S, Feng L, Graettinger G et al. (2015). Natural and unnatural oil slicks in the Gulf of Mexico. J Geophys Research 120: 8364–8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor BJ, Biddle JF, Siebert JR, Staunton E, Hegg EL, Matthysse AG et al. (2013). Why orange Guaymas Basin Beggiatoa spp. are orange: single-filament-genome-enabled identification of an abundant octaheme cytochrome with hydroxylamine oxidase, hydrazine oxidase, and nitrite reductase activities. App Environ Microbiol 79: 1183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY et al. (2015). CDD: NCBI's conserved domain database. Nucleic Acids Research 43: D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Chen IM, Chu K, Szeto E, Palaniappan K, Grechkin Y et al. (2012). IMG/M: the integrated metagenome data management and comparative analysis system. Nucleic Acids Res 40: D123–D129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KE, Ozsvar J, Coleman NV. (2014). SmoXYB1C1Z of Mycobacterium sp. strain NBB4: a soluble methane monooxygenase (sMMO)-like enzyme, active on C2 to C4 alkanes and alkenes. Appl Environ Microbiol 80: 5801–5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J 71: 10–12. [Google Scholar]
- Mason OU, Han J, Woyke T, Jansson JK. (2014. a). Single-cell genomics reveals features of a Colwellia species that was dominant during the Deepwater Horizon oil spill. Front Microbiol 5: 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason OU, Hazen TC, Borglin S, Chain PS, Dubinsky EA, Fortney JL et al. (2012). Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J 6: 1715–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason OU, Scott NM, Gonzalez A, Robbins-Pianka A, Baelum J, Kimbrel J et al. (2014. b). Metagenomics reveals sediment microbial community response to Deepwater Horizon oil spill. ISME J 8: 1464–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CS, Baker BJ, Thomas BC, Singer SW, Banfield JF. (2011). EMIRGE: reconstruction of full-length ribosomal genes from microbial community short read sequencing data. Genome Biol 12: R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Nielsen AK, Gerdes K, Murrell JC. (1997). Copper-dependent reciprocal transcriptional regulation of methane monooxygenase genes in Methylococcus capsulatus and Methylosinus trichosporium. Mol Microbiol 25: 399–409. [DOI] [PubMed] [Google Scholar]
- O'Neill JG, Wilkinson JF. (1977). Oxidation of ammonia by methane-oxidizing bacteria and the effects of ammonia on methane oxidation. J General Microbiol 100: 407–412. [Google Scholar]
- Patel RN, Hou CT, Laskin AI, Felix A, Derelanko P. (1979). Microbial oxidation of gaseous hydrocarbons. II. Hydroxylation of alkanes and epoxidation of alkenes by cell-free particulate fractions of methane-utilizing bacteria. J Bacteriol 139: 675–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Leung HC, Yiu SM, Chin FY. (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28: 1420–1428. [DOI] [PubMed] [Google Scholar]
- Peterson CH, Anderson SS, Cherr GN, Ambrose RF, Anghera S, Bay S et al. (2012). A tale of two spills: Novel science and policy implications of an emerging new oil spill model. BioScience 461: 461–469. [Google Scholar]
- Pritchard PH, Mueller JG, Rogers JC, Kremer FV, Glaser JA. (1992). Oil spill bioremediation: experiences, lessons and results from the Exxon Valdez oil spill in Alaska. Biodegradation 3: 315–335. [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond MC, Valentine DL. (2012). Natural gas and temperature structured a microbial community response to the Deepwater Horizon oil spill. Proc Natl Acad Sci USA 109: 20292–20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers AR, Sharma S, Tringe SG, Martin J, Joye SB, Moran MA. (2013). Transcriptional response of bathypelagic marine bacterioplankton to the Deepwater Horizon oil spill. ISME J 7: 2315–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RL, Overholt WA, Hagan C, Huettel M, Kostka JE, Konstantinidis KT. (2015). Microbial community successional patterns in beach sands impacted by the Deepwater Horizon oil spill. ISME J 9: 1928–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz HN, Jorgensen BB. (2001). Big bacteria. Annu Rev Microbiol 55: 105–137. [DOI] [PubMed] [Google Scholar]
- Scott NM, Hess M, Bouskill NJ, Mason OU, Jansson JK, Gilbert JA. (2014). The microbial nitrogen cycling potential is impacted by polyaromatic hydrocarbon pollution of marine sediments. Front Microbiol 5: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon I, Morowitz MJ, Thomas BC, Costello EK, Relman DA, Banfield JF. (2013). Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res 23: 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spier C, Stringfellow WT, Hazen TC, Conrad M. (2013). Distribution of hydrocarbons released during the 2010 MC252 oil spill in deep offshore waters. Environ Pollut 173: 224–230. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USCG. (2012). Polluting Incidents In and Around US Waters. A Spill/Release Compendium: 1969–2011.
- Valentine DL, Fisher GB, Bagby SC, Nelson RK, Reddy CM, Sylva SP et al. (2014). Fallout plume of submerged oil from Deepwater Horizon. Proc Natl Acad Sci USA 111: 15906–15911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beilen JB, Funhoff EG, van Loon A, Just A, Kaysser L, Bouza M et al. (2006). Cytochrome P450 alkane hydroxylases of the CYP153 family are common in alkane-degrading eubacteria lacking integral membrane alkane hydroxylases. Appl Environ Microbiol 72: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beilen JB, Kingma J, Witholt B. (1994). Substrate specificity of the alkane hydroxylase system of Pseudomonas oleovorans GPo1. Enzyme Microbial Technol 16: 904–911. [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel M, Salman-Carvalho V, Woyke T, Richter M, Schulz-Vogt HN, Flood BE et al. (2016). Single-cell sequencing of thiomargarita reveals genomic flexibility for adaptation to dynamic redox conditions. Front Microbiol 7: 964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Scott AJ. (2012). Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 28: 1033–1034. [DOI] [PubMed] [Google Scholar]
- Yan B, Passow U, Chanton JP, Nothig EM, Asper V, Sweet J et al. (2016). Sustained deposition of contaminants from the Deepwater Horizon spill. Proc Natl Acad Sci USA 113: E3332–E3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S, Cruz-Garcia C, Sanford R, Ritalahti KM, Loffler FE. (2015). Denitrification versus respiratory ammonification: environmental controls of two competing dissimilatory NO3(-)/NO2(-) reduction pathways in Shewanella loihica strain PV-4. ISME J 9: 1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zedelius J, Rabus R, Grundmann O, Werner I, Brodkorb D, Schreiber F et al. (2011). Alkane degradation under anoxic conditions by a nitrate-reducing bacterium with possible involvement of the electron acceptor in substrate activation. Environ Microbiol Rep 3: 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zukunft P. (2010). Operational Science Advisory Team Report (OSAT).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.