Abstract
Aerobic methanotrophic bacteria have evolved a specialist lifestyle dependent on consumption of methane and other short-chain carbon compounds. However, their apparent substrate specialism runs contrary to the high relative abundance of these microorganisms in dynamic environments, where the availability of methane and oxygen fluctuates. In this work, we provide in situ and ex situ evidence that verrucomicrobial methanotrophs are mixotrophs. Verrucomicrobia-dominated soil communities from an acidic geothermal field in Rotokawa, New Zealand rapidly oxidised methane and hydrogen simultaneously. We isolated and characterised a verrucomicrobial strain from these soils, Methylacidiphilum sp. RTK17.1, and showed that it constitutively oxidises molecular hydrogen. Genomic analysis confirmed that this strain encoded two [NiFe]-hydrogenases (group 1d and 3b), and biochemical assays revealed that it used hydrogen as an electron donor for aerobic respiration and carbon fixation. While the strain could grow heterotrophically on methane or autotrophically on hydrogen, it grew optimally by combining these metabolic strategies. Hydrogen oxidation was particularly important for adaptation to methane and oxygen limitation. Complementary to recent findings of hydrogenotrophic growth by Methylacidiphilum fumariolicum SolV, our findings illustrate that verrucomicrobial methanotrophs have evolved to simultaneously utilise hydrogen and methane from geothermal sources to meet energy and carbon demands where nutrient flux is dynamic. This mixotrophic lifestyle is likely to have facilitated expansion of the niche space occupied by these microorganisms, allowing them to become dominant in geothermally influenced surface soils. Genes encoding putative oxygen-tolerant uptake [NiFe]-hydrogenases were identified in all publicly available methanotroph genomes, suggesting hydrogen oxidation is a general metabolic strategy in this guild.
Introduction
Aerobic methane-oxidising bacteria (methanotrophs) consume the potent greenhouse gas methane (CH4) (Kirschke et al., 2013). They serve as the primary biological sink of atmospheric methane (~30 Tg annum−1) (Hanson and Hanson, 1996) and, together with anaerobic methane-oxidising archaea, also capture the majority of biologically and geologically produced CH4 before it enters the atmosphere (Oremland and Culbertson, 1992). Relative to their global impact as greenhouse gas mitigators, aerobic methanotrophs exhibit low phylogenetic diversity and are presently limited to 26 genera in the Alphaproteobacteria and Gammaproteobacteria (Euzéby, 1997), two candidate genera in the phylum Verrucomicrobia (Op den Camp et al., 2009; van Teeseling et al., 2014), and two representatives of candidate phylum NC10 (Ettwig et al., 2010; Haroon et al., 2013). Reflecting their aerobic methylotrophic lifestyle, methanotrophs thrive in oxic–anoxic interfaces where CH4 fluxes are high, including peat bogs, wetlands, rice paddies, forest soils and geothermal habitats (Singh et al., 2010; Knief, 2015). However, they also exist within soil and marine ecosystems where CH4 and oxygen (O2) are more variable (Knief et al., 2003; Tavormina et al., 2010; Knief, 2015).
Based on current paradigms, aerobic methanotrophs are thought to primarily grow on one-carbon (C1) compounds in the environment (Dedysh et al., 2005). All species can grow by oxidising CH4 to methanol via particulate or soluble methane monooxygenase. They subsequently oxidise methanol to carbon dioxide (CO2), yielding reducing equivalents (e.g. NADH) for respiration and biosynthesis. Proteobacterial methanotrophs generate biomass by assimilating the intermediate formaldehyde via the ribulose monophosphate or serine pathways (Hanson and Hanson, 1996). In contrast, verrucomicrobial methanotrophs oxidise methanol directly to formate (Keltjens et al., 2014) and generate biomass by fixing CO2 via the Calvin–Benson–Bassham cycle (Khadem et al., 2011). While these specialist C1-based metabolisms are thought to be the primary growth strategy under optimal conditions (i.e. CH4 and O2 replete conditions), they would presumably be less effective in dynamic environments where CH4 and oxidant availability are likely to fluctuate. To add to this complexity, the methane monooxygenase reaction (CH4+O2+[NAD(P)H+H+]/QH2→CH3OH+NAD(P)+/Q+H2O) (Hakemian and Rosenzweig, 2007) is metabolically demanding, given it requires simultaneous sources of CH4, endogenous reductant (NAD(P)H or quinol) and exogenous O2 to proceed. Methanotrophs therefore must carefully allocate resources to meet carbon, energy and reductant demands (Hanson and Hanson, 1996). This complex balancing act provokes that, in order to be viable in environments limited for CH4 and O2 gases (Knief et al., 2003; Tavormina et al., 2010), methanotrophs should be able to supplement C1 usage with other energy-yielding strategies.
Recent pure culture studies have provided evidence that CH4-oxidising bacteria are indeed more metabolically versatile than previously thought. A minority of conventional methanotrophs can meet energy demands by oxidising the trace concentrations of CH4 (1.8 ppmv) found in the atmosphere (Kolb et al., 2005; Ho et al., 2013; Cai et al., 2016). Contrary to the long-held paradigm that methanotrophs are obligate methylotrophs, species from three alphaproteobacterial genera have been shown to grow on simple organic acids, alcohols and short-chain alkane gases (Dedysh et al., 2005; Crombie and Murrell, 2014). Most recently, it has been shown that some methanotrophs are not exclusive heterotrophs: the verrucomicrobium Methylacidiphilum fumariolicum SolV can sustain chemolithoautotrophic growth on molecular hydrogen (H2) through the activity of two [NiFe]-hydrogenases (Mohammadi et al., 2016). Proteobacterial methanotrophs can also consume H2, though to date this process has only been reported as providing reductant to supplement methanotrophic growth (Chen and Yoch, 1987; Shah et al., 1995; Hanczár et al., 2002). Our recent findings demonstrating a widespread distribution and diversity of hydrogenases in aerobic bacteria, in specific methanotrophs (Greening et al., 2014a,2014b, 2015, 2016), led us to surmise that H2 metabolism could serve a multifaceted role in adaptation of methanotrophic bacteria to their environment. Specifically, H2 may serve as an important electron donor for the organism to meet carbon, energy and reductant demands in response to fluctuations in CH4 and oxidant availability.
In this work, we addressed this hypothesis by conducting an interdisciplinary investigation of the role of H2 in defining the physiology and ecology of verrucomicrobial methanotrophs. Evidence obtained from in situ field studies indicate that Verrucomicrobia simultaneously oxidised CH4 and H2 in geothermally heated soils in Rotokawa, New Zealand, suggesting they are mixotrophic with respect to energy metabolism. Pure culture studies on a verrucomicrobium representative isolated from this site confirmed that the microorganism grew most efficiently through a mixotrophic lifestyle and depended on H2 consumption to acclimate to fluctuations in CH4 and O2 availability. Integrating these findings with genome surveys, we propose that H2 oxidation expands the ecological niche of methanotrophs, enabling them to meet energy and biomass demands in dynamic environments where O2 and CH4 concentrations are variable. We provide evidence that, while methanotrophic bacteria are often pervasively viewed as C1 specialists, their niche space is likely broader than previously recognised. Combining heterotrophic and lithotrophic electron donors allows for a more flexible growth/survival strategy with clear ecological benefits (Semrau et al., 2011).
Materials and methods
Environmental sampling
Soil samples (~50 g) were collected every 10 cm from the surface of the Rotokawa sampling site (38°37′30.8″S, 176°11′55.3″E) to a maximum depth of 50 cm. Soil temperatures were measured in the field using a 51II single input digital thermometer (Fluke, Everett, WA, USA). The pH of the soil (1 g in 10 ml of dH2O) was measured upon returning to the laboratory using a model HI11310 pH probe (Hanna Instruments, Woonsocket, RI, USA). Soil gas samples were collected every 10 cm using a custom-built gas-sampling probe equipped with a 1 l gas-tight syringe (SGE Analytical Science, Melbourne, VIC, Australia). Gas samples were collected and stored at 25 °C in 50 ml Air & Gas Sampling Bags (Calibrated Instruments, McHenry, MD, USA) and were processed within 48 h on a 490 MicroGC (Agilent Technologies, Santa Clara, CA, USA) equipped with Molecular Sieve 5A with a heated injector (50 °C, back-flush at 5.10 s, column at 90 °C, 150 kPa), a PoraPak Q column with a heated injector (50 °C, no back-flush, column at 70 °C, 50 kPa) and a 5CB column with a heated injector (50 °C, no back-flush, column at 80 °C, 150 kPa). CH4 and H2 gas consumption by soil microbial communities was determined by incubating 1 g of soils collected from the Rotokawa sampling site (depth <10 cm) in 112 ml gas-tight serum bottles at 37 and 50 °C. The serum bottle headspaces were amended with 300 ppmv H2 or 400 ppmv CH4. Headspace CH4 and H2 mixing ratios were measured with a PeakPerformer gas chromatograph (Peak Laboratories, Mountain View, CA, USA) equipped with a flame ionising detector (FID: CH4) and a PP1 Gas Analyzer (Peak Laboratories, Mountain View, CA, USA) equipped with a reducing compound photometer (RCP: H2).
Isolation and cultivation of Methylacidiphilum sp. RTK17.1
Soil samples (1 g) from the first 10 cm of soil at the Rotokawa sampling site were inoculated into serum bottles containing 50 ml media (pH 2.5). All cultivations were performed in a V4 mineral medium as described previously (Dunfield et al., 2007) but with the addition (0.2 μm) of rare earth elements lanthanum and cerium (Pol et al., 2014). CH4 (10% v/v) and CO2 (1%) were added to an air headspace and samples were incubated at 60 °C with shaking 150 r.p.m. CH4 in the headspace was monitored with a PeakPerformer gas chromatograph (Peak Laboratories) equipped with an FID. Following several passages (10% v/v) into liquid media, enrichments were transferred onto solid media. Following several weeks incubation (60 °C) single colonies were re-streaked before being transferred back into liquid media. Isolate identity was confirmed via sequencing of the 16S rRNA gene (Macrogen, South Korea) using bacterial 9f/1492r primers (Weisburg et al., 1991).
Bioreactor and batch cultivation
Methylacidiphilum sp. RTK17.1 was cultivated to the stationary phase for subsequent hydrogenase activity and oxygen respiration measurements in a semi-continuous static-liquid fed-batch bioreactor (New Brunswick; volume 1 l, pH control 2.5, temp 50 °C, agitation 100 r.p.m.) in an artificial headspace composed of 10% CH4, 10% H2, 20% O2, 40% CO2 (v/v, balance N2; flow rate 60 ml min−1) equipped with headspace recirculation and automated sampling via a 490 MicroGC (Agilent Technologies). Gas mixtures were supplied for 2 min every hour at a rate of 60 ml min−1. Gas feeds for the bioreactor, batch and chemostat cultivation-based experimental work are presented as headspace compositions. For batch culture experiments, 350 ml cultures (in triplicate) were incubated in 1 l rubber-stoppered Schott bottles. The headspace of the bottles was amended with different mixing ratios of H2, CH4, O2 and CO2 gas, as described in figure legends. Acetylene gas was added in some experiments (4% v/v) to inhibit MMO activity as previously described (Bédard and Knowles, 1989). Finally, to determine if growth was enhanced in the presence of H2, 20 paired cultures (n=40) of Methylacidiphilum sp. RTK17.1 were incubated with or without 1% (v/v) H2 in an air headspace supplemented with (v/v) 10% CH4 and 1% CO2. Cultures were incubated in a custom test tube oscillator (agitation 1.2 Hz; Terratec, Hobart, TAS, Australia). Following 7 days incubation, total protein was determined by the Bradford assay (Bradford, 1976). Statistical significance of observed differences of growth yields was determined using a Student’st-test (α=0.05). Headspace mixing ratios of H2 and CH4 were monitored throughout batch experiments by GC as described above.
Chemostat cultivation
Chemostat cultivation of Methylacidiphilum sp. RTK17.1 was performed to investigate the influence of H2 on growth under O2-replete and O2-limiting conditions. A 1 l bioreactor (BioFlo 110; New Brunswick Scientific, Edison, NJ, USA) was used for these studies. Cultures were incubated at 50 °C and pH 2.5 with continuous stirring (800 r.p.m.). Bioreactor volume was kept constant at 0.5 l by automatic regulation of the culture level. V4 mineral media was supplied at a constant flow rate of 10 ml h−1 (D=0.02 h−1). Dissolved O2 was monitored using an InPro 6810 Polarographic Oxygen Sensor (Mettler-Toledo, Columbus, OH, USA). Custom gas mixtures were prepared in a compressed gas cylinder and supplied to the chemostat at a rate of 10 ml min−1 using a mass flow controller (El-flow, Bronkhorst, Netherlands). Gas mixtures contained approximately (v/v) 3% CH4 and 26% CO2 for all experiments. For O2-replete and O2-limiting conditions, influent O2 was supplied at (v/v) 14.1% and 3.5%, respectively. Within respiring cultures, these values corresponded to 57.5% and 0.17% oxygen saturation. High, medium and low H2 experiments consisted of (v/v) 1.9%, 0.7% and 0.4% H2 additions. The balance of all gas mixtures was made up with N2. Cell density in liquid samples was monitored by measuring turbidity at 600 nm using a Ultrospec 10 cell density meter (Amersham Bioscience, UK). One unit of OD600 was found to be equivalent to 0.43 g l−1 cell dry weight for Methylacidiphilum sp. RTK17.1. After achieving a steady-state condition as determined by OD600, influent and effluent gas concentrations were monitored over several days using a 490 MicroGC (Agilent Technologies). Biomass cell dry weight was used to calculate growth rate and specific gas consumption rate.
Whole-cell biochemical assays
Hydrogenase activity of Methylacidiphilum sp. RTK17.1 was measured in stationary-phase cultures harvested from the bioreactor. For amperometric measurements, whole cells were concentrated 5-, 10-, 20- and 30-fold by centrifugation followed by resuspension in V4 mineral medium (pH 3.0). Rate of H2 oxidation was measured at 50 °C using an H2-MR microsensor (Unisense, Denmark) as previously described (Berney et al., 2014b; Greening et al., 2015). For colourimetric assays, 500 ml culture was harvested by centrifugation (15 min, 5000 × g, 4 °C) and treated as previously described (Greening et al., 2014a, 2015) to prepare crude, cytosolic and membrane fractions for analysis. To test for hydrogenase activity, samples (20 μg protein) from each cell fraction were incubated with 1 ml 50 mm potassium phosphate buffer (pH 7.0) and 50 μm benzyl viologen for 8 h in an anaerobic chamber (5% H2, 10% CO2, 85% N2 (v/v)). Debris was removed by centrifugation (15 min, 10 000 × g, 4 °C) and the absorbance of the supernatants was read at 604 nm in a Jenway 6300 spectrophotometer (Cole Palmer, UK). O2 consumption experiments were performed on cell suspensions of Methylacidiphilum sp. RTK17.1 to determine the influence of endogenous glycogen catabolism (Khadem et al., 2012a) on O2-dependent hydrogenase measurements. For these experiments, 2 ml cells (OD600 1.0) were added to a Clarke-type oxygen electrode and incubated at 50 °C for up to 12 min without the addition of exogenous energy sources. Cell suspensions were treated with the protonophore 1 μm, carbonyl cyanide m-chlorophenyl hydrazine (CCCP), 1 mm iodoacetamide (an inhibitor of glycolysis) and 1 mm potassium cyanide (KCN) to determine whether observed rates of O2 consumption were a consequence of glycogen catabolism. An oxygen solubility value of 220 nmol ml−1 was used for calculations. Values were expressed as nmol O2 min−1 (mg protein)−1. Protein was determined, from lysed cell pellets, using the BCA assay (Thermo Fisher Scientific, Waltham, MA, USA), with bovine serum albumin as a standard.
CO2 fixation was measured by incubating cultures with 14C-labelled HCO3− (as CO2 at medium pH of 2.3). Triplicate cultures were initially grown at 50 °C with a headspace of CH4 (20%), CO2 (10%), H2 (10%) in air. At late exponential stage growth, the headspaces of cultures (and heat-killed controls) were replaced by sparging with N2 (10 min) and then amended with H2 (8%), CO2 (8%) and O2 (1%). 15 μCi of 14C-HCO3− (51 mCi mmol−1; American Radiolabeled Chemical, St Louis, MO, USA) were added to each culture. Given that the pKa for HCO3− at 50 °C is ~6.8 (Amend and Shock, 2001), it was assumed that all added radiolabelled substrate equilibrated with unlabelled CO2 pools immediately. Subsamples (1 ml) were harvested by centrifugation at regular time points, washed with sterile medium and subjected to liquid scintillation counting using Cytoscint cocktail as previously described (Urschel et al., 2015). Disintegrations per minute were converted to rates of CO2 fixed using previously described approaches (Urschel et al., 2015).
Details of nucleic acid extraction, amplification, genome sequencing, environmental quantitative PCR and soil microbial community composition determination methodologies are presented in the Supplementary Information.
Results and discussion
Verrucomicrobia-dominated surface soils serve as a sink of geothermally derived H2 and CH4 in Rotokawa geothermal field
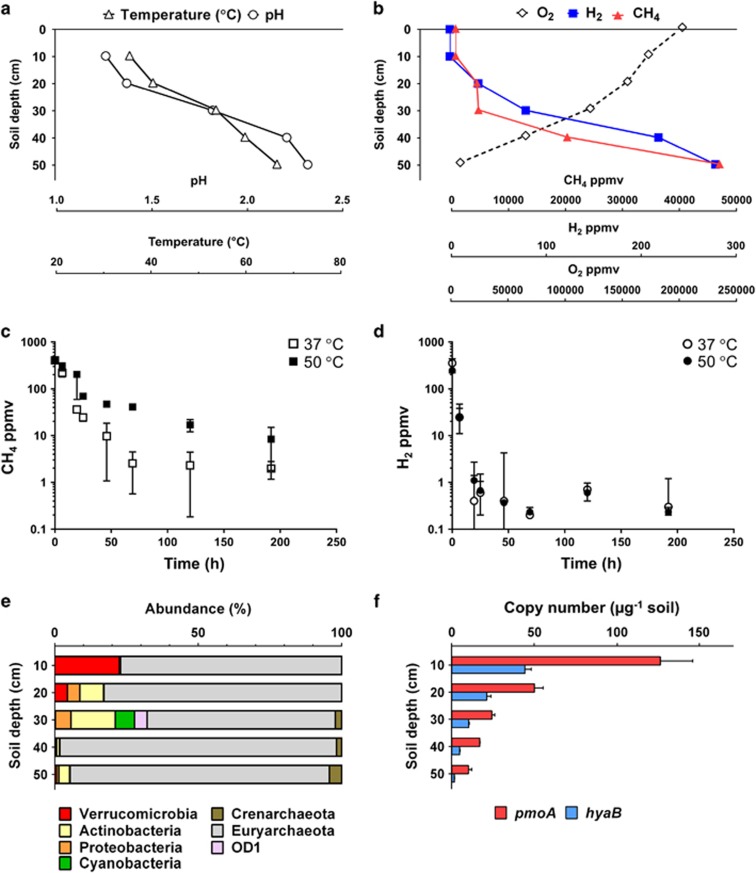
We performed a geochemical, molecular and biochemical survey of CH4 and H2 metabolism in an acidic geothermal soil in Rotokawa, New Zealand. The Rotokawa geothermal field is a predominately steam-driven system dominated by acidic and sulphurous springs and heated soil features. We selected a geothermally heated and acidic soil where previous studies have indicated methanotrophic activity (Sharp et al., 2014). Substantial vertical gradients in temperature, pH and mixing ratios of CH4, H2 and O2 were observed in the soil profile (Figure 1a). Consistent with the geothermal activity at the site, high soil mixing ratios of CH4 (47 000 ppmv) and H2 (280 ppmv) were detectable at the deepest soil depths sampled. The levels of both gases decreased in the upper 30 cm of soil and, in the case of H2, dropped towards atmospheric levels by 10 cm depth (Figure 1b). These sharp decreases suggested that there were active methanotrophs and hydrogenotrophs in the oxic zone of the soil that consume most geothermally derived gas before it is emitted into the atmosphere. Indeed, microcosm incubations containing surface soils and associated communities rapidly consumed H2 and CH4 introduced into ambient air headspaces. Rates of H2 oxidation exceeded that of CH4, suggesting that H2 serves as a major energy source for this geothermal soil community (Figures 1c and Figures 1d).
Figure 1.
Geochemical, biochemical and molecular profile of CH4 and H2 oxidation at a geothermal field in Rotokawa, New Zealand. (a) Temperature and pH of the soils at different depths. (b) Soil mixing ratios of CH4, H2 and O2 at different depths. (c) Oxidation of CH4 by surface soils. (d) Oxidation of H2 by surface soils. In both (c) and (d), soil samples of 1 g were collected from the first 10 cm of soil from the profile and incubated in serum vials containing a CH4- or H2-supplemented ambient air headspaces. The average and standard deviation of triplicate samples are shown. (e) Community structure of the study site at different soil depths. Illumina 16S rRNA gene sequencing was performed on total genomic DNA extracted from samples taken at 10–50 cm soil depth. Non-rarefied abundance results (%) are shown for all OTUs (>100 reads) from 130 289 total sequence reads (with an average of 26 058 per sample depth). Consistent with a methanotrophic lifestyle, all verrucomicrobial OTUs were further classified into the family Methylacidiphilaceae. (f) Abundance of genes encoding Verrucomicrobia-type particulate methane monooxygenase (pmoA) and aerobic uptake hydrogenase (hyaB) plotted as a function of soil depth. Error bars represent the standard deviation of triplicate measurements on each extract. Differences in the copy number between pmoA and hyaB are attributable to the multiple isoforms of pmoA encoded in Methylacidiphilum spp. genomes.
To infer the microorganisms responsible for CH4 and H2 uptake, we determined the microbial community structure of the soil profile. Consistent with findings in other acidic soil ecosystems (Golyshina, 2011; Sharp et al., 2014; Lee et al., 2016), the euryarchaeotal order Thermoplasmatales was dominant at all depths. Methanotrophic verrucomicrobial genera, specifically Methylacidiphilum spp. and Methylacidimicrobium spp., were the dominant bacterial OTUs in surface soils and accounted for 47% of all bacterial 16S rRNA gene sequences in the soil profile (Figure 1e). Bacteria from these genera have been previously isolated in acidic geothermal soils in New Zealand (Dunfield et al., 2007; Sharp et al., 2014), Kamchatka (Islam et al., 2008) and Italy (Pol et al., 2007; van Teeseling et al., 2014). As the only known acidophilic methanotrophs (Op den Camp et al., 2009), it is probable that the verrucomicrobial phylotypes were solely responsible for CH4 consumption in this ecosystem. Moreover, given putative uptake [NiFe]-hydrogenases have been detected in the genomes of Methylacidiphilales but not in Thermoplasmatales (Hou et al., 2008; Khadem et al., 2012b; Greening et al., 2016), it was likely that the Verrucomicrobia detected in these soils serve as major sinks of H2 in this ecosystem. To test this possibility, we designed PCR primers to detect the presence of genes encoding the large subunits of the three particulate methane monooxygenases (pmoA) and a single oxygen-tolerant uptake hydrogenase (hyaB) encoded in the genome of Methylacidiphilum infernorum V4 (Hou et al., 2008) (Supplementary Table S1). These primers were applied in qPCRs on DNA extracts from the Rotokawa soil profile. Both the hydrogenase and methane monoxygenase genes were detected at all depths, with the most abundant templates detected in the top 10 cm of soil (Figure 1f), corresponding to the zones with the highest relative abundance of Verrucomicrobia-affiliated sequences and where the lowest CH4 and H2 soil gas concentrations were detected (Figure 1b).
A verrucomicrobial strain isolated from Rotokawa constitutively oxidises CH4 and H2 gas
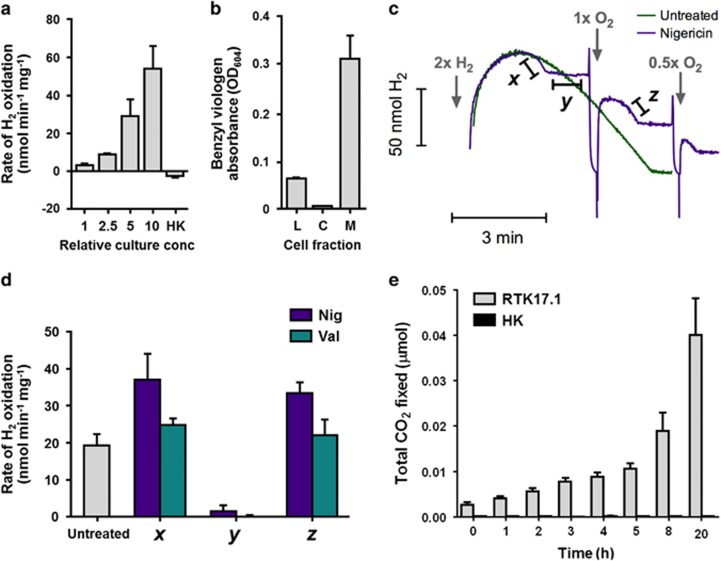
To gain insight into the metabolic strategies that Verrucomicrobia use to dominate bacterial assemblages in geothermally heated acidic soil ecosystems, we isolated a thermotolerant methanotroph from surface soils. The strain, Methylacidiphilum sp. RTK17.1 (Supplementary Table S2), grew optimally at pH 2.5, 50 °C (Tmax 60 °C) and shared 99% 16S rRNA gene sequence identity with Methylacidiphilum infernorum V4 (Dunfield et al., 2007). Bacteriological characterisation confirmed that the strain, in common with other verrucomicrobial methanotrophs (Khadem et al., 2012a, 2011), oxidised CH4, fixed CO2 and accumulated glycogen. In addition, cultures rapidly consumed exogenous H2 (Supplementary Figures S2 and S3). Real-time amperometric measurements confirmed that the strain oxidised H2 under oxic conditions at rates proportional to increases in cell density (Figure 2a). H2 oxidation occurred in all batch culture conditions tested, including when CH4 was absent (Supplementary Figure S1A), when CH4 was in excess (Supplementary Figure S1C), and following inhibition of CH4 oxidation with acetylene (Supplementary Figure S2). This suggests that H2 and CH4 are oxidised independently serving to energise the respiratory chain through the reduction of the quinone pool. Moreover, this expands the role of hydrogenases in aerobic methanotrophs beyond their previously suggested role of providing reductant for pMMO (Shah et al., 1995; Hanczár et al., 2002). The observation that RTK17.1 can constitutively oxidise H2 and CH4 parallels results from the soil study showing that both H2 and CH4 are simultaneously oxidised (Figure 1). Considering that Verrucomicrobia are dominant among taxa putatively capable of oxidising H2 or CH4, this provides further evidence that verrucomicrobial methanotrophs adopt a mixotrophic lifestyle with respect to their energy metabolism.
Figure 2.
H2 oxidation drives aerobic respiration and CO2 fixation in Methylacidiphilum sp. RTK17.1. (a) Real-time oxidation of H2 by bioreactor-cultivated whole cells. Rates of H2 uptake were measured amperometrically using a H2 microsensor. Density dependence and heat sensitivity (HK) of the process are shown. (b) Localisation of hydrogenase activity in cell membranes. Activity was measured colourimetrically by incubating cell fractions in an anaerobic chamber in the presence of H2 and the artificial electron acceptor/redox dye benzyl viologen. The protein concentration-normalised absorbance of activity in cell lysates (L), cytosols (C) and membranes (M) are shown. (c) Aerobic respiratory dependence of H2 uptake in whole cells. Real-time traces in untreated cells and nigericin-treated cells are shown. The relative amounts of H2 and O2 added at specific time points are shown. (d) Rates of hydrogen oxidation of untreated, nigericin-treated and valinomycin-treated cells. For the uncoupler-treated cultures, the initial (x), O2-limiting (y) and O2-restored (z) rates of H2 oxidation are shown, which correspond to the rates highlighted in panel (c). Endogenous glycogen catabolism likely contributed to oxygen limitation (y) observed in nigericin-treated cells (Supplementary Figure S5B). (e) CO2 fixation by batch-cultivated whole cells cultivated under microoxic growth conditions with H2 and O2 as the sole reductant and oxidant (Supplementary Figure S2A). 14C-labelled CO2 is incorporated into biomass in live but not heat-killed (HK) cultures. CO2 fixed per mol of biomass in live and heat-killed cells is presented as a function of time.
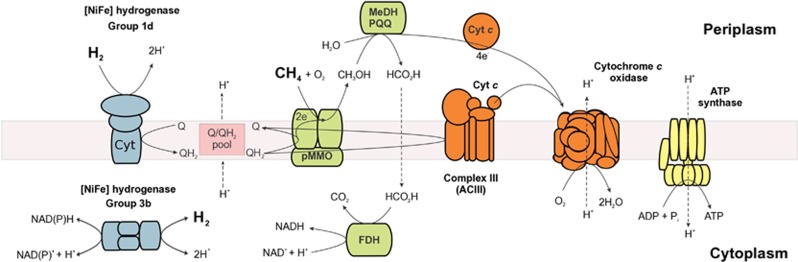
We sequenced the genome of Methylacidiphilum sp. RTK17.1 to obtain further insights into the potential functionality of this taxon (Supplementary Table S2). Genes encoding key enzymes and pathways for CH4 oxidation to CO2, CO2 fixation through the Calvin–Benson–Bassham pathway, and aerobic respiration (Figure 3) were highly conserved with those identified in other Methylacidiphilum strains (Hou et al., 2008; Khadem et al., 2012b; Erikstad and Birkeland, 2015). We also detected two [NiFe]-hydrogenase-encoding gene clusters in the genome (Supplementary Figure S3) and confirmed their expression during aerobic growth with CH4 and H2 by RT-PCR (Supplementary Figure S4). The gene clusters were classified as groups 1d (hyaABC) and 3b (hyhBGSL) [NiFe]-hydrogenases based on phylogenetic affiliation with biochemically characterised enzymes (Supplementary Figure S3; Greening et al., 2016; Søndergaard et al., 2016). Biochemically characterised group 1d [NiFe]-hydrogenase are H2-uptake multimeric proteins that are membrane-bound via their cytochrome b subunit (hyaC) and function by transferring electrons into the respiratory chain via the quinone pool (Fritsch et al., 2011). Consistent with the observed activity of the [NiFe]-hydrogenase in the presence of O2 (Figure 2a), enzymes of this class are predicted to be O2-tolerant due to the presence of a novel [4Fe3S] cluster that protects the O2-sensitive active site from oxidative damage (Fritsch et al., 2011; Shomura et al., 2011). Indeed, the six cysteine residues required to ligate such a cluster were conserved in the deduced RTK17.1 HyaB protein sequence. In comparison, biochemically characterised group 3b hydrogenases are reversible cytosolic enzymes that are relatively O2-sensitive (Kwan et al., 2015); they directly couple NAD(P)H oxidation to H2 formation during fermentation (Berney et al., 2014a) and, in some cases, H2 oxidation by these enzymes supports CO2 fixation through the production of reduced electron carriers (Yoon et al., 1996). The RTK17.1 [NiFe]-hydrogenase combination differs from Methylacidiphilum fumariolicum SolV, where group 1 h/5 (hhyLH; a putative high-affinity H2 uptake hydrogenase) and group 1d [NiFe]-hydrogenases (annotated as hupSLZ) were reported (Mohammadi et al., 2016), with our previous survey (Greening et al., 2016) also showing SolV encodes a group 3b enzyme (Supplementary Figure S3).
Figure 3.
Proposed model of methane (CH4) and hydrogen (H2) oxidation in Methylacidiphilum sp. RTK17.1. During mixotrophic growth, the oxidation of both H2 and CH4 yields reducing equivalents in the form of reduced quiones (QH2). A large proton-motive force is generated and sufficient ATP is produced for growth via an H+-translocating F1Fo-ATP synthase. Some of the quinol generated through H2 oxidation provides the electrons necessary for pMMO catalysis. Following CH4 oxidation by pMMO, ensuing reactions catalysed by an XoxF-type methanol dehydrogenase (MeDH) and formate dehydrogenase (FDH) contribute additional reductant (cyt c and NADH) into the respiratory chain for ATP production and growth (Keltjens et al., 2014). NADH reduced through the actions of the formate dehydrogenase and H2-dependent group 3b [NiFe]-hydrogenase is used to support CO2 fixation through the Calvin–Benson–Bassham cycle. Respiratory complexes I and II are not shown but are encoded in the genome of Methylacidiphilum sp. RTK17.1 (Supplementary Table S1).
H2 oxidation supports aerobic respiration and CO2 fixation in the verrucomicrobial isolate
The observation that RTK17.1 encodes and utilises [NiFe]-hydrogenases prompted biochemical studies to investigate the role of H2 in the metabolism of this bacterium. Biochemical assays targeting the group 1d [NiFe]-hydrogenase demonstrated that it is a membrane-bound uptake hydrogenase linked to the aerobic respiratory chain, consistent with our genome-based predictions. Fractionation experiments confirmed the activity was membrane-localised, as shown by the 31-fold increase in activity in membranes when compared to the cytosolic fraction (Figure 2b). We next tested the effect of the ionophores nigericin and valinomycin on rates of H2 oxidation on whole cells (in the absence of CH4). These compounds (nigericin and valinomycin) dissipate components of the electrochemical gradient used for ATP synthesis (pH and charge gradient, respectively) and the cellular response is to increase respiration to replenish the electrochemical gradient (Cook et al., 2014) in a phenomenon known as uncoupling. H2 oxidation increased upon treatment with these ionophores (Figures 2c and d), showing hydrogenase activity behaves as expected from a component of the energy-conserving respiratory chain. This uncoupled activity rapidly ceased, due to O2 consumption by the cells suspended in the sealed chamber, but could be restored by further supplementation with O2. These results show hydrogenase is a bona fide component of this microorganism’s respiratory chain and is coupled to the activity of terminal cytochrome oxidases. Under these conditions, the onset of O2-limitation was likely exacerbated by the catabolism of endogenous glycogen reserves (Supplementary Figure S5). Collectively, these findings demonstrate that this group 1d [NiFe]-hydrogenase is a membrane-bound, respiratory-linked, O2-tolerant/dependent enzyme that drives ATP synthesis as has been observed in other aerobic hydrogenotrophs.
To test whether H2 oxidation coupled to O2 reduction could support CO2 fixation in RTK17.1, we transferred mixotrophically grown, log phase cultures into a new microoxic headspace (O2 1% v/v) in which H2 (8% v/v of the headspace) was present as the sole exogenous electron donor and CO2 (8% v/v) as the sole carbon source. Trace 14CO2 (0.1% of total CO2 supplied) was added and the amount fixed into biomass sampled over time was determined by measuring disintegrations per minute (DPM) via liquid scintillation counting. We observed systematic increases in DPMs associated with cells sampled over a 20 h period relative to controls, indicating that CO2 was rapidly incorporated into biomass in a time-dependent manner. The number of DPMs associated with cells after 20 h of incubation were 200-fold greater in live than heat-killed cells (Figure 2e), showing that biological CO2 fixation occurs in cultures supplied with H2 as the sole reductant and O2 as the sole oxidant. This activity was not observed in the absence of exogenous H2, indicating that H2 serves as the source of reductant for CO2 fixation under these conditions.
H2 oxidation supports adaptation of verrucomicrobial methanotrophs to CH4 and O2 limitation
The finding that Methylacidiphilum sp. RTK17.1 couples H2 oxidation to aerobic respiration and carbon fixation suggests that it can grow chemolithoautotrophically. Consistent with this hypothesis, we observed a small but significant increase in biomass in cultures grown under microoxic conditions when H2, CO2 and O2 (1% headspace concentration) were supplied as the sole electron donor, carbon source and electron acceptor, respectively (Supplementary Figure S2A). This biomass increase was concomitant with increased amounts of CO2 fixed (Figure 2e), and the amount of carbon fixed per cell (40 to 80 fmol cell−1) produced over this time period was consistent with biomass yields from other studies (Maestrini et al., 2000). However, the observed growth rates (0.005 h−1) were substantially lower than observed when RTK17.1 was supplied with CH4, CO2 and H2 (0.037 h−1) and when Methylacidiphilum fumariolicum SolV was grown autotrophically in similar conditions (0.047 h−1) (Mohammadi et al., 2016). We also observed that autotrophic growth was only sustained when RTK17.1 was incubated under microoxic conditions (1% O2) rather than in an oxic (20% O2) headspace. We speculate that, under such microoxic conditions, sufficient O2 is available to drive hydrogenotrophic aerobic respiration through activity of the group 1d [NiFe]-hydrogenase. Simultaneously, it is likely that the O2-sensitive group 3b [NiFe]-hydrogenase can remain active and is able to supply reducing equivalents required for CO2 fixation. In support of this notion, H2 oxidation sustained energy-conservation via the group 1d hydrogenase of non-growing CH4-limited cultures in the presence of ambient O2 (Supplementary Figure S2C) and enhanced growth yields in CH4-replete cultures (Supplementary Figure S2D).
To better understand the role of H2 and CH4 oxidation during mixotrophic growth, we compared growth and gas consumption kinetics of the cells cultivated in a chemostat under six different conditions (Table 1). We observed that H2 addition into the feedgas of Methylacidiphilum sp. RTK17.1 increased growth yields under O2-replete and O2-limiting conditions. Whereas CH4 oxidation predominated under O2-replete conditions, the specific consumption rate of H2 increased 80-fold and exceeded rates of CH4 oxidation under O2-limiting conditions. In combination, these results show that Methylacidiphilum sp. RTK17.1 grows mixotrophically and modulates rates of H2 and CH4 consumption in response to the availability of O2 in order to balance energy-generation and carbon fixation.
Table 1. H2 oxidation by Methylacidiphilumsp. RTK17.1 during chemostat cultivation.
| Conditiona,b | Growth ratec |
H2
consumption |
CH4
consumption |
Biomass yieldd | ||
|---|---|---|---|---|---|---|
| (mg l−1 h−1)c | (mmol l−1 h−1)c | (mmol gCDW−1 h−1)c | (mmol l−1 h−1)c | (mmol gCDW−1 h−1)c | (gCDW molCH4−1)c | |
| O2-replete, medium H2 | 9.33 (±0.52) | 0.01 (±0.01) | 0.03 (±0.03) | 1.06 (±0.12) | 2.40 (±0.38) | 8.90 (±1.51)e |
| O2-replete, no H2 | 8.17 (±0.82) | 0.00 | 0.00 | 1.22 (±0.05) | 2.85 (±0.18) | 6.68 (±0.41) |
| O2-limiting, high H2 | 5.57 (±0.16) | 0.78 (±0.02) | 2.66 (±0.11) | 0.55 (±0.03) | 1.89 (±0.11) | 10.11 (±0.64)f |
| O2-limiting, medium H2 | 4.99 (±0.17) | 0.33 (±0.01) | 1.34 (±0.06) | 0.70 (±0.02) | 2.80 (±0.13) | 7.16 (±0.34) |
| O2-limiting, low H2 | 5.60 (±0.31) | 0.19 (±0.00) | 0.73 (±0.04) | 0.71 (±0.01) | 2.68 (±0.16) | 7.87 (±0.45)f |
| O2-limiting, no H2 | 5.40 (±0.26) | 0.00 | 0.00 | 0.80 (±0.05) | 3.11 (±0.27) | 6.79 (±0.55) |
Feedgas was continuously supplied at a rate of 10 ml min−1 with the following composition % (v/v): O2-replete, 14.1% O2-limiting, 3.5%. High, medium and low H2 experiments consisted of 1.9%, 0.7% and 0.4% H2 supply. For all experiments excess CH4 was supplied at 3%, CO2 at 26% with the balance made up with N2. O2 saturations in the medium were 57.5% and 0.17% for the O2-replete and O2-limiting conditions, respectively.
A dilution rate of 0.02 h−1 was maintained for all experiments.
Gram cell dry weight (gCDW) measurements were determined by correlating OD600 measurements to an existing standard curve (OD600=1 corresponds to 0.43 gCDW l−1).
Standard deviation shown in brackets are calculated from a minimum of triplicate measurements.
Denotes significant increase (P-value <0.05) in yield from O2-replete, no H2 result; unpaired t-test, one tailed.
Denotes significant increase (P-value <0.05) in yield from O2-limiting, no H2 result; unpaired t-test, one tailed.
H2 oxidation may be a general ecological strategy for verrucomicrobial and proteobacterial methanotrophs
In this work, we demonstrated that a verrucomicrobial methanotroph adopts a mixotrophic lifestyle both in situ and ex situ. The environmental isolate Methylacidiphilum sp. RTK17.1 sustains aerobic respiration and carbon fixation by using organic (CH4) and inorganic (H2) electron donors either in concert or separately depending on substrate availability. Through the dual use of both electron donors, the bacterium is able to more flexibly adjust its metabolism to meet energy and carbon demands in response to simulated environmental change. A model of how CH4 and H2 metabolism is integrated into the physiology of this microorganism, based in part on genomic information (Supplementary Table S2), is shown in Figure 3. Integrating our genomic, physiological and biochemical findings, we conclude that the group 1d [NiFe]-hydrogenase is a membrane-bound uptake hydrogenase that is directly linked to the aerobic respiratory chain and supplements the quinone pool to power the methane monoxygenase reaction or feed directly into Complex III. The organism is also capable of using H2 as a reductant to support CO2 fixation through the Calvin–Benson-Bassham pathway, likely through the cytosolic NAD(P)-coupled group 3b [NiFe]-hydrogenase. The [NiFe]-hydrogenase combination (group 1d and group 3b) in this RTK17.1 only supports weak autotrophic growth, but provides multiple layers of support for a mixotrophic lifestyle. In addition to supporting growth and survival during periods of CH4 limitation, our data show that H2 is the preferred electron donor during O2-limiting conditions. Under these conditions, rates of H2 consumption increased by 77-fold and exceeded observed rates of CH4 oxidation (Table 1). This is likely to be a consequence of two factors. Firstly, some hydrogenases such as the group 3b [NiFe]-hydrogenase are inhibited at high O2 concentrations (Kwan et al., 2015). Secondly, methanotrophy is more resource-intensive than canonical aerobic hydrogenotrophy, given it requires O2 both as a substrate for methane monooxygenase and as the terminal electron acceptor for respiration (Hakemian and Rosenzweig, 2007).
Comparison of our independent findings with those made by Mohammadi et al. (2016) suggests that verrucomicrobial methanotrophs have evolved a range of strategies to integrate H2 metabolism into their physiology. Both Methylacidiphilum sp. RTK17.1 and Methylacidiphilum fumariolicum SolV are capable of sustaining chemolithoautotrophic growth on H2/CO2 under microoxic conditions. However, the strains grow at drastically different rates of 0.005 and 0.047 h−1 (Mohammadi et al., 2016), respectively, under optimal conditions. These differences may reflect that, while both strains possess group 1d (hyaABC/hupLSZ) and group 3b (hyhBGSL) hydrogenases, SolV has also acquired a group 1 h enzyme (hhyLH) (Greening et al., 2016) with surprisingly fast whole-cell kinetics (Mohammadi et al., 2016). It is possible that, with this enhanced hydrogenase suite, SolV may be able to more efficiently partition electrons derived from H2 oxidation between respiration and carbon fixation. In addition to supporting hydrogenotrophic growth, both organisms also modulate hydrogenase expression and H2 oxidation rates in response to simulated environmental change, such as CH4 and O2 availability (Mohammadi et al., 2016). While the physiological significance of this regulation was not explored in SolV, our studies inferred that H2 co-oxidation with CH4 enhanced yields during CH4 surplus and sustained survival during CH4 limitation in RTK17.1. Further differences between the strains are reflected in the regulatory profile, with the group 1d enzyme constitutively expressed in RTK17.1, but repressed in favour of the group 1 h enzyme under oxic conditions in SolV (Mohammadi et al., 2016). Overall, our findings suggest that SolV may fulfil a similar ecological niche to classical Knallgas bacteria (e.g. Ralstonia eutropha), switching between efficient heterotrophic and autotrophic growth dependent on energy availability. In contrast, H2 metabolism appears to be more important for optimising growth and survival of RTK17.1 in response to energy and O2 availability. In this regard, this organism’s metabolism more closely resembles the mixotrophic strategy employed by Mycobacterium smegmatis (Berney et al., 2014a; Greening et al., 2014b). Future studies would benefit from side-by-side comparisons of these strains under equivalent conditions and further exploration of the physiological role and biochemical features of the group 3b and group 1 h [NiFe]-hydrogenase enzymes.
More generally, we predict that H2 oxidation is likely to support the majority of aerobic methanotrophic bacteria. Whereas only a few methanotrophic genera appear to be capable of heterotrophic generalism (Crombie and Murrell, 2014), genomic surveys (Peters et al., 2015; Greening et al., 2016) show that all 31 publicly available aerobic methanotrophic genomes harbour the capacity to metabolise H2 (Supplementary Figure S3). As with SolV (Mohammadi et al., 2016) and RTK17.1, most of the surveyed methanotroph genomes that were found to encode for [NiFe]-hydrogenases have been shown to support aerobic respiration (groups 1d, 1h, 2a) and carbon fixation (groups 3b, 3d, 1 h) (Greening et al., 2016). Reports showing H2 oxidation by several proteobacterial strains further support the classification of these enzymes as uptake hydrogenases (Chen and Yoch, 1987; Shah et al., 1995; Hanczár et al., 2002). H2 is likely to be a particularly attractive energy source for methanotrophs because of its relative ubiquity when compared to C1 compounds. H2 is biologically produced by diverse organisms across the three domains of life as a result of fermentation, photobiological processes and nitrogen fixation (Peters et al., 2013; Schwartz et al., 2013; Poudel et al., 2016). Moreover, verrucomicrobial and proteobacterial methanotrophs harbouring the recently described group 1 h [NiFe]-hydrogenases (Greening et al., 2014a, 2015) may be capable of scavenging atmospheric H2 to survive CH4 starvation. Considering these observations in concert, it seems likely that hydrogenases in aerobic methanotrophs function to supplement the energetic and reductant requirements in environments where CH4 and O2 gases are limiting or variable. Thus, while methanotrophic bacteria are often pervasively viewed as C1 specialists, we propose that, via the utilisation of hydrogenases as part of a mixotrophic strategy, the niche space of methanotrophs is much broader than previously recognised. Combining heterotrophic and lithotrophic electron donors allows for a more flexible growth/survival strategy, with clear ecological benefits (Semrau et al., 2011). We therefore predict that most methanotrophs will be able to use H2 to support either autotrophic growth, mixotrophic growth or long-term persistence/maintainence.
Finally, our geochemical and microbial community diversity investigation of the Rotokawa geothermal field provides ecological support to our assertion that the metabolic flexibility of methanotrophs enhances niche expansion in situ. We provide genetic and biochemical evidence that methanotrophic Verrucomicrobia inhabiting the near-surface soils co-metabolised CH4 and H2 gas (Figure 1) and in doing so adopt a clear mixotrophic strategy. In acidic geothermal soils, we demonstrated that verrucomicrobial methanotrophs have grown to be the dominant bacterial taxon by simultaneously consuming gases primarily of geothermal and atmospheric origin, that is, CH4 and H2 as energy sources, respectively, CO2 as a carbon source and O2 as oxidant. Their metabolic flexibility also ensures resilience to temporal and spatial variations in the availability of key substrates allowing for CH4 oxidation via the monooxygenase reaction. More generally, the prevalent narrative that methanotrophic bacteria are methylotrophic specialists is based on studies under optimal growth conditions and ignores the requirement of these organisms to adapt to environmental variations requiring a certain level of metabolic versatility. Intimate evolutionary and ecological interactions are likely to have selected for a spectrum of different lifestyles across methanotrophic lineages, ranging from strict C1 specialism to broad substrate generalism, depending on the environment. However, based on the presence of [NiFe]-hydrogenase in numerous methanotroph genomes (Supplementary Figure S3) and the data presented here, we contend it is likely that most methanotrophs depend on H2 oxidation to some extent to support either growth and/or survival. This finding has broad implications for future investigations on the ecology of methanotrophs as well as the biogeochemical cycles of H2 and CH4.
Acknowledgments
This work was supported by Geothermal Resources of New Zealand (CRC, MBS, KMH and JFP) and Environmental Technologies (CRC, DG, BM and CC) research programmes at GNS Science and Scion, respectively. CRC was further supported by Genomes Canada, the Manitoba Research Innovation Fund (MIRF) and The Royal Society of New Zealand (Marsden Grant GNS1601). CG was supported by a CSIRO Office of the Chief Executive Postdoctoral Fellowship and an ARC DECRA Fellowship (DE170100310). KH was supported by a University of Otago Postgraduate Scholarship. ESB acknowledges support for this work from the NASA Astrobiology Insititute (Grant #NNA15BB02A). Ngāti Tahu-Ngāti Whaoa are acknowledged as the iwi having mana whenua (customary rights) over the Rotokawa geothermal field and we thank the Runanga Trust for their support of our research. Relevant gene sequences (accession numbers KU509367-KU509352, KY820885) have been deposited into GenBank for archival storage.
Author contributions
CRC, CG and MBS conceived the study. CRC, MBS, CG, GC, KMH, KH, CC, DJG, RS, ESB and BM contributed to experimental design. CRC, BM, CC, DJG and MHK conducted bioreactor and wet lab experiments. CRC and MBS performed fieldwork, JFP determined microbial community structure, and CRC, MBS and CG conducted genomic analyses. CRC, KH and CG undertook cell respiratory analysis, and ESB and MBS performed the radioactive tracing experiments. CRC, CG, MBS, KH and GC wrote the manuscript.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Amend JP, Shock EL. (2001). Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiol Rev 25: 175–243. [DOI] [PubMed] [Google Scholar]
- Bédard C, Knowles R. (1989). Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol Rev 53: 68–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berney M, Greening C, Conrad R, Jacobs WR, Cook GM. (2014. a). An obligately aerobic soil bacterium activates fermentative hydrogen production to survive reductive stress during hypoxia. Proc Natl Acad Sci USA 111: 11479–11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berney M, Greening C, Hards K, Collins D, Cook GM. (2014. b). Three different [NiFe] hydrogenases confer metabolic flexibility in the obligate aerobe Mycobacterium smegmatis. Environ Microbiol 16: 318–330. [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Cai Y, Zheng Y, Bodelier PLE, Conrad R, Jia Z. (2016). Conventional methanotrophs are responsible for atmospheric methane oxidation in paddy soils. Nat Commun 7: 11728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YP, Yoch DC. (1987). Regulation of two nickel-requiring (inducible and constitutive) hydrogenases and their coupling to nitrogenase in Methylosinus trichosporium OB3b. J Bacteriol 169: 4778–4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook GM, Greening C, Hards K, Berney M (2014). Energetics of pathogenic bacteria and opportunities for drug development. In: Poole RK (ed.). Advances in Bacterial Pathogen Biology, vol. 65. Elsevier Academic Press: Cambridge, MA, USA, pp 1–62.. [DOI] [PubMed]
- Crombie AT, Murrell JC. (2014). Trace-gas metabolic versatility of the facultative methanotroph Methylocella silvestris. Nature 510: 148–151. [DOI] [PubMed] [Google Scholar]
- Dedysh SN, Knief C, Dunfield PF. (2005). Methylocella species are facultatively methanotrophic. J Bacteriol 187: 4665–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunfield PF, Yuryev A, Senin P, Smirnova AV, Stott MB, Hou S et al. (2007). Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450: 879–882. [DOI] [PubMed] [Google Scholar]
- Erikstad H-A, Birkeland N-K. (2015). Draft genome sequence of Candidatus Methylacidiphilum kamchatkense Strain Kam1, a thermoacidophilic methanotrophic Verrucomicrobium. Genome Announc 3: e00065–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM et al. (2010). Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464: 543–548. [DOI] [PubMed] [Google Scholar]
- Euzéby JP. (1997). List of bacterial names with standing in nomenclature: a folder available on the Internet. Int J Syst Evol Microbiol 47: 590–592. [DOI] [PubMed] [Google Scholar]
- Fritsch J, Scheerer P, Frielingsdorf S, Kroschinsky S, Friedrich B, Lenz O et al. (2011). The crystal structure of an oxygen-tolerant hydrogenase uncovers a novel iron-sulphur centre. Nature 479: 249–252. [DOI] [PubMed] [Google Scholar]
- Golyshina OV. (2011). Environmental, biogeographic, and biochemical patterns of archaea of the family Ferroplasmaceae. Appl Environ Microbiol 77: 5071–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening C, Berney M, Hards K, Cook GM, Conrad R. (2014. a). A soil actinobacterium scavenges atmospheric H2 using two membrane-associated, oxygen-dependent [NiFe] hydrogenases. Proc Natl Acad Sci USA 111: 4257–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB et al. (2016). Genome and metagenome surveys of hydrogenase diversity indicate H2 is a widely-utilised energy source for microbial growth and survival. ISME J 10: 761–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening C, Carere CR, Rushton-Green R, Harold LK, Hards K, Taylor MC et al. (2015). Persistence of the dominant soil phylum Acidobacteria by trace gas scavenging. Proc Natl Acad Sci USA 112: 10497–10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening C, Villas-Bôas SG, Robson JR, Berney M, Cook GM. (2014. b). The growth and survival of Mycobacterium smegmatis is enhanced by co-metabolism of atmospheric H2. PLoS One 9: e103034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakemian AS, Rosenzweig AC. (2007). The biochemistry of methane oxidation. Annu Rev Biochem 76: 223–241. [DOI] [PubMed] [Google Scholar]
- Hanczár T, Csáki R, Bodrossy L, Murrell CJ, Kovács KL. (2002). Detection and localization of two hydrogenases in Methylococcus capsulatus (Bath) and their potential role in methane metabolism. Arch Microbiol 177: 167–172. [DOI] [PubMed] [Google Scholar]
- Hanson RS, Hanson TE. (1996). Methanotrophic bacteria. Microbiol Rev 60: 439–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P et al. (2013). Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500: 567–570. [DOI] [PubMed] [Google Scholar]
- Ho A, Kerckhof F, Luke C, Reim A, Krause S, Boon N et al. (2013). Conceptualizing functional traits and ecological characteristics of methane‐oxidizing bacteria as life strategies. Environ Microbiol Rep 5: 335–345. [DOI] [PubMed] [Google Scholar]
- Hou S, Makarova KS, Saw JHW, Senin P, Ly BV, Zhou Z et al. (2008). Complete genome sequence of the extremely acidophilic methanotroph isolate V4, Methylacidiphilum infernorum, a representative of the bacterial phylum Verrucomicrobia. Biol Direct 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam T, Jensen S, Reigstad LJ, Larsen Ø, Birkeland N-K. (2008). Methane oxidation at 55 C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc Natl Acad Sci USA 105: 300–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keltjens JT, Pol A, Reimann J, den Camp HJMO. (2014). PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl Microbiol Biotechnol 98: 6163–6183. [DOI] [PubMed] [Google Scholar]
- Khadem AF, Pol A, Wieczorek A, Mohammadi SS, Francoijs K-J, Stunnenberg HG et al. (2011). Autotrophic methanotrophy in Verrucomicrobia: Methylacidiphilum fumariolicum SolV uses the Calvin-Benson-Bassham cycle for carbon dioxide fixation. J Bacteriol 193: 4438–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadem AF, van Teeseling MCF, van Niftrik L, Jetten MSM, Op den Camp HJM, Pol A. (2012. a). Genomic and physiological analysis of carbon storage in the verrucomicrobial methanotroph Ca. Methylacidiphilum fumariolicum SolV. Front Microbiol 3: 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadem AF, Wieczorek AS, Pol A, Vuilleumier S, Harhangi HR, Dunfield PF et al. (2012. b). Draft genome sequence of the volcano-inhabiting thermoacidophilic methanotroph Methylacidiphilum fumariolicum strain SolV. J Bacteriol 194: 3729–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschke S, Bousquet P, Ciais P, Saunois M, Canadell JG, Dlugokencky EJ et al. (2013). Three decades of global methane sources and sinks. Nat Geosci 6: 813–823. [Google Scholar]
- Knief C. (2015). Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front Microbiol 6: 1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knief C, Lipski A, Dunfield PF. (2003). Diversity and activity of methanotrophic bacteria in different upland soils. Appl Environ Microbiol 69: 6703–6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb S, Knief C, Dunfield PF, Conrad R. (2005). Abundance and activity of uncultured methanotrophic bacteria involved in the consumption of atmospheric methane in two forest soils. Environ Microbiol 7: 1150–1161. [DOI] [PubMed] [Google Scholar]
- Kwan P, McIntosh CL, Jennings DP, Hopkins RC, Chandrayan SK, Wu C-H et al. (2015). The [NiFe]-Hydrogenase of Pyrococcus furiosus exhibits a new type of oxygen tolerance. J Am Chem Soc 137: 13556–13565. [DOI] [PubMed] [Google Scholar]
- Lee KC, Stott MB, Dunfield PF, Huttenhower C, McDonald IR, Morgan XC. (2016). The Chthonomonas calidirosea genome is highly conserved across geographic locations and distinct chemical and microbial environments in New Zealand’s Taupō Volcanic Zone. Appl Environ Microbiol 82: 3572–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestrini S, Bechemin C, Grzebyk D, Hummert C. (2000). Phosphorus limitation might promote more toxin content in the marine invader dinoflagellate Alexandrium minutum. Plankt Biol Ecol 47: 7–11. [Google Scholar]
- Mohammadi S, Pol A, van Alen TA, Jetten MSM, Op den Camp HJM. (2016). Methylacidiphilum fumariolicum SolV, a thermoacidophilic ‘Knallgas’ methanotroph with both an oxygen-sensitive and -insensitive hydrogenase. ISME J. 11: 945–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Op den Camp HJM, Islam T, Stott MB, Harhangi HR, Hynes A, Schouten S et al. (2009). Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ Microbiol Rep 1: 293–306. [DOI] [PubMed] [Google Scholar]
- Oremland RS, Culbertson CW. (1992). Importance of methane-oxidizing bacteria in the methane budget as revealed by the use of a specific inhibitor. Nature 356: 421–423. [Google Scholar]
- Peters JW, Boyd ES, D’Adamo S, Mulder DW, Therien J, Posewitz MC. (2013) Hydrogenases, nitrogenases, anoxia, and H2 production in water-oxidizing phototrophs. In: Algae for Biofuels and Energy. Springer Netherlands, pp 37–75. [Google Scholar]
- Peters JW, Schut GJ, Boyd ES, Mulder DW, Shepard EM, Broderick JB et al. (2015). [FeFe]- and [NiFe]-hydrogenase diversity, mechanism, and maturation. Biochim Biophys Acta 1853: 1350–1369. [DOI] [PubMed] [Google Scholar]
- Pol A, Barends TRM, Dietl A, Khadem AF, Eygensteyn J, Jetten MSM et al. (2014). Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ Microbiol 16: 255–264. [DOI] [PubMed] [Google Scholar]
- Pol A, Heijmans K, Harhangi HR, Tedesco D, Jetten MSM, den Camp HJMO. (2007). Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 450: 874–878. [DOI] [PubMed] [Google Scholar]
- Poudel S, Tokmina-Lukaszewska M, Colman DR, Refai M, Schut GJ, King PW et al. (2016). Unification of [FeFe]-hydrogenases into three structural and functional groups. Biochim Biophys Acta 1860: 1910–1921. [DOI] [PubMed] [Google Scholar]
- Schwartz E, Fritsch J, Friedrich B. (2013) H2-Metabolizing Prokaryotes. Springer Berlin Heidelberg: Berlin, Heidelberg. [Google Scholar]
- Semrau JD, DiSpirito AA, Vuilleumier S. (2011). Facultative methanotrophy: false leads, true results, and suggestions for future research. FEMS Microbiol Lett 323: 1–12. [DOI] [PubMed] [Google Scholar]
- Shah NN, Hanna ML, Jackson KJ, Taylor RT. (1995). Batch cultivation of Methylosinus trichosporium OB3B: IV. Production of hydrogen‐driven soluble or particulate methane monooxygenase activity. Biotechnol Bioeng 45: 229–238. [DOI] [PubMed] [Google Scholar]
- Sharp CE, Smirnova AV, Graham JM, Stott MB, Khadka R, Moore TR et al. (2014). Distribution and diversity of Verrucomicrobia methanotrophs in geothermal and acidic environments. Environ Microbiol 16: 1867–1878. [DOI] [PubMed] [Google Scholar]
- Shomura Y, Yoon K-S, Nishihara H, Higuchi Y. (2011). Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase. Nature 479: 253–256. [DOI] [PubMed] [Google Scholar]
- Singh BK, Bardgett RD, Smith P, Reay DS. (2010). Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat Rev Microbiol 8: 779–790. [DOI] [PubMed] [Google Scholar]
- Søndergaard D, Pedersen CNS, Greening C. (2016). HydDB: a web tool for hydrogenase classification and analysis. Sci Rep 6: 34212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina PL, Ussler W, Joye SB, Harrison BK, Orphan VJ. (2010). Distributions of putative aerobic methanotrophs in diverse pelagic marine environments. ISME J 4: 700–710. [DOI] [PubMed] [Google Scholar]
- van Teeseling MCF, Pol A, Harhangi HR, van der Zwart S, Jetten MSM, den Camp HJMO et al. (2014). Expanding the verrucomicrobial methanotrophic world: description of three novel species of Methylacidimicrobium gen. nov. Appl Environ Microbiol 80: 6782–6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urschel MR, Kubo MD, Hoehler TM, Peters JW, Boyd ES. (2015). Carbon source preference in chemosynthetic hot spring communities. Appl Environ Microbiol 81: 3834–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. (1991). 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173: 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K-S, Ueda Y, Ishii M, Igarashi Y, Kodama T. (1996). NADH: ferredoxin reductase and NAD-reducing hydrogenase activities in Hydrogenobacter thermophilus strain TK-6. FEMS Microbiol Lett 139: 139–142. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.