Abstract
Chemosynthetic Fe-oxidizing communities are common at diffuse-flow hydrothermal vents throughout the world’s oceans. The foundational members of these communities are the Zetaproteobacteria, a class of Proteobacteria that is primarily associated with ecosystems fueled by ferrous iron, Fe(II). We report here the discovery of two new isolates of Zetaproteobacteria isolated from the Mid-Atlantic Ridge (TAG-1), and the Mariana back-arc (SV-108), that are unique in that they can utilize either Fe(II) or molecular hydrogen (H2) as sole electron donor and oxygen as terminal electron acceptor for growth. Both strains precipitated Fe-oxyhydroxides as amorphous particulates. The cell doubling time on H2 vs Fe(II) for TAG-1 was 14.1 vs 21.8 h, and for SV-108 it was 16.3 vs 20 h, and it appeared both strains could use either H2 or Fe(II) simultaneously. The strains were close relatives, based on genomic analysis, and both possessed genes for the uptake NiFe-hydrogenase required for growth on H2. These two strains belong to Zetaproteobacteria operational taxonomic unit 9 (ZetaOTU9). A meta-analysis of public databases found ZetaOTU9 was only associated with Fe(II)-rich habitats, and not in other environments where known H2-oxidizers exist. These results expand the metabolic repertoire of the Zetaproteobacteria, yet confirm that Fe(II) metabolism is the primary driver of their physiology and ecology.
Introduction
Ferrous iron is an important driver of chemosynthetic or chemolithoautotrophic microbial ecosystems at both deep and shallow marine hydrothermal vents, and it is now well documented that specialized communities adapted for microaerobic growth on Fe(II) thrive at these vents (Kato et al., 2012; Hoshino et al., 2016). A number of Fe-oxidizing bacteria (FeOB) produce organo-metallic filamentous Fe-oxides to construct a woven fabric mat that provides enough structural integrity for colonization by other microbes (Chan et al., 2016). The population structure of active marine iron mats is dominated by members of the Zetaproteobacteria, one of the seven officially recognized classes of Proteobacteria (Makita et al., 2016). In general, the Proteobacteria are well known for the remarkable breadth of their metabolic diversity, and the different classes of Proteobacteria occupy a wide variety of both oxic and anoxic niches, where they are often conspicuous for their high relative abundances. The Zetaproteobacteria are unique in this sense, as they are only found associated with marine systems, or ancient marine sediments, where Fe(II) is a prevalent electron donor (McAllister et al., 2011; Scott et al., 2015). Thus, the Zetaproteobacteria are more metabolically specialized than the other classes of Proteobacteria, with the exception of the class Acidithiobacillia, which are obligate iron or sulfur utilizers, but restricted to acidic environments (Williams and Kelly, 2013). Consistent with the Zetaproteobacteria being specialists for Fe(II)-based lithotrophy, thus far all the known isolates are obligate Fe-oxidizers that prefer microaerobic conditions. This makes the Zetaproteobacteria a compelling group of organisms for understanding how aspects of phylogenetic diversity contribute to functional unity.
The Fe(II)-rich ecosystems of these FeOB inhabit are almost exclusively associated with lower temperature (<100 °C) diffuse vents. The cumulative contribution of diffuse hydrothermalism to the biogeochemical cycling of elements at the seafloor may be as great, or greater than at high-temperature vents (Wankel et al., 2011; Resing et al., 2015). Nonetheless, the microbiology and geochemistry of diffuse-flow systems is not as well studied as high temperature, focused flow vents most typified by black smokers. In theory, lower temperature vents offer greater niche-space for colonization by diverse microbial communities. The thermal gradients in diffuse-flow systems are less extreme; therefore, they encompass a spatially larger habitable zone that can include the subsurface (Orcutt et al., 2011). As a result, they may offer heterogeneous gradients of electron donors and acceptors to support diverse microbial ecosystems. Previous studies have shown that the diversity of Zetaproteobacteria phylotypes is as great, or greater, within a particular vent site than it is between vent sites separated by large geographical distances (Davis and Moyer, 2008; McAllister et al., 2011; Scott et al., 2015). Other recent studies showed that physicochemical differences in temperature, oxygen and Fe(II) concentrations, as well as hydrodynamics played an important role in shaping the community structure of microbial iron mats (Fullerton et al., 2017; Scott et al., 2017). What is not known is how metabolic differences—specifically the ability to utilize alternative electron donors to iron or for that matter alternative electron-acceptors—drives community diversity, and whether Zetaproteobacteria are all obligate Fe oxidizers. Here, we describe two new isolates of Zetaproteobacteria that come from quite different hydrothermal systems at nearly opposite sides of the Earth. These two strains are close phylogenetically and share many phenotypic attributes including the ability to use H2 in addition to Fe(II) as their sole electron donor, thus expanding the metabolic repertoire of the Zetaproteobacteria.
Materials and methods
Source and isolation of strains
TAG-1 and SV-108 were isolated from marine hydrothermal vents on the Mid-Atlantic Ridge (MAR) in the Atlantic Ocean and the Southern Mariana Trough in the Pacific Ocean, respectively. At the MAR an iron mat sample was collected from the Trans-Atlantic Geotraverse (TAG) vent site (3626 mbsl; 26°9′45.9″N, 044°46′38.5″W) using a microbial mat sampler (described in Breier et al., 2012) capable of collecting samples with sub-centimeter vertical resolution. The sample was associated with a small (approximately 10 cm2), fluffy yellow mat at a diffuse vent site with a temperature of 5 °C at the vent orifice. Upon return to the ship a subsample of this material was used to initiate an enrichment culture for FeOB using an artificial seawater medium (ASW) amended with zero valent iron powder (ASW/ZVI) as the Fe(II) source (Emerson and Floyd, 2005; Emerson et al., 2007). An initial set of serial dilutions from 10−2 to 10−7 was done in Petri plates each containing 15 ml of ASW/ZVI and incubated in an anaerobic jar under a microoxic atmosphere created using a Campypak (Becton Dickinson, Franklin Lakes, NJ, USA) as described previously (Emerson and Floyd, 2005). After 10 days there was significant growth at the 10−2 dilution, as well as some cells present at 10−3 and 10−4 dilutions. A subsequent dilution series (10−2–10−7) was carried out in ASW/ZVI and within 6 days, the original 10−2 dilution had growth at 10−6, while there was no growth from the original 10−4 dilution. This second 10−6 dilution culture was placed in a serum bottle under a N2 atmosphere and returned to Bigelow Laboratory where three subsequent dilutions to extinction (10−3 to 10−8) were carried out, again under microaerobic growth conditions. In each case the highest dilution that yielded growth after 1 week was either 10−6 or 10−7, in no case was there growth at 10−8. Observations by phase contrast and epifluorescence microscopy showed a uniform, rod-shaped cell morphology, but no distinctive morphology for the iron oxides.
At the Mariana, an iron mat sample was collected at the Snail Vents (also known as the Fryer Site), near Marker 108 (2850 mbsl, 12°57′166″N, 143°37′142″W). Snail Vents are on the Malaguana–Gadao Ridge axis of the back-arc spreading ridge in the Southern Mariana Trough. The sample was from flocculant yellow iron mat associated with a diffuse vent with an orifice temperature of 27 °C. The same procedure was followed in the isolation as used for TAG-1, including initial growth in a 10−2 dilution aboard the ship followed by four series of dilution to extinction. The final culture also yielded a rod-shaped cell that precipitated Fe-oxides, but did not produce a distinctive iron oxide morphology. Neither of these strains grew on marine R2A medium, indicating the absence of heterotrophic bacteria. Both strains were grown in batch culture on ASW/ZVI for DNA extraction and sequencing of the small subunit (SSU) rRNA gene. The SSU rRNA sequences of both strains indicated a single phylotype was present that was most closely related to the Zetaproteobacteria. The MAR isolate was designed TAG-1, and the Mariana isolate was designed SV-108.
Both bacterial strains were deposited in the culture collections as following number: strain TAG-1, DSMZ 103937, JCM 31637, NCMA B5; strain SV-108, NCMA B6.
Cultivation of TAG-1 and SV-108.
Both strains were cultivated in ASW that contained either 0.5 g l−1 (9.35 mm) ammonium chloride or 0.8 g l−1 (9.41 mm) sodium nitrate as the N-source. The pH was adjusted and buffered to 6.5 with 20 mm sodium bicarbonate and bubbling with carbon dioxide. Cultivation was conducted in 20 ml ASW medium in 30-ml sealed glass bottles under N2/CO2 headspace with regular feeding of 200 μl sterilized air every 48 h to maintain microoxic conditions and 1 mm ferrous chloride (FeCl2) or 700 μl H2 (approximately 4% v/v of headspace) as the electron donor, or in a 15 ml of ASW/ZVI in a Petri plate under a microoxic atmosphere created using a Campypak. Bacterial growth on FeCl2 and hydrogen, and on ammonia and nitrate in bottles, was compared respectively through microscopic cell-counting using SYTO13 staining (Thermo Scientific, Waltham, MA, USA) at different time points of cultivation and establishing the growth curves as has been described previously (Emerson et al., 2007). Concentrations of Fe(II) in the culture were monitored using the phenanthroline method (Tamura et al., 1974). The temperature and pH optima for growth was determined at different temperatures (5–37 °C) and pH values (5.5–7.5, buffered by 20 mm MES or HEPES).
Monitoring of hydrogen oxidation by TAG-1 and SV-108
Bacterial cells of strain TAG-1 and SV-108 were inoculated into 20 ml of ASW medium in a 30-ml bottle, with a headspace filled with N2/CO2 and supplied with additional 700 μl hydrogen (final approx. 4% v/v) and 300 μl sterilized air. The H2 concentration was measured each day by taking a 100 μl of headspace gas with a sterile syringe. Gas pressure in the culture vials was measured using a digital pressure meter (GMH 3111, Greisinger Electronic, Regenstauf, Germany) to calculate the amount of H2 consumed. To replenish the supply of oxygen, 200 μl sterilized air was supplied to the culture every 2 days. An aliquot of the medium (approximately 0.1 ml) was removed from the culture bottle each day for cell counts. All incubations were prepared as biological triplicates, and included a cell-free control. The H2 concentration in the headspace of the TAG-1 and SV-108 incubations was measured with a Hewlett-Packard Co. 5980 series II gas chromatograph (Hewlett-Packard, Palo Alto, CA, USA) coupled to a thermal conductivity detector (TCD-GC) equipped with a molecular sieve 13X column (3 m × 1/8″ Alltech, Unterhaching, Germany). The conditions for analysis were as follows: oven temperature, 40 °C; injection temperature, 150 °C; detection temperature, 175 °C; carrier gas (argon) flow rate, 30 ml min−1 (27 psi). The concentration of H2 was monitored and quantified on 32 Karat software (Beckmann Coulter, Krefeld, Germany). A standard curve for H2 was prepared for every measurement using 0.04–3.74% (v/v) H2 standards prepared in serum bottles.
Whole-genome sequencing of TAG-1 and SV-108
For genome sequencing both TAG-1 and SV-108 were grown in approximately 100 ml of a H2-containing medium. After 5 days of growth, cells were concentrated by centrifugation (16 000 g for 10 min), and DNA was extracted using a PowerMax Soil kit (Mo Bio Laboratories, Solana Beach, CA, USA). The genome of TAG-1 was sequenced at the Department of Energy’s, Joint Genome Institute (JGI) as part of their microbial sequencing program, using Illumina technology as previously described (Emerson et al., 2013). The genome of SV-108 was sequenced and assembled by the sequencing facility at the Bigelow Laboratory Single Cell Genomics Center using Illumina (Nextseq) technology. The assembled genomes of both organisms were uploaded to JGI’s Integrated Microbial Genome and Microbiome (IMG/MER) for annotation. Subsequent analysis of both genomes was done using tools provided on the IMG/MER website (https://img.jgi.doe.gov/cgi-bin/mer/main.cgi; Markowitz et al., 2009, 2014). The IMG Taxon ID for the genome of TAG-1 is 2852580733 and for SV-108 it is 2617270712.
Phylogenetic analyses
To understand the phylogeny of TAG-1 and SV-108, phylogenetic trees based on maximum-likelihood using the program MEGA (ver. 7.0.21; Kumar et al., 2016) were created with SSU rRNA gene sequences, concatenated sequences of housekeeping genes, and some key functional gene sequences, respectively. The housekeeping genes used for building concatenated trees were as follows: DNA gyrase subunit B (gyrB), isoleucyl-tRNA synthase (ileS), leucyl-tRNA synthase (leuS), CTP synthase (pyrG), leader peptidase A (lepA), DNA recombination and repair protein (recA), elongation factor P (efp), RNA polymerase α-subunit (rpoA), RNA polymerase major σ factor (rpoD), Hsp70 cochaperone (dnaJ) and tmRNA-binding protein (smpB) (Gil et al., 2004; Santos and Ochman, 2004). For all analyses, the relevant nucleic acid sequences were aligned using ClustalW in MEGA and the trees were created using Tamura-Nei model with 1000 bootstrap iterations.
Whole-genome comparisons
Similarities among whole genomes of TAG-1 and SV-108 with other Zetaproteobacteria isolates (Mariprofundus ferrooxydans PV-1, JV-1 and M34, Mariprofundus spp. EKF-M39, and DIS-1) were investigated by calculating average amino-acid identities (AAIs) and nucleic acid identities (ANIs). AAIs were calculated by CompareM (ver. 0.0.21; https://github.com/dparks1134/CompareM). ANIs were calculated using JSpeciesWS Online Service (http://jspecies.ribohost.com/jspeciesws/; Richter and Rosselló-Móra, 2009; Richter et al., 2015). The synteny between the genomes of TAG-1 and SV-108 was conducted using Mauve (ver. 2.4.0; Darling et al., 2010).
Biogeography and ecological analysis
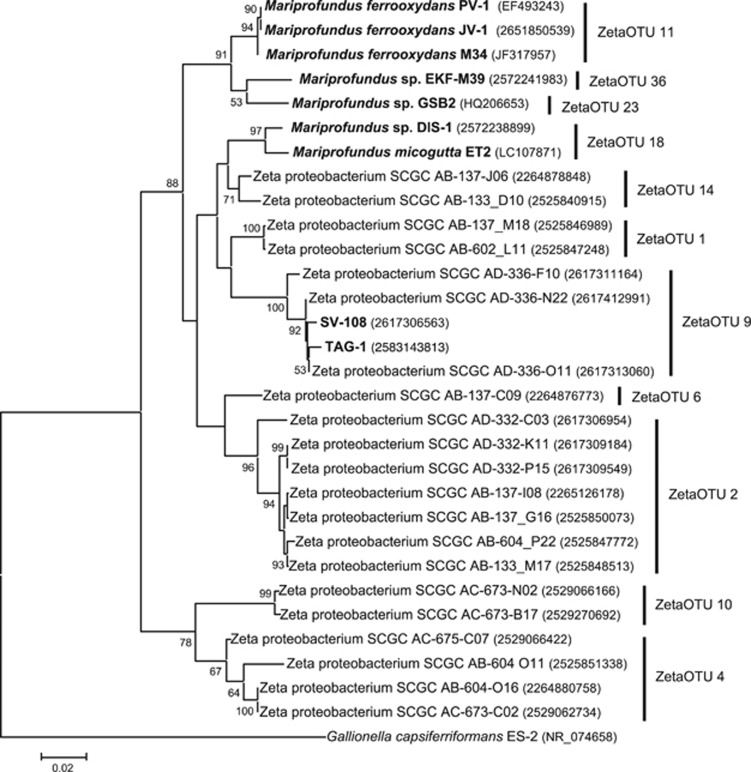
Initial phylogenetic and classification analysis using ZetaHunter (https://github.com/mooreryan/ZetaHunter, last accessed 01.01.2017) to assess SSU rRNA gene sequences showed that TAG-1 and SV-108 belong to Zetaproteobacteria ZetaOTU9 (McAllister et al., 2011; Figure 4). To better understand the global distribution of these isolates and ZetaOTU9, we used the Integrated Microbial Next Generation Sequencing (IMNGS, https://www.imngs.org, last accessed 04.01.2017; Lagkouvardos et al., 2016) server. IMNGS is an online platform that allows users to conduct comprehensive searches of SSU rRNA gene sequences against raw sequence read archives retrieved from the International Nucleotide Sequence Database Collaboration. The current build (accessed January 2017) has roughly 88 500 samples, 32 890 of which are from human-associated studies and the remainder from a wide array of environmental and host-associated habitats. IMNGS offers two types of queries—a taxonomy query and a similarity query. To gain a general sense of Zetaproteobacteria distribution, we first ran the taxonomy query. Because there are no lower taxonomic ranks for Zetaproteobacteria, we screened the entire class. We then conducted a similarity search of all near full-length SSU rRNA single amplified genome (SAG) and pure culture sequences classified at ZetaOTU9 (by ZetaHunter) against the IMNGS database using a similarity threshold of 99% and a minimum length of 200 bp.
Results
Growth studies of TAG-1 and SV-108
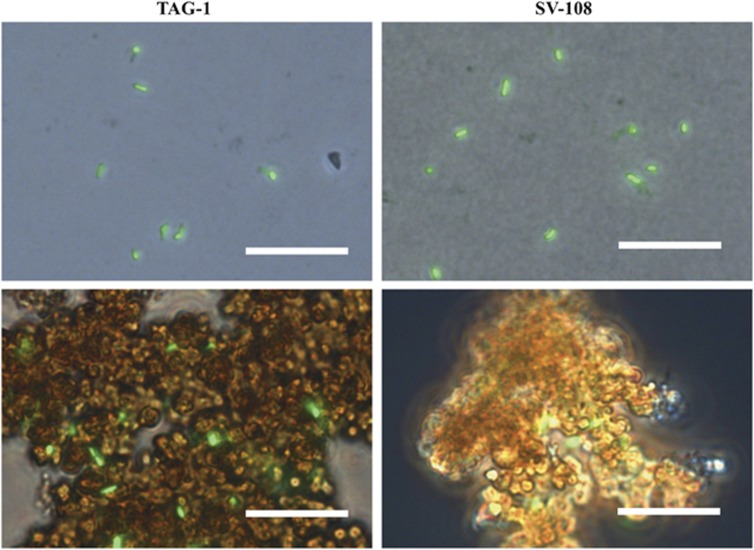
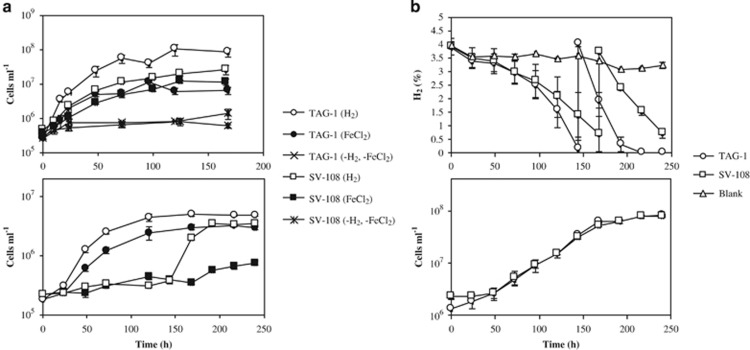
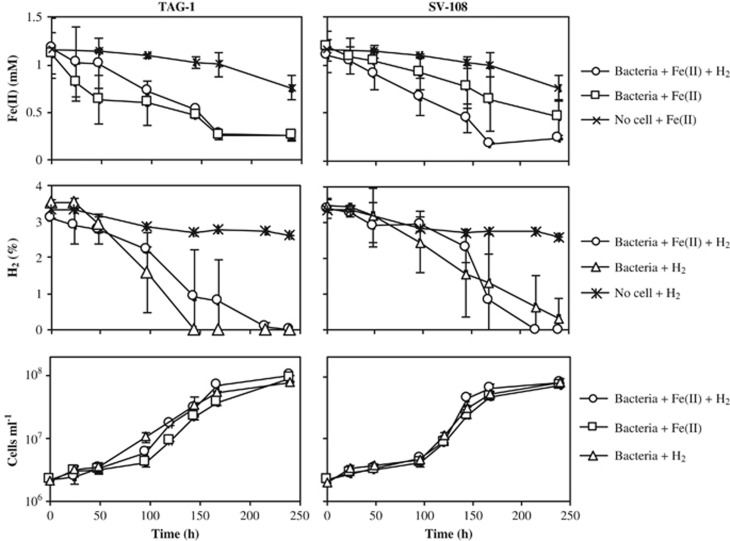
TAG-1 and SV-108 are both rod-shaped cells with dimensions of 0.3 × 1.5–2 μm (Figure 1). Motility was often, but not always, observed, and both strains contained a complement of flagellar synthesis genes and chemotaxis genes (Supplementary Tables S1 and S2). When cultivated with Fe(II), Fe-oxyhydroxides precipitated that had no discernible morphology, although the cells were often associated with the oxides. Both strains grew on either FeCl2 or H2 with ammonium chloride as a N source (Figure 2a). Growth on H2 was faster than growth on FeCl2 especially in TAG-1, with calculated doubling times of 14.1 and 21.8 h (TAG-1) and 16.3 and 20.0 h (SV-108) for H2 and Fe(II), respectively. These doubling times were calculated based on the rapid growth phases of the growth curves in Figure 2, thus represent an optimal doubling time. For both strains consumption of H2 supplied in the headspace corresponded with cell growth, and the addition of H2 to week-old cultures led to rapid H2 consumption (Figure 2b). There was no consumption of H2 in the cell-free controls, indicating no abiotic H2 consumption. TAG-1 consumed H2 in the headspace with a rate of 4.83 μmol day−1 during the first 6 days of incubation, and 12.27 μmol day−1 following a second addition of H2 (Supplementary Figure S1). Cell yield of TAG-1 was 2.36 × 107 cells per μmol H2 consumed, which is a comparable value to the calculated cell biomass yield of Cupriavidus necator, a model H2-oxidizing chemolithoautotrophic bacterium (Bongers, 1970). When TAG-1 was grown with both Fe(II) and H2, or either substrate alone, the consumption of Fe(II) and H2 co-occurred (Figure 3). SV-108, on the other hand, showed slower H2 oxidation in the presence of Fe(II) than cultures without Fe(II) during first 5–6 days, while Fe(II) disappeared more quickly in the presence of H2 (Figure 3), suggesting this organism may have a growth preference for Fe(II) over H2.
Figure 1.
Microscopic images of TAG-1 (left) and SV-108 (right) cells grown on H2 (above) and FeCl2 (below). Cells were stained with SYTO13. Scale bars indicate 10 μm.
Figure 2.
(a) Growth curves of TAG-1 (○, ●) and SV-108 (□, ▪) on H2 (○, □) or FeCl2 (●, ▪) with ammonium chloride (upper, n=3) or sodium nitrate (lower, n=2). Hydrogen and Fe(II) sources were regularly fed. Growth curves with neither H2 nor Fe(II) source (with ammonium chloride) were also shown. (b) Monitoring of hydrogen concentrations (upper) and cell numbers (lower) in strain TAG-1 (○) and SV-108 (□) cultures (n=3). Additional H2 was fed after 144 h (TAG-1) and 168 h (SV-108) of incubation. Error bars indicate standard deviations.
Figure 3.
Monitoring of Fe(II) (upper) and hydrogen (middle) concentrations and cell numbers (lower) in strain TAG-1 (left column) and SV-108 (right) cultures which were supplied with simultaneous use of Fe(II) and hydrogen (○) or only Fe(II) (□) or hydrogen (Δ). Error bars indicate SD (n=3).
Growth studies revealed that nitrate had an inhibitory or bacteriostatic effect on the growth of SV-108, since there was a pronounced lag phase of 3–6 days for cultures grown on either Fe(II) or H2 in the presence of 9.41 mm sodium nitrate, but not on 9.35 mm ammonium chloride (Figure 2a). Nitrate-grown cells of SV-108 that were sub-cultured into new nitrate-containing medium exhibited a similar lag phase (data not shown), indicating this lag was not strictly due to an adaptation to nitrate. TAG-1 did not show any differential response in growth based on nitrogen source.
The temperature (20 °C) and pH (6.5–7.0) optima of both strains are summarized in Supplementary Table S3. Neither strain grew on heterotrophic R2A medium under microoxic conditions. It was discovered that organic compounds could inhibit the growth of both strains. Both strains can be cryo-preserved in 10% glycerol (w/v final concentration); however, upon recovery, the glycerol stocks had to be diluted 1:100 or 1:1000 in order to initiate growth. This phenomenon was tested further by adding either glycerol or glucose to either TAG-1 or SV-108 grown on ZVI; neither strain grew in the presence of 0.5% glycerol or 1% glucose; however, at lower concentrations (0.1% glycerol or 0.5% glucose) the cells were able to grow so long as H2 was present.
Phylogenetic analysis of TAG-1 and SV-108
The respective genome sizes of TAG-1 and SV-108 were 2.16 and 2.14 Mb; other pertinent genome information is shown in Supplementary Table S4. Phylogenetic analysis confirmed the morphological and physiological observations that these two strains are close relatives, sharing 99% homology between their respective SSU rRNA genes. The SSU rRNA gene phylogeny showed the two strains clustered closely together within the Zetaproteobacteria (Figure 4). More detailed phylogenetic trees that used concatenated functional genes gyrB-ileS-leuS-efp-lepA-pyrG-recA-rpoA-rpoD-dnaJ-smpB confirmed this relationship (Supplementary Figure S2). TAG-1 and SV-108 clustered most closely with three SAGs that originated from the MAR, and formed a small phylogenetic clade (Figure 4). The ANIs and AAIs between isolates of Zetaproteobacteria with sequenced genomes (PV-1, JV-1, M34, EKF-M39, DIS-1, TAG-1, and SV-108) are shown in Supplementary Figure S3, and provided further confirmation of the relatedness of TAG-1 and SV-108 to one another, and distance from the other Zetaproteobacteria isolates. As shown in Supplementary Figure S4 there was considerable synteny between the genomes of TAG-1 and SV-108. Based on their relatedness to one another, but genetic, physiological, and morphological differences from other Zetaproteobacteria, it is reasonable to propose TAG-1 and SV-108 are strains of a novel genus and species.
Figure 4.
Maximum-likelihood phylogenetic tree of SSU rRNA gene of TAG-1, SV-108 and other Zetaproteobacteria isolates and SAGs. Isolated strains of Zetaproteobacteria are in bold, and GenBank accession no. or Gene IDs on IMG are given in parentheses. The tree was rooted with SSU rRNA gene of G. capsiferriformans ES-2. The tree was created with 1000 bootstrap iteration and the values below 50 are not reported.
Genome analysis of TAG-1 and SV-108
The mechanism of iron oxidation in the Zetaproteobacteria remains largely unknown; however, several candidate genes or gene clusters have been identified either through comparative genomics or proteomics (Singer et al., 2011; Emerson et al., 2013; Barco et al., 2015; Fullerton et al., 2017). Thus far, a gene in M. ferrooxydans identified as a distant homolog of Cyc2, an outer membrane protein in Acidithiobacillus that has iron oxidation activity, and designated Cyc2PV-1 has been found in the genomes of all marine and freshwater lithotrophic FeOB (Barco et al., 2015). TAG-1 (IMG locus tag DM09DRAFT_0629) and SV-108 (Ga0073143_10672) both encode Cyc2PV-1 homologs according to BLAST analysis. The alternative respiratory complex III (AC-III) including the actB gene that encodes for a molybdopterin oxidoreductase is another important component in the genomes of a number of marine and freshwater FeOB. TAG-1 has this complex, but also contains a second gene encoding a molybdopterin oxidoreductase. This second gene is in a cluster that includes the large and small subunits of the assimilatory nitrate reductase. This same gene cluster is conserved in the genomes of several Zetaproteobacteria (Field et al., 2014; Fullerton et al., 2017). Interestingly, SV-108 did not have the AC-III complex, but does encode nitrite and nitrate assimilatory reductases (Supplementary Figure S5).
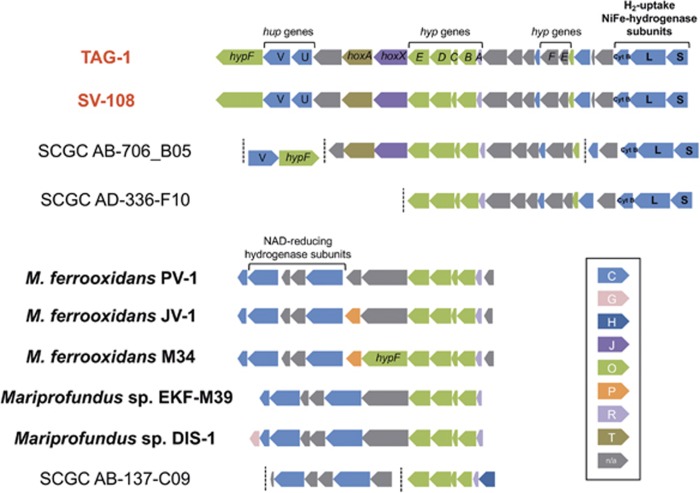
Consistent with their ability to grow on H2, both TAG-1 and SV-108 possessed nearly identical gene clusters that consisted of the subunits and assembly proteins of the H2-uptake NiFe-hydrogenase (Figure 5; Greening et al., 2016). The assembly protein genes (hypA, B, C, D, E and F) were also detected in genomes of other isolates of Zetaproteobacteria; however, these strains do not have subunit genes of the uptake NiFe-hydrogenase, but instead had the genes for a NAD-reducing hydrogenase.
Figure 5.
Gene clusters of NiFe-hydrogenase encoded by TAG-1, SV-108 and other Zetaproteobacteria isolates and SAGs. Genes are shown in different colors according to COG functional category as listed in a box. Genes annotated as subunits of H2-uptake NiFe-hydrogenase are labeled with bold letters (L: large subunit, S: small subunit, and Cyt B: cytochrome b subunit). Dash lines indicate end of the contig.
In terms of autotrophic potential, TAG-1 and SV-108 had gene clusters consistent with CO2 fixation via the Calvin–Benson–Bassham pathway, including the large subunit cbbM gene of the Form II ribulose-1,5-bisphosphate carboxylase (RubisCO) and the phosphoribulokinase gene (Berg, 2011). A phylogenetic comparison of the cbbM gene of TAG-1 and SV-108 showed a 92% similarity of nucleic acid sequences between these two genes, and lower (66–79%) similarities with other Zetaproteobacteria isolates and SAGs (Supplementary Figure S6). Neither TAG-1 or SV-108 encoded Form I RubisCO, implying they may have a preference for high CO2 and low oxygen conditions (Tabita et al., 2008).
For nitrogen acquisition, both TAG-1 and SV-108 have a suite of genes involved in nitrogen metabolism, as listed in Supplementary Table S5. These include an assimilatory nitrate reductase, and nitrite reductase that, as noted above, contain a molybdopterin oxidoreductase. They also have a nitric oxide reductase as well as ammonium transporters. TAG-1 and SV-108 share all nitrogen-related genes checked in this study. The reason for nitrate-sensitivity of SV-108 is still unclear.
Ecological analysis
A previous study found that the Zetaproteobacteria are primarily associated with high iron environments in the ocean (Scott et al., 2015). ZetaOTU9, the clade containing TAG-1 and SV-108, was present at diffuse-flow Fe(II)-rich vents at Rainbow and Snakepit hydrothermal vent fields, in addition to TAG, located on the MAR, although a previous study, based on an earlier phylogenetic analysis, identified ZetaOTU9 as OTU3 at TAG, Rainbow and Snakepit (Scott et al., 2015). Somewhat surprisingly, at the Snail Vent site in the Southern Mariana Trough, analysis of community abundance by SSU rRNA amplicon sequencing found that although the relative abundance of Zetaproteobacteria was 10–12% of community, ZetaOTU9 only accounted for 0.05% of the Zetaproteobacteria reads (Hager et al., 2017). This may help explain why only a low dilution (10−2) enrichment was successful. A comprehensive taxonomic search of SSU rRNA raw sequencing read archives using IMNGS found Zetaproteobacteria were present (mostly at abundances of <0.01%) in only 700 of over 55 500 available non-human samples. A similarity search of representative sequences classified as ZetaOTU9 against IMNGS found hits in only 17 of these samples (Table 1). The two samples in the IMNGS where ZetaOTU9 had substantial abundance, accounting for 22–37% of the total community, was from freshly synthesized basalt chips incubated on abyssal plain of the Atlantic Ocean (Henri et al., 2016). These were colonized by Zetaproteobacteria that were presumed to be growing on Fe(II) released from the basalt. Remarkably ZetaOTU9 accounted for >90% of the Zetaproteobacteria reads from these samples.
Table 1. Distribution of SSU rRNA gene of ZetaOTU9 SAGs and pure culture isolates.
| SRA IDs | Size | Zeta reads | ZetaOTU9 reads |
|---|---|---|---|
| ERR313278a | 12769 | 11 | 1 |
| ERR313285a | 12765 | 11 | 1 |
| ERR313301a | 159368 | 127 | 19 |
| SRR1573430b | 31256 | 3417 | 3403 |
| SRR1573432b | 16973 | 3330 | 3112 |
| SRR2130144c | 37846 | 4 | 4 |
| SRR2130147c | 36968 | 3 | 3 |
| SRR2130148c | 53226 | 1 | 1 |
| SRR2130149c | 45262 | 1 | 1 |
| SRR2130151c | 30321 | 6 | 6 |
| SRR2130152c | 49326 | 18 | 9 |
| SRR2130153c | 32058 | 11 | 9 |
| SRR2130155c | 20423 | 7 | 7 |
| SRR592604d | 5902 | 1 | 1 |
| SRR592631d | 4566 | 1 | 1 |
Size refers to the total reads in each library, and ZetaOTU9 reads are a subset of the Zeta reads.
Marine sediments of the Barents Sea.
MAR Basaltic glass.
Po river Prodelta and Mar Piccolo of Taranto surface sediments bacterial communities.
Logatchev field in the MAR.
Discussion
Previous isolates of marine FeOB have all been obligate Fe-oxidizers, thus TAG-1 and SV-108 are the first Zetaproteobacteria shown to grow microaerobically on H2 as an alternative substrate. Phylogenetic analysis of conserved genes as well as comparative genomics clearly shows these two strains are not closely related to other isolates belonging to the Zetaproteobacteria. Furthermore, they do not produce any identifiable extracellular organo-metallic structures such as stalks, sheaths or organized filaments that are characteristic of other known marine FeOB. Consistent with a lack of stalk production, TAG-1 and SV-108 do not possess the putative genes for stalk-formation (xag operon), although these are conserved in the genomes of several Mariprofundus species (Kato et al., 2015). Presumably these two strains produce a different type of exopolymer that prevents them from becoming encrusted in Fe-oxides, as has been proposed for several, non-stalk forming freshwater FeOB (Emerson et al., 2013). Despite coming from hydrothermal vents on opposite sides of the world, the two isolates share most of their genes in common and their genomes have substantial regions of synteny. Together these results demonstrate TAG-1 and SV-108 represent a novel genus within the Zetaproteobacteria, and the name ‘Ghiorsea bivora’ is proposed.
Metabolism
Thermodynamically H2 is a better energy source than Fe(II) with a moderately higher free energy, ΔG°=−237 kJ mol−1 (−474 kJ per mol O2 reduced) for H2 vs ΔG°=−90 kJ mol−1 for Fe(II) (−360 kJ per mol O2 reduced) (Emerson et al., 2010). The doubling times of TAG-1 and SV-108 were 55.0% and 23.2% more rapid for H2-grown cells compared to Fe(II)-grown cells, respectively, and cell yields were marginally higher on H2 compared to Fe(II) (Figure 2a). While we could not totally account for auto-oxidation of Fe(II) in the substrate utilization experiments, it appeared both substrates were consumed simultaneously, indicating these organisms are not adapted to preferentially use either Fe(II) or H2, despite H2 being the more energetically favorable electron donor.
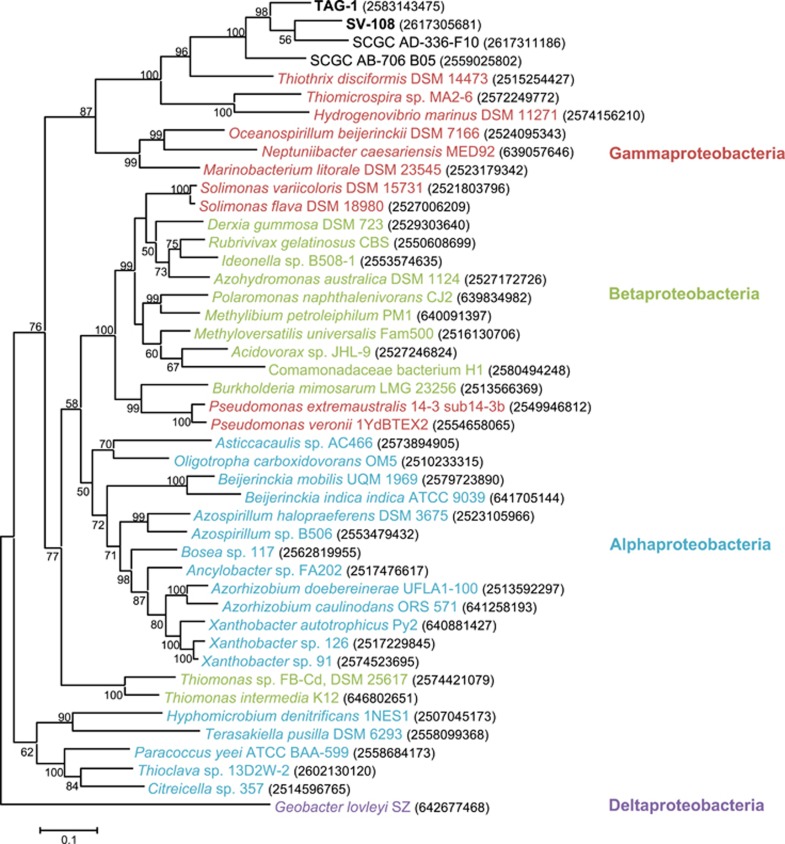
Both strains share nearly identical suites of genes involved in H2 metabolism that include the canonical genes for the oxygen-tolerant, membrane-bound H2-uptake NiFe-hydrogenase (Greening et al., 2016). These proteins contain signal sequences and are presumably located in the periplasm where they can initiate H2 oxidation coupled to energy conservation via the electron transport chain. The phylogenetic analysis of the large subunit of the hydrogenase revealed that two SAG genomes belonging to ZetaOTU9 (SCGC AB-706_B05 and SCGC AD-336-F10) also contain uptake NiFe-hydrogenase (Figure 5). Together these genes from the Zetaproteobacteria fall into a diverse cluster of related genes in the Gammaproteobacteria, with the closest relatives coming from lithotrophic sulfur-oxidizing bacteria (Figure 6). This raises the possibility that H2 oxidation was acquired through different horizontal gene transfers.
Figure 6.
Maximum-likelihood phylogenetic trees of H2-uptake NiFe-hydrogenase large subunit gene of TAG-1, SV-108, Zetaproteobacteria SAGs and related bacterial species. Gene IDs on IMG are given in parentheses, and bacterial classes (Alpha-, Beta-, Gamma- and Deltaproteobacteria) are shown in different colors. The tree was rooted with the gene of G. lovleyi (Deltaproteobacteria). The tree was created with 1000 bootstrap iteration and the values below 50 are not reported.
Regarding iron oxidation, we still lack definitive evidence as to how FeOB conserve energy from Fe(II) oxidation (White et al., 2016); however, these two strains illustrate the diversity of specific pathways that are likely involved in this process. Both strains have a gene homolog to Cyc2PV-1 that a recent proteomic analysis indicated could be an important outer membrane cytochrome that may initiate iron oxidation. The Cyc2 homologs encoded by TAG-1, SV-108 and an SAG belonging to ZetaOTU9 (SCGC AD-336-F10) show high gene homologies (Supplementary Figure S7). Neither strain however, possesses a gene homologous to the Cyc1PV-1, a soluble cytochrome that has been suggested to play a role in shuttling electrons across the periplasm in M. ferrooxydans. Both strains have the capacity to produce the different complexes of the electron transport chain; however, TAG-1 does not have a cytochrome bc1 complex (complex III), although it does have the AC-III, while SV-108 does not have AC-III but does have the cytochrome bc1 complex III. Unlike most of the other FeOB genomes, neither strain has the cytochrome bd complex that encodes a terminal oxidase with high oxygen affinity that can couple directly with the quinone pool. In both strains, complex IV, the terminal oxidase of the electron transport chain, is encoded by a single gene cluster ccb3-type cytochrome oxidase. This is in contrast to M. ferrooxydans that has this same gene cluster, but also has a second cluster of genes that encode a ccb3-type cytochrome oxidase. Proteomic analysis showed that this second cytochrome oxidase was the most highly expressed in M. ferrooxydans (Barco et al., 2015). Somewhat surprisingly, TAG-1, but not SV-108, also has a gene cluster that includes the coxA (DM09DRAFT_0178) and coxB (DM09DRAFT_0179) genes encoding an aa3-type terminal cytochrome oxidase with lower affinity for oxygen. Thus far, TAG-1 is only genome from an FeOB isolate that has this cytochrome oxidase, although it is present in SAGs and metagenomes of Zetaproteobacteria (Fullerton et al., 2017).
Based on this genomic analysis, we hypothesize that TAG-1 may exhibit more flexibility regarding its ability to grow and survive under higher or more dynamic oxygen concentrations. A related aspect is the capacity for these organisms to defend against reactive oxygen species such as hydrogen peroxide (H2O2) and super-oxides. This is especially relevant given that Fenton chemistry involves the reaction of H2O2 with Fe(II) to produce hydroxyl radicals that are extremely damaging to cellular organic matter (Imlay, 2008). Somewhat surprising then is that in the genome of SV-108, neither catalase or superoxide dismutase was found, while TAG-1 had a catalase-peroxidase gene (DM09DRAFT_0584) but also lacked superoxide dismutase (Supplementary Table S6). Superoxide dismutase is a ubiquitous gene in aerobic organisms, and is even found in many anaerobes (Hewitt and Morris, 1975; Imlay, 2008). Thus far, the sequenced genomes of all other FeOB have contained superoxide dismutase (Mumford et al., 2016). For defense against H2O2 production, both TAG-1 and SV-108 contain two copies of a cytochrome c-peroxidase gene. Cytochrome c-peroxidase is excreted to the periplasm, and may act as a defense mechanism against exogenously produced H2O2. The most abundant genes involved in reactive oxygen species protection for both strains were peroxiredoxins (TAG-1=4 copies; SV-108=5 copies; Supplementary Table S6) that utilize a thiol-based mechanism to react with H2O2 in the cytoplasm. Presumably it is this mechanism that is the primary defense for reactive oxygen species in TAG-1 and SV-108. It should be noted, however, that our results cannot eliminate the possibility of either mis-annotation of functional genes, or that a gene is missing from these near complete, but not finished genomes. Some physiological capabilities of TAG-1 and SV-108, such as oxygen tolerance and utilization of nitrogen-species, still have to be evaluated via further cultivation-based tests.
Ecological implications
All previous isolates of Zetaproteobacteria have been obligate Fe-oxidizers, so the discovery of these two H2-utilizing strains adds a new dimension to the physiology of this group. Because the Zetaproteobacteria are phylogenetically differentiated from other Proteobacteria at the class level, we are able to effectively track individual OTUs within them (McAllister et al., 2011). TAG-1 and SV-108, as well as the two SAGs that also had uptake hydrogenase genes (Figure 5) belong to ZetaOTU9; however, none of the other genomes of Zetaproteobacteria, either from pure cultures, SAGs, or metagenomes have these hydrogenase genes, suggesting that growth on H2 may be a trait only found in ZetaOTU9. When analyzed relative to the dominant ZetaOTUs in a variety of chemosynthetic iron communities, the overall abundance of ZetaOTU9 is often at background levels of <0.1% of the community. However, in samples where its abundance is >1%, it is often a dominant phylotype of the Zetaproteobacteria. For example, among vent sites at the MAR, ZetaOTU9 was prevalent in five samples (two from TAG, two from Snakepit, one from Rainbow), accounting for 60–95% of Zetaproteobacteria reads, but made-up less than 5% of the reads in three other samples (one each from TAG, Snakepit and Rainbow; Scott et al., 2015). At the Mariana, as mentioned in the results, ZetaOTU9 was in low abundance at the Snail Vents where SV-108 was isolated. However, in a broader survey of chemosynthetic iron mats along the Mariana Arc and back-arc, ZetaOTU9 accounted for >5% of the Zetaproteobacteria reads in 6 of 21 discrete samples from 5 different sites. At one of these sites ZetaOTU9 accounted for 75% of the total Zetaproteobacteria reads (Hager et al., 2017). An extensive survey of iron mats at L-ō‘ihi Seamount, found ZetaOTU9 was only present in veil-like mats that are structurally dominated by sheath-forming Zetaproteobacteria (Scott et al., 2017).
In addition to being abundant on fresh basalts (Henri et al., 2016) as noted above, other sites where ZetaOTU9 is abundant are mild steel coupons incubated in coastal seawater (McBeth and Emerson, 2016), and in the subsurface of the ocean crust (Kato et al., 2009). These latter habitats are locales where H2 may be present. On steel, ZVI, given the right conditions, can react with seawater to produce H2, and in the subsurface water–rock reactions may generate H2. The capacity to use H2 as a sole electron donor would certainly give strains like TAG-1 and SV-108 a competitive advantage over obligate Fe(II)-oxidizers. Unfortunately, it has not been possible to obtain H2 measurements from the iron mats where ZetaOTU9 was abundant. It is worth pointing out, however, that samples from the MAR, L-ō‘ihi, and the Mariana were all collected at high (sub-centimeter) spatial resolution, thus the corresponding presence and absence of ZetaOTU9 with specific samples that in some cases were only a few centimeters apart, indicates that there may be chemical (e.g., the presence of H2), mineralogical and/or physical drivers that may be controlling their population dynamics. This underscores the importance of being able to collect relevant physicochemical data at spatial scales that can be co-registered with community analysis, so fundamental drivers of microbial diversity are revealed.
The IMNGS meta-analysis included a number of sites where H2 is likely present as a potential energy source, such as sediments and hydrothermal vent communities, but where the abundance of Fe(II) is insufficient to support Fe(II)-fueled chemosynthesis. This indicates that despite having the metabolic flexibility of being able to use H2 in addition to Fe(II), TAG-1 and SV-108, and by extension ZetaOTU9, are still limited to Fe(II)-rich habitats. Thus the adaptations required to grow on Fe(II) likely play a stronger role in selection than the capacity to grow on H2. There can be appreciable fluxes of H2-associated diffuse venting systems, and the dynamics of these fluxes indicate there is significant microbial metabolism of H2 (Wankel et al., 2011). The discovery that this particular clade of FeOB that grow as well or better on H2 than Fe(II) supports this finding, and indicates the Zetaproteobacteria contribute to chemosynthesis in these systems via H2 utilization in addition to iron oxidation. A similar role has been suggested for chemolithoautotrophic sulfur-oxidizing bacteria like Thiomicrospira (Hansen and Perner, 2015).
Taxonomy
TAG-1 and SV-108 represent the most unique isolates within the Zetaproteobacteria discovered to date. We propose they form a novel genus and species, thus representing the second genus and third species within this class.
Description of Ghiorsea gen. nov.
Ghiorsea gen. nov. (Ghi.or.se.a. N. L. fem. n. Ghiorsea, named after contemporary American microbiologist William C Ghiorse, in recognition of his important contributions to the field of geomicrobiology.
Gram-negative rod-shaped cells. Growth is obligately lithotrophic, oxygen-dependent, and requires marine salts; habitat, marine microbial iron mats. The type species is: Ghiorsea bivora gen. nov. sp. nov.
Description of Ghiorsea bivora sp. nov.
G. bivora (bi.vor.a, L. prefix bi two; L. comb. –vora, ones that eat; N.L. n. bivora one that eats two substrates).
Displays the following properties in addition to those given by the genus description. The cells are rods, approximately 0.3 × 1.5–2 μm. Motility is often observed. Lithotrophic growth on either ferrous iron or on hydrogen gas under microaerobic conditions. Result of growth on ferrous iron is the precipitation of iron-oxyhydroxides with no determinant structure. No observable growth on organic compounds; elevated concentrations of organics may be bacteriostatic. The optimum growth temperature is 20 °C. The pH range for growth is 6.0–7.0. The G+C content of the DNA is 43.7%. The type strain is TAG-1 T (=DSMZ 103937; =JCM 31637;=NCMA B5; IMG genome identification number 2582580733) isolated from an iron-rich microbial mat associated with diffuse hydrothermal venting at the TAG hydrothermal vent site on the MAR.
Acknowledgments
We are indebted to Captains and crew of the R/V Knorr and R/V Roger Revelle, and the team of ROV Jason II engineers who were instrumental in sample collection. We thank Dr George Luther for the opportunity to participate in SNAPMORE cruise to the MAR. We thank Anna Leavitt and Sarabeth George for help with culturing and maintenance of TAG-1 and SV-108, and Dr Roman Barco for his input on the manuscript. We also thank the anonymous reviewers for helpful comments. Sequencing and assembly of the genome of SV-108 was carried out by the Single Cell Genomics Center at Bigelow Laboratory (supported in part by NSF Major Research Infrastructure grant OCE-1335810), and we especially thank Liz Fergusson and Joe Brown for their assistance. Sequencing of TAG-1 by JGI (supported by US Department of Energy Office of Science Contract No. DE-AC02-05CH11231) was funded through JGI’s Community Sequencing Program, Project 560. Funding for this work was provided by the NSF Biological Oceanography program OCE-1155754, NASA Exobiology NNX15AM11G, as well as OCE-1155756 along with funding from Western Washington University's Office of Research and Sponsored Programs (to CLM), and the Center for Dark Energy Biosphere Investigations (C-DEBI). This is C-DEBI contribution no.379.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Barco RA, Emerson D, Sylvan JB, Orcutt BN, Jacobson Meyers ME, Ramírez GA et al. (2015). New insight into microbial iron oxidation as revealed by the proteomic profile of an obligate iron-oxidizing chemolithoautotroph. Appl Environ Microbiol 81: 5927–5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IA. (2011). Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol 77: 1925–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongers L. (1970). Energy generation and utilization in hydrogen bacteria. J Bacteriol 104: 145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breier JA, Gomez-Ibanez D, Reddington E, Huber JA, Emerson D. (2012). A precision multi-sampler for deep-sea hydrothermal microbial mat studies. Deep Sea Res I 70: 83–90. [Google Scholar]
- Chan CS, McAllister SM, Leavitt AH, Glazer BT, Krepski ST, Emerson D. (2016). The architecture of iron microbial mats reflects the adaptation of chemolithotrophic iron oxidation in freshwater and marine environments. Front Microbiol 7: 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5: e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RE, Moyer CL. (2008). Extreme spatial and temporal variability of hydrothermal microbial mat communities along the Mariana Island Arc and southern Mariana back-arc system. J Geophys Res 113: B8S15. [Google Scholar]
- Emerson D, Floyd MM. (2005). Enrichment and isolation of iron-oxidizing bacteria at neutral pH. Methods Enzymol 397: 112–123. [DOI] [PubMed] [Google Scholar]
- Emerson D, Rentz JA, Lilburn TG, Davis RE, Aldrich H, Chan C et al. (2007). A novel lineage of Proteobacteria involved in formation of marine Fe-oxidizing microbial mat communities. PLoS One 2: e667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson D, Fleming EJ, McBeth JM. (2010). Iron-oxidizing bacteria: an environmental and genomic perspective. Annu Rev Microbiol 64: 561–583. [DOI] [PubMed] [Google Scholar]
- Emerson D, Field EK, Chertkov O, Davenport KW, Goodwin L, Munk C et al. (2013). Comparative genomics of freshwater Fe-oxidizing bacteria: implications for physiology, ecology, and systematics. Front Microbiol 4: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field EK, Sczyrba A, Lyman AE, Harris CC, Woyke T, Stepanauskas R et al. (2014). Genomic insights into the uncultivated marine Zetaproteobacteria at Loihi Seamount. ISME J 9: 857–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton H, Hager KW, McAllister SM, Moyer CL. (2017). Hidden diversity revealed by genome-resolved metagenomics of iron-oxidizing microbial mats from L-ō'ihi Seamount, Hawai’i. ISME J 11: 1900–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil R, Silva FJ, Peretó J, Moya A. (2004). Determination of the core of a minimal bacterial gene set. Microbiol Mol Biol Rev 68: 518–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB et al. (2016). Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J 10: 761–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager KW, Fullerton H, Butterfield DA, Moyer CL. (2017). Community structure of lithotrophically-driven microbial mats from the Mariana Arc and back-arc. Front Microbiol (in press). [DOI] [PMC free article] [PubMed]
- Hansen M, Perner M. (2015). A novel hydrogen oxidizer amidst the sulfur-oxidizing Thiomicrospira lineage. ISME J 9: 696–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henri PA, Rommevaux-Jestin C, Lesongeur F, Mumford A, Emerson D, Godfroy A et al. (2016). Structural iron (II) of basaltic glass as an energy source for Zetaproteobacteria in an abyssal plain environment, off the Mid Atlantic Ridge. Front Microbiol 6: 1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt J, Morris J. (1975). Superoxide dismutase in some obligately anaerobic bacteria. FEBS Lett 50: 315–318. [DOI] [PubMed] [Google Scholar]
- Hoshino T, Kuratomi T, Morono Y, Hori T, Oiwane H, Kiyokawa S et al. (2016). Ecophysiology of Zetaproteobacteria associated with shallow hydrothermal iron-oxyhydroxide deposits in Nagahama Bay of Satsuma Iwo-Jima, Japan. Front Microbiol 6: 1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA. (2008). Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77: 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Yanagawa K, Sunamura M, Takano Y, Ishibashi J, Kakegawa T et al. (2009). Abundance of Zetaproteobacteria within crustal fluids in back-arc hydrothermal fields of the Southern Mariana Trough. Environ Microbiol 11: 3210–3222. [DOI] [PubMed] [Google Scholar]
- Kato S, Nakamura K, Toki T, Ishibashi J, Tsunogai U, Hirota A et al. (2012). Iron-based microbial ecosystem on and below the seafloor: a case study of hydrothermal fields of the Southern Mariana Trough. Front Microbiol 3: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Ohkuma M, Powell DH, Krepski ST, Oshima K, Hattori M et al. (2015). Comparative genomic insights into ecophysiology of neutrophilic, microaerophilic iron oxidizing bacteria. Front Microbiol 6: 1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagkouvardos I, Joseph D, Kapfhammer M, Giritli S, Horn M, Haller D et al. (2016). IMNGS: a comprehensive open resource of processed 16 S rRNA microbial profiles for ecology and diversity studies. Sci Rep 6: 33721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makita H, Tanaka E, Mitsunobu S, Miyazaki M, Nunoura T, Uematsu K et al. (2016). Mariprofundus micogutta sp. nov., a novel iron-oxidizing zetaproteobacterium isolated from a deep-sea hydrothermal field at the Bayonnaise knoll of the Izu-Ogasawara arc, and a description of Mariprofundales ord. nov. and Zetaproteobacteria classis nov. Arch Microbiol 199: 335–346. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, Mavromatis K, Ivanova NN, Chen I-MA, Chu K, Kyrpides NC. (2009). IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25: 2271–2278. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Pillay M et al. (2014). IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res 42: D560–D567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister SM, Davis RE, McBeth JM, Tebo BM, Emerson D, Moyer CL. (2011). Biodiversity and emerging biogeography of the neutrophilic iron-oxidizing Zetaproteobacteria. Appl Environ Microbiol 77: 5445–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBeth JM, Emerson D. (2016). In situ microbial community succession on mild steel in estuarine and marine environments: exploring the role of iron-oxidizing bacteria. Front Microbiol 7: 767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumford AC, Adaktylou IJ, Emerson D. (2016). Peeking under the iron curtain: development of a microcosm for imaging colonization of steel surfaces by Mariprofundus sp. DIS-1, an oxygen tolerant Fe-oxidizing bacterium. Appl Environ Microbiol 82: 6799–6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orcutt BN, Sylvan JB, Knab NJ, Edwards KJ. (2011). Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol Mol Biol Rev 75: 361–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resing JA, Sedwick PN, German CR, Jenkins WJ, Moffett JW, Sohst BM et al. (2015). Basin-scale transport of hydrothermal dissolved metals across the South Pacific Ocean. Nature 523: 200–203. [DOI] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106: 19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. (2015). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32: 929–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos SR, Ochman H. (2004). Identification and phylogenetic sorting of bacterial lineages with universally conserved genes and proteins. Environ Microbiol 6: 754–759. [DOI] [PubMed] [Google Scholar]
- Scott JJ, Breier JA, Luther G III, Emerson D. (2015). Microbial iron mats at the Mid-Atlantic Ridge and evidence that Zetaproteobacteria may be restricted to iron-oxidizing marine systems. PLoS One 10: e0119284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JJ, Glazer BT, Emerson D. (2017). Bringing microbial diversity into focus: high-resolution analysis of iron mats from the L-ō‘ihi Seamount. Environ Microbiol 19: 301–316. [DOI] [PubMed] [Google Scholar]
- Singer E, Emerson D, Webb EA, Barco RA, Kuenen JG, Nelson WC et al. (2011). Mariprofundus ferrooxydans PV-1 the first genome of a marine Fe(II) oxidizing Zetaproteobacterium. PLoS One 6: e25386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabita FR, Satagopan S, Hanson TE, Kreel NE, Scott SS. (2008). Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J Exp Bot 59: 1515–1524. [DOI] [PubMed] [Google Scholar]
- Tamura H, Goto K, Yotsuyanagi T, Nagayama M. (1974). Spectrophotometric determination of iron(II) with 1,10-phenanthroline in presence of large amounts of iron(III). Talanta 21: 314–318. [DOI] [PubMed] [Google Scholar]
- Wankel SD, Germanovich LN, Lilley MD, Genc G, DiPerna CJ, Bradley AS et al. (2011). Influence of subsurface biosphere on geochemical fluxes from diffuse hydrothermal fluids. Nat Geosci 4: 461–468. [Google Scholar]
- White G, Edwards M, Gomez-Perez L, Richardson D, Butt J, Clarke T. (2016). Mechanisms of bacterial extracellular electron exchange. Adv Microb Physiol 68: 87–138. [DOI] [PubMed] [Google Scholar]
- Williams KP, Kelly DP. (2013). Proposal for a new class within the phylum ProteobacteriaAcidithiobacillia classis nov., with the type order Acidithiobacillales, and emended description of the class Gammaproteobacteria. Int J Syst Evol Microbiol 63: 2901–2906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.