Abstract
Background
Engineering of the antibody's fragment crystallizable (Fc) by modifying the amino acid sequence (Fc protein engineering) or the glycosylation pattern (Fc glyco-engineering) allows enhancing effector functions of tumor targeting antibodies. Here, we investigated whether complement-dependent cytotoxicity (CDC) and antibody-dependent cell-mediated cytotoxicity (ADCC) of CD20 antibodies could be improved simultaneously by combining Fc protein engineering and glyco-engineering technologies.
Methods and Results
Four variants of the CD20 antibody rituximab were generated: a native IgG1, a variant carrying the EFTAE modification (S267E/H268F/S324T/G236A/I332E) for enhanced CDC as well as glyco-engineered, non-fucosylated derivatives of both to boost ADCC. The antibodies bound CD20 specifically with similar affinity. Antibodies with EFTAE modification were more efficacious in mediating CDC, irrespective of fucosylation, than antibodies with wild-type sequences due to enhanced C1q binding. In contrast, non-fucosylated variants had an enhanced affinity to FcγRIIIA and improved ADCC activity. Importantly, the double-engineered antibody lacking fucose and carrying the EFTAE modification mediated both CDC and ADCC with higher efficacy than the native CD20 IgG1 antibody.
Conclusion
Combining glyco-engineering and protein engineering technologies offers the opportunity to simultaneously enhance ADCC and CDC activities of therapeutic antibodies. This approach may represent an attractive strategy to further improve antibody therapy of cancer and deserves further evaluation.
Keywords: Antibody therapy, Fc engineering, ADCC, CDC, CD20
Introduction
Therapeutic antibodies represent potent treatment options in cancer therapy [1,2]. In particular, CD20 antibodies are well established in the treatment of B-cell lymphomas and leukemias, and several CD20 antibodies, including rituximab, ofatumumab and obinutuzumab, are approved for clinical use [3]. However, monoclonal antibodies rarely cure patients as monotherapy, not all patients benefit from this generally well-tolerated therapeutic option, and relapses still remain a serious problem. Thus, further improving antibody therapy is a major issue in current translational research.
Deeper insights into antibody effector functions provided the basis for the generation of ‘fit-for-purpose’ antibodies by rational design [1,2,4,5]. In vitro tumor targeting antibodies like rituximab can eliminate malignant cells by different means, including induction of cell death, complement-dependent cytotoxicity (CDC), and recruitment of effector cells for antibody-dependent cell-mediated cytotoxicity (ADCC) or antibody-dependent cellular phagocytosis (ADCP) by engagement of activating Fcγ receptors (FcγR). However, antibodies may vary in effector functions depending on the isotype, the target antigen and its expression levels, or the recognized epitope [6,7,8,9,10]. Traditionally, CD20 antibodies are grouped into type I or type II antibodies [10], which both trigger ADCC effectively, but differ in their capacities to trigger CDC or direct cell death. Thus, type I antibodies (e.g. rituximab) strongly mediate CDC, but weakly elicit direct cell death, while type II antibodies (e.g. obinutuzumab) efficiently induce direct cell death but exert poor CDC activity. Yet, in vivo the situation is more complex, and the relative contribution of different antibody functions is not fully understood. Animal models have suggested that functions mediated by the fragment crystalizable (Fc) such as CDC or effector cell recruitment are crucial in CD20 antibody therapy [11,12,13,14]. Clinically, improved responses to rituximab or other therapeutic antibodies were observed in patients with homozygous expression of the FcγRIIIA-158V allelic variant, which binds the antibody Fc domain with higher affinity, in comparison to patients carrying the low-affinity FcγRIIIA-158F allele [15,16,17,18,19], pointing to a role of FcγRIIIA-expressing natural killer (NK) cells, macrophages, or monocytes. Moreover, activation of NK cells upon rituximab infusion was demonstrated in patients with the high-affinity FcγRIIIA polymorphism [20]. Whereas these results indicate a pivotal role for FcγR engagement and effector cell activation, a contribution of CDC in antibody therapy has not been proven [21]. However, regarding CD20 antibody therapy, a role of CDC has been supported by studies showing that complement is consumed upon rituximab infusion, that patients may benefit from infusion of plasma as a source of complement, and that post-rituximab treatment expression levels of inhibitory membrane-bound complement regulatory protein (mCRP) CD59 were increased in antibody-resistant chronic lymphocytic leukemia (CLL) patients [22,23,24,25].
Fc engineering strategies represent a promising approach to further enhance the efficacy of antibody therapy. Considering ADCC and CDC as important antibody functions, Fc modifications enhancing affinity to activating FcγR or C1q have gained peculiar interest. Two different technologies, either modification of the glycosylation pattern (Fc glyco-engineering) or alteration of the amino acid sequence (Fc protein engineering), have been established. Fc glyco-engineering was applied in particular to enhance ADCC. Thus, glyco-engineered antibodies, now lacking fucosylation of the N297-linked oligosaccharide, had a selectively enhanced affinity to FcγRIIIA and exerted improved efficacy in inducing ADCC by NK cells [26,27,28]. With obinutuzumab, a first glyco-engineered CD20 antibody has been approved for treatment of CLL [29,30,31]. Fc protein engineering approaches were employed to promote either FcγR or C1q binding [4]. A number of amino acid exchanges were identified, which markedly increased affinity to activating FcγR and substantially enhanced ADCC and ADCP [32,33]. In other studies amino acid alterations were found to specifically enhance CDC [34,35,36]. Alternatively, CDC activity was enhanced by generation of mixed-isotype IgG1/IgG3 variants of rituximab or by conversion of IgG1 into IgG3 antibodies [37,38]. In another attempt, CDC was augmented by introducing distinct amino acid exchanges favoring antibody hexamer assembly [39,40]. However, although distinct Fc modifications were identified that either promoted ADCC or CDC, simultaneous enhancement of both effector functions by amino acid alteration remains difficult, probably due to an overlap in the putative binding site for C1q [41] and the binding site for classical FcγR [42,43]. Actually, some CDC-optimized antibody variants had a drop in ADCC activity, why additional rescue modifications were required [34].
In an attempt to engineer antibodies for both enhanced ADCC and CDC, we investigated whether both functions could be improved simultaneously by combining protein engineering and glyco-engineering technologies. Therefore, five amino acid exchanges (S267E/H268F/S324T/G236A/I332E, referred to as EFTAE), which in combination were shown to enhance CDC while maintaining the ADCC activity of native IgG1 antibodies [34], were introduced into the Fc domain of the CD20 antibody rituximab. The antibody was then expressed in a fully fucosylated form or as a glyco-engineered, non-fucosylated derivative, and ADCC and CDC activities of these differentially modified antibodies were analyzed in comparison to the corresponding native IgG1 molecule.
Material and Methods
Cell Culture
Daudi (Burkitt lymphoma) and baby hamster kidney BHK-21 cells (American Type Culture Collection (ATCC), Manassas, VA, USA) were cultured in RPMI 1640 Glutamax-I medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal calf serum (FCS; Thermo Fisher Scientific), 100 U/ml penicillin, and 100 µg/ml streptomycin (Thermo Fisher Scientific; R10+ medium). MEC-2 cells (CLL; German Collection of Microorganism and Cell Cultures (DSMZ), Braunschweig, Germany) were maintained in Iscove's MDM medium (Thermo Fisher Scientific) containing 20% FCS, 100 U/ml penicillin, and 100 µg/ml streptomycin. GRANTA-519 (mantle cell lymphoma; DSMZ) and Chinese hamster ovary CHO-K1 cells (DSMZ) were kept in DMEM medium (Thermo Fisher Scientific) supplemented with 10% FCS, 100 U/ml penicillin and 100 µg/ml streptomycin. CHO glycosylation-mutant Lec13 cells [44,45] were grown in MEM alpha medium containing nucleosides (Thermo Fisher Scientific) and supplemented with 10% dialyzed FCS (Thermo Fisher Scientific), 100 U/ml penicillin and 100 µg/ml streptomycin. Medium of transfected CHO-K1 and Lec13 cells was supplemented with 500 μg/ml hygromycin B (Thermo Fisher Scientific). BHK-21 cells stably transfected with expression vectors encoding FcεRI γ chain and either human FcγRIIIA 158V (BHK-CD16-158V) or FcγRIIIA 158F (BHK-CD16-158F) allelic variants were cultured in medium supplemented with 10 μmol/l methotrexate (Sigma-Aldrich, Munich, Germany) and 500 μg/ml geneticin (Thermo Fisher Scientific) [46].
Antibodies
For generation of antibody expression vector sequences encoding variable light (VL) and heavy (VH) chains of rituximab were synthesized de novo (Eurofins, Ebersberg, Germany) according to published sequences [47]. VL was ligated in frame into antibody κ light (LC) chain expression vector pSectag2-LC [48]. The sequence encoding VH was inserted in heavy chain (HC) expression vectors pSectag2-HC (encoding a native IgG1 Fc domain [48]) and pSectag2-HC-EFTAE (encoding the engineered Fc domain with amino acid substitutions S267E/H268F/S324T/G236A/I332E [34]; unpublished data). Similarly, expression vectors for corresponding HER2 antibody variants were constructed using VL and VH sequences from antibody trastuzumab [49]. Correctness of cloned sequences was confirmed by Sanger sequencing of final constructs. For expression, CHO-K1 or Lec13 cells were stably transfected with antibody LC and HC expression constructs with the Amaxa Nucleofectior System (Lonza, Cologne, Germany) using transfection kit V according to the manufacturer's recommendations as described previously [50]. After 48 h, medium was exchanged by culture medium containing 500 μg/ml hygromycin B. Stably transfected production lines were established by selection with hygromycin B (500 μg/ml). After establishing single-cell subclones by limiting dilution, single clones with moderate to high production rates were identified by flow cytometry analysis of supernatants. Antibodies were purified from cell culture supernatant with CaptureSelect™ IgG-CH1 Affinity Matrix (Thermo Fisher Scientific) and affinity chromatography using gravity flow columns (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer's recommendations. Antibody concentration and integrity were determined by quantitative capillary electrophoresis using Experion™ Pro260 technology (Bio-Rad Laboratories) in accordance with the manufacturer's protocol. Trastuzumab was purchased from Roche (Penzberg, Germany).
Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE), Western Blot, and Lectin Blot Analysis
SDS-PAGE under reducing and non-reducing conditions was performed according to standard procedures [50]. Briefly, 1–2 μg of purified antibodies was loaded on 6% or 12% polyacrylamide gels. Gels were either stained directly with colloidal Coomassie brilliant blue staining solution (Carl Roth GmbH, Karlsruhe, Germany) or blotted to PVDF membranes. Human IgG Fc was detected using goat-anti-human-IgG-HRP conjugate (Sigma Aldrich) as previously described [50]. Lectin blots using biotinylated Aleuria aurantia lectin (Vector Laboratories, Burlingame, CA, USA) and HRP-conjugated NeutrAvidin (Thermo Fisher Scientific) were performed as previously described [50].
Flow Cytometry
For indirect immunofluorescence staining, 3 × 105 cells were washed in phosphate-buffered saline supplemented with 1% bovine serum albumin (Sigma-Aldrich) and 0.1% sodium-azide (PBA buffer). Cells were incubated with antibodies at the indicated concentrations on ice for 30 min, washed two times with 500 μl PBA buffer, and stained with FITC-conjugated anti-human IgG Fc F(ab‘)2 fragments of polyclonal goat antibodies (DAKO, Glostrup, Denmark) or FITC-labeled goat anti-mouse IgG Fc F(ab‘)2 antibodies (Sigma-Aldrich). After a final wash, cells were analyzed on a Navios flow cytometer (Beckman Coulter, Brea, CA, USA). 10,000 events were counted, and dead cells and cellular debris were excluded by using appropriate forward and side scatter gates. To analyze C1q deposition 3 × 105 Daudi cells were first incubated with antibodies at 25 µg/ml in 50 µl R10+ medium on ice for 20 min. Human serum was added to R10+ medium to a final concentration of 2% and incubated for neutralization of C5 with eculizumab (Alexion Pharma GmbH, Munich, Germany) at a concentration of 200 µg/ml at room temperature for 20 min. Then 50 µl were added to antibody-coated cells. Cells were incubated at 37 °C for 10 min and then washed three times. Finally, cells were incubated with a murine FITC-conjugated anti-C1q antibody (DAKO) for 1 h; cells were washed three times, re-suspended in cold PBA, and analyzed for cell-bound C1q by flow cytometry. Expression of mCRPs was determined using mouse anti-human CD46 IgG1 (Thermo Fisher Scientific), CD55 IgG1 (BioRad), and CD59 IgG2a antibodies (EXBIO, Vestec, Czech Republic) at a concentration of 50 µg/ml. As isotypes purified murine hybridoma anti-myc IgG1 antibody 9E10 (ATCC) and anti-keyhole limpet hemocyanin IgG2a antibody (R&D Systems, Minneapolis, MN, USA) were used.
Cytotoxicity Assay
CDC and ADCC were determined in standard 51Cr release experiments as described [50]. Human mononuclear cells (MNCs) and plasma, which were separated from citrate-anticoagulated blood from healthy volunteers by density gradient centrifugation using Easycoll (Biochrom, Berlin, Germany), served as a source of effector cells and complement, respectively. In CDC assays, plasma was used at 25%, and recombinant hirudin (Refludan®, Bayer HealthCare Pharmaceuticals, Wayne, NJ, USA) was added to a concentration of 10 µg/ml as anticoagulant. In ADCC experiments MNCs were applied at an effector-to-target cell ratio of 40:1.
Statistical Analysis
Graphical and statistical analyses were performed using GraphPad Prism 5.0 software. P values were calculated using repeated measures ANOVA and Bonferroni post-tests. The null hypothesis was rejected for p < 0.05.
Results
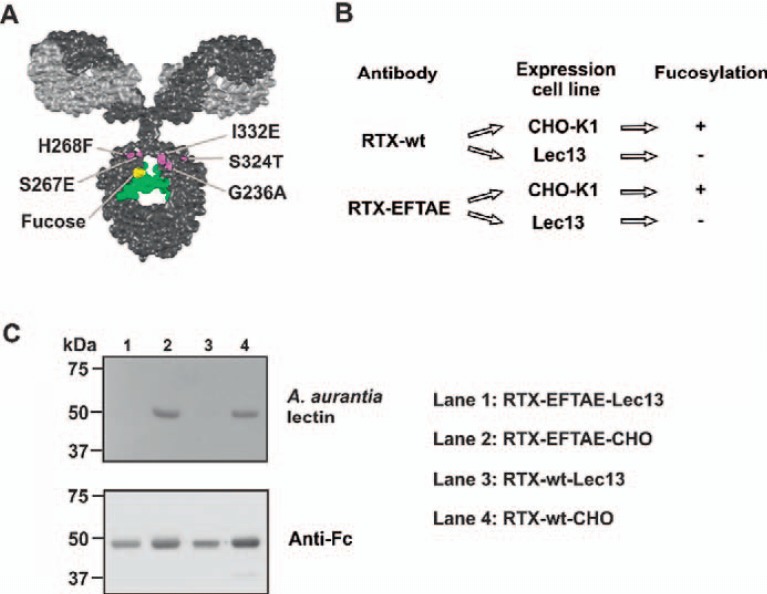
With the aim to enhance CDC and ADCC simultaneously, the Fc domain of the CD20 antibody rituximab was double-engineered by combining Fc protein engineering and Fc glyco-engineering technologies (fig. 1). Thus, the amino acid substitutions S267E/H268F/S324T/G236A/I332E (EFTAE modification), which previously have been shown to enhance CDC while preserving ADCC activity, were introduced into the antibody constant heavy region 2 (fig. 1A). To also increase its ADCC activity, the antibody was glyco-engineered by expression in Lec13 cells, which produce IgG1 molecules lacking Fc fucosylation (fig. 1B). By expression in CHO-K1 cells, a fucosylated EFTAE-modified derivative was generated as a control. Similarly, corresponding native wild-type CD20 antibody sequences were expressed in CHO-K1 or Lec13 cells to generate corresponding antibody variants lacking the EFTAE modification (fig. 1B). This resulted in four different CD20 antibodies, which were referred to as RTX-EFTAE-Lec13 (double-engineered Fc domain for enhanced CDC and ADCC), RTX-wt-CHO (unmodified IgG1 Fc domain), RTX-EFTAE-CHO (protein-engineered Fc) or RTX-wt-Lec13 (glyco-engineered Fc). The antibodies were purified by affinity chromatography from cell culture supernatants of stably transfected cell lines. Integrity and purity of antibody preparations were confirmed by reducing or non-reducing SDS-PAGE and subsequent Coomassie blue staining (unpublished data). To determine the fucosylation status of antibody variants expressed in different host cell lines, lectin blots using biotinylated Aleuria aurantia lectin were performed (fig. 1C). In agreement with previous findings [50], antibodies expressed in CHO-K1 cells (i.e., RTX-wt-CHO and RTX-EFTAE-CHO) were fucosylated, whereas their derivatives produced in Lec13 cells (i.e., RTX-wt-Lec13 and RTX-EFTAE-Lec13) lacked fucosylation.
Fig. 1.
Generation of Fc engineered variants of antibody rituximab. A Illustration of positions of amino acid substitutions S267E/H268F/S324T/G236A/I332E (EFTAE-modification; in magenta) within the constant heavy chain (CH) 2 domain, which enhance CDC activity, and the fucose residue (in yellow), which is critical for FcγRIII binding and ADCC. The VL chain is depicted in light grey and the heavy chain in dark grey. The N 297-linked glycan is colored in green. The IgG model structure is based on the pdb file provided by Dr. Mike Clark [57] and was modified using Discovery Studio Visualizer (Biovia, San Diego, CA, USA). B The EFTAE modification was introduced into Fc domain sequences of antibody rituximab (RTX-EFTAE). Both RTX-EFTAE and a variant with a wild-type Fc domain (RTX-wt) sequence were expressed in CHO-K1 and Lec13 cells to generate fucosylated antibodies (RTX-wt-CHO and RTX-EFTAE-CHO) and corresponding non-fucosylated derivatives (RTX-wt-Lec13 and RTX-EFTAE-Lec13), respectively. C After purification by affinity chromatography fucosylation of antibodies was analyzed by lectin blot using biotinylated A. aurantia lectin and HRP-conjugated neutrAvidin protein showing that antibodies produced in Lec13 cells lacked fucose in contrast to antibodies expressed in CHO-K1 cells. As a control antibody heavy chains were detected by Western blot analysis using HRP-conjugated anti-human IgG Fc antibody. Data from one representative experiment out of two performed are presented.
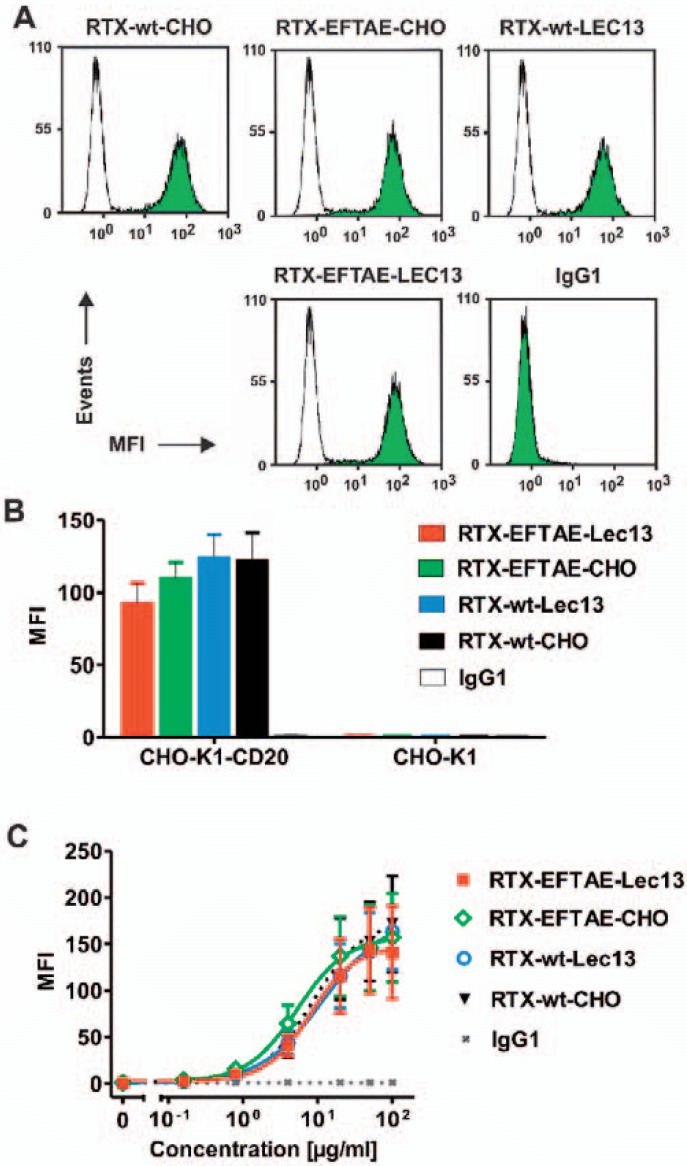
Analysis of CD20 binding by flow cytometry revealed that antigen specificity was not altered by expression in different cell lines or Fc modifications. Thus, all rituximab variants bound both CD20-positive MEC-2 cells (fig. 2A) and CHO-K1 cells that were stably transfected with human CD20 (CHO-K1-CD20; fig. 2B). In contrast, no binding to non-transfected CHO-K1 cells (fig. 2B) and CD20-negative tumor cell lines (e.g. SK-BR-3 cells) was observed (data not shown). Importantly, all antibodies exerted similar affinity to CD20 irrespective of their Fc domain modification as revealed by comparison of dose-dependent binding curves using CHO-K1-CD20 cells and flow cytometry analysis (fig. 2C). Thus, CD20 specificity and binding avidity was maintained despite different Fc manipulations.
Fig. 2.
CD20 binding analysis. A CD20-positive MEC-2 cells were incubated in buffer alone (white peaks) or in the presence of the indicated antibodies at 50 µg/ml (green peaks), then reacted with FITC-conjugated anti-human IgG Fc F(ab')2 and analyzed by flow cytometry. B RTX-wt-CHO, RTX-EFTAE-CHO, RTX-wt-Lec13 and RTX-EFTAE-Lec13 (concentration: 50 µg/ml) specifically bound to CHO-K1-CD20 cells but did not react with non-transfected CHO-K1 cells. Bars indicate mean values ± SEM (n = 2). Antibodies were detected with FITC-conjugated anti-human IgG Fc F(ab')2 fragments and flow cytometry. Trastuzumab was used as control antibody (MFI, mean fluorescence intensity). C Antibody variants were analyzed for binding to CHO-K1-CD20 cells at varying concentrations using secondary FITC-conjugated anti-human IgG Fc F(ab')2 fragments for detection and flow cytometry. Data points represent mean values ± SEM (n = 4).
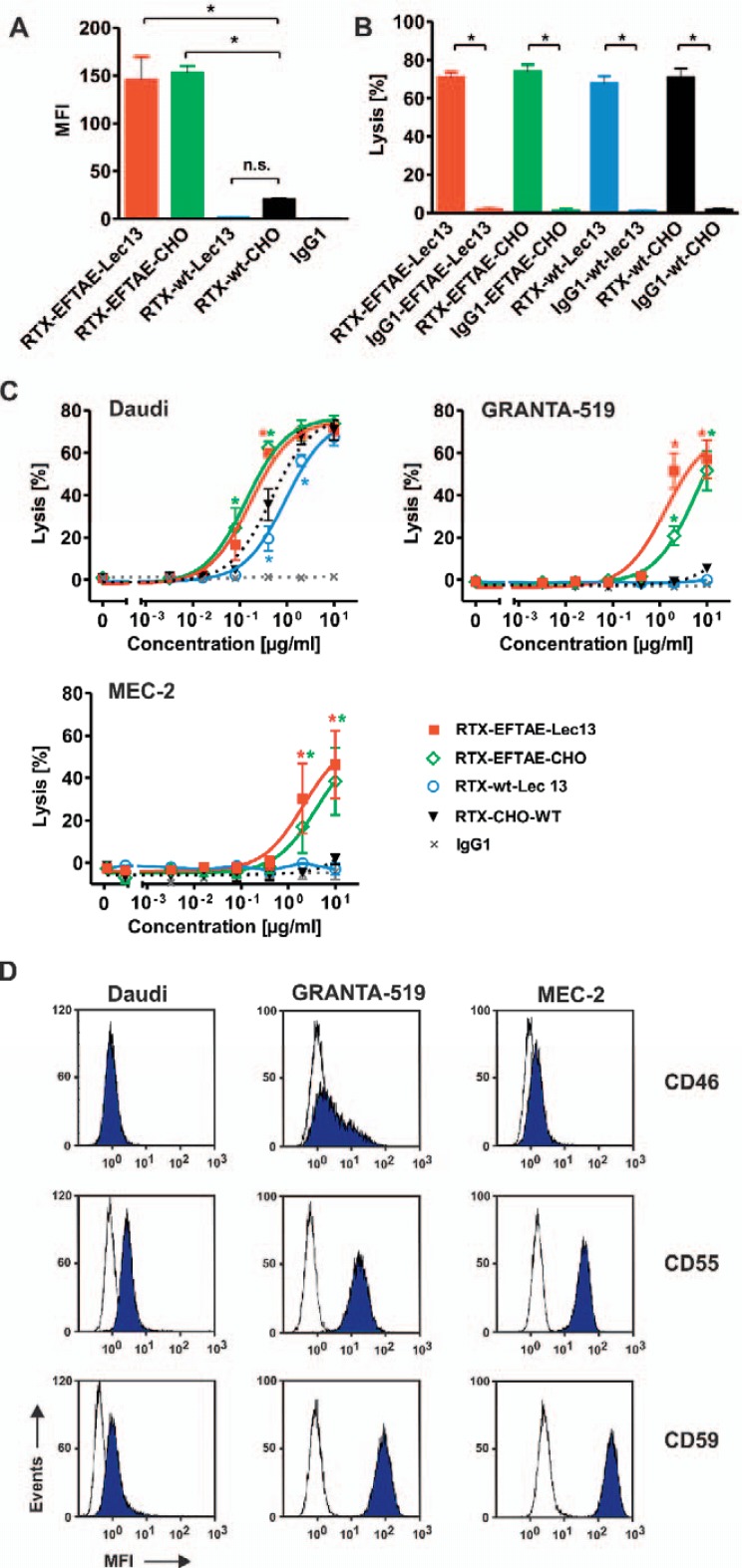
Efficient deposition of C1q on target cells is a prerequisite for induction of CDC via the classical pathway. Therefore, it was analyzed whether the EFTAE modification enhanced the abilities of the antibodies to fix C1q on CD20-positive lymphoma cells and, if this strategy was applicable, to non-fucosylated antibodies (fig. 3A). To this, CD20-positive Daudi cells were opsonized with RTX-wt-CHO, RTX-EFTAE-CHO, RTX-wt-Lec13, or RTX-EFTAE-Lec13, then incubated with human serum as a source of C1q in the presence of the C5 neutralizing antibody eculizumab to block CDC, and finally reacted with a C1q-specific antibody. Flow cytometry analysis revealed that higher amounts of C1q were bound by target cells coated with RTX-EFTAE-CHO or RTX-EFTAE-Lec13, presumably due to an increased gain in affinity to C1q achieved by the EFTAE modification. Obviously, C1q binding efficacy was similar for the protein-engineered RTX-EFTAE-CHO antibody variant and the double-engineered antibody RTX-EFTAE-Lec13.
Fig. 3.
Induction of CDC by rituximab antibody variants. A Daudi cells were coated with RTX-wt-CHO, RTX-EFTAE-CHO, RTX-wt-Lec13 or RTX-EFTAE-Lec13 (concentration: 50 µg/ml) and then incubated in the presence of human serum (1%) as a source of C1q. Eculizumab was added to block CDC. Deposition of C1q was analyzed with a FITC-coupled mouse anti-human C1q antibody by flow cytometry. Bars represent mean values ± SEM (n = 3). B CDC by rituximab variants in comparison to corresponding HER2-specific control antibodies was analyzed by 51Cr release experiments using Daudi cells as targets in the presence of 25% human plasma. Antibodies were applied at 10 µg/ml. Mean values ± SEM are depicted. Significant differences between CD20 antibodies and similarly designed control proteins are indicated (*, p ≤ 0.05; n = 3). C Dose-dependent induction of CDC against Daudi (n = 3), GRANTA-519 (n = 4) and MEC-2 (n = 4) cells by rituximab variants. Human plasma (25%) was added as a source of complement. Statistically significant differences in CDC between engineered antibodies and the native CD20 IgG1 molecule are indicated (*, P ≤ 0.05). D Daudi, GRANTA-519 and MEC-2 cells were incubated with specific antibodies against mCRPs CD46, CD55 or CD59 (blue peaks) or isotype matched control antibodies (white peaks), which were subsequently detected with secondary FITC-conjugated goat anti-mouse IgG Fc F(ab')2 fragments, and expression levels were analyzed by flow cytometry. Results from one representative experiment are shown (n = 3; MFI, mean fluorescence intensity).
To analyze the ability of rituximab variants to kill lymphoma cells by CDC, 51Cr release experiments were performed using human plasma and Daudi Burkitt's lymphoma cells (fig. 3B). All CD20 antibodies triggered efficient CDC against Daudi cells, whereas similarly constructed control antibodies directed against HER2 and harboring the corresponding Fc modifications were not effective. Thus, RTX-wt-CHO, RTX-EFTAE-CHO, RTX-wt-Lec13, and RTX-EFTAE-Lec13 triggered CDC in a target antigen-dependent manner. When dose-dependent induction of CDC against Daudi cells was analyzed, CD20 antibody variants were all effective at nanomolar concentrations but, importantly, differed markedly in their potency (fig. 3C). Thus, RTX-EFTAE-CHO and RTX-EFTAE-Lec13 had equal activity and were most effective showing an approximately 4- to 5-fold lower half-maximum effective concentration than RTX-wt-CHO and RTX-wt-Lec13. The observed differences in the CDC activity between antibodies were even more pronounced, when GRANTA-519 mantle cell lymphoma or MEC-2 CLL cells were used as target cells. Here, antibodies lacking the EFTAE modification hardly triggered CDC, whereas RTX-EFTAE-CHO and RTX-EFTAE-Lec13 induced substantial target cell lysis, although higher concentrations were required in comparison to experiments with Daudi cells.
One explanation for the observed differences in the susceptibility of these cell lines to CDC may be variation in the expression of mCRPs CD46, CD55, and CD59. Therefore cell lines were analyzed for surface levels of CD46, CD55, and CD59 by flow cytometry (fig. 3D). Interestingly, MEC-2 and GRANTA-519 cells expressed significantly higher levels of all three mCRPs than the CDC-sensitive Daudi cells. Thus, expression of complement defense proteins may contribute to the observed differences between cell lines in CDC assays.
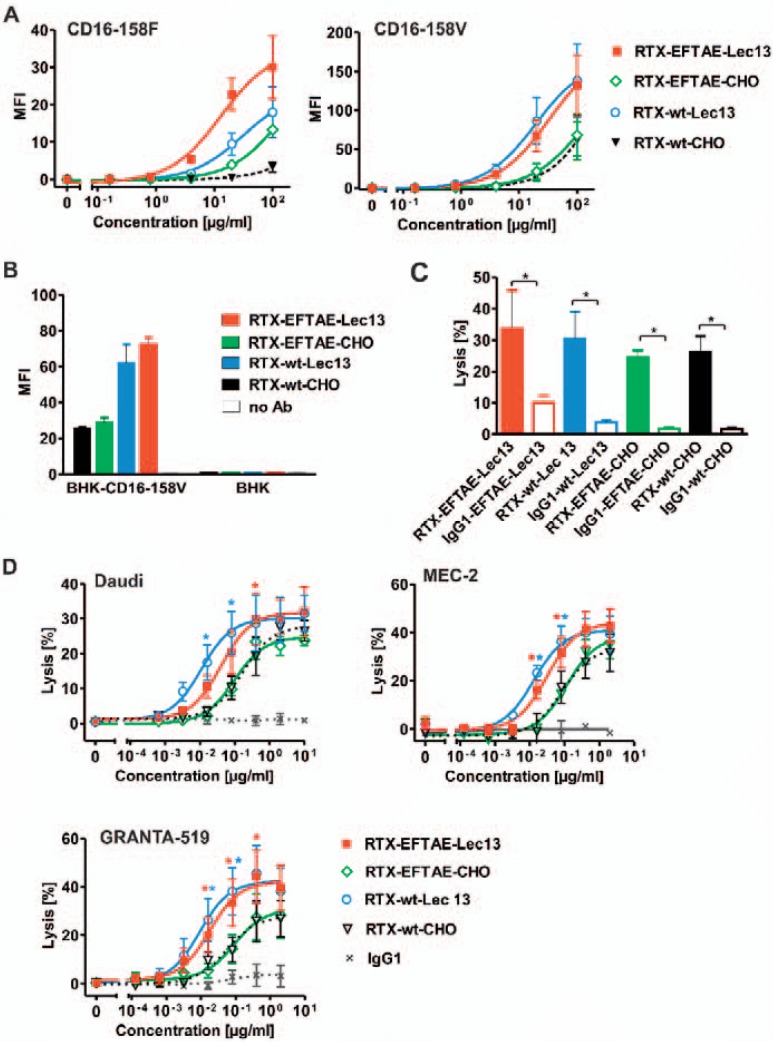
Next, to examine the affinity of different antibody constructs to FcγRIIIA, dose-dependent binding to BHK cells stably transfected with either FcγRIIIA-158V or FcγRIIIA-158F expression constructs was analyzed (fig. 4A). We found that both non-fucosylated antibody variants had a higher affinity to both FcγRIIIA allelic variants relative to RTX-wt-CHO and RTX-EFTAE-CHO. None of the antibodies bound to non-transfected BHK cells, confirming that measured differences in the fluorescence intensities were due to altered FcγRIIIA binding (fig. 4B). The ability of the antibody derivatives to induce ADCC was examined in 51Cr release experiments using Daudi lymphoma cells as targets and human MNCs as effector cells (fig. 4C). All rituximab variants triggered ADCC, whereas corresponding control antibodies targeting HER2 were not effective, further confirming the antigen-specific mode of action. To compare the potency of the CD20 antibody variants, dose-dependent induction of ADCC was analyzed using the cell lines Daudi, GRANTA-519, and MEC-2 (fig. 4D). Importantly, the observed gain in the affinity to FcγRIIIA exerted by non-fucosylated antibody variants (fig. 4A) resulted in a higher potency to trigger ADCC (fig. 4D). Thus, RTX-EFTAE-Lec13 and RTX-wt-Lec13 were more efficacious in inducing ADCC against all three cell lines tested than the fucosylated CHO antibodies, irrespective of the EFTAE mutation. Of note, antibody RTX-wt-Lec13 was only slightly more effective than the double-engineered RTX-EFTAE-Lec13 antibody, and the observed differences did not reach statistical significance.
Fig. 4.
FcγRIIIA binding and induction of ADCC by rituximab antibody variants. A Dose-dependent binding to BHK cells transfected with expression vectors encoding FcεRI γ chain and either human FcγRIIIA-158V (BHK-CD16-158V) or FcγRIIIA-158V (BHK-CD16-158F) was analyzed by flow cytometry using FITC-coupled anti-human IgG Fc F(ab')2 fragments. B Antigen-specific binding was verified by analyzing binding to BHK-CD16-158V vs. un-transfected BHK cells. Antibodies were applied at 50 µg/ml and detected as described above. C ADCC by rituximab variants in comparison to corresponding HER2-specific control antibodies was analyzed by 51Cr release experiments with Daudi cells as targets and human MNCs as effector cells. Antibodies were applied at 10 µg/ml. Mean values ± SEM are depicted. Significant differences between CD20 antibodies and similarly designed control antibodies are indicated (*, p ≤ 0.05; n = 3). D Dose-dependent induction of ADCC against Daudi (n = 3), MEC-2 (n = 4) and GRANTA-519 cells (n = 4) by rituximab variants using MNC effector cells. Statistically significant differences in target cell lysis between Fc-engineered antibodies and the native CD20 IgG1 molecule are indicated (*, p ≤ 0.05).
In conclusion, combining glyco-engineering and protein engineering technologies allows enhancing both CDC and ADCC activities of therapeutic antibodies simultaneously. The Fc double-engineering approach may represent an attractive strategy to further improve antibody therapy of cancer and may deserve further evaluation towards clinical testing.
Discussion
In an attempt to enhance CDC and ADCC antibody functions simultaneously, an Fc double-engineered variant of the CD20 antibody rituximab was generated by combining protein engineering and glyco-engineering technologies. The resulting non-fucosylated CD20 antibody with the EFTAE -modification [34] was more efficacious in mediating CDC and ADCC against lymphoma or leukemia cells than the corresponding native IgG1. These results suggest that glyco-engineering and protein engineering technologies can be applied to the same antibody molecule, which offers the opportunity to generate antibodies with both enhanced ADCC and CDC activity.
Fc-engineered antibodies are increasingly gaining importance in antibody therapy of cancer [1,4]. Whereas antibodies optimized for CDC to our knowledge have not been evaluated in patients, to date two Fc glyco-engineered antibodies (i.e., mogamulizumab and obinutuzumab) with enhanced FcγRIIIA binding and ADCC activity have been approved for clinical use. However, it still remains unclear, whether these Fc modifications indeed translate into higher therapeutic efficacy in patients since direct comparisons between Fc-engineered antibodies and their corresponding native IgG1 counterparts in patients are still lacking [51].
Different murine models have suggested that both complement and effector cell recruitment represent important in vivo effector functions for antibodies targeting CD20 or other tumor-associated antigens, suggesting that enhancement of both effector functions may be beneficial. Of note, the relative contribution of complement and FcγR engagement varied between different murine models: in some models the therapeutic efficacy of the antibody largely depended on CDC, whereas in other models the antibody strictly required FcγR engagement [11,12,13]. In patients, tumor cell characteristics, such as target antigen expression levels or cell surface expression of antigens that regulate susceptibility of tumor cells to CDC or ADCC, may determine which killing mechanism is available to the therapeutic antibody. Thus, expression of antigens inhibiting effector cell activation (e.g. human leukocyte antigens or CD47 [5,52]) or receptors promoting cellular cytotoxicity (e.g. NKG2D [53]) may play a role. Likewise expression of mCRPs may protect tumor cells from CDC and thus lower the relative contribution of this elimination mechanism [54]. However, inhibitory effects may be overcome with Fc-engineered antibodies, as also suggested in the current study. Thus, rituximab variants with the EFTAE modification triggered CDC against MEC-2 and GRANTA-519 cells, which abundantly expressed mCRPs and were almost resistant to CDC by the native IgG1 antibody.
More recent animal data suggest that in vivo mechanisms of CD20 antibodies are affected by additional factors such as tumor burden or the anatomic location [14]. Whereas low tumor load was eradicated by CDC, in the situation of high tumor load both complement and FcγR engagement were required. In addition, an impact of the tumor microenvironment on antibody functions has been suggested [55]. Thus, in human CD20 transgenic mice, depletion of distinct B-cell compartments were dependent on different mechanisms [55]. While CDC was the underlying elimination mechanism in killing of marginal-zone B cells, FcγR-dependent mechanisms were required for elimination of blood B cells as well as eradication of lymph node and follicular B cells in the spleen. Thus, in certain situations both CDC and effector cell-mediated killing mechanisms may be required for sufficient target cell depletion, suggesting that particularly in such situations double-engineered antibodies with both enhanced ADCC and CDC activity may have advantages over native antibodies, or antibodies optimized only for one effector function.
Enhancing of CDC and ADCC simultaneously is difficult to achieve by amino acid alterations alone. In one approach, Fc glyco-engineering was applied to an IgG1/IgG3 mixed-isotype antibody, which resulted in enhanced CDC and ADCC activities [37]. Results of the current study provide profound evidence that augmented ADCC and CDC activity can also be achieved by combining Fc protein engineering and Fc glyco-engineering technology. Type I CD20 antibodies such as rituximab are typically characterized by strong potency to trigger CDC and ADCC, which at least in part is attributed to favorable characteristics of the target antigen and the recognized epitope [25]. Whether this double-engineering approach is applicable to other CD20 antibodies or antibodies targeting other antigens still needs to be investigated. The observed higher ADCC activity with MNC effector cells presumably reflects NK cell activity in short-time 51Cr release experiments. If double-engineered, non-fucosylated antibodies endowed with the EFTAE modification also have a higher activity in the activation of myeloid effector cells for ADCC or ADCP remains to be determined. The influence of the EFTAE modification may be more pronounced with myeloid effector cells than with NK cells since this modification affects affinity to both activating FcγRIIA and inhibitory FcγRIIB receptors [34] which are both expressed by macrophages and monocytes, but not by NK cells [56].
In conclusion, ADCC and CDC activities of therapeutic antibodies may be enhanced simultaneously by combining Fc glyco-engineering and protein engineering technologies as exemplified here for non-fucosylated CD20 antibodies harboring the EFTAE modification, which exerted significantly improved effector functions. Thus, this double-engineering approach may represent an attractive strategy to further improve antibody therapy of tumors and may deserve further evaluation towards clinical testing.
Disclosure Statement
The authors declare no competing financial interests.
Acknowledgements
Professor Pamela Stanley (Albert Einstein College of Medicine of Yeshiva University, New York, NY, USA) is kindly acknowledged for providing the cell line Lec13. M. P. is supported by the Mildred-Scheel professorship program by the Deutsche Krebshilfe e. V. C. K and M. P. are supported by a research grant from the Wilhelm Sander Foundation (2014.134.1). M.P. and T.V. receive research support by the Deutsche Forschungsgemeinschaft (PE1425/5-1 and VA124/9-1). We thank Anja Muskulus and Britta von Below for excellent technical assistance.
References
- 1.Weiner GJ. Building better monoclonal antibody-based therapeutics. Nat Rev Cancer. 2015;15:361–370. doi: 10.1038/nrc3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott AM, Wolchok JD, Old LJ. Antibody therapy of cancer. Nat Rev Cancer. 2012;12:278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- 3.Lim SH, Levy R. Translational medicine in action: anti-CD20 therapy in lymphoma. J Immunol. 2014;193:1519–1524. doi: 10.4049/jimmunol.1490027. [DOI] [PubMed] [Google Scholar]
- 4.Kellner C, Derer S, Valerius T, Peipp M. Boosting ADCC and CDC activity by Fc engineering and evaluation of antibody effector functions. Methods. 2014;65:105–113. doi: 10.1016/j.ymeth.2013.06.036. [DOI] [PubMed] [Google Scholar]
- 5.Ferris RL, Jaffee EM, Ferrone S. Tumor antigen-targeted, monoclonal antibody-based immunotherapy: clinical response, cellular immunity, and immunoescape. J Clin Oncol. 2010;28:4390–4399. doi: 10.1200/JCO.2009.27.6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niwa R, Sakurada M, Kobayashi Y, Uehara A, Matsushima K, Ueda R, Nakamura K, Shitara K. Enhanced natural killer cell binding and activation by low-fucose IgG1 antibody results in potent antibody-dependent cellular cytotoxicity induction at lower antigen density. Clin Cancer Res. 2005;11:2327–2336. doi: 10.1158/1078-0432.CCR-04-2263. [DOI] [PubMed] [Google Scholar]
- 7.van Meerten T, van Rijn RS, Hol S, Hagenbeek A, Ebeling SB. Complement-induced cell death by rituximab depends on CD20 expression level and acts complementary to antibody-dependent cellular cytotoxicity. Clin Cancer Res. 2006;12:4027–4035. doi: 10.1158/1078-0432.CCR-06-0066. [DOI] [PubMed] [Google Scholar]
- 8.Beers SA, Glennie MJ, White AL. Influence of immunoglobulin isotype on therapeutic antibody function. Blood. 2016;127:1097–1101. doi: 10.1182/blood-2015-09-625343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niederfellner G, Lammens A, Mundigl O, Georges GJ, Schaefer W, Schwaiger M, Franke A, Wiechmann K, Jenewein S, Slootstra JW, Timmerman P, Brannstrom A, Lindstrom F, Mossner E, Umana P, Hopfner KP, Klein C. Epitope characterization and crystal structure of GA101 provide insights into the molecular basis for type I/II distinction of CD20 antibodies. Blood. 2011;118:358–367. doi: 10.1182/blood-2010-09-305847. [DOI] [PubMed] [Google Scholar]
- 10.Cragg MS, Glennie MJ. Antibody specificity controls in vivo effector mechanisms of anti-CD20 reagents. Blood. 2004;103:2738–2743. doi: 10.1182/blood-2003-06-2031. [DOI] [PubMed] [Google Scholar]
- 11.Di Gaetano N, Cittera E, Nota R, Vecchi A, Grieco V, Scanziani E, Botto M, Introna M, Golay J. Complement activation determines the therapeutic activity of rituximab in vivo. J Immunol. 2003;171:1581–1587. doi: 10.4049/jimmunol.171.3.1581. [DOI] [PubMed] [Google Scholar]
- 12.Clynes RA, Towers TL, Presta LG, Ravetch JV. Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets. Nat Med. 2000;6:443–446. doi: 10.1038/74704. [DOI] [PubMed] [Google Scholar]
- 13.de Haij S, Jansen JH, Boross P, Beurskens FJ, Bakema JE, Bos DL, Martens A, Verbeek JS, Parren PW, van de Winkel JG, Leusen JH. In vivo cytotoxicity of type I CD20 antibodies critically depends on Fc receptor ITAM signaling. Cancer Res. 2010;70:3209–3217. doi: 10.1158/0008-5472.CAN-09-4109. [DOI] [PubMed] [Google Scholar]
- 14.Boross P, Jansen JH, de Haij S, Beurskens FJ, van der Poel CE, Bevaart L, Nederend M, Golay J, van de Winkel JG, Parren PW, Leusen JH. The in vivo mechanism of action of CD20 monoclonal antibodies depends on local tumor burden. Haematologica. 2011;96:1822–1830. doi: 10.3324/haematol.2011.047159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, Watier H. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood. 2002;99:754–758. doi: 10.1182/blood.v99.3.754. [DOI] [PubMed] [Google Scholar]
- 16.Weng WK, Levy R. Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J Clin Oncol. 2003;21:3940–3947. doi: 10.1200/JCO.2003.05.013. [DOI] [PubMed] [Google Scholar]
- 17.Persky DO, Dornan D, Goldman BH, Braziel RM, Fisher RI, Leblanc M, Maloney DG, Press OW, Miller TP, Rimsza LM. Fc gamma receptor 3a genotype predicts overall survival in follicular lymphoma patients treated on SWOG trials with combined monoclonal antibody plus chemotherapy but not chemotherapy alone. Haematologica. 2012;97:937–942. doi: 10.3324/haematol.2011.050419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Musolino A, Naldi N, Bortesi B, Pezzuolo D, Capelletti M, Missale G, Laccabue D, Zerbini A, Camisa R, Bisagni G, Neri TM, Ardizzoni A. Immunoglobulin G fragment C receptor polymorphisms and clinical efficacy of trastuzumab-based therapy in patients with HER-2/neu-positive metastatic breast cancer. J Clin Oncol. 2008;26:1789–1796. doi: 10.1200/JCO.2007.14.8957. [DOI] [PubMed] [Google Scholar]
- 19.Bibeau F, Lopez-Crapez E, Di Fiore F, Thezenas S, Ychou M, Blanchard F, Lamy A, Penault-Llorca F, Frebourg T, Michel P, Sabourin JC, Boissiere-Michot F. Impact of Fc{gamma}RIIa-Fc{gamma}RIIIa polymorphisms and KRAS mutations on the clinical outcome of patients with metastatic colorectal cancer treated with cetuximab plus irinotecan. J Clin Oncol. 2009;27:1122–1129. doi: 10.1200/JCO.2008.18.0463. [DOI] [PubMed] [Google Scholar]
- 20.Veeramani S, Wang SY, Dahle C, Blackwell S, Jacobus L, Knutson T, Button A, Link BK, Weiner GJ. Rituximab infusion induces NK activation in lymphoma patients with the high-affinity CD16 polymorphism. Blood. 2011;118:3347–3349. doi: 10.1182/blood-2011-05-351411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weng WK, Levy R. Expression of complement inhibitors CD46, CD55, and CD59 on tumor cells does not predict clinical outcome after rituximab treatment in follicular non-Hodgkin lymphoma. Blood. 2001;98:1352–1357. doi: 10.1182/blood.v98.5.1352. [DOI] [PubMed] [Google Scholar]
- 22.Bannerji R, Kitada S, Flinn IW, Pearson M, Young D, Reed JC, Byrd JC. Apoptotic-regulatory and complement-protecting protein expression in chronic lymphocytic leukemia: relationship to in vivo rituximab resistance. J Clin Oncol. 2003;21:1466–1471. doi: 10.1200/JCO.2003.06.012. [DOI] [PubMed] [Google Scholar]
- 23.Kennedy AD, Beum PV, Solga MD, DiLillo DJ, Lindorfer MA, Hess CE, Densmore JJ, Williams ME, Taylor RP. Rituximab infusion promotes rapid complement depletion and acute CD20 loss in chronic lymphocytic leukemia. J Immunol. 2004;172:3280–3288. doi: 10.4049/jimmunol.172.5.3280. [DOI] [PubMed] [Google Scholar]
- 24.Klepfish A, Schattner A, Ghoti H, Rachmilewitz EA. Addition of fresh frozen plasma as a source of complement to rituximab in advanced chronic lymphocytic leukaemia. Lancet Oncol. 2007;8:361–362. doi: 10.1016/S1470-2045(07)70106-7. [DOI] [PubMed] [Google Scholar]
- 25.Lim SH, Beers SA, French RR, Johnson PW, Glennie MJ, Cragg MS. Anti-CD20 monoclonal antibodies: historical and future perspectives. Haematologica. 2010;95:135–143. doi: 10.3324/haematol.2008.001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrara C, Stuart F, Sondermann P, Brunker P, Umana P. The carbohydrate at FcgammaRIIIa Asn-162. An element required for high affinity binding to non-fucosylated IgG glycoforms. J Biol Chem. 2006;281:5032–5036. doi: 10.1074/jbc.M510171200. [DOI] [PubMed] [Google Scholar]
- 27.Ferrara C, Grau S, Jager C, Sondermann P, Brunker P, Waldhauer I, Hennig M, Ruf A, Rufer AC, Stihle M, Umana P, Benz J. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose. Proc Natl Acad Sci U S A. 2011;108:12669–12674. doi: 10.1073/pnas.1108455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shields RL, Lai J, Keck R, O'Connell LY, Hong K, Meng YG, Weikert SH, Presta LG. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J Biol Chem. 2002;277:26733–26740. doi: 10.1074/jbc.M202069200. [DOI] [PubMed] [Google Scholar]
- 29.Mossner E, Brunker P, Moser S, Puntener U, Schmidt C, Herter S, Grau R, Gerdes C, Nopora A, van Puijenbroek E, Ferrara C, Sondermann P, Jager C, Strein P, Fertig G, Friess T, Schull C, Bauer S, Dal Porto J, Del Nagro C, Dabbagh K, Dyer MJ, Poppema S, Klein C, Umana P. Increasing the efficacy of CD20 antibody therapy through the engineering of a new type II anti-CD20 antibody with enhanced direct and immune effector cell-mediated B-cell cytotoxicity. Blood. 2010;115:4393–4402. doi: 10.1182/blood-2009-06-225979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goede V, Fischer K, Busch R, Engelke A, Eichhorst B, Wendtner CM, Chagorova T, de la Serna J, Dilhuydy MS, Illmer T, Opat S, Owen CJ, Samoylova O, Kreuzer KA, Stilgenbauer S, Dohner H, Langerak AW, Ritgen M, Kneba M, Asikanius E, Humphrey K, Wenger M, Hallek M. Obinutuzumab plus chlorambucil in patients with CLL and coexisting conditions. N Engl J Med. 2014;370:1101–1110. doi: 10.1056/NEJMoa1313984. [DOI] [PubMed] [Google Scholar]
- 31.Illidge T, Klein C, Sehn LH, Davies A, Salles G, Cartron G. Obinutuzumab in hematologic malignancies: lessons learned to date. Cancer Treat Rev. 2015;41:784–792. doi: 10.1016/j.ctrv.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Lazar GA, Dang W, Karki S, Vafa O, Peng JS, Hyun L, Chan C, Chung HS, Eivazi A, Yoder SC, Vielmetter J, Carmichael DF, Hayes RJ, Dahiyat BI. Engineered antibody Fc variants with enhanced effector function. Proc Natl Acad Sci U S A. 2006;103:4005–4010. doi: 10.1073/pnas.0508123103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horton HM, Bernett MJ, Pong E, Peipp M, Karki S, Chu SY, Richards JO, Vostiar I, Joyce PF, Repp R, Desjarlais JR, Zhukovsky EA. Potent in vitro and in vivo activity of an Fc-engineered anti-CD19 monoclonal antibody against lymphoma and leukemia. Cancer Res. 2008;68:8049–8057. doi: 10.1158/0008-5472.CAN-08-2268. [DOI] [PubMed] [Google Scholar]
- 34.Moore GL, Chen H, Karki S, Lazar GA. Engineered Fc variant antibodies with enhanced ability to recruit complement and mediate effector functions. MAbs. 2010;2:181–189. doi: 10.4161/mabs.2.2.11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Idusogie EE, Wong PY, Presta LG, Gazzano-Santoro H, Totpal K, Ultsch M, Mulkerrin MG. Engineered antibodies with increased activity to recruit complement. J Immunol. 2001;166:2571–2575. doi: 10.4049/jimmunol.166.4.2571. [DOI] [PubMed] [Google Scholar]
- 36.Lee CH, Romain G, Yan W, Watanabe M, Charab W, Todorova B, Lee J, Triplett K, Donkor M, Lungu OI, Lux A, Marshall N, Lindorfer MA, Goff OR, Balbino B, Kang TH, Tanno H, Delidakis G, Alford C, Taylor RP, Nimmerjahn F, Varadarajan N, Bruhns P, Zhang YJ, Georgiou G. IgG Fc domains that bind C1q but not effector Fcgamma receptors delineate the importance of complement-mediated effector functions. Nat Immunol. 2017;18:889–898. doi: 10.1038/ni.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Natsume A, In M, Takamura H, Nakagawa T, Shimizu Y, Kitajima K, Wakitani M, Ohta S, Satoh M, Shitara K, Niwa R. Engineered antibodies of IgG1/IgG3 mixed isotype with enhanced cytotoxic activities. Cancer Res. 2008;68:3863–3872. doi: 10.1158/0008-5472.CAN-07-6297. [DOI] [PubMed] [Google Scholar]
- 38.Rosner T, Derer S, Kellner C, Dechant M, Lohse S, Vidarsson G, Peipp M, Valerius T. An IgG3 switch variant of rituximab mediates enhanced complement-dependent cytotoxicity against tumour cells with low CD20 expression levels. Br J Haematol. 2013;161:282–286. doi: 10.1111/bjh.12209. [DOI] [PubMed] [Google Scholar]
- 39.Diebolder CA, Beurskens FJ, de Jong RN, Koning RI, Strumane K, Lindorfer MA, Voorhorst M, Ugurlar D, Rosati S, Heck AJ, van de Winkel JG, Wilson IA, Koster AJ, Taylor RP, Saphire EO, Burton DR, Schuurman J, Gros P, Parren PW. Complement is activated by IgG hexamers assembled at the cell surface. Science. 2014;343:1260–1263. doi: 10.1126/science.1248943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tammen A, Derer S, Schwanbeck R, Rosner T, Kretschmer A, Beurskens FJ, Schuurman J, Parren PW, Valerius T. Monoclonal antibodies against epidermal growth factor receptor acquire an ability to kill tumor cells through complement activation by mutations that selectively facilitate the hexamerization of IgG on opsonized cells. J Immunol. 2017;198:1585–1594. doi: 10.4049/jimmunol.1601268. [DOI] [PubMed] [Google Scholar]
- 41.Schneider S, Zacharias M. Atomic resolution model of the antibody Fc interaction with the complement C1q component. Mol Immunol. 2012;51:66–72. doi: 10.1016/j.molimm.2012.02.111. [DOI] [PubMed] [Google Scholar]
- 42.Radaev S, Motyka S, Fridman WH, Sautes-Fridman C, Sun PD. The structure of a human type III Fcgamma receptor in complex with Fc. J Biol Chem. 2001;276:16469–16477. doi: 10.1074/jbc.M100350200. [DOI] [PubMed] [Google Scholar]
- 43.Sondermann P, Huber R, Oosthuizen V, Jacob U. The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature. 2000;406:267–273. doi: 10.1038/35018508. [DOI] [PubMed] [Google Scholar]
- 44.Ripka J, Adamany A, Stanley P. Two Chinese hamster ovary glycosylation mutants affected in the conversion of GDP-mannose to GDP-fucose. Arch Biochem Biophys. 1986;249:533–545. doi: 10.1016/0003-9861(86)90031-7. [DOI] [PubMed] [Google Scholar]
- 45.Patnaik SK, Stanley P. Lectin-resistant CHO glycosylation mutants. Methods Enzymol. 2006;416:159–182. doi: 10.1016/S0076-6879(06)16011-5. [DOI] [PubMed] [Google Scholar]
- 46.Glorius P, Baerenwaldt A, Kellner C, Staudinger M, Dechant M, Stauch M, Beurskens FJ, Parren PW, Winkel JG, Valerius T, Humpe A, Repp R, Gramatzki M, Nimmerjahn F, Peipp M. The novel tribody [(CD20)(2)xCD16] efficiently triggers effector cell-mediated lysis of malignant B cells. Leukemia. 2013;27:190–201. doi: 10.1038/leu.2012.150. [DOI] [PubMed] [Google Scholar]
- 47.Anderson DR, Hanna N, Leonard JE, Newman RA, Rastetter WH, Reff ME.Chimeric anti-CD20 antibody. Patent EP2000149A1 https://docs.google.com/viewer?url=patentimages.storage.googleapis.com/pdfs/162dad44e5fd9390cee1/EP2000149A1.pdf (last accessed August 24, 2017).
- 48.Schewe DM, Alsadeq A, Sattler C, Lenk L, Vogiatzi F, Cario G, Vieth S, Valerius T, Rosskopf S, Meyersieck F, Alten J, Schrappe M, Gramatzki M, Peipp M, Kellner C. An Fc engineered CD19 antibody eradicates MRD in patient-derived MLL-rearranged acute lymphoblastic leukemia xenografts. Blood. 2017 doi: 10.1182/blood-2017-01-764316. doi: 101182/blood-2017-01-764316. [DOI] [PubMed] [Google Scholar]
- 49.Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, Rowland AM, Kotts C, Carver ME, Shepard HM. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci U S A. 1992;89:4285–4289. doi: 10.1073/pnas.89.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Repp R, Kellner C, Muskulus A, Staudinger M, Nodehi SM, Glorius P, Akramiene D, Dechant M, Fey GH, van Berkel PH, van de Winkel JG, Parren PW, Valerius T, Gramatzki M, Peipp M. Combined Fc-protein- and Fc-glyco-engineering of scFv-Fc fusion proteins synergistically enhances CD16a binding but does not further enhance NK-cell mediated ADCC. J Immunol Methods. 2011;373:67–78. doi: 10.1016/j.jim.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 51.van Oers MH. CD20 antibodies: type II to tango? Blood. 2012;119:5061–5063. doi: 10.1182/blood-2012-04-420711. [DOI] [PubMed] [Google Scholar]
- 52.Chao MP, Alizadeh AA, Tang C, Myklebust JH, Varghese B, Gill S, Jan M, Cha AC, Chan CK, Tan BT, Park CY, Zhao F, Kohrt HE, Malumbres R, Briones J, Gascoyne RD, Lossos IS, Levy R, Weissman IL, Majeti R. Anti-CD47 antibody synergizes with rituximab to promote phagocytosis and eradicate non-Hodgkin lymphoma. Cell. 2010;142:699–713. doi: 10.1016/j.cell.2010.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Inagaki A, Ishida T, Yano H, Ishii T, Kusumoto S, Ito A, Ri M, Mori F, Ding J, Komatsu H, Iida S, Ueda R. Expression of the ULBP ligands for NKG2D by B-NHL cells plays an important role in determining their susceptibility to rituximab-induced ADCC. Int J Cancer. 2009;125:212–221. doi: 10.1002/ijc.24351. [DOI] [PubMed] [Google Scholar]
- 54.Meyer S, Leusen JH, Boross P. Regulation of complement and modulation of its activity in monoclonal antibody therapy of cancer. MAbs. 2014;6:1133–1144. doi: 10.4161/mabs.29670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gong Q, Ou Q, Ye S, Lee WP, Cornelius J, Diehl L, Lin WY, Hu Z, Lu Y, Chen Y, Wu Y, Meng YG, Gribling P, Lin Z, Nguyen K, Tran T, Zhang Y, Rosen H, Martin F, Chan AC. Importance of cellular microenvironment and circulatory dynamics in B cell immunotherapy. J Immunol. 2005;174:817–826. doi: 10.4049/jimmunol.174.2.817. [DOI] [PubMed] [Google Scholar]
- 56.Nimmerjahn F, Ravetch JV. Antibodies, Fc receptors and cancer. Curr Opin Immunol. 2007;19:239–245. doi: 10.1016/j.coi.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 57.Clark MR. IgG effector mechanisms. Chem Immunol. 1997;65:88–110. [PubMed] [Google Scholar]