Abstract
Recent Hsp104 structural studies have reported both planar and helical models of the hexameric structure. The conformation of Hsp104 monomers within the hexamer is affected by nucleotide ligation. After nucleotide-driven hexamer formation, Hsp104-catalyzed disruption of protein aggregates requires binding to the peptide substrate. Here, we examine the oligomeric state of Hsp104 and its peptide binding competency in the absence of nucleotide and in the presence of ADP, ATPγS, AMPPNP, or AMPPCP. Surprisingly, we found that only ATPγS facilitates avid peptide binding by Hsp104. We propose that the modulation between high-and low-peptide affinity states observed with these ATP analogues is an important component of the disaggregation mechanism of Hsp104.
Saccharomyces cerevisiae Hsp104 is a member of the Hsp100 (heat shock protein) family of AAA+ (ATPases associated with diverse cellular activities) motor proteins that function broadly to help organisms survive and adapt to environmental stress.1,2 Accumulation of unfolded or misfolded proteins into aggregates can occur when protein homeostasis systems are overwhelmed. Hsp104 promotes cell survival by resolving these aggregates.3 Such aggregation in humans is implicated in neurodegenerative diseases, including Parkinson’s disease and amyotrophic lateral sclerosis. Remarkably, no cytoplasmic homologue to Hsp104 exists in humans. Consequently, there is great interest in understanding how Hsp104 resolves protein aggregates, with a vision of developing it for therapeutic applications.4–6
Like other members of the Hsp100 family, Hsp104 is biologically active as a hexameric ring but may exist in a dynamic equilibrium of oligomers.7 ClpB, the Escherichia coli homologue of Hsp104 populates a distribution of oligomers, affected by protein concentration, salt concentration, and ligand (nucleotide).8,9 The nonhydrolyzable ATP analogue β, γ-imidoadenosine 5'-triphosphate (AMPPNP) was used to populate hexamers for crystallographic and cryoelectron microscopy studies of Thermus thermophilus ClpB (TClpB).10,11 While ClpB is structurally similar to Hsp104 and both proteins function to resolve amorphous protein aggregates, Hsp104 has unique amyloid remodeling12 and prion propagating13 activities that ClpB does not share. Furthermore, complementary cochaperones from the prokaryotic and eukaryotic systems are not interchangeable, highlighting a species-specific interface.14,15 Thus, investigations of Hsp104 provide information that is not available from the investigations of ClpB.
Interestingly, although the biologically active form of these motor proteins is a hexameric ring, Hsp104, ClpB, and closely related AAA+ family member ClpA have crystallized in extended spiral filaments.10,16,17 In contrast, a study of AAA+ protein ClpC with AMPPNP revealed planar ring hexamers in the crystallographic unit.18 Many electron microscopy (EM) studies of Hsp104 have also shown planar hexameric rings with a variety of bound nucleotides.3,17,19–21 The poorly hydrolyzable ATP analogue, adenosine 5'-(3-thiotriphosphate) (ATPγS), and ADP have been used to investigate conformation, oligomerization, and protein remodeling activity of wild-type Hsp104, whereas ATP has been used with Hsp104 variants that are deficient in ATP hydrolysis.22,23
Each monomer of Hsp104, ClpB, ClpA, and ClpC has two nucleotide binding and hydrolysis domains that, in most reconstructions and models, form stacked rings in the planar hexamer.3,10,17–21,24 Monomers adopt different conformations within the ring when unligated or ligated with different nucleotides. These studies led to proposed mechanisms of conformational changes throughout the ATP binding and hydrolysis cycle that are mechanistically linked to disaggregase activity.19 In contrast to these planar structures, a recent EM study showed Hsp104 bound by AMPPNP in a novel helical hexameric spiral with an asymmetric seam.25
Given the abundance of structural models for Hsp104 and the influence of nucleotide binding on conformation, we sought to address the question of whether the oligomers bound by various nucleotides in solution differ structurally and display different polypeptide binding activity.
We have shown that Hsp104 binds FITC-casein in the presence of ATPγS.5 Previously, Bösl et al. demonstrated that Hsp104 binds to another model substrate (RCMLa) in the presence of ATPγS. ADP did not facilitate polypeptide binding because of the lower affinity of Hsp104 for the polypeptide in the ADP-bound state. Surprisingly, ATP also failed to facilitate peptide binding, because of rapid hydrolysis of ATP resulting in the low-affinity ADP-bound state.22 Here, we use a polypeptide consisting of the first 50 amino acids of RepA, a soluble substrate used to examine Hsp104 protein remodeling activity.5,26–29
We extend our investigation to include AMPPNP, which was used to examine both the TClpB crystal structure and the Hsp104 spiral hexamer. In addition, we include β, γ-methyl-eneadenosine 5'-triphosphate (AMPPCP), which has been used for structural studies of Hsp104 and E. coli ClpB.10,25,30 Because ATP is rapidly hydrolyzed, resulting in ADP and inorganic phosphate, ATP is not included in this study. We found that only ATPγS promotes the formation of an Hsp104 structure that is competent for stable polypeptide binding under the conditions tested.
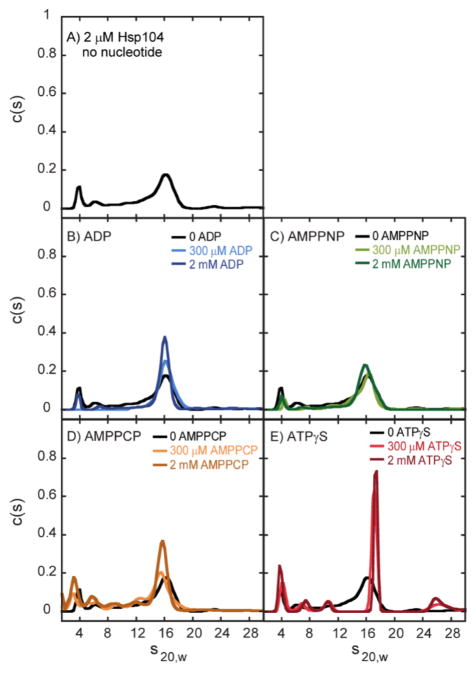
As a necessary precursor to investigating peptide binding, we first addressed two questions with respect to the quaternary structure of Hsp104. First, is nucleotide binding required for the formation of hexameric rings of Hsp104? Second, if nucleotide binding is required, which nucleotides can fulfill this role? To address these questions, we performed sedimentation velocity experiments to examine Hsp104 assembly. Experiments were performed on 2 μM Hsp104 in the absence of any nucleotide. Figure 1A shows the resulting c(s) distribution, where multiple peaks are observed, indicating that various oligomeric states are populated in the absence of a nucleotide. However, one peak with an s20,w of ~16 S dominates the c(s) distribution, suggesting that one oligomeric state is predominantly populated under these conditions.
Figure 1.
Sedimentation velocity c(s) distributions of Hsp104 in the absence and presence of nucleotides. Sedimentation velocity experiments were performed with 2 μM Hsp104 (A) in the absence and in the presence of (B) ADP, (C) AMPPNP, (D) AMPPCP, or (E) ATPγS.
Experiments were then performed with 2 μM Hsp104 in the presence of 300 μM ADP, AMPPNP, AMPPCP, or ATPγS. Panels B–E of Figure 1 show the resulting c(s) distributions from analysis of the sedimentation boundaries. In all cases, a dominant peak between ~15.5 and 17 S was observed. Strikingly, the ATPγS-bound structure exhibits a dominant population of oligomers with a sedimentation coefficient that is larger than those of the other nucleotide-bound structures (see Table 1).
Table 1.
Hsp104 Predominant c(s) Distributions and Frictional Ratiosa
| condition | s̄20,w (S) | f/f0 |
|---|---|---|
| no nucleotide | 15.8 ± 0.3 | 1.35 ± 0.03 |
| ADP | 16.1 ± 0.3 | 1.35 ± 0.02 |
| AMPPNP | 15.9 ± 0.1 | 1.36 ± 0.01 |
| AMPPCP | 15.6 ± 0.1 | 1.39 ± 0.01 |
| ATPγS | 17.0 ± 0.2 | 1.28 ± 0.02 |
Standard deviation based on the average of two replicates at each of two nucleotide concentrations (see the Supporting Information).
To assess whether 300 μM nucleotide was a sufficiently high concentration to saturate Hsp104 and maximize oligomerization, sedimentation velocity experiments were repeated with 2 μM Hsp104 in the presence of 2 mM nucleotide (Figure 1B–E). In all cases, increasing the nucleotide concentration did not shift the dominant peak in the c(s) distribution. Moreover, the magnitude of that peak, which reflects the relative concentration of that oligomer in solution, did not significantly change. Both E. coli ClpB and Hsp104 share structural domain organization, including the presence of an M domain. Our previous work indicates an s20,w of (17.6 ± 0.6) S for the ClpB hexamer, consistent with the dominant peaks observed in Figure 1A–E.8 Thus, while definitive assignment of peaks to specific oligomers requires a multifaceted investigation beyond the scope of this study, the observed dominant peaks are likely hexameric Hsp104.
Table 1 shows the weight-average sedimentation coefficient, s̄20,w, for the dominant peak observed in the absence and presence of the nucleotides tested. Most strikingly, ATPγS binding clearly populates a hexamer that is hydrodynamically different from the hexamer populated with ADP, AMPPNP, AMPPCP, or no nucleotide.
The potentiated variant Hsp104A503S has enhanced disaggregation activity in numerous biological assays.5 The mechanistic basis for this gain of function is unknown. To test whether Hsp104A503S has nucleotide-linked oligomerization properties different from those of wild-type Hsp104, we subjected the variant to sedimentation velocity experiments. As with wild-type Hsp104, hexamers populated in the presence of ATPγS are hydrodynamically different from those populated in the absence of nucleotide or in the presence of ADP, AMPPNP, or AMPPCP (Supporting Figure 1A–E and Supporting Table 1), though the difference between the ATPγS-bound and AMPPNP-bound states is smaller than that observed for the wild type. A similar observation was made for E. coli ClpB (Supporting Figure 2A–E and Supporting Table 1).
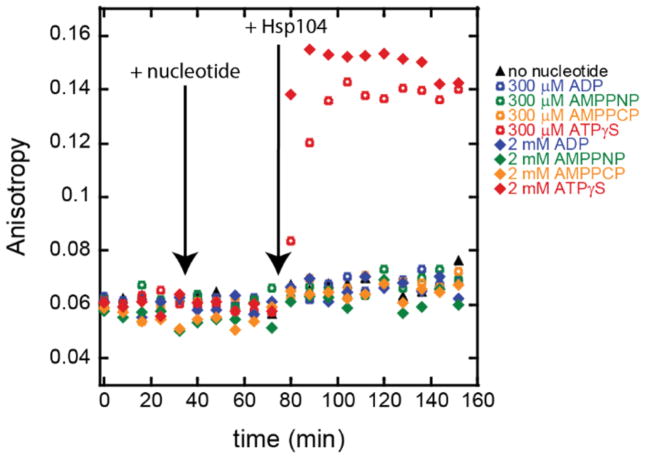
The analysis of the sedimentation velocity experiments presented in Figure 1 indicates that higher-order oligomers, consistent with hexamers, are observed under all conditions tested. The remaining question is whether these oligomers exhibit avid peptide binding. To test for peptide binding activity, we used a fluorescein-modified model polypeptide substrate termed FluNCysRepA50mer. Figure 2 shows the fluorescence anisotropy of a peptide solution upon addition of various nucleotides and Hsp104. The experiment begins with each sample containing only FluNCysRepA50mer. All time points of <40 min in Figure 2 report the anisotropy of the peptide alone, where r = 0.060 ± 0.004 (replicate, r = 0.069 ± 0.004).
Figure 2.
Fluorescence anisotropy measurements of FluNCysRe-pA50mer before and after subsequent additions of nucleotide and Hsp104 as indicated.
Next, as shown in Figure 2 by an arrow indicating nucleotide addition, ADP, AMPPNP, AMPPCP, or ATPγS was added to each sample to a final nucleotide concentration of 300 μM. The anisotropy of the peptide in the presence of each nucleotide was within one standard deviation of the anisotropy of FluNCysRepA50mer alone, indicating that the nucleotide did not influence the observed anisotropy (Table 2).
Table 2.
Fluorescence Anisotropy for FluNCysRepA50mera
| ADP (300 μM) | AMPPNP (300 μM) | AMPPCP (300 μM) | ATPγS (300 μM) | ADP (2 mM) | AMPPNP (2 mM) | AMPPCP (2 mM) | ATPγS (2 mM) | |
|---|---|---|---|---|---|---|---|---|
| + nucleotide | 0.061 ± 0.002 | 0.062 ± 0.003 | 0.060 ± 0.002 | 0.059 ± 0.001 | 0.059 ± 0.003 | 0.055 ± 0.003 | 0.055 ± 0.003 | 0.059 ± 0.002 |
| replicate | 0.072 ± 0.001 | 0.071 ± 0.004 | 0.063 ± 0.003 | 0.070 ± 0.003 | 0.073 ± 0.004 | 0.066 ± 0.002 | 0.064 ± 0.003 | 0.065 ± 0.002 |
| + Hsp104 | 0.068 ± 0.003 | 0.067 ± 0.004 | 0.068 ± 0.003 | 0.139 ± 0.002 | 0.066 ± 0.003 | 0.062 ± 0.004 | 0.065 ± 0.002 | 0.150 ± 0.005 |
| replicate | 0.076 ± 0.002 | 0.076 ± 0.002 | 0.070 ± 0.002 | 0.190 ± 0.004 | 0.076 ± 0.002 | 0.073 ± 0.003 | 0.070 ± 0.002 | 0.178 ± 0.003 |
The average and standard deviations were calculated as described in the Supporting Information.
Hsp104 was then added to each sample to a final concentration of 2 μM (Figure 2, arrow indicating Hsp104 addition). Strikingly, the anisotropy increased to 0.139 ± 0.002 for the sample containing 300 μM ATPγS. The increase in anisotropy indicates slower tumbling of FluNCysRepA50mer, consistent with peptide bound by the Hsp104 oligomer. In contrast, the anisotropy measurement did not significantly increase upon addition of Hsp104 in the presence of other nucleotides. As summarized in Table 2, the anisotropy measurements of the peptide in the presence of Hsp104 and ADP, AMPPNP, or AMPPCP were within one standard deviation of each other, and within one standard deviation of that of FluNCysRepA50mer in the presence of Hsp104 with no nucleotide, where r = 0.068 ± 0.004 (replicate, r = 0.072 ± 0.002). Inspection of the plot also reveals a minor upward trend in the anisotropy measurements taken after addition of Hsp104 under all conditions except ATPγS, indicating that there may be some small population of Hsp104 binding peptide under those conditions, consistent with substantially weaker affinity. These observations indicate that the ATPγS-bound Hsp104 oligomer has a unique competency for interactions with the polypeptide, not conferred by other nucleotides.
An alternative explanation is that 300 μM nucleotide is not sufficient to saturate Hsp104 binding. To test this possibility, the experiment was repeated using 2 mM nucleotide. The filled diamonds in Figure 2 illustrate that at 2 mM nucleotide, the anisotropy of the peptide remained constant (from ~40 to ~80 min). Table 2 lists the anisotropy measurements for the peptide under each of the 2 mM nucleotide conditions. Note that each measurement is within one standard deviation of the anisotropy value for the free peptide, where r = 0.060 ± 0.004 (replicate, r = 0.069 ± 0.004). Upon addition of Hsp104 to a final concentration of 2 μM, again only the ATPγS condition exhibited an increased anisotropy of 0.150 ± 0.005 that is consistent with binding polypeptide (Figure 2). Though a single representative time course is shown in Figure 2, the results are reproducible. The replicate time course is shown in Supporting Figure 3, while anisotropy values from the replicate are included in Table 2 for comparison. Both Hsp104A503S and ClpB were tested at nucleotide concentrations of 300 μM and 2 mM; the same trends were observed as for wild-type Hsp104 (Supporting Figure 4 and Supporting Tables 2 and 3).
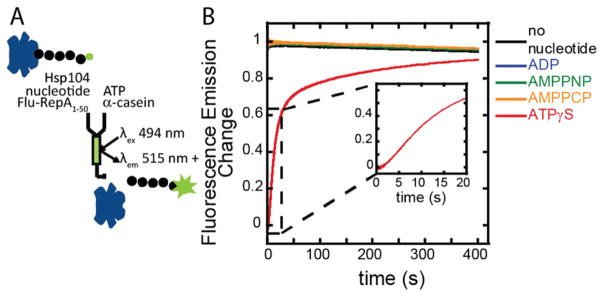
We next sought to determine what conditions form a peptide-Hsp104 complex that is poised for ATP-dependent polypeptide translocation. We determined these conditions using a fluorescence stopped-flow assay, developed by our lab to study polypeptide translocation by ClpA and ClpB.31–33 As schematized in Figure 3A, Hsp104, nucleotide, polypeptide solutions were rapidly mixed with ATP and a protein trap to allow a single turnover of ATP-dependent translocation. The polypeptide substrate bound by Hsp104 exhibits quenched fluorescence (Supporting Figure 5) that is relieved upon Hsp104 dissociation.
Figure 3.
(A) Fluorescence stopped-flow reaction scheme and (B) time courses. Hsp104 and FluNCysRepA50mer were incubated in the presence of various nucleotides and then rapidly mixed with ATP and α-casein to test for the presence of a translocation-ready complex.
As indicated in Figure 3B, the time courses for the samples incubated in the absence of nucleotide or with ADP, AMPPNP, or AMPPCP show an insignificant decrease in the intensity of the fluorescence signal over time, not indicative of ATP-driven events associated with a bound Hsp104-polypeptide complex. The time course for the ATPγS condition shows a low initial fluorescence, caused by fluorescence quenching in the bound complex (Supporting Figure 5), followed by an increase in fluorescence upon rapid mixing with ATP and protein trap. Note the presence of a lag phase (inset) indicating that assembled Hsp104 bound to the polypeptide in the presence of ATPγS takes at least two repeating kinetic steps before dissociating from the substrate, which is distinctly different from what we have reported for ClpB.33 (This finding has prompted a thorough investigation of the Hsp104 translocation mechanism that is currently under way.) The stopped-flow results, like the fluorescence-anisotropy results, show that of the nucleotides tested only ATPγS supports avid binding of the polypeptide by Hsp104.
A prerequisite for resolving protein aggregates is binding a polypeptide substrate. Our sedimentation velocity work shows that there is a structural difference, resulting in a larger sedimentation coefficient and a smaller frictional coefficient for the ATPγS-bound Hsp104 hexamer compared to those of the apo, ADP-bound, AMPPNP-bound, or AMPPCP-bound Hsp104 hexamers (Table 1). The smaller frictional ratio, calculated in relation to a hydrated sphere of equivalent mass, indicates that the ATPγS-bound structure is more spherical than the other structures. The larger frictional ratio of the AMPPNP-bound structure reflects a less spherical shape, which is consistent with the asymmetric spiral hexamer observed by Yokom et al.25 However, on the basis of the results reported here, that structure is not competent for avid polypeptide binding. The same general shape as that seen for the AMPPNP-bound Hsp104 oligomer is expected for the apo, ADP-bound, and AMPPCP-bound oligomers from our results.
Our anisotropy experiments show that, under the conditions tested, Hsp104 binds the unstructured polypeptide substrate only in the presence of ATPγS. Additionally, only ATPγS-bound Hsp104 forms a translocation-ready complex with a polypeptide in our stopped-flow experiments. Thus, only the unique structural conformation of ATPγS-bound Hsp104 is competent for avidly binding a polypeptide. Our findings are consistent with those of Lee et al., who showed that, because of differences in quaternary structure, AMPPNP, ADP, and the absence of a nucleotide do not promote binding of TClpB to the polypeptide substrate, while ATPγS does.11
Studying ClpA, Farbman et al. described two possible peptide substrate-processing mechanisms as either an “alternating affinity” or a “constant affinity” model.34 As each NBD cycles through ATP hydrolysis, the resulting conformational changes are associated with either a high or low affinity for the peptide. The coordination of these hydrolysis-driven conformational changes within the hexameric ring allows individual subunits to release a peptide substrate while the hexameric ring retains the substrate through multiple rounds of ATP hydrolysis.
Here, and in previous work, nucleotide analogues are used to approximate the structure of hydrolyzable ATP or a transition state along the reaction pathway from ATP to ADP-Pi. A single monomer within a hexamer that is bound by a transition state analogue likely reflects a physiologically relevant conformation along the reaction pathway. However, it is unlikely that all 12 NBDs within a hexamer would be synchronized with respect to hydrolysis, so a hexameric structure in which all protomers are ligated by the transition state analogue is not likely to be a conformational structure observed in the biological activity of the motor. Here, we see that when Hsp104 is saturated with ATPγS, a high-peptide affinity state is achieved. In contrast, when the motor is saturated by AMPPNP, AMPPCP, ADP, or no nucleotide, a low-peptide affinity state is induced.
Possible mechanistic implications proposed by Yokom et al. and Heuck et al. merit further investigation.17,25 Hsp104 monomers within a hexamer may alternate between the conformations that give rise to planar and helical hexamers. Moreover, it has been reported that a mixture of ATP and ATPγS yields activity similar to that observed in the presence of cochaperone Hsp70.26 Taken with our findings, this may indicate that binding by cochaperone Hsp70 is part of the mechanism of modulation between high- and low-peptide affinity states. Alternatively, a composite of the protomer conformation used to build up the planar and hexameric rings may be used in concert as the active hexamer cycles through ATP while engaged with a peptide substrate. Aggregates may also present multiple binding sites unlike soluble peptides that might increase binding affinity.
Supplementary Material
Acknowledgments
Funding
This work was supported by National Science Foundation Grant MCB-1412624 to A.L.L. and National Institutes of Health Grant R01GM099836 to J.S.
We thank Peter Prevelige for confirmation of the ClpB sequence by mass spectrometry, Jingzhi Li and Bingdong Sha for protein preparation discussions, Amber Tariq for assistance with Hsp104 preparations, Woody Robbins and Kim Hardy for use of the French press, and Ethan Cagle for assistance with peptide labeling.
Footnotes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio-chem.7b00225.
Experimental procedures and additional supporting data (PDF)
References
- 1.Schirmer EC, Glover JR, Singer MA, Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289. [PubMed] [Google Scholar]
- 2.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A Class of Chaperone-Like ATPases Associated with the Assembly, Operation, and Disassembly of Protein Complexes. Genome Res. 1999;9:27. [PubMed] [Google Scholar]
- 3.Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hspl04. Nature. 1994;372:475. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 4.Shorter J. Hsp104: A Weapon to Combat Diverse Neurodegenerative Disorders. Neurosignals. 2008;16:63. doi: 10.1159/000109760. [DOI] [PubMed] [Google Scholar]
- 5.Jackrel ME, DeSantis ME, Martinez BA, Castellano LM, Stewart RM, Caldwell KA, Caldwell GA, Shorter J. Potentiated Hsp104 Variants Antagonize Diverse Proteotoxic Misfolding Events. Cell. 2014;156:170. doi: 10.1016/j.cell.2013.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackrel ME, Shorter J. Potentiated Hsp104 variants suppress toxicity of diverse neurodegenerative disease-linked proteins. Dis Models & Mech. 2014;7:1175. doi: 10.1242/dmm.016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hattendorf DA, Lindquist SL. Cooperative kinetics of both Hsp104 ATPase domains and interdomain communication revealed by AAA sensor-1 mutants. EMBO J. 2002;21:12. doi: 10.1093/emboj/21.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin J, Lucius AL. Examination of the dynamic assembly equilibrium for E. coli ClpB. Proteins: Struct, Funct, Genet. 2015;83:2008. doi: 10.1002/prot.24914. [DOI] [PubMed] [Google Scholar]
- 9.Lin J, Lucius AL. Examination of ClpB Quaternary Structure and Linkage to Nucleotide Binding. Biochemistry. 2016;55:1758. doi: 10.1021/acs.biochem.6b00122. [DOI] [PubMed] [Google Scholar]
- 10.Lee S, Sowa ME, Watanabe YH, Sigler PB, Chiu W, Yoshida M, Tsai FT. The Structure of ClpB: A Molecular Chaperone that Rescues Proteins from an Aggregated State. Cell. 2003;115:229. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 11.Lee S, Choi JM, Tsai FT. Visualizing the ATPase Cycle in a Protein Disaggregating Machine: Structural Basis for Substrate Binding by ClpB. Mol Cell. 2007;25:261. doi: 10.1016/j.molcel.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeSantis ME, Leung EH, Sweeny EA, Jackrel ME, Cushman-Nick M, Neuhaus-Follini A, Vashist S, Sochor MA, Knight MN, Shorter J. Operational Plasticity Enables Hsp104 to Disaggregate Diverse Amyloid and Nonamyloid Clients. Cell. 2012;151:778. doi: 10.1016/j.cell.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shorter J, Lindquist S. Hsp104 Catalyzes Formation and Elimination of Self-Replicating Sup35 Prion Conformers. Science. 2004;304:1793. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 14.Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: A Novel Chaperone System that Rescues Previously Aggregated Proteins. Cell. 1998;94:73. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 15.Schlee S, Beinker P, Akhrymuk A, Reinstein J. A Chaperone Network for the Resolubilization of Protein Aggregates: Direct Interaction of ClpB and DnaK. J Mol Biol. 2004;336:275. doi: 10.1016/j.jmb.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 16.Guo F, Maurizi MR, Esser L, Xia D. Crystal Structure of ClpA, an Hsp100 Chaperone and Regulator of ClpAP Protease. J Biol Chem. 2002;277:46743. doi: 10.1074/jbc.M207796200. [DOI] [PubMed] [Google Scholar]
- 17.Heuck A, Schitter-Sollner S, Suskiewicz MJ, Kurzbauer R, Kley J, Schleiffer A, Rombaut P, Herzog F, Clausen T. Structural basis for the disaggregase activity and regulation of Hsp104. eLife. 2016;5:e21516. doi: 10.7554/eLife.21516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F, Mei Z, Qi Y, Yan C, Hu Q, Wang J, Shi Y. Structure and mechanism of the hexameric MecA-ClpC molecular machine. Nature. 2011;471:331. doi: 10.1038/nature09780. [DOI] [PubMed] [Google Scholar]
- 19.Wendler P, Shorter J, Snead D, Plisson C, Clare DK, Lindquist S, Saibil HR. Motor Mechanism for Protein Threading through Hsp104. Mol Cell. 2009;34:81. doi: 10.1016/j.molcel.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee S, Sielaff B, Lee J, Tsai FT. CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc Natl Acad Sci U S A. 2010;107:8135. doi: 10.1073/pnas.1003572107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroni M, Kummer E, Oguchi Y, Wendler P, Clare DK, Sinning I, Kopp J, Mogk A, Bukau B, Saibil HR. Head-to-tail interactions of the coiled-coil domains regulate ClpB activity and cooperation with Hsp70 in protein disaggregation. eLife. 2014;3:e02481. doi: 10.7554/eLife.02481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bösl B, Grimminger V, Walter S. Substrate Binding to the Molecular Chaperone Hsp104 and Its Regulation by Nucleotides. J Biol Chem. 2005;280:38170. doi: 10.1074/jbc.M506149200. [DOI] [PubMed] [Google Scholar]
- 23.Klosowska A, Chamera T, Liberek K. Adenosine diphosphate restricts the protein remodeling activity of the Hsp104 chaperone to Hsp70 assisted disaggregation. eLife. 2016;5:e15159. doi: 10.7554/eLife.15159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Effantin G, Ishikawa T, De Donatis GM, Maurizi MR, Steven AC. Local and Global Mobility in the ClpA AAA+ Chaperone Detected by Cryo-Electron Microscopy: Functional Connotations. Structure. 2010;18:553. doi: 10.1016/j.str.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yokom AL, Gates SN, Jackrel ME, Mack KL, Su M, Shorter J, Southworth DR. Spiral architecture of the Hsp104 disaggregase reveals the basis for polypeptide translocation. Nat Struct Mol Biol. 2016;23:830. doi: 10.1038/nsmb.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doyle SM, Shorter J, Zolkiewski M, Hoskins JR, Lindquist S, Wickner S. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat Struct Mol Biol. 2007;14:114. doi: 10.1038/nsmb1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doyle SM, Hoskins JR, Wickner S. Collaboration between the ClpB AAA+ remodeling protein and the DnaK chaperone system. Proc Natl Acad Sci U S A. 2007;104:11138. doi: 10.1073/pnas.0703980104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doyle SM, Shastry S, Kravats AN, Shih YH, Miot M, Hoskins JR, Stan G, Wickner S. Interplay between E. coli DnaK, ClpB and GrpE during Protein Disaggregation. J Mol Biol. 2015;427:312. doi: 10.1016/j.jmb.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee J, Kim JH, Biter AB, Sielaff B, Lee S, Tsai FT. Heat shock protein (Hsp) 70 is an activator of the Hsp104 motor. Proc Natl Acad Sci U S A. 2013;110:8513. doi: 10.1073/pnas.1217988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeymer C, Barends TR, Werbeck ND, Schlichting I, Reinstein J. Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor. Acta Crystallogr, Sect D: Biol Crystallogr. 2014;70:582. doi: 10.1107/S1399004713030629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajendar B, Lucius AL. Molecular Mechanism of Polypeptide Translocation Catalyzed by the Escherichia coli ClpA Protein Translocase. J Mol Biol. 2010;399:665. doi: 10.1016/j.jmb.2010.03.061. [DOI] [PubMed] [Google Scholar]
- 32.Miller JM, Lin J, Li T, Lucius AL. E. coli ClpA Catalyzed Polypeptide Translocation Is Allosterically Controlled by the Protease ClpP. J Mol Biol. 2013;425:2795. doi: 10.1016/j.jmb.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li T, Weaver CL, Lin J, Duran EC, Miller JM, Lucius AL. Escherichia coli ClpB is a non-processive polypeptide translocase. Biochem J. 2015;470:39. doi: 10.1042/BJ20141457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farbman ME, Gershenson A, Licht S. Single-Molecule Analysis of Nucleotide-Dependent Substrate Binding by the Protein Unfoldase ClpA. J Am Chem Soc. 2007;129:12378. doi: 10.1021/ja074168x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.