Abstract
The p38 MAPK family is composed of four kinases of which p38α/MAPK14 is the major proinflammatory member. These kinases contribute to many inflammatory diseases, but the currently available p38 catalytic inhibitors (e.g., SB203580) are poorly effective and cause toxicity. We reasoned that the failure of catalytic p38 inhibitors may derive from their activity against noninflammatory p38 isoforms (e.g., p38β/MAPK11) and loss of all p38α-dependent responses, including anti-inflammatory, counterregulatory responses via mitogen- and stress-activated kinase (MSK) 1/2 and Smad3. We used computer-aided drug design to target small molecules to a pocket near the p38α glutamate–aspartate (ED) substrate-docking site rather than the catalytic site, the sequence of which had only modest homology among p38 isoforms. We identified a lead compound, UM101, that was at least as effective as SB203580 in stabilizing endothelial barrier function, reducing inflammation, and mitigating LPS-induced mouse lung injury. Differential scanning fluorimetry and saturation transfer difference–nuclear magnetic resonance demonstrated specific binding of UM101 to the computer-aided drug design–targeted pockets in p38α but not p38β. RNA sequencing analysis of TNF-α–stimulated gene expression revealed that UM101 inhibited only 28 of 61 SB203580-inhibited genes and 7 of 15 SB203580-inhibited transcription factors, but spared the anti-inflammatory MSK1/2 pathway. We provide proof of principle that small molecules that target the ED substrate-docking site may exert anti-inflammatory effects similar to the catalytic p38 inhibitors, but their isoform specificity and substrate selectivity may confer inherent advantages over catalytic inhibitors for treating inflammatory diseases.
The p38 MAPK family of stress- and cytokine-activated kinases contribute to the pathogenesis of many human diseases, including cancer (1), rheumatoid arthritis (2), cardiovascular disease (3), multiple sclerosis (4), inflammatory bowel disease (5), chronic obstructive pulmonary disease and asthma (6), and acute lung injury (ALI) (7). Among the many important biological processes regulated by p38 MAPKs, regulation of endothelial and epithelial barrier function (8), leukocyte trafficking (9), and cytokine expression (2) are central to the pathogenesis of acute and chronic inflammatory disorders. Although preclinical studies strongly support the pharmacologic targeting of p38 as treatment for inflammatory diseases, p38 inhibitors have had very limited success in clinical testing because of dose-limiting toxicity and lack of efficacy. Of the 36 phase II clinical trials of p38 inhibitors listed on ClinicalTrials.gov (https://www.clinicaltrials.gov), the results of only eight studies have been published or listed on this site and showed little clinical benefit (10–13) and/or moderate toxicity (12).
All available p38 inhibitors block catalytic activity either by directly competing for ATP binding or by allosterically causing conformational changes that preclude access of ATP to the catalytic site (14). Davidson et al. (15) identified a purported p38α substrate-selective inhibitor, CMPD1, which selectively inhibited MAPK-activated protein kinase-2 (MK2) phosphorylation in in vitro kinase assays, but CMPD1 bound near the p38α active site and was subsequently shown to lack substrate-selectivity when tested in cells (16). Almost all available inhibitors are active against both p38α and p38β (17), and some are active against additional p38 isoforms. Yet genetic and pharmacologic studies have identified p38α as the proinflammatory isoform (18, 19), whereas other studies have demonstrated p38β signaling to be cytoprotective (20, 21). Therefore, inhibition of p38β may contribute to both lack of efficacy and toxicity of non–isoform-selective p38 inhibitors. However, the extensive structural conservation of the catalytic module across most protein kinases presents a challenge to developing catalytic inhibitors with high selectivity, especially for individual p38 isoforms (17).
Even if the catalytic inhibitors were absolutely selective for p38α, by design these compounds would block all p38α signaling events, many of which are essential for reestablishing and maintaining homeostasis. For example, p38α not only activates expression of proinflammatory cytokines, it also activates anti-inflammatory cytokines and counterregulatory dual-specificity protein phosphatase-2 (DUSP2) through the p38α substrate, mitogen- and stress-activated kinase (MSK) 1/2 (22, 23). The transient decrease and subsequent rebound of serum C-reactive protein (CRP) levels seen in clinical trials of p38 catalytic inhibitors (12, 13, 24) might be caused by the loss of the MSK1/2-dependent anti-inflammatory signaling.
As an alternative to the catalytic inhibitors, we targeted the substrate binding groove of p38α, which stretches between two acidic patches, the common docking (CD) and glutamate–aspartate (ED) domains (25, 26), and is distinct from the DEF substrate-binding pocket (27). Downstream substrates, upstream activating kinases, and possibly scaffolding molecules all interact with p38 through these sites (25). We used computer-aided drug design (CADD) to target low m.w. compounds to a pocket near the p38α ED substrate binding site, which binds MK2 (28), a p38α substrate known to mediate endothelial permeability and neutrophil transendothelial migration (TEM) in vitro and pulmonary edema in a mouse lung injury model (7), whereas anti-inflammatory MSK1/2 appears to bind to the CD site (26). Using this algorithm, we identified p38α-binding compounds with high efficiency, including a lead compound, 4-chloro-N-(4-[(1,1-dioxo-1λ~6~,4-thiazinan-4-yl) methyl]phenyl)benzamide (UM101), that selectively bound p38α and not p38β, stabilized endothelial barrier function in human lung microvascular endothelial cells (HMVECLs), inhibited LPS-induced proinflammatory gene expression in THP1 cells, and was well tolerated and more potent than SB203580 in mitigating experimental ALI.
Materials and Methods
Chemicals, recombinant proteins, and Abs
Mouse anti-human p38α and rabbit anti-phospho-MK2 (T222) and phospho-STAT-1 (S727) were obtained from Cell Signaling Tehcnologies (Danvers, MA). The coding sequences for human p38α variant 2 and p38β (with N-terminal hemagglutinin tag) were amplified by PCR and cloned into pRSetA (Thermo Fisher Scientific, Waltham, MA). Mutations were introduced into p38α using Quikchange (Stratagene) and confirmed by bidirectional sequencing. To generate activated dual-phosphorylated p38α for in vitro kinase assays, p38α variant 2 was amplified by PCR and cloned into the first multicloning site of pETDuet (EMD-Millipore) in-frame with an N-terminal His tag sequence. A gene block containing the optimized sequence for the constitutively active human MKK6 S207G/T211G mutant (29) was synthesized (Genscript, Piscataway, NJ) and cloned into the second multicloning site of pETDuet-p38α. Plasmids were transformed in Escherichia coli BL21 and proteins were purified using cobalt columns (TALON; Clontech Laboratories, Mountain View, CA), and confirmed by SDS-PAGE and immunoblotting. The p38α protein expressed from the pETDuet plasmid was confirmed to be >80% dual-phosphorylated by MALDI-TOF in the University of Maryland School of Pharmacy Proteomics Core. The compounds identified in the CADD screen were purchased from Maybridge Chemical (Belgium). Recombinant MK2, STAT-1α, and activating transcription factor (ATF) 2 protein were purchased from Thermo Fisher Scientific (PV3317), SignalChem (Richmond, BC, Canada) (S52-50G), and Origene (Rockville, MD) (TP710017), respectively.
CADD identification of lead compounds
Based on the x-ray crystal structure of mouse p38α (PDB ID: 1P38), we used a step-wise iterative CADD process to screen an in silico database of small molecule compounds available from Maybridge Chemical Screening Collection for the potential to bind in a pocket near the ED substrate binding site (Fig. 1A, 1B) using the same methods as were used to identify novel ERK inhibitors (30). In silico preparation of p38α conformation was performed using CHARMM36 protein and CHARMM general force field (31, 32) with the Nanoscale Molecular Dynamics program (33), to identify local potential ligand-binding pockets (34). Protein structures were subjected to clustering (35) to identify 20 representative protein conformations to account for protein flexibility. Screening was performed in the following stages: 1) potential inhibitor binding sites were identified (36); 2) compounds were ranked based on their van der Waals and electrostatic interaction energies with the protein binding pockets using the program DOCK (37–39) with size-based score normalization (40); 3) the top 50,000 compounds were subjected to a second in silico screen with additional relaxation of the ligands during simulated binding and the top 1000 compounds were selected based on total interaction energy including score normalization based on ligand size; and 4) chemical fingerprint-based cluster analysis (41, 42) of the top scoring compounds using the program MOE (Chemical Computing Group) was performed to identify chemically diverse compounds and the final list of potential p38α-interacting compounds were selected based on a scalar bioavailability metric, 4DBA (43), which accounts for the physiochemical descriptors in Lipinski’s Rule of Five (44) (Fig. 1D).
FIGURE 1.
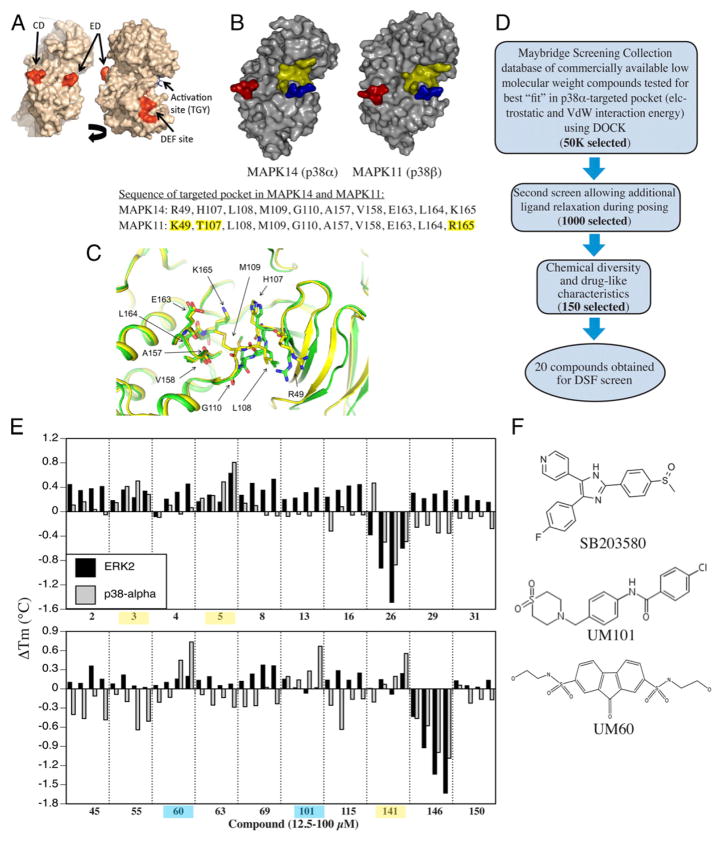
Design of substrate-selective p38 inhibitors. (A) Structure of p38α showing CD, ED, DEF, and activation site. (B) Comparison of p38α and β structure. CD and ED sites colored red and blue and the CADD target yellow. The sequence comprising the CADD target on p38α and the corresponding site on p38β differ in only 3 of 10 aa (highlighted yellow). (C) Overlap of CADD target structure in apo- (PDB:1P38; green) and dual-phosphorylated (PDB:3PY3; yellow) mouse p38α. (D) Overview of CADD screening strategy. (E) DSF screening of compounds added at 10, 25, 50, or 100 μM to recombinant p38α or ERK2 with binding indicated by increase in melting temperature. Compounds binding ERK2 and p38 α highlighted yellow. Those only binding to p38 α are highlighted in blue. (F) Chemical structure of UM60, UM101, and SB203580.
Differential scanning fluorimetry
Binding of CADD-selected compounds to p38α and β isoforms was tested experimentally using differential scanning fluorimetry (DSF), which evaluates changes in the target protein melting temperature (ΔTm) due to interactions with the test compound (45). SYPRO orange (Thermo Fisher Scientific) diluted 1:1000 in 10 mM HEPES, 150 mM NaCl (pH 7.5), and 1 μM unphosphorylated recombinant human p38α were added to 96-well PCR plates. Then 50 nM to 200 μM test compound dissolved in 100% DMSO (2% final DMSO concentration) was added, the plates were mixed, sealed, centrifuged at 1000 rpm for 1 min, and melting curves were performed using an Applied Biosystems StepOne real-time PCR instrument. The melting point was determined from the first derivative curve.
Cell culture
HMVECLs were purchased from Promocell, Heidelberg, DE, maintained in Endothelial Cell Growth Medium MV2, used at passage 3–10, and studied at postconfluence according to the supplier’s protocol as we have previously described (8, 9, 46, 47). The THP1 human monocyte cell line (American Type Culture Collection no. TIB202) was maintained in RPMI 1640 medium supplemented with 2 mM L-glutamine, 1 mM sodium pyruvate, 10 mM HEPES buffer (pH 7.3), penicillin, streptomycin, 0.05 mM β-mercaptoethanol, and 10% defined FBS (Life Technologies, Grand Island, NY). HeLa cells (American Type Culture Collection no. CCL-2) were cultured in DMEM with 4.5 g/l glucose, 1 mM sodium pyruvate, 2 mM L-glutamine, penicillin, streptomycin, and 10% FBS. Prior to experimental exposure, THP1 cells were differentiated by treating with 5 ng/ml PMA (Sigma-Aldrich) for 24 h, washing with PBS, and culturing at 37°C in PMA-free media for an additional 24 h.
Endothelial permeability assay
Permeability of HMVECL monolayers was assessed by measuring transendothelial flux of 10 kDa dextran conjugated to Cascade blue fluorescent dye for 30 min at 37°C in Matrigel-coated 3 μm pore size Transwell plates as we have described (8).
TEM assay
Isolation of neutrophils from heparinized venous blood collected from healthy volunteers, calcein labeling, and measurement of IL-8–directed TEM through HMVECLs was measured as previously described (9, 48, 49). The study was approved by the University of Maryland Baltimore Institutional Review Board.
Mouse acute lung injury model
Male CD-1 mice weighing 25–30 g were purchased from Charles River and housed in the Baltimore Veterans Administration Medical Center Animal Care Facility under American Association of Laboratory Animal Care–approved conditions. All protocols were approved by the University of Maryland Baltimore Institutional Animal Care and Use Committee. Inhibitors were tested in a mouse intratracheal (i.t.) LPS/febrile-range hyperthermia–induced ALI model as we have previously described (50). Mice were pre-treated with SB203580 or putative p38 inhibitors in ≤2%DMSO via 0.5 ml i.p. injection 1 h prior to i.t. instillation of 50 μg LPS and switch to a 37°C incubator, which increases core temperature to ~39.5°C. The dose of SB203580 used, 1 mg (40 mg/kg body weight), was previously shown to be maximally effective in animal models of septic shock (51). Mice were euthanized after 24 h, the lungs were lavaged with a total of 2 ml PBS, the cells were counted, and the cell-free lavage fluid was analyzed for protein content using the Bradford method (Bio-Rad), as previously described (50).
Inhibition of substrate phosphorylation
A functional analysis of UM101 to block p38-dependent phosphorylation of MK2 and STAT-1 was performed in HeLa cells. The cells were pretreated with SB203580 or UM101 for 30 min and then activated with 25 μg/ml anisomycin for 10–60 min. Cell were lysed in radioimmunoprecipitation buffer containing protease and phosphatase inhibitors and lysates were resolved by SDS-PAGE, transferred to polyvinylidene fluoride membrane, blocked with 5% nonfat dry milk, and probed with primary Abs against phosphorylated MK2 and STAT-1, and total p38α as a loading control. Bands were detected using secondary Abs conjugated to infrared fluorophores and infrared fluorescence imaging (Odyssey; LI-COR) and the data were quantified using the LI-COR image analysis software. The inhibitor effect on p38α phosphorylation of specific substrates was further analyzed by in vitro kinase assay in 20 μl reaction volume containing 75 ng of active p38α (expressed from pETDuet), substrate proteins (100–150 ng), 5 μM SB203580 or 10–50 μM UM101, and 5 μCi of ATP [γ-32P] in kinase buffer [50 mM Tris-HCl, 10 mM MgCl2, 0.1 mM EDTA, 2 mM DTT, 0.01% Brij 35, (pH 7.5)]. Reactions were performed at 37°C for 10–120 min, then terminated by adding and equal volume of 2× SDS-PAGE sample buffer. The reaction mixtures were separated by SDS-PAGE. The gels were dried and the radioactive phosphate incorporation into substrates was measured by phosphorimaging on a Typhoon FLA 7000 (GE Healthcare Life Sciences). The data were quantified using ImageQuant TL (GE Healthcare Life Sciences) and expressed as relative values.
Cytotoxicity assay
Cytotoxicity was monitored in parallel HMVEC-L monolayers established in 96-well culture plates using a colorimetric assay that measured the reduction of 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium to a formazan dye (CellTiter 96; Promega, Madison, WI) according to the manufacturer’s protocol and quantifying product formation by measuring absorption at 490 nm.
Gene expression
RNA integrity was confirmed by Agilent Bioanalyzer 2100 and all samples were confirmed to have RNA integrity scores of 10 prior to further analysis. Poly(A)-enriched samples were reverse transcribed and sequenced using the Illumina HiSeq platform to generate at least 90 million reads per sample. Intergenic sequence accounted for <0.7% of all reads, indicating minimal genomic DNA contamination. Raw data were analyzed using the TopHat read alignment tool and Homo sapiens genomic reference sequence (Ensembl version GRCh38.78). Differential gene expression was analyzed using the DESeq R package (Bioconductor) and the negative binomial model. Criteria for significant differences in gene expression were: 1) false discovery rate <0.05; 2) expression level >10th percentile; and 3) ≥2-fold change. The differential gene expression patterns were further analyzed using the PathwayNet (Troyanskaya Lab, Princeton) and Ingenuity Pathway Analysis (Qiagen) tools. Cytokine gene expression in THP1 cells was analyzed by quantitative RT-PCR using primers in a commercially available PCR array (HCA-II array; Real Time Primers, Elkins Park, PA) and SYBR-green reaction mix (Bio-Rad), and a Bio-Rad iCycler IQ Optical Module according to the supplier’s protocol. Data were quantified using the gene expression Ct difference method (52) and standardized to the levels of the housekeeping gene, GAPDH, using Ct values automatically determined by the thermocycler as we have previously described (53). The RNA sequencing (RNASeq) data have been deposited in the Gene Expression Omnibus database (accession number GSE93330; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93330).
Saturation transfer difference–nuclear magnetic resonance
A 40 mM stock solution of UM101 was made in d6-DMSO. Saturation transfer difference–nuclear magnetic resonance (STD-NMR) samples contained 150 mM NaCl, 50 mM phosphate (pH 7), 200 μM UM101, and 5 μM p38 protein in D2O. Spectra were recorded on an Agilent DD2 500-MHz spectrometer equipped with a 5-mm inverse proton–fluorine–carbon–nitrogen probe head at 27°C. During each transient, the protein was saturated with a series of 58 GAUSSIAN-shaped pulses (50 ms with 1 ms delay between pulses) using the vendor-supplied STD–excitation-sculpting pulse sequence, for a total saturation time of 3 s. The on-resonance irradiation of protein was done at 0.5 ppm and off-resonance irradiation at 30 ppm. The vendor-supplied WATERGATE pulse sequence was used to suppress the water signal in the STD spectrum. The on-resonance and off-resonance pulse sequences were subtracted internally. A total of 16,384 transients were collected for each STD experiment with a 1 s delay between acquisitions, 6000 Hz spectral width, and 1.3 s acquisition time.
Statistical methods
Data are presented as mean ± SE. Differences among >2 groups were analyzed by applying a Tukey Honestly Significant Difference test to a one-way ANOVA. Differences between dose-response curves was analyzed by multivariate ANOVA (MANOVA). Differences with p < 0.05 were considered significant.
Results
CADD modeling of p38 MAPK substrate-docking site and compound identification
We used a CADD-based strategy to identify low m.w. compounds predicted to bind near the ED substrate-docking site of mouse unphosphorylated p38α (MAPK14 variant-1; PDB:1P38), which is >99% identical to human p38α (variant-2) (Fig. 1A). The ED and CD sites in p38α are located at either end of a substrate-binding cleft located on the opposite side of the protein from the catalytic site (Fig. 1A). We identified a pocket near the ED binding site comprising 10 aa, only seven of which were identical in p38α and p38β (Fig. 1B). Overlay of structures of mouse unphosphorylated (PDB:1P38) and dual-phosphorylated p38α (PDB:3PY3) revealed near-superposition of the targeted pocket in the two forms (Fig. 1C).
An overview of the CADD screening and compound testing protocols is shown in Fig. 1D. The compounds in the Maybridge Screening Collection were analyzed for binding to the targeted p38α pocket based on van der Waals and electrostatic interaction energies (40), chemical diversity by chemical fingerprint-based cluster analysis (41, 42), solubility, m.w., and number of hydrogen-bonding functional groups that maximize bioavailability (43). A panel of 150 diverse compounds was selected for potential biological testing; 20 of these structurally dissimilar compounds were obtained for functional analysis (Supplemental Fig. 1, Table I).
Table I.
CADD-identified p38α ED site-binding candidates screened for p38α binding
| CADD no. | Compound IDa | m.w. | logPb | p38α ΔTm (°C) at 100 μMc | ERK2 ΔTm (°C) at 100 μMc |
|---|---|---|---|---|---|
| 2 | SEW 06373 | 417 | 3.19 | −0.05 | 0.412 |
| 3 | HTS 02798 | 415 | 0.67 | 0.282 | 0.337 |
| 4 | HTS 13333 | 312 | −1.10 | 0.065 | 0.452 |
| 5 | SCR 00846 | 418 | 2.22 | 0.808 | 0.628 |
| 8 | AW 00509 | 317 | 1.13 | −0.07 | 0.531 |
| 13 | SEW 06264 | 309 | 0.28 | 0.005 | 0.390 |
| 16 | SCR 00610 | 339 | 1.69 | −0.052 | 0.444 |
| 23 | SCR 01200 | 378 | 2.79 | −0.488 | −0.598 |
| 29 | BTB 05645 | 350 | 3.07 | −0.353 | 0.342 |
| 31 | KM 04113 | 304 | 1.83 | −0.278 | 0.153 |
| 43 | CD 11992 | 300 | 1.16 | −0.485 | 0.151 |
| 55 | SP 01164 | 2.11 | 1.92 | −0.506 | 0.022 |
| 60 | BTB 13869 | 426 | 0.28 | 0.735 | 0.195 |
| 63 | PD 00612 | 294 | 0.61 | −0.287 | 0.075 |
| 69 | KM 00081 | 345 | 1.68 | −0.233 | 0.361 |
| 101 | HTS 05732 | 378 | 2.31 | 0.667 | 0.0175 |
| 115 | NRB 03986 | 278 | 3.88 | −0.156 | 0.246 |
| 141 | SEW 02182 | 318 | 2.46 | 0.554 | 0.238 |
| 146 | KM 10445 | 313 | 2.55 | −1.084 | −1.632 |
| 150 | HTS 03239 | 341 | 1.68 | −0.171 | 0.133 |
Compound ID from Maybridge portfolio.
logP is the logarithm of the estimated octanol/water partition coefficient, a measure of compound solubility.
Change in melting temperature relative to DMSO control in DSF assay.
Screening compounds for direct, selective interaction with p38α
Test compounds at 10–100 μM were screened for binding to recombinant p38α and ERK2 using DSF (45) (Fig. 1E, Table I). Five compounds caused concentration-dependent stabilization of p38α, indicating binding. Three of these also stabilized ERK2 (3, 5, and 141, highlighted yellow in Fig. 1E) and two, N2,N7-di(2-hydroxyethyl)-9-oxo-9H-2,7-fluorenedisulfonamide (UM60) and UM101, highlighted blue, stabilized p38α but not ERK2. These two structurally dissimilar compounds (Fig. 1F), added at 100 μM, increased the melting temperature of p38α by ~0.7°C compared with a 6°C increase with SB203580.
Effects of compounds on endothelial barrier functions
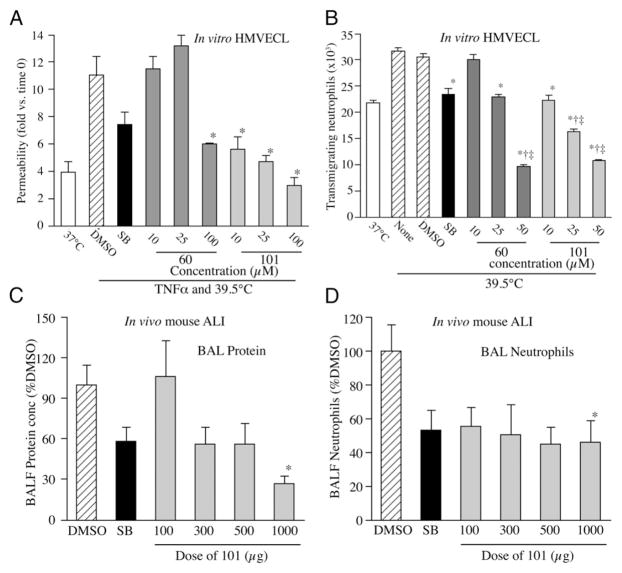
We tested the capacity of UM60 and UM101 to stabilize endothelial barriers to macromolecules and neutrophils in TNF-α– and hyperthermia-stressed HMVECL monolayers (8, 9) (Fig. 2). Combined exposure to 1 ng/ml TNF-α and hyperthermia (39.5°C) for 6 h increased HMVECL permeability for 10 kDa dextran 2.8-fold compared with untreated 37°C cells. Pretreating with 10 μM SB203580 for 30 min reduced TNF-α–/hyperthermia-induced permeability by 50% (Fig. 2A). Pretreatment with UM60 at 10 and 25 μM had no effect on permeability, but 100 μM UM60 reduced the TNF-α–/hyperthermia-induced permeability increase by 71%, whereas UM101 at 10, 25, and 100 μM reduced the TNF-α–/hyperthermia-induced permeability increase by 74, 89, and >100%, respectively.
FIGURE 2.
Biological effects of p38 inhibitors. (A and B) Effect of 10 μM SB203580 (SB), or indicated concentration of UM60 or 101 on HMVECL permeability (A) and capacity for IL-8–directed neutrophil TEM (B). Cells were pretreated with DMSO or compounds for 1 h, then incubated with 10 ng/ml TNF-α for 6 h at 39.5°C prior to permeability assay (A) or at 39.5°C without additional stimulus for 6 h prior to TEM assay (B). Mean ± SE. *p < 0.0001 versus DMSO, †p < 0.0001 versus SB, ‡p < 0.005 versus 37°C. (C and D) Male CD1 mice were pretreated with 1 mg SB or 0.1–1 mg UM101 prior to i.t. instillation of 50 μg LPS and hyperthermia exposure. *p < 0.05 versus DMSO.
Preincubating HMVECLs at 39.5°C for 6 h increased subsequent IL-8–directed neutrophil TEM from 22.8 ± 0.45 × 103 to 31.8 ± 0.54 × 103 neutrophils (Fig. 2B). Pretreatment with 10 μM SB203580 reduced hyperthermia-augmented neutrophil TEM by 84%. UM60 at 10 and 25 μM and UM101 at 10 μM reduced hyperthermia-augmented increase in TEM by 18, 89, and 95%. UM60 at 50 μM and UM101 at 25 and 50 μM reduced TEM to less than baseline levels. Neither compound was toxic in lactate dehydrogenase release and 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium assays when added to HMVECLs at 100 μM for 48 h (data not shown).
Comparing effectiveness of SB203580 and UM101 in mouse ALI
We compared the effectiveness of UM60, UM101, and SB203580 in mitigating transalveolar protein and neutrophil extravasation in a mouse model of LPS-/hyperthermia-induced ALI (50) (Fig. 2C, 2D). Mice received a single i.p. injection of 100, 300, 500, or 1000 μg UM101, 1000 μg UM60, or 1000 μg SB203580 in 0.5 ml 2% DMSO 30 min prior to i.t. instillation of 50 μg LPS and transfer to hyperthermic chambers. Control mice received DMSO. Four of six UM60-treated, one of six SB203580-treated, and 1 of 11 DMSO-treated control mice died within 24 h. All 16 UM101-pretreated mice survived. Lung lavage from DMSO-pretreated, LPS-/hyperthermia-challenged mice contained 1.09 ± 0.19 mg/ml protein, and 3.97 ± 1.07 × 106 neutrophils. Compared with DMSO-treated controls, lavage protein concentration and neutrophil content in mice pretreated with 1000 μg SB203580 were reduced by 42 and 46.8%, respectively. Lavage protein concentration in mice pretreated with 100, 300, 500, and 1000 μg UM101 was reduced by 0, 44.1, 43.9, and 92.9%, respectively, and lavage neutrophil content was reduced by 44.4, 49.5, 55.3, and 54%, respectively.
Effect of SB203580 and UM101 on LPS-induced gene expression in human THP1 promonocytes
The effects of UM101 and SB203580 on inflammatory cytokine expression were compared by pretreating PMA-differentiated THP1 cells with 25 μM SB203580 or 10, 25, or 100 μM UM101 for 30 min, then stimulating with 100 ng/ml LPS, and harvesting RNA 4 h later for analysis by PCR-based cytokine array. Of 16 LPS-stimulated genes in the array, SB203580 inhibited expression of seven: IL-1α, IL-8, TNFSF8 (CD30 ligand), TNFSF9 (CD137 ligand), CXCL5, CCL7, and CCL17 (Table II). UM101 inhibited expression of all SB203580-inhibited genes except TNFSF9, and inhibited four SB203580-insensitive genes, IL-1β, CXCL1, TNFSF15, and CCL5.
Table II.
Effects of SB203580 and UM101 on LPS-induced cytokine expression in THP1 cells
| Gene | DMSOa | ANOVAb | SB20358025 μM | p versus LPSc | UM101 10 μM | p versus LPSc | UM101 25 μM | p versus LPS | UM101 100 μM | p versus LPSc |
|---|---|---|---|---|---|---|---|---|---|---|
| IL-1A | 453 ± 24 | <0.0001 | 141 ± 9.2 | <0.0001 | 424 ± 22.6 | 0.74 | 339 ± 13.5 | 0.041 | 88 ± 3.33 | <0.0001 |
| IL-8 | 56.5 ± 3.3 | 0.0026 | 9.6 ± 0.1 | 0.002 | 35.6 ± 0.7 | 0.40 | 26.7 ± 4.1 | 0.564 | 19.7 ± 1.8 | 0.015 |
| TNFSF8 | 60.5 ± 5.5 | 0.0073 | 20.6 ± 8.8 | 0.024 | 23.5 ± 8.3 | 0.37 | 10.5 ± 3.9 | 0.006 | 20.7 ± 9.5 | 0.025 |
| CXCL5 | 49.7 ± 2.9 | <0.0001 | 3.2 ± 1.0 | <0.0001 | 23.2 ± 3.7 | 0.0002 | 8.7 ± 2.9 | <0.0001 | 3.1 ± 0.2 | <0.0001 |
| CCL7 | 12.8 ± 1.2 | <0.0001 | 4.2 ± 0.3 | <0.0001 | 7.7 ± 0.3 | 0.0036 | 6.2 ± 0.9 | 0.0036 | 4 ± 0.4 | <0.0001 |
| CCL17 | 56.9 ± 6.1 | <0.0001 | 21.5 ± 3.7 | 0.001 | 30.4 ± 4.7 | 0.008 | 11 ± 1.0 | 0.0004 | 2.5 ± 0.33 | <0.0001 |
| TNFSF9 | 50.8 ± 6.1 | 0.0046 | 20.7 ± 3.1 | 0.0054 | 48 ± 2.1 | 0.99 | 38.2 ± 6.9 | 0.334 | 32 ± 1.12 | 0.086 |
| IL-1B | 171 ± 9.0 | 0.0089 | 187 ± 7.4 | 0.988 | 104 ± 21 | 0.382 | 88 ± 9.0 | 0.204 | 51.6 ± 5.2 | 0.033 |
| CXCL1 | 24.5 ± 0.5 | <0.0001 | 28.2 ± 1.9 | 0.577 | 19.8 ± 1.8 | 0.36 | 12.8 ± 2.5 | 0.005 | 5.2 ± 1.0 | <0.0001 |
| TNFSF15 | 9.6 ± 1.1 | 0.0012 | 10 ± 1.1 | 0.998 | 7.6 ± 0.9 | 0.544 | 5.4 ± 0.8 | 0.053 | 2.9 ± 0.6 | 0.003 |
| CCL5 | 7.6 ± 0.9 | 0.0045 | 3.6 ± 0.8 | 0.26 | 3 ± 0.5 | 0.018 | 2.7 ± 0.2 | 0.008 | 2.6 ± 1.2 | 0.006 |
| CCL4 | 188 ± 12 | 0.9519 | 188 ± 16 | NS | 174 ± 41 | NS | 191 ± 57 | NS | 217 ± 51 | NS |
| CCL20 | 82.5 ± 27.8 | 0.1189 | 106 ± 15.1 | NS | 63 ± 3.1 | NS | 63.4 ± 1.0 | NS | 42.7 ± 12.7 | NS |
| CXCL2 | 122 ± 11.0 | 0.9887 | 125 ± 4.6 | NS | 128 ± 20.0 | NS | 132 ± 22.9 | NS | 130 ± 6.4 | NS |
| TNF | 115 ± 13/1 | 0.6112 | 66.4 ± 9.6 | NS | 87 ± 12.4 | NS | 95.9 ± 21.2 | NS | 80 ± 14.5 | NS |
| BMP6 | 8.1 ± 1.8 | 0.1195 | 4.1 ± 1.1 | NS | 8.9 ± 1.7 | NS | 7.8 ± 1.1 | NS | 3.9 ± 0.5 | NS |
All values are fold-change mRNA levels versus unstimulated PMA-differentiated THP1 cells.
Cells were preincubated with 0.4% DMSO or inhibitors for 1 h, then stimulated with 100 ng/ml LPS for 2 h.
p values from one-way ANOVA.
p values from the Tukey Honestly Significant Difference post hoc test.
Comparing effects of SB203580 and UM101 on TNF-α–induced gene expression in HMVECLs
We compared the effects of UM101 and SB203580 on TNF-α–induced gene expression in HMVECLs using RNASeq. We pre-treated HMVECLs for 1 h with 10 μM SB203580 or 100 μM UM101, and then stimulated with 10 ng/ml TNF-α for 3 h. We used a UM101 concentration 10-fold higher than its biologically effective dose in HMVECL barrier assays to ensure identifying any partial overlap with SB203580. The TNF-α concentration and duration of stimulation used were based on published studies (54, 55) and confirmed by preliminary quantitative RT-PCR analysis of IL-8 and IL-1β mRNA expression (data not shown). After filtering the RNASeq results for genes having ≥10 reads in at least one sample per experiment, we found 511 genes that were upregulated and 520 downregulated by ≥2-fold by TNF-α treatment (Supplemental Table I). SB203580 inhibited expression of 61 TNF-α–induced genes, 28 of which were also inhibited by UM101 (Supplemental Fig. 2, Supplemental Table I, Table III). SB203580 increased expression of 38 genes, 10 of which were also increased by UM101. Of the 28 genes inhibited by both SB203580 and UM101, 22 coded for known proteins, including IL-1β, CCL17, MMP9, IDO1, CXCL5, 10 and 11, hyaluronan synthase-3, MUC4, and PLA2 (Table III). Of the 33 genes inhibited by SB203580 but not UM101, 24 coded for known proteins, including GM-CSF, IL-1α, TNF-α, IL-12 receptor-β1, and hyaluronan synthase-2 (Table III). Quantitative RT-PCR confirmed the RNASeq results for a subset of these genes and showed that UM101, but not SB203580, increased expression of DUSP2 and 10 (Supplemental Fig. 3).
Table III.
Effect of SB203580 and UM101 in HMVECLs on TNFα-induced genes
| Gene Symbol | Gene Name | Log Fold-Change Versus DMSO
|
|
|---|---|---|---|
| SB203580 | UM101 | ||
| Genes inhibited by both SB203580 and UM101 | |||
| PRRG4 | Proline rich Gla 4 | −1.782580534 | −1.198488233 |
| TSLP | Thymic stromal lymphopoietin | −1.651439594 | −1.336652511 |
| CCL17 | Chemokine (C-C motif) ligand 17 | −1.834143455 | −2.730309773 |
| EXOC3L4 | Exocyst complex component 3-like 4 | −1.479163179 | −1.160471021 |
| MMP9 | Matrix metallopeptidase 9 | −1.157091348 | −1.091179627 |
| IDO1 | Ido 1 | −3.510632932 | −3.567987354 |
| CXCL10 | Chemokine (C-X-C motif) ligand 10 | −3.100915562 | −4.369836708 |
| CD200 | Cd200 | −1.729285649 | −1.538155406 |
| SLC15A3 | Solute carrier family 15, member 3 | −1.00338842 | −1.73105887 |
| VDR | Vitamin D receptor | −1.16718631 | −1.19731694 |
| IL1B | Il-1β | −1.401586926 | −1.172530543 |
| GPR88 | G protein-coupled receptor 88 | −1.397083754 | −2.150599176 |
| CD207 | CD207 (langerin) | −1.547757288 | −3.382437255 |
| TCHH | Trichohyalin | −1.504958085 | −1.547665316 |
| HAS3 | Hyaluronan synthase 3 | −1.43377734 | −1.124339564 |
| GBP1P1 | Guanylate binding protein 1 | −1.363287203 | −1.755706078 |
| MUC4 | Mucin-4 | −2.859491876 | −1.057315692 |
| ELOVL7 | ELOVL fatty acid elongase 7 | −1.340933369 | −1.381063226 |
| CXCL11 | Chemokine (C-X-C motif) ligand 11 | −1.377905942 | −4.136354868 |
| GBP4 | Guanylate binding protein 4 | −1.259283076 | −2.835947907 |
| PLA1A | Phospholipase A1 member A | −1.27452433 | −1.500633356 |
| CXCL5 | Chemokine (C-X-C motif) ligand 5 | −1.017427849 | −1.468307731 |
| Genes inhibited by SB203580 but not UM101 | |||
| CSF2 | GM-CSF | −1.634082807 | NS |
| RND1 | Rho family gtpase 1 | −1.15027449 | NS |
| SLC26A9 | Solute carrier family 26, member 9 | −2.092734866 | NS |
| HAS2 | Hyaluronan synthase 2 | −2.646197932 | NS |
| CD69 | Cd69 | −1.467068659 | NS |
| ANKRD55 | Ankyrin repeat domain 55 | −1.204412851 | NS |
| NKX3-1 | NK3 homeobox 1 | −1.105417452 | NS |
| EBI3 | EBV induced 3 | −1.393947741 | NS |
| NKD2 | Naked cuticle homolog 2 | −1.418158287 | NS |
| F2RL3 | Coagulation factor II receptor-like 3 | −2.642662346 | NS |
| IL1A | IL-1alpha | −2.152720278 | NS |
| TP63 | Tumor protein 63 | −1.312154792 | NS |
| HAND1 | Heart/neural crest derivatives expressed 1 | −1.617984328 | NS |
| IL12RB1 | IL 12 receptor, β 1 | −1.409364824 | NS |
| ERVFRD-1 | Endogenous retrovirus group FRD, member 1 | −1.234925956 | NS |
| TMEM52B | Transmembrane protein 52B | −1.109879612 | NS |
| BPGM | 2,3-bisphosphoglycerate mutase | −1.204500786 | NS |
| TOX | Thymocyte selection-associated HMGB | −1.422546092 | NS |
| HPCA | Hippocalcin | −2.219945733 | NS |
| TNF | Tnf-α | −1.478211598 | NS |
| FAM101A | Family with sequence similarity 101, member A | −1.712019627 | NS |
| OLR1 | Oxidized low density lipoprotein receptor 1 | −1.013271327 | NS |
| HEPACAM | Hepatic and glial cell adhesion molecule | −1.211648309 | NS |
| P2RX2 | Purinergic receptor P2X | −2.244463614 | NS |
HMVECL treated 1 h with 0.4% DMSO, 10 μM SB20350, or 100 μM UM101, then 4 h with 10 ng/ml TNF-α.
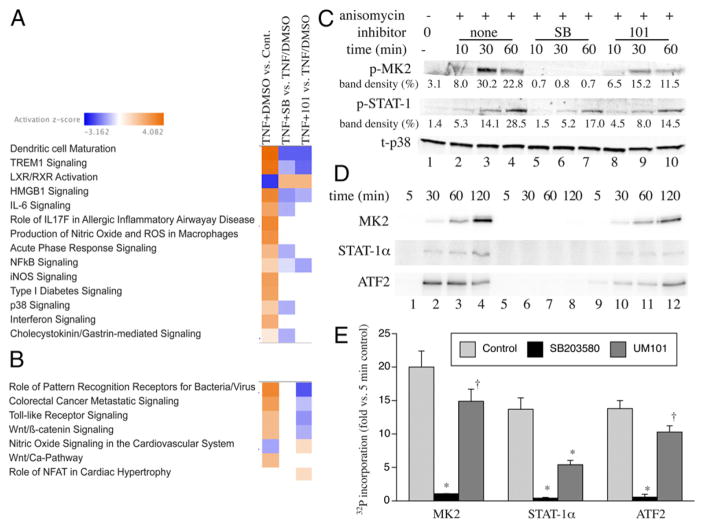
The differentially expressed genes were further analyzed using PathwayNet and Ingenuity tools to identify the transcription factors and biological pathways regulated by the two inhibitors. PathwayNet analysis suggested that UM101 inhibits some of the SB203580-inhibited transcription factors (STAT-1, c-Fos, c-Jun, NF-κB, p53, PPARγ, and Sp1), but not others (ATF1, ATF2, Elk1, c/EBPβ, USF1, SMAD3, FOXO1, and CREB via MSK1/2). Ingenuity analysis suggested that both SB203580 and UM101 inhibit dendritic cell maturation, triggering receptors expressed on myeloid cells-1, high mobility group box 1, and NF-κB pathways and both increase liver X-receptor/retinoid X-receptor activation, whereas only SB203580 inhibits IL-6, acute phase, and cholecystokinin/gastrin-mediated pathways (Fig. 3A). UM101 at 100 μM reduced expression of 115 genes and increased expression of 119 genes that were not modified by SB203580 (Supplemental Table I), which ingenuity pathway analysis suggested reduced TLR and Wnt/β-catenin signaling and increased NO in cardiovascular disease pathways (Fig. 3B).
FIGURE 3.
Biochemical effects of substrate-selective p38 inhibitors. (A and B) Heat maps from RNASeq showing IPA pathways inhibited by SB203580 alone or SB203580 and UM101 or (A) and those only inhibited by UM101 (B). (C) HeLa cells were pretreated with 50 μM UM101 or 10 μM SB203580 (SB) for 30 min, then treated with anisomycin for 10–60 min, and immunoblotted for phospho-MK2, phospho-STAT-1, and total p38. The means of the band densities from two experiments expressed as percent of total density for all bands are shown below each corresponding band. (D and E) In vitro kinase reaction containing activated p38α, ATP [γ-32P], 5 μM SB203580, or 50 μM UM101, and the indicated substrate were incubated at 37°C for indicated time, separated by SDS-PAGE, and analyzed by phosphorimaging. A representative of three gels (D) and means ± SE of the 120-min band densities expressed as fold-change versus the 5-min control reaction band are shown. *p < 0.0001, †p < 0.01 versus control.
Comparing effects of SB203580 and UM101 on p38 MAPK substrate phosphorylation profile
To assess whether UM101 selectively inhibits phosphorylation consistent with its target, HeLa cells were pretreated for 30 min with 10 μM SB203580, 50 μM UM101, or 0.1% DMSO vehicle control, then with the p38 activator, anisomycin (25 μg/ml), and phosphorylated MK2, and STAT-1 were analyzed by immunoblotting (Fig. 3C). Anisomycin-stimulated phosphorylation of MK2 and STAT-1 were reduced by both 10 μM SB203580 and 50 μM UM1010, but to a greater extent with SB203580. The in vivo phosphorylation studies were complemented by in vitro kinase assays, comparing the effects of 5 μM SB203580 with 50 μM UM101 on phosphorylation of MK2, STAT-1α, and ATF2 by p38α (Fig. 3D, 3E). Whereas SB203580 abrogated phosphorylation of all three substrates, UM101 partially reduced phosphorylation of the three substrates by varying degrees with the greatest effect on STAT-1α.
Analyzing specific binding of UM101 to p38α
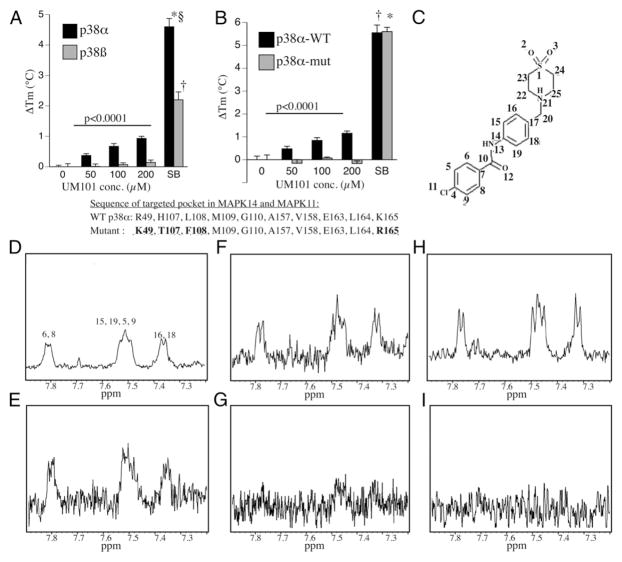
We used DSF to analyze concentration-specific binding of UM101 to p38α and p38β. Whereas SB203580 stabilized both p38α and p38β, UM101 only stabilized p38α (Fig. 4A). To confirm that UM101 bound the CADD-targeted pocket, we used DSF to compare binding of UM101 and SB203580 to wild-type p38α and a p38α mutant with substitutions of 4 of the 10 amino acids defining the CADD-targeted pocket (R49K/HL107-8TF/K165R) (Fig. 4B). The mutant exhibited SB203580-binding that was identical to wild-type p38α, but no detectable UM101 binding.
FIGURE 4.
Binding specificity of UM101 and SB203580. (A) DSF analysis of indicated concentration of UM101 and 10 μM SB203580 (SB) binding to recombinant p38α and p38β. Mean ± SE of four experiments. *,†,§p < 0.0001 versus p38α with DMSO, p38β with DMSO, and p38β with SB203580, respectively. p < 0.0001 for difference between UM101 binding to p38α and p38β by MANOVA. (B) DSF analysis of indicated concentration of UM101 and 10 μM SB203580 (SB) binding to recombinant wild-type p38α and a p38α mutant with four mutations in CADD-targeted pocket (highlighted and bolded). Mean ± SE of four experiments. *,†p < 0.0001 versus wild-type with DMSO and mutant with DMSO, respectively. p < 0.0001 for difference between UM101 binding to wild-type and mutant p38α by MANOVA. (C–I) STD-NMR was performed with UM101 and p38α (D and E), p38β (F and G), and the p38α mutant (H and I). The 1D (D, F, and H) and STD spectra (E, G, and I) from the same sample are shown. The tentative peak assignments are indicated in (D). The structure of UM101 with the protons labeled is shown in (C).
We confirmed selective binding of UM101 to the CADD-targeted pocket in p38α using STD-NMR (Fig. 4C–I). A 1D spectrum of UM101 in the presence of p38α is shown in Fig. 4D and the STD spectrum of the same sample is shown in Fig. 4E. The peaks in the 1D spectrum are labeled according to tentative peak assignments of UM101 in aqueous form, based on assignments of UM101 in 2 mM d6-DMSO, which were obtained from the use of 1D proton and [13C] and two-dimensional hetero-nuclear multiple bond correlation experiments (data not shown). The shifts of the peaks in the STD spectrum correspond well to those of the 1D spectrum, thus indicating that protons in both aromatic rings of UM101 interact with p38α. In contrast, although the 1D spectra for UM101 with p38β and mutated p38α were similar to that of UM101/p38α (Fig. 4F, 4H), the interaction of UM101 with p38β and mutated p38α is much weaker, as indicated by the barely discernible peaks of the aromatic protons in the STD spectrum of UM101 with p38β (Fig. 4G) and mutated p38α (Fig. 4I).
Discussion
In targeting a pocket near the ED substrate–docking site of p38α, we theoretically avoided interfering with CD-specific substrates, including MSK1/2 (25, 26), which limits inflammation through expression of IL-10 and DUSP2 (22). Among the effects of MSK1/2 deletion in mice is increased and prolonged LPS-induced expression of the CRP regulator, IL-6, suggesting a possible mechanism of the rebound in serum CRP observed in some clinical trials of catalytic p38 inhibitors (12, 13, 24).
Mouse unphosphorylated p38α/MAPK14 variant-1 differs from its human variant-2 by only 2 aa, H48L and A263T, and from mouse variant-2 and human variant-1 by only 14 aa between residues 230 and 254. Neither these amino acid differences nor the phosphorylation state of p38α (Fig. 1C) are predicted to significantly alter the structure of the CD or ED sites of our CADD targeted pocket, thereby validating our use of mouse unphosphorylated p38α variant-1 for the CADD search and unphosphorylated recombinant human p38α variant-2 protein for the DSF screen. Although DSF is less sensitive than other assays of ligand:protein binding (56), it is low cost and has relatively high throughput. DSF detected p38α binding by 25% of the CADD-identified compounds screened and selective p38α binding by 10%, demonstrating good efficiency of both the CADD and DSF screening strategies. The 10% hit rate of our CADD search for substrate-selective p38α inhibitors was similar to our prior experience with CADD-identified substrate-selective ERK inhibitors (30) and much greater than the usual 0.1–0.01% hit rate using experimental screening alone (57).
We identified a lead compound that has favorable biological effects in human cell culture models and in a mouse model of inflammatory lung injury. The CADD-targeted pocket in p38α differed from the corresponding pocket in p38β in 3 of 10 aa, which provided an opportunity for p38α selectivity. We confirmed selective binding of UM101 to p38α using complementary technologies. DSF, which detects ligand-induced protein stabilization (45), showed UM101 to cause a concentration-dependent increase in melting temperature of p38α but not p38β (Fig. 3D). The smaller effect of UM101 compared with SB203580 on p38α melting suggests lower p38α binding affinity of substrate-selective versus catalytic inhibitors, which is similar to our prior experience with substrate-selective ERK inhibitors (30). The smaller effect of SB203580 on p38β than p38α is consistent with the known ~10-fold higher binding affinity of SB203580 for p38α (58). STD-NMR, which measures low-affinity protein:ligand binding via nonscalar magnetization transfer from protein to ligand protons (59), confirmed specific UM101 binding to p38α and localized the interaction to its aromatic rings. We indirectly confirmed that UM101 bound to its CADD target by showing that mutating 4 of 10 aa in the targeted pocket abrogated UM101 binding whereas SB203580 binding was preserved.
In vitro kinase assays illustrate how UM101 and SB203580 differ in how they modify p38α activity. Whereas SB203580 abrogated phosphorylation of all three substrates tested, MK2, ATF2, and STAT-1α, UM101 only partially reduced substrate phosphorylation. It exerted the greatest effect on STAT-1α and the least on MK2. Although x-ray crystallographic data showing that MK2 binds the ED site of p38α led us to predict that UM101 would block its phosphorylation, MK2 simultaneously interacts with the CD site (28), which may explain why UM101 exerted only modest effects on its phosphorylation in HeLa cells (Fig. 3C) and in in vitro kinase assays (Fig. 3D, 3E).
Both of the selective p38α binding compounds, UM60 and UM101, exerted SB203580-like in vitro and in vivo endothelial-barrier–stabilizing and macrophage-cytokine–modifying effects, thereby validating our ED-targeting strategy. We note that the in vivo dose response for UM101 differs for transalveolar protein leak and neutrophil migration, suggesting that different UM101-inhibitible p38-dependent pathways may contribute to endothelial protein leak and neutrophil transmigration. Alternatively, UM101 may exert its effects on neutrophil transmigration in vivo by affecting both endothelial cells and neutrophils as p38 activation in both cells contributes to neutrophil transendothelial transmigration (9). Because UM101 more effectively stabilized endothelial barriers than SB203580 (Fig. 2A, 2B) despite having less effect on MK2 phosphorylation (Fig. 3C), we evaluated additional molecular actions by comparing the effects of UM101 and SB203580 on global gene expression using RNASeq in TNF-α–treated HMVECLs. TNF-α increased expression of 511 genes by ≥2-fold, of which 61 were reduced and 38 increased by pretreatment with 10 μM SB203580. Despite using a concentration of UM101 that was >10-fold higher than required to stabilize HMVECL barrier functions (Fig. 2A, 2B), UM101 modified expression of only 38 of the 99 SB203580-modified genes.
PathwayNet analysis showed UM101 blocked only seven of the 15 SB203580-blocked transcription factors. MSK1/2 was among those spared by UM101, which is consistent with the targeting strategy for UM101 to the ED site. Because p38 MAPK exerts transcriptional and posttranscriptional control of gene expression through phosphorylation of multiple protein substrates (60), substrate-selective inhibitors like UM101 are likely to impact gene expression and cell function differently than catalytic inhibitors. For example, sparing of MSK1/2 phosphorylation by UM101 preserves expression of counterregulatory genes, including DUSP2 and 10 (22, 23), which were increased by treatment with UM101 but not by treatment with SB203580.
The partial functional overlap of UM101 and SB203580 revealed by RNASeq is consistent with the design of UM101 as a noncatalytic substrate-selective inhibitor, but might also be the result of off-target effects of SB203580, which include receptor-interacting protein kinase-2, cyclin G-associated kinase, and casein kinase-1δ (58). However, none of the SB203580-inhibited transcription factors identified by the PathwayNet analysis are known substrates for these kinases as analyzed with PhosphoNetworks (61).
Although the high concentration of UM101 used in this initial analysis may have caused some p38-independent actions, we believe the following data support a conclusion that UM101 exerts its biological effects predominantly by modifying p38α: 1) DSF and STD-NMR show p38α-specific binding of UM101; 2) p38α binding of UM101 was abrogated by mutating four of 10 target pocket amino acids; 3) UM60 and 101 both bind p38α and exert effects on endothelial function similar to those of SB203580; 4) UM101 partially blocked phosphorylation of the p38 substrates MK2 and STAT-1 in TNF-α–stimulated HeLa cells; and 5) UM101 inhibited the expression of about half the genes inhibited by SB203580. UM101 may be more effective than SB203580 in stabilizing the endothelial barrier because of its selective sparing of potential counter-regulatory genes, such as GM-CSF (62), MSK1/2-dependent anti-inflammatory genes (22, 23), and p38β-dependent prosurvival genes (20, 21). Although the apparent affinity of UM101 for p38α is much greater than for p38β, additional work is required to definitively address whether it is functionally specific for the p38α isoform.
In summary, we have shown that a CADD strategy that targets the ED substrate–docking site in p38α efficiently identifies low m.w. compounds with favorable biological effects relevant to inflammation and organ injury. We have identified a novel lead compound, UM101, which is at least as effective as the prototypical p38 catalytic inhibitor, SB203580, in stabilizing the endothelial barrier to macromolecules and neutrophils in vitro and in vivo. These results demonstrate the feasibility of our strategy to efficiently identify new therapeutic agents that favorably modify rather than ablate pathogenic signaling pathways by targeting MAPK substrate–binding sites, and show that such agents express favorable biological effect profiles and therapeutic indexes.
Supplementary Material
Acknowledgments
This work was supported by National Heart, Lung, and Blood Institute Grant R01HL69057 (to J.D.H. and P.S.) and U.S. Department of Veterans Affairs Grant BX002143 (to J.D.H.).
We thank Sean Daugherty for help with the RNASeq data analysis.
Abbreviations used in this article
- ALI
acute lung injury
- ATF
activating transcription factor
- CADD
computer-aided drug design
- CD
common docking
- CRP
C-reactive protein
- DSF
differential scanning fluorimetry
- DUSP2
dual-specificity protein phosphatase-2
- ED
glutamate–aspartate
- HMVECL
human lung microvascular endothelial cell
- i.t
intratracheal
- MANOVA
multivariate ANOVA
- MK2
MAPK-activated protein kinase-2
- MSK
mitogen- and stress-activated kinase
- RNASeq
RNA sequencing
- STD-NMR
saturation transfer difference–nuclear magnetic resonance
- TEM
transendothelial migration
- UM60
N2,N7-di(2-hydroxyethyl)-9-oxo-9H-2,7-fluorenedisulfonamide
- UM101
4-chloro-N-(4-[(1,1-dioxo-1λ~6~,4-thiazinan-4-yl)methyl]phenyl)benzamide
Footnotes
The sequences presented in this article have been deposited in the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93330) under accession number GSE93330.
The online version of this article contains supplemental material.
Disclosures
A.D.M. is cofounder and Chief Scientific Officer of SilcsBio LLC. The other authors have no financial conflicts of interest.
References
- 1.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 2.Thalhamer T, McGrath MA, Harnett MM. MAPKs and their relevance to arthritis and inflammation. Rheumatology (Oxford) 2008;47:409–414. doi: 10.1093/rheumatology/kem297. [DOI] [PubMed] [Google Scholar]
- 3.Fisk M, Gajendragadkar PR, Mäki-Petäjä KM, Wilkinson IB, Cheriyan J. Therapeutic potential of p38 MAP kinase inhibition in the management of cardiovascular disease. Am J Cardiovasc Drugs. 2014;14:155–165. doi: 10.1007/s40256-014-0063-6. [DOI] [PubMed] [Google Scholar]
- 4.Krementsov DN, Thornton TM, Teuscher C, Rincon M. The emerging role of p38 mitogen-activated protein kinase in multiple sclerosis and its models. Mol Cell Biol. 2013;33:3728–3734. doi: 10.1128/MCB.00688-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng YJ, Li YY. The role of p38 mitogen-activated protein kinase in the pathogenesis of inflammatory bowel disease. J Dig Dis. 2011;12:327–332. doi: 10.1111/j.1751-2980.2011.00525.x. [DOI] [PubMed] [Google Scholar]
- 6.Chung KF. p38 mitogen-activated protein kinase pathways in asthma and COPD. Chest. 2011;139:1470–1479. doi: 10.1378/chest.10-1914. [DOI] [PubMed] [Google Scholar]
- 7.Damarla M, Hasan E, Boueiz A, Le A, Pae HH, Montouchet C, Kolb T, Simms T, Myers A, Kayyali US, et al. Mitogen activated protein kinase activated protein kinase 2 regulates actin polymerization and vascular leak in ventilator associated lung injury. PLoS One. 2009;4:e4600. doi: 10.1371/journal.pone.0004600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah NG, Tulapurkar ME, Damarla M, Singh IS, Goldblum SE, Shapiro P, Hasday JD. Febrile-range hyperthermia augments reversible TNF-α-induced hyperpermeability in human microvascular lung endothelial cells. Int J Hyperthermia. 2012;28:627–635. doi: 10.3109/02656736.2012.690547. [DOI] [PubMed] [Google Scholar]
- 9.Tulapurkar ME, Almutairy EA, Shah NG, He JR, Puche AC, Shapiro P, Singh IS, Hasday JD. Febrile-range hyperthermia modifies endothelial and neutrophilic functions to promote extravasation. Am J Respir Cell Mol Biol. 2012;46:807–814. doi: 10.1165/rcmb.2011-0378OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damjanov N, Kauffman RS, Spencer-Green GT. Efficacy, pharmacodynamics, and safety of VX-702, a novel p38 MAPK inhibitor, in rheumatoid arthritis: results of two randomized, double-blind, placebo-controlled clinical studies. Arthritis Rheum. 2009;60:1232–1241. doi: 10.1002/art.24485. [DOI] [PubMed] [Google Scholar]
- 11.Watz H, Barnacle H, Hartley BF, Chan R. Efficacy and safety of the p38 MAPK inhibitor losmapimod for patients with chronic obstructive pulmonary disease: a randomised, double-blind, placebo-controlled trial. Lancet Respir Med. 2014;2:63–72. doi: 10.1016/S2213-2600(13)70200-5. [DOI] [PubMed] [Google Scholar]
- 12.MacNee W, Allan RJ, Jones I, De Salvo MC, Tan LF. Efficacy and safety of the oral p38 inhibitor PH-797804 in chronic obstructive pulmonary disease: a randomised clinical trial. Thorax. 2013;68:738–745. doi: 10.1136/thoraxjnl-2012-202744. [DOI] [PubMed] [Google Scholar]
- 13.Schreiber S, Feagan B, D’Haens G, Colombel JF, Geboes K, Yurcov M, Isakov V, Golovenko O, Bernstein CN, Ludwig D, et al. BIRB 796 Study Group. Oral p38 mitogen-activated protein kinase inhibition with BIRB 796 for active Crohn’s disease: a randomized, double-blind, placebo-controlled trial. Clin Gastroenterol Hepatol. 2006;4:325–334. doi: 10.1016/j.cgh.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 14.Pargellis C, Tong L, Churchill L, Cirillo PF, Gilmore T, Graham AG, Grob PM, Hickey ER, Moss N, Pav S, Regan J. Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat Struct Biol. 2002;9:268–272. doi: 10.1038/nsb770. [DOI] [PubMed] [Google Scholar]
- 15.Davidson W, Frego L, Peet GW, Kroe RR, Labadia ME, Lukas SM, Snow RJ, Jakes S, Grygon CA, Pargellis C, Werneburg BG. Discovery and characterization of a substrate selective p38alpha inhibitor. Biochemistry. 2004;43:11658–11671. doi: 10.1021/bi0495073. [DOI] [PubMed] [Google Scholar]
- 16.Hendriks BS, Seidl KM, Chabot JR. Two additive mechanisms impair the differentiation of ‘substrate-selective’ p38 inhibitors from classical p38 inhibitors in vitro. BMC Syst Biol. 2010;4:23. doi: 10.1186/1752-0509-4-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marber MS, Molkentin JD, Force T. Developing small molecules to inhibit kinases unkind to the heart: p38 MAPK as a case in point. Drug Discov Today Dis Mech. 2010;7:e123–e127. doi: 10.1016/j.ddmec.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beardmore VA, Hinton HJ, Eftychi C, Apostolaki M, Armaka M, Darragh J, McIlrath J, Carr JM, Armit LJ, Clacher C, et al. Generation and characterization of p38beta (MAPK11) gene-targeted mice. Mol Cell Biol. 2005;25:10454–10464. doi: 10.1128/MCB.25.23.10454-10464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Keefe SJ, Mudgett JS, Cupo S, Parsons JN, Chartrain NA, Fitzgerald C, Chen SL, Lowitz K, Rasa C, Visco D, et al. Chemical genetics define the roles of p38alpha and p38beta in acute and chronic inflammation. J Biol Chem. 2007;282:34663–34671. doi: 10.1074/jbc.M704236200. [DOI] [PubMed] [Google Scholar]
- 20.Ferrari G, Terushkin V, Wolff MJ, Zhang X, Valacca C, Poggio P, Pintucci G, Mignatti P. TGF-β1 induces endothelial cell apoptosis by shifting VEGF activation of p38(MAPK) from the prosurvival p38β to proapoptotic p38α. Mol Cancer Res. 2012;10:605–614. doi: 10.1158/1541-7786.MCR-11-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu H, Yanamandala M, Lee TC, Kim JK. Mitochondrial p38β and manganese superoxide dismutase interaction mediated by estrogen in cardiomyocytes. PLoS One. 2014;9:e85272. doi: 10.1371/journal.pone.0085272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ananieva O, Darragh J, Johansen C, Carr JM, McIlrath J, Park JM, Wingate A, Monk CE, Toth R, Santos SG, et al. The kinases MSK1 and MSK2 act as negative regulators of Toll-like receptor signaling. Nat Immunol. 2008;9:1028–1036. doi: 10.1038/ni.1644. [DOI] [PubMed] [Google Scholar]
- 23.Kim C, Sano Y, Todorova K, Carlson BA, Arpa L, Celada A, Lawrence T, Otsu K, Brissette JL, Arthur JS, Park JM. The kinase p38 alpha serves cell type-specific inflammatory functions in skin injury and coordinates pro- and anti-inflammatory gene expression. Nat Immunol. 2008;9:1019–1027. doi: 10.1038/ni.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheriyan J, Webb AJ, Sarov-Blat L, Elkhawad M, Wallace SM, Mäki-Petäjä KM, Collier DJ, Morgan J, Fang Z, Willette RN, et al. Inhibition of p38 mitogen-activated protein kinase improves nitric oxide-mediated vasodilatation and reduces inflammation in hypercholesterolemia. Circulation. 2011;123:515–523. doi: 10.1161/CIRCULATIONAHA.110.971986. [DOI] [PubMed] [Google Scholar]
- 25.Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 26.Tanoue T, Maeda R, Adachi M, Nishida E. Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J. 2001;20:466–479. doi: 10.1093/emboj/20.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzarum N, Komornik N, Ben Chetrit D, Engelberg D, Livnah O. DEF pocket in p38α facilitates substrate selectivity and mediates autophosphorylation. J Biol Chem. 2013;288:19537–19547. doi: 10.1074/jbc.M113.464511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ter Haar E, Prabhakar P, Liu X, Lepre C. Crystal structure of the p38 alpha-MAPKAP kinase 2 heterodimer. [Published erratum appears in 2007 J. Biol. Chem. 282: 14684.] J Biol Chem. 2007;282:9733–9739. doi: 10.1074/jbc.M611165200. [DOI] [PubMed] [Google Scholar]
- 29.Raingeaud J, Gupta S, Rogers JS, Dickens M, Han J, Ulevitch RJ, Davis RJ. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 30.Hancock CN, Macias A, Lee EK, Yu SY, Mackerell AD, Jr, Shapiro P. Identification of novel extracellular signal-regulated kinase docking domain inhibitors. J Med Chem. 2005;48:4586–4595. doi: 10.1021/jm0501174. [DOI] [PubMed] [Google Scholar]
- 31.Best RB, Zhu X, Shim J, Lopes PE, Mittal J, Feig M, Mackerell AD., Jr Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J Chem Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD., Jr CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foster TJ, MacKerell AD, Jr, Guvench O. Balancing target flexibility and target denaturation in computational fragment-based inhibitor discovery. J Comput Chem. 2012;33:1880–1891. doi: 10.1002/jcc.23026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karpen ME, Tobias DJ, Brooks CL., III Statistical clustering techniques for the analysis of long molecular dynamics trajectories: analysis of 2.2-ns trajectories of YPGDV. Biochemistry. 1993;32:412–420. doi: 10.1021/bi00053a005. [DOI] [PubMed] [Google Scholar]
- 36.Zhong S, MacKerell AD., Jr Binding response: a descriptor for selecting ligand binding site on protein surfaces. J Chem Inf Model. 2007;47:2303–2315. doi: 10.1021/ci700149k. [DOI] [PubMed] [Google Scholar]
- 37.DesJarlais RL, Sheridan RP, Dixon JS, Kuntz ID, Venkataraghavan R. Docking flexible ligands to macromolecular receptors by molecular shape. J Med Chem. 1986;29:2149–2153. doi: 10.1021/jm00161a004. [DOI] [PubMed] [Google Scholar]
- 38.Kuntz ID, Blaney JM, Oatley SJ, Langridge R, Ferrin TE. A geometric approach to macromolecule-ligand interactions. J Mol Biol. 1982;161:269–288. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- 39.Makino S, I, Kuntz D. Automated flexible ligand docking method and its application for database search. J Comput Chem. 1997;18:1812–1825. [Google Scholar]
- 40.Pan Y, Huang N, Cho S, MacKerell AD., Jr Consideration of molecular weight during compound selection in virtual target-based database screening. J Chem Inf Comput Sci. 2003;43:267–272. doi: 10.1021/ci020055f. [DOI] [PubMed] [Google Scholar]
- 41.Butina D. Unsupervised data base clustering on daylight’s fingerprint and tanimoto similarity: a fast and automated way to cluster small and large data sets. J Chem Inf Comput Sci. 1999;39:747–750. [Google Scholar]
- 42.Godden JW, Stahura FL, Bajorath J. Anatomy of fingerprint search calculations on structurally diverse sets of active compounds. J Chem Inf Model. 2005;45:1812–1819. doi: 10.1021/ci050276w. [DOI] [PubMed] [Google Scholar]
- 43.Oashi T, Ringer AL, Raman EP, Mackerell AD. Automated selection of compounds with physicochemical properties to maximize bio-availability and druglikeness. J Chem Inf Model. 2011;51:148–158. doi: 10.1021/ci100359a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lipinski CA. Drug-like properties and the causes of poor solubility and poor permeability. J Pharmacol Toxicol Methods. 2000;44:235–249. doi: 10.1016/s1056-8719(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 45.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 46.Gong P, Angelini DJ, Yang S, Xia G, Cross AS, Mann D, Bannerman DD, Vogel SN, Goldblum SE. TLR4 signaling is coupled to SRC family kinase activation, tyrosine phosphorylation of zonula adherens proteins, and opening of the paracellular pathway in human lung microvascular endothelia. J Biol Chem. 2008;283:13437–13449. doi: 10.1074/jbc.M707986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu A, Gong P, Hyun SW, Wang KZ, Cates EA, Perkins D, Bannerman DD, Puché AC, Toshchakov VY, Fang S, et al. TRAF6 protein couples Toll-like receptor 4 signaling to Src family kinase activation and opening of paracellular pathway in human lung microvascular endothelia. J Biol Chem. 2012;287:16132–16145. doi: 10.1074/jbc.M111.310102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hasday JD, Bannerman D, Sakarya S, Cross AS, Singh IS, Howard D, Drysdale BE, Goldblum SE. Exposure to febrile temperature modifies endothelial cell response to tumor necrosis factor-alpha. J Appl Physiol. 2001;90:90–98. doi: 10.1152/jappl.2001.90.1.90. [DOI] [PubMed] [Google Scholar]
- 49.Sakarya S, Rifat S, Zhou J, Bannerman DD, Stamatos NM, Cross AS, Goldblum SE. Mobilization of neutrophil sialidase activity desialylates the pulmonary vascular endothelial surface and increases resting neutrophil adhesion to and migration across the endothelium. Glycobiology. 2004;14:481–494. doi: 10.1093/glycob/cwh065. [DOI] [PubMed] [Google Scholar]
- 50.Rice P, Martin E, He JR, Frank M, DeTolla L, Hester L, O’Neill T, Manka C, Benjamin I, Nagarsekar A, et al. Febrile-range hyperthermia augments neutrophil accumulation and enhances lung injury in experimental gram-negative bacterial pneumonia. J Immunol. 2005;174:3676–3685. doi: 10.4049/jimmunol.174.6.3676. [DOI] [PubMed] [Google Scholar]
- 51.Badger AM, Bradbeer JN, Votta B, Lee JC, Adams JL, Griswold DE. Pharmacological profile of SB 203580, a selective inhibitor of cytokine suppressive binding protein/p38 kinase, in animal models of arthritis, bone resorption, endotoxin shock and immune function. J Pharmacol Exp Ther. 1996;279:1453–1461. [PubMed] [Google Scholar]
- 52.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 53.Gupta A, Cooper ZA, Tulapurkar ME, Potla R, Maity T, Hasday JD, Singh IS. Toll-like receptor agonists and febrile range hyperthermia synergize to induce heat shock protein 70 expression and extracellular release. J Biol Chem. 2013;288:2756–2766. doi: 10.1074/jbc.M112.427336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang MZ, Tsukahara H, Hayakawa K, Todoroki Y, Tamura S, Ohshima Y, Hiraoka M, Mayumi M. Effects of antioxidants and NO on TNF-alpha-induced adhesion molecule expression in human pulmonary microvascular endothelial cells. Respir Med. 2005;99:580–591. doi: 10.1016/j.rmed.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 55.Viemann D, Goebeler M, Schmid S, Nordhues U, Klimmek K, Sorg C, Roth J. TNF induces distinct gene expression programs in microvascular and macrovascular human endothelial cells. J Leukoc Biol. 2006;80:174–185. doi: 10.1189/jlb.0905530. [DOI] [PubMed] [Google Scholar]
- 56.Vivoli M, Novak HR, Littlechild JA, Harmer NJ. Determination of protein-ligand interactions using differential scanning fluorimetry. J Vis Exp. 2014;(91):51809. doi: 10.3791/51809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burkhard K, Smith S, Deshmukh R, MacKerell AD, Jr, Shapiro P. Development of extracellular signal-regulated kinase inhibitors. Curr Top Med Chem. 2009;9:678–689. doi: 10.2174/156802609789044416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Godl K, Wissing J, Kurtenbach A, Habenberger P, Blencke S, Gutbrod H, Salassidis K, Stein-Gerlach M, Missio A, Cotten M, Daub H. An efficient proteomics method to identify the cellular targets of protein kinase inhibitors. Proc Natl Acad Sci USA. 2003;100:15434–15439. doi: 10.1073/pnas.2535024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mayer M, Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J Am Chem Soc. 2001;123:6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 60.Cuadrado A, Nebreda AR. Mechanisms and functions of p38 MAPK signalling. Biochem J. 2010;429:403–417. doi: 10.1042/BJ20100323. [DOI] [PubMed] [Google Scholar]
- 61.Hu J, Rho HS, Newman RH, Hwang W, Neiswinger J, Zhu H, Zhang J, Qian J. Global analysis of phosphorylation networks in humans. Biochim Biophys Acta. 2014;1844(Pt B):224–231. doi: 10.1016/j.bbapap.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao J, Chen L, Shu B, Tang J, Zhang L, Xie J, Liu X, Xu Y, Qi S. Granulocyte/macrophage colony-stimulating factor attenuates endothelial hyperpermeability after thermal injury. Am J Transl Res. 2015;7:474–488. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.