Abstract
Horizontal gene transfer mediated by broad-host-range plasmids is an important mechanism of antibiotic resistance spread. While not all bacteria maintain plasmids equally well, plasmid persistence can improve over time, yet no general evolutionary mechanisms have emerged. Our goal was to identify these mechanisms, and to assess if adaptation to one plasmid affects the permissiveness to others. We experimentally evolved Pseudomonas sp. H2 containing multi-drug resistance plasmid RP4, determined plasmid persistence and cost using a joint experimental-modeling approach, resequenced evolved clones, and reconstructed key mutations. Plasmid persistence improved in fewer than 600 generations because the fitness cost turned into a benefit. Improved retention of naive plasmids indicated that the host evolved towards increased plasmid permissiveness. Key chromosomal mutations affected two accessory helicases and the RNA polymerase β-subunit. Our and other findings suggest that poor plasmid persistence can be caused by a high cost involving helicase-plasmid interactions that can be rapidly ameliorated.
Keywords: Multidrug resistance, Pseudomonas, horizontal gene transfer, experimental evolution, adaptation
Analyses of the genome sequences of prokaryotes have clearly revealed that horizontal gene transfer (HGT) plays an important role in their evolution (1). It mediates wholesale acquisition of genes encoding traits that can be advantageous. A prime example of great concern to human health is the plasmid mediated spread of antibiotic resistance and virulence genes, such as the recent worldwide dissemination of resistance to colistin, an antibiotic of last resort (2). Plasmids are very common in bacteria (3), and those with a broad host-range play a critical role in gene spread among phylogenetically distinct bacteria (4). The same plasmid types found before the antibiotic era now often contain multiple antibiotic resistance genes (5), and are present in both clinical and environmental isolates (6).
Not all bacteria maintain plasmids equally well, with great differences even between members of the same genus (7). To curb the spread of plasmid-mediated antibiotic resistance and virulence it is vital that we understand the basis of these differences, and how so-called ‘plasmid permissiveness’ can evolve over time. Persistence of a plasmid in a bacterial population hinges upon correctly balancing efficient replication, accurate segregation, plasmid cost, conjugative transfer, post-segregational killing and selection on beneficial accessory traits (8, 9). While these processes are entirely or partially encoded by the plasmid, they also require host housekeeping functions. Plasmids and chromosomes thus interact in ways we do not yet understand (10, 11, 12), resulting in host-dependent plasmid persistence (7).
Poor bacteria-plasmid relationships can improve due to evolution of the plasmid (13, 14), the host (10, 15, 16) or both (17, 18). Plasmid-encoded mutations tend to affect replication, cost, or inheritance (14, 18, 19, 20) while those in the host often affect global gene expression and plasmid cost (10, 11). However, no general mechanism for plasmid-host stabilization has yet emerged. It is also unknown if a host that adapts to one plasmid can be pre-adapted to others, thereby facilitating persistence and spread of a range of multidrug resistance (MDR) plasmids.
To determine evolutionary patterns of plasmid stabilization in bacteria, several combinations of plasmids and hosts should be experimentally evolved and analyzed. Here we report on the experimental evolution of an environmental strain, Pseudomonas sp. nov. H2, carrying plasmid RP4. Strain H2 poorly maintains this and other plasmids of the incompatibility (Inc) group IncP-1, in contrast to close relatives such as P. putida (7, 21). This makes it an attractive host to determine the molecular mechanisms of bacteria-plasmid evolution. RP4 is a highly promiscuous, multidrug resistance prototype IncP-1α plasmid that was found in P. aeruginosa isolated from a burn wound (22, 23). We show that its persistence evolved rapidly and resulted in plasmid addiction, due to chromosomal mutations in genes encoding accessory helicases and the β-subunit of the RNA polymerase, rpoB. More importantly, we demonstrate for the first time that a host adapted to one plasmid, can become generally more permissive to plasmid carriage. Understanding how these bacterial genes affect MDR plasmid retention could aide alternative drug therapies needed to combat the spread of antibiotic resistance (24).
RESULTS
An unstable plasmid-host pair stabilized due to host adaptation
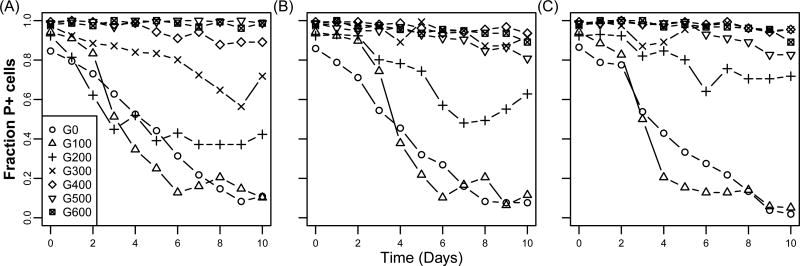
To experimentally evolve a bacterium-plasmid association, three replicate populations of Pseudomonas sp. nov. H2 containing the promiscuous MDR plasmid RP4 were grown for ca. 600 generations (gen.) by serial batch transfer in Tryptic Soy Broth (TSB) and in the presence of the host- and plasmid-selective antibiotics, rifampicin (Rif) and tetracycline (Tet), respectively. At 100 gen. intervals plasmid persistence in the absence of Tet was assessed and shown to improve rapidly in all three replicate populations (Fig. 1 and Supplemental Fig. 1).
Figure 1. Plasmid persistence profiles measured at 100 gen. intervals during the 600 gen. evolution experiment for replicate populations A to C (panels A to C, respectively).
Each data point represents the mean fraction of plasmid-containing (P+) cells (n = 3).
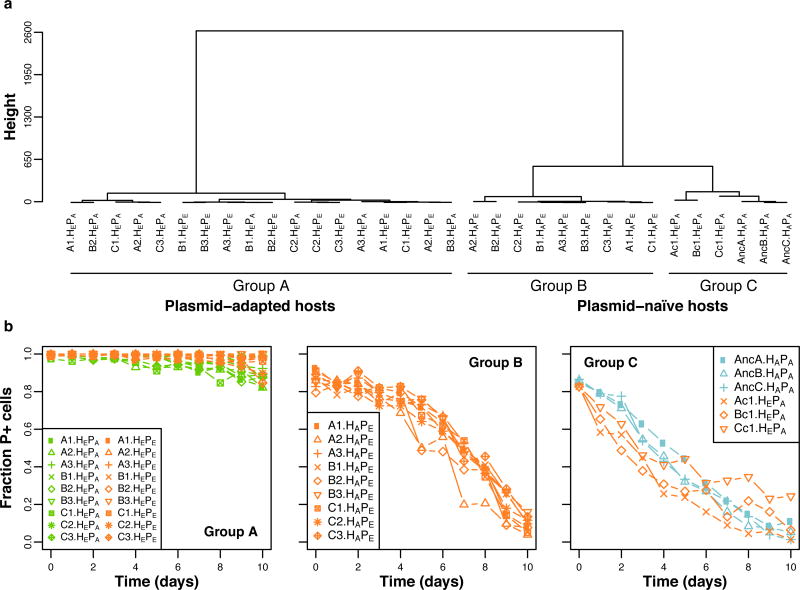
To establish if plasmid persistence increased due to evolution of the plasmid, the host, or both, it was measured for three clones (numbered 1–3) from each replicate population, and for all their possible bacteria-plasmid permutations: the ancestral plasmid (PA) and plasmids from nine evolved clones (PE) in both ancestral (HA) and evolved hosts (HE). Since host adaptation to its environment may affect plasmid persistence, we also determined the persistence of PA in one clone from each of three evolved plasmid-free populations (populations Ac-Cc, clones Ac1-Cc1, hereafter named ‘control’ populations or clones). The ‘plasmid persistence profiles’, representing the plasmid persistence dynamics over time, were analyzed and compared using a recently described segregation and selection (SS) plasmid population dynamics model, combined with linkage cluster analysis (18). The difference between two plasmid persistence profiles is reflected in the magnitude of the Bayesian Information Criteria (BIC) generated by the pairwise comparisons, which permits the complex time series data to be clustered objectively. The plasmid persistence profiles clustered into three distinct groups, A–C (Fig. 2A). The group that contained all the RP4-adapted HE hosts with either PA or PE (Group A) was clearly separated from the group containing the ancestral hosts with evolved plasmids, HAPE (Group B) (ΔBIC: −13,134 the more negative ΔBIC, the larger the difference between groups). Fig. 2B and Supplemental Fig. 1 show this clustering was due to improved persistence of both PE and PA in HE (Group A). The mean predicted time for the plasmid-bearing fraction to reach 1% (T1%) was 34 days for Group A and only 18 days for Group B (Supplemental Table 1). This improved persistence was not due to adaptation to the environment, since the control clones clustered separately (Fig. 2A, Group C) due to very poor retention of PA, much like HAPA clones (Fig. 2B, Supplemental Fig. 1; ΔBIC −18,439 and −1,343, respectively). These findings indicate that the populations evolved with RP4 improved their ability to retain this plasmid due to plasmid-adaptive chromosomal mutations.
Figure 2. The mutations responsible for increased plasmid persistence are located in the host chromosome.
(a) Based on complete linkage distance analysis of the plasmid persistence profiles, permutations of all the RP4-adapted hosts, HEPA or HEPE, clustered together (Group A), whereas all permutations of evolved plasmids in ancestral host (HAPE) clustered as Group B, and all ancestral or control hosts with the ancestral plasmids in Group C (HAPA and HEPA, respectively). (b) Plasmid persistence profiles for the individual ancestral, evolved and control clones and their different permutations, grouped (A to C) according to their clustering. Each data point represents the mean fraction of plasmid-containing (P+) cells (n = 3). The different host-plasmid permutations are: Ancestral host-ancestral plasmid (HAPA); evolved host-evolved plasmid (HEPE); ancestral host-evolved plasmid (HAPE) and evolved host-ancestral plasmid (HEPA). See also Fig. S1 and Table S1 for the modeled predictions.
Plasmid stabilization was due to plasmid addiction
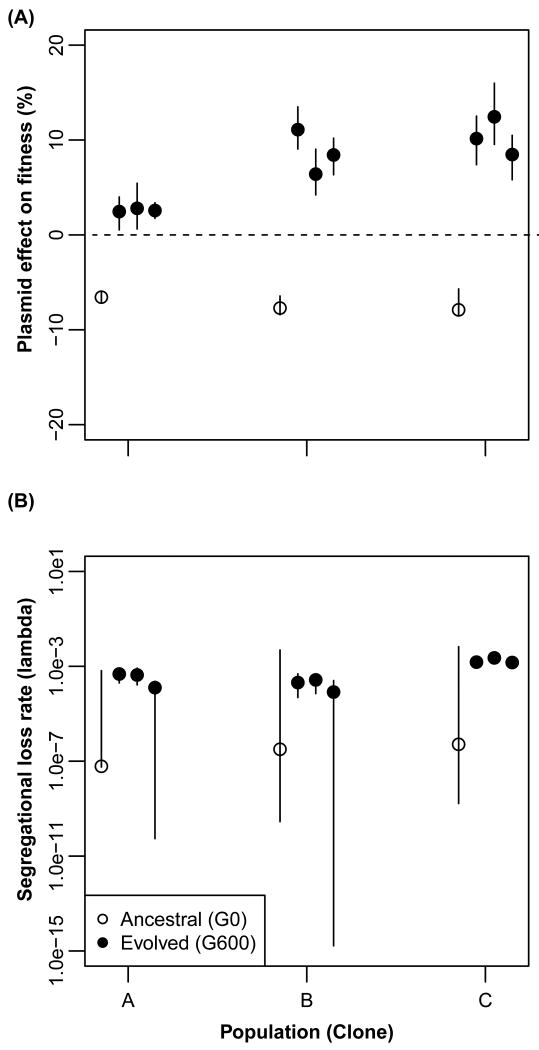
To test if the improved persistence of RP4 was due to a change in conjugation rate (γ), segregation rate (λ), or plasmid cost (σ), we estimated model-based parameters and performed conjugation and competition experiments. First, to determine the best fitting model we fit the plasmid persistence profiles to both the segregation and selection (SS) and the horizontal transfer (HT) models described previously (7, 25). Since the SS model provided a better fit, conjugation was considered not important for persistence of RP4 (Supplemental Table 2). This was supported by the low empirically measured transfer rate in liquid cultures (Supplemental Fig. 2). Estimates of λ and σ were obtained by jointly analyzing data from plasmid persistence and competition experiments. For unstable bacteria-plasmid pairs the traditional estimations of plasmid cost through competition assays are confounded by the high plasmid loss rate during the assay. Moreover, that loss rate is often not reported. For highly persistent bacteria-plasmid associations, the plasmid persistence data do not contain enough information about the dynamics as plasmid-free hosts are rare, and they can be confounded by evolutionary changes during the course of the persistence assay. Our joint analysis provides the benefit of more accurate estimation of both parameters while taking into account the growth dynamics in serial batch culture (See Methods and Supplemental Methods). The most likely estimates (MLEs) for λ ranged from 6.16 × 10−8 to 2.29 × 10−3, thus negligibly low. The plasmid cost σ was however drastically different between ancestral and evolved clones. While plasmid carriage imposed a fitness cost σ of 6.5 – 7.9 % in the HAPA clones, it conferred a clear fitness benefit of 2.4 – 8.5 % in all HEPE clones (Fig. 3; Supplemental Table 3). Thus, in line with the first study on so-called addiction of a small non-conjugative plasmid by Lenski et al. (26), these results show that persistence of a self-transmissible MDR plasmid can improve solely due to chromosomal mutations that turned a plasmid cost into a benefit, resulting in plasmid addiction. So far as we know, no other study has shown the cost of a self-transmissible MDR plasmid to evolve to a benefit via chromosomal mutations only. It provides another explanation for how antibiotic resistance can spread and persist in bacterial populations.
Figure 3. Evolution of plasmid cost into benefit rather than a change in segregational loss frequency facilitated improved plasmid persistence.
The effect of the plasmid on host fitness, i.e. plasmid cost (a), and segregational loss frequency (b) were jointly estimated using conjugated data from plasmid persistence assays and competition experiments using the SS model for the ancestral host-plasmid pairs HAPA and their respective evolved HEPE clones from replicate populations A to C. Evolved clones in each population are ordered sequentially from 1 to 3. For each conjugated dataset n = 3. The vertical lines represent deviations in the model output. The large deviations indicate that a wide range of maximum likelihood estimates are possible for that parameter. The asymmetry of the lines is due to the date being plotted on a log scale.
Adaptation to one plasmid improved general plasmid permissiveness
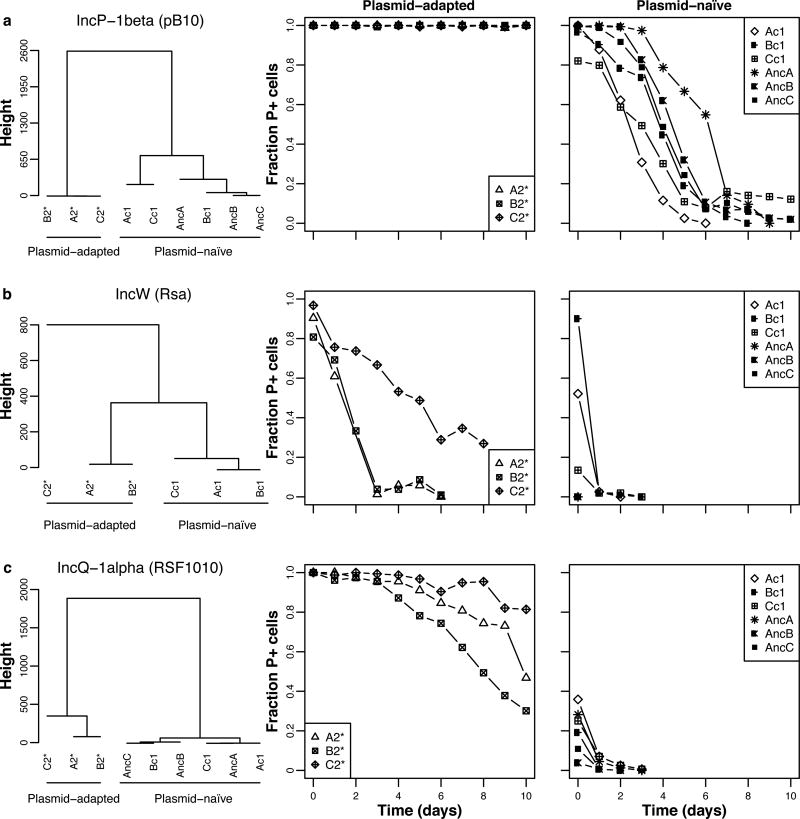
To determine whether host adaptation to RP4 was plasmid-specific, or allowed for generally improved plasmid permissiveness, we tested the persistence of three naive plasmids in three plasmid-free segregants of RP4-adapted hosts (A2*, B2*, C2*), their corresponding ancestral hosts, and three control hosts (Ac1-Cc1). These plasmids were a closely related IncP-1β plasmid, and two plasmids that belong to different incompatibility groups: an IncW plasmid with a distantly related replication system, and an IncQ plasmid with an unrelated replication machinery. Strikingly, all three plasmids showed higher persistence in the RP4-adapted clones than in the ancestral and control strains (Fig. 4 and Supplemental Fig. 3; Supplemental Table 4). Thus by adapting to one particular MDR plasmid. Pseudomonas sp. nov. H2 became more permissive towards distantly related antibiotic resistance plasmids. Bacterial communities and populations are known to be diverse in their plasmid permissiveness, but the underlying genetics are not understood (27, 28). These findings also suggest that the plasmid-addiction observed for RP4 was not due to the plasmid’s Tet resistance (TetR) gene, as previously shown (26), since the TetR gene is not present on two of the three other plasmids. Addiction of our Pseudomonas strain to RP4 must thus be due to other genes shared by these three plasmids.
Figure 4. Pseudomonas sp. nov. H2 evolved to be more permissive towards both related and unrelated plasmids.
The plasmid persistence profiles in three clones each of RP4-adapted, ancestral and control hosts are shown for three naive plasmids: (A) the closely related IncP-1 β plasmid pB10, (B) the distantly related IncW plasmid Rsa, and (C) the unrelated IncQ-1α plasmid RSF1010. They always clustered into two distinct groups: Group A contained the three RP4-adapted hosts and Group B the ancestral and control hosts, naïve to the plasmid. Note that due to the extremely low persistence of plasmids Rsa and RSF1010, the initial fractions of plasmid-containing (P+) ancestral cells were not close to 1, even though the pre-culture contained plasmid-selective antibiotics. This is due to survival of plasmid-free cells in the pre-cultures and additional plasmid loss in the colonies during replica-plating. Moreover, the persistence of Rsa in the ancestral host was so poor that no temporal data could be collected to compare to the other Rsa persistence profiles. Each data point represents the mean fraction of P+ cells (n = 3). See also Fig. S3 and Table S4 for the modeled predictions.
Genetic solutions towards improved plasmid cost and persistence
To identify mutations that could explain the improved persistence of plasmid RP4 we determined the complete genome sequences of all nine evolved clones, three control and three ancestral clones. Fourteen mutated chromosomal loci were identified among the nine evolved clones after excluding mutations also found in the ancestral and control clones (Fig. 5, Supplemental Table 5). No mutations were found in any of the plasmid genomes, thus confirming that their improved persistence was due to host evolution.
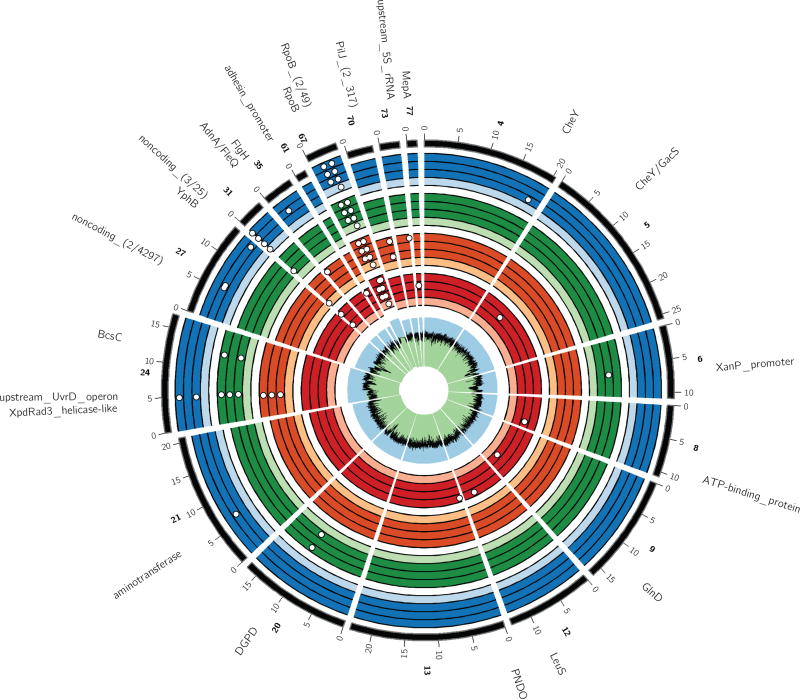
Figure 5. SNPs identified in the chromosome of each sequenced clone, as compared to the Rif sensitive reference strain Pseudomonas sp. nov. H2.
White dots represent SNPs. The black line within the innermost green (low) and blue (high) ring indicates GC-content. Each set of four colored rings represents four clones from the following populations: control (red) and the three plasmid-containing populations A–C (orange, green, blue, respectively). Within these are shown, again from inside to outside: the ancestral (light-colored) and three evolved (dark-colored) clones. The latter correspond to control clones Ac1, Bc1, Cc1, and RP4-adapted clones A1-3, B1-3, and C1-3. The outermost black line represents the reference genome. Only contigs with at least 1 SNP in comparison to the reference genome are displayed. Excluded are three small contigs (84, 86 and 132) possibly representing different regions of the same lapA gene and which contains multiple SNPs present in either the ancestral or various evolved strains. However, due to highly variable sequencing coverage, most SNPs are likely sequencing errors. Numbered ticks indicate nucleotide position (× 1000) along the contig and the contig numbers are indicated in bold. When two or more SNPs cannot be resolved in the figure the numbers in parentheses behind the gene or locus names indicate the number of unique variants followed by the number of nucleotides that separate those variants. The radius of contigs 24 and 67 is enlarged by 105%, and nucleotides 10,700 to 11,000 on contig 67 are zoomed 100-fold to highlight and resolve the helicase and rpoB mutations, respectively. Importantly, there were three rpoB mutations on contig 67: one in all strains including the RifR ancestors, causing Rif resistance, and two in all strains evolved with and without RP4. For details on the mutations and a full description of gene names, see Supplemental Table 5.
Only two mutations were common among multiple clones from one or two plasmid-containing populations. One was a non-synonymous transversion in a gene encoding a Xpd/Rad3-like helicase protein (64% of the residues modeled at >90% accuracy on the Phyre2 protein fold recognition portal; 29). This SNP, hereafter named Xpd/Rad3D672A, was found in three sequenced clones from populations A and two from population C (Supplemental Table 5). Moreover, the mutation was present in half of the sequenced target-specific amplicons obtained from these populations, but below the detection limit in population B, the ancestral- and the control populations (Supplemental Fig. 4). In clones A2 and C3 it was the only mutation after excluding the control SNPs. Finally, based on analysis using SNAP2 (ProteinPredict; 30) the altered residue may have a detrimental effect on protein structure and hence function of the accessory helicase (Supplemental Fig. 5). Together these findings suggest that the mutation was selected by the presence of the plasmid.
The only other common SNP was found in all three clones from population B. It was an A to C transversion 32-bp upstream of an operon encoding an rRNA-binding protein (40% of the residues modeled at >91% accuracy), a DNA repair protein (65% of the residues modeled at 100% accuracy) and an UvrD helicase (94% of the residues modeled at 100% accuracy). This atypical UvrD operon is located 2,007 bp downstream of the Xpd/Rad3-like ORF and both are within a low GC island that was likely acquired by horizontal gene transfer from an unknown source (Supplemental Fig. 6). This SNP was present in 30% of the sequenced amplicons from population B, and below the detection limit in all other populations, again suggesting it was adaptive to the plasmid (Supplemental Fig. 4). Although no consensus promoter elements could be identified in the vicinity of the SNP, we still refer to it as PuvrD_A-32C. Real-Time qPCR transcription analyses on one ancestral and evolved strain with and without plasmid show that the presence of the plasmid downregulated transcription of uvrD in the ancestor but not in the evolved strain (Fig. S9). This suggests that fine-tuning of UvrD levels was important for plasmid cost amelioration. The remaining SNPs were located in CDSs involved in metabolism, chemotaxis and response, or upstream of a gene encoding the 5S rRNA. Since they were never found in more than two clones from one population, we assumed they were not under strong selection.
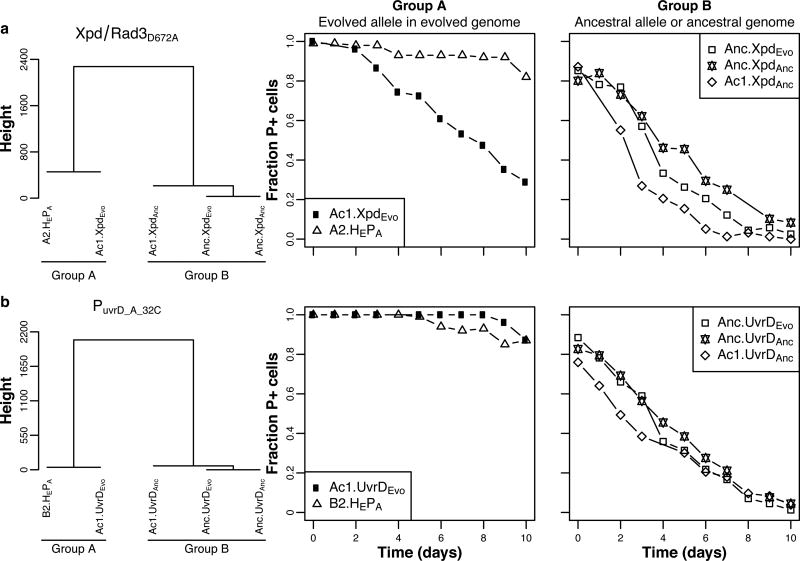
To determine whether the accessory helicase SNPs were required for increased plasmid persistence, we separately reconstructed Xpd/Rad3D672A and PuvrD_A-32C in an ancestral and control host. In the ancestral host neither of the evolved alleles improved persistence of RP4. In contrast, these same two alleles, especially PuvrD_A-32C, improved persistence in control host Ac1, albeit not quite to the same level as the corresponding evolved clones (Fig. 6; Supplemental Fig. 7 and Supplemental Tables 6, 7, 8). Moreover, the alternate plasmids pB10, Rsa and RSF1010 were also much more persistent in control host Ac1 with evolved helicase alleles than in host Ac1 with ancestral allele. These results strongly suggest that epistasis between these accessory helicase mutations and at least one other mutation was responsible for general plasmid stabilization.
Figure 6. At least two mutations were required for full plasmid persistence, one plasmid-adaptive and one environment-adaptive mutation.
Complete linkage cluster analysis (left) of the persistence profiles (shown to the right) of plasmid-adapted clones A2 (A2.HEPA in panel A) and B2 (B2.HEPA in panel B), and control clone Ac1 and ancestral clone Anc, each containing either the evolved or ancestral allele of Xpd/Rad3D672A (denoted as XpdEvo or XpdAnc) (A), and PuvrD_A-32C (UvrDEvo or UvrDanc) (B). See also Supplemental Fig. 7 and Supplemental Tables 6, 7, 8 for the modeled predictions. Each data point represents the mean fraction of plasmid-containing cells (n = 3).
To identify mutations that may interact with the accessory helicase mutations, we screened for CDSs that had mutated in both the plasmid-adapted and plasmid-free control host Ac1. Only one such CDS was found and encodes the β-subunit of RNA polymerase (RpoB). Eleven of the 12 evolved strains contained the same secondary SNP in rpoB, whereas the remaining strain, Cc1, had a different SNP in the same gene (Fig. 5 and Supplemental Table 5). Since the SNP in RpoB also arose in strains that did not carry RP4, they were likely adaptations to the growth environment. Thus, in control clone Ac1::PuvrD_A-32C, which retained the plasmid almost as well as the corresponding RP4-adapted host, the gene that most likely interacted with PuvrD_A-32C was rpoB.
DISCUSSION
The role of helicases in plasmid stabilization
Strikingly, our and at least three other studies that evolved different host-plasmid pairs now suggest that maladapted interactions between plasmids and host-encoded helicases adversely affect plasmid cost and persistence. Moreover, these interactions can often be improved by single mutations, suggesting we are zooming in on a potential general mechanism of bacterial adaptation to plasmids. First, in P. aeruginosa PAO1, loss-of-function mutations in a putative accessory helicase with a UvrD-like helicase C-terminal domain ameliorated the cost of a small non-mobilizable plasmid (16). This initial cost was due to the plasmid’s replication initiation (Rep) protein triggering an SOS response in the ancestral host, and the helicase knockout mutation reduced Rep expression (11). Second, experimental evolution of an IncP-1β mini-replicon in Shewanella oneidensis MR-1 improved plasmid cost and persistence through loss of a helicase (DnaB) binding domain in the plasmid’s Rep protein, reducing the protein’s affinity for DnaB (14). This likely avoided an SOS response that may explain the high cost of the ancestral plasmid (20). Third, when we evolved that same plasmid in another host, it stabilized in two clones due to a SNP in either the dnaB promoter or a uvrD gene (18). Finally, in the present study SNPs affecting two accessory helicases again compensated for the cost of RP4, and improved the persistence of this and three other BHR plasmids.
Helicases are involved in many aspects of DNA and RNA metabolism, such as replication (replicative helicases), and DNA repair, recombination, translocation, transcription, translation, and resolution of replication-transcription conflicts (accessory helicases) (31, 32, 33). Accessory helicases such as Xpd/Rad3 and UvrD, generally have variable C- and N-terminal accessory domains which determine their physiological specificity (34). Interestingly, UvrD and the Xpd/Rad3-like helicase DinG, have been shown to be upregulated as part of the SOS response induced by plasmid entrance and replication in a naïve host (35, 36, 37, 38). In E. coli, the UvrD homologue Rep helicase has been shown to interact with DnaB, acting as a second motor that improves replication fork movement on the chromosome (39). UvrD helicases have also been shown to ‘backtrack’ the RNA polymerase complex to slow down transcription, thus preventing the complex from colliding with the replication fork and causing dsDNA breaks (32, 38).
In our study, host adaptation to plasmid carriage was facilitated by two different mutations; one that likely changed the Xpd/Rad3-like structure, and one that likely affected UvrD abundance. It was interesting to see that our plasmid decreased uvrD transcript levels in the ancestor where it imposed a high cost, but not in the evolved strains, where it had become beneficial. This suggests that higher UvrD levels are needed for plasmid persistence. The mechanisms by which these mutations affected plasmid cost and persistence are currently not understood but the topic of future work. A simple explanation like a change in plasmid copy number can probably be excluded based on very similar plasmid sequence coverage for the ancestral and evolved genomes (data not shown). We postulate three not necessarily mutually exclusive models: the accessory helicases interact with DnaB (i) or the plasmid replication initiation protein TrfA (ii) to modulate plasmid replication efficiency, or (iii) mutations in the helicases ameliorate the fitness cost of plasmid RP4 through their regulatory function.
Whatever the mechanism, further research should confirm that accessory helicases are involved in plasmid persistence across pathogens. The two accessory helicase genes in our Pseudomonas strain were likely acquired by HGT, consistent with previous findings for P. aeruginosa PAO1 (11), where it was proposed they caused genetic conflict with the plasmid. We intend to test whether these helicases hamper or improve persistence of various resident MDR plasmids in other strains, as this could aid the development of strategies aimed at slowing down the spread of antibiotic resistance in bacterial pathogens.
Potential epistasis between helicase and RpoB mutations
Stabilization of plasmid RP4 required not only mutations in loci selected in the presence of the plasmid, but also at least one mutation that seemed adaptive to the growth environment. The gene rpoB, which encodes the β-subunit of RNA polymerase (RNAP), was mutated across all sequenced plasmid-containing and control clones. It is thus the most likely candidate for epistatic interactions with the accessory helicases. The RNAP holoenzyme, consisting of five subunits, αI, αII, β, β′ and ω, together with the σ factor, is responsible for transcription (40). The β-subunit specifically, in addition to DNA binding, is involved in the modulation of transcription through interaction with σ factors (41) and DNA helicases (38). The rpoB mutation in question was secondary after the initial rpoB mutation that resulted in Rif resistance (RifR) in our ancestral strain. In the absence of the helicase mutations, these rpoB mutations did not improve plasmid persistence at all (Fig. 2). Thus, they most likely affect it only through epistatic interaction with the helicase mutations.
We propose two possible mechanisms of epistasis between the rpoB and helicase mutations that are not mutually exclusive. First, based on their location, the SNPs in rpoB likely compensated for the cost imposed by the initial RifR rpoB mutation (Table S5) by ameliorating transcription efficiency (42, 43). It is possible that without this compensatory mutation the helicase mutations were unable to significantly improve plasmid cost and persistence. Second, it is striking that accessory helicases can bind to the RNAP complex, in particular RpoB, to slow down transcription and regulate backtracking (32, 38). Was there a need for mutations in RpoB to modify this physical interaction? Our secondary RpoB mutations are closer to the active site than to the helicase-binding residues, suggesting they may not affect helicase binding. They are also not close to the rpoB mutations that were shown to rescue viability of strains without accessory helicases (34). Future studies are needed to determine the mechanism by which the helicase and possibly rpoB mutations can transform a plasmid cost into a benefit for its bacterial host.
In conclusion, to combat the spread and persistence of plasmid-mediated antibiotic resistance, novel therapeutic approaches are needed that target mechanisms that affect stable retention of MDR plasmids (24). To do so we need to understand which chromosomal gene products stabilize or destabilize MDR plasmids across bacterial species and how. Our study led to at least three important conclusions that may impact the way we tackle MDR plasmid spread: (i) bacteria can adapt to conjugative MDR plasmids by changing plasmid cost into benefit, resulting in greatly improved plasmid persistence; (ii) this can be due to mutations affecting helicases that initially impaired plasmid persistence, a recurring evolutionary pattern that may lead to new antimicrobial therapies; and (iii) bacterial adaptation to one plasmid can lead to generally improved plasmid permissiveness, enabling future retention of MDR plasmids. So far as we know, this is the first time that antibiotic exposure is shown to select for bacterial mutants with increased general permissiveness toward transmissible drug resistance plasmids. These mutations may threaten the efficacy of traditional antibiotic treatments even more than single drug resistance mutations, as adaptation of a pathogen to one plasmid may result in improved retention of other plasmid-encoded antibiotic resistance determinants.
MATERIALS AND METHODS
Bacterial strains, plasmids and media
The bacterial strains and plasmids used in this study are listed in Supplemental Table 9. Unless otherwise stated, the RifR Pseudomonas sp. H2 was cultured in TSB or in Tryptic Soy Agar (TSA) supplemented with Rif (50 µg.ml−1) and, when necessary for selection of plasmid RP4, tetracycline (Tc, 10 µg.ml−1). The various strains of E. coli were cultured in LB broth or on LB agar supplemented with diaminopimelic acid (DAP; 10 µg.ml−1), ampicillin (100 µg.ml−1), kanamycin (50 µg.ml−1) and Tc (10 µg.ml−1) as required. All strains were grown at 30 ℃.
Plasmids RP4, pB10 and pRSA were introduced into H2 by conjugation from E. coli AT1036 while plasmid RSF1010.Km was introduced by mobilization from E. coli S17.1pir+. Briefly, 1 ml of donor and recipient cultures were harvested by centrifugation and resuspended in 100 µl phosphate buffered saline (PBS, pH 7.4); 50 µl of each cell type was then mixed together and spotted onto a TSA surface and incubated at 30 ℃. After 1 h the cell mixture was scraped off the agar surface using a sterile inoculation loop, suspended in 1 ml PBS and a dilution series spread onto the appropriate donor-, recipient- and transconjugant-selective TSA media. The remainder of the donor and recipient cultures were also spread onto transconjugant-selective TSA to verify media selectivity. Evolved RP4 plasmids were introduced into the ancestral H2 host by electroporation using standard methods (44).
Molecular methods
Conventional plasmid isolation and DNA manipulation techniques were used as described in Sambrook and Russell (44). Sanger sequencing was sourced to Elim Biopharm (California) and reaction composition were as per their instruction. Restriction enzymes and high-fidelity Phusion DNA polymerase were obtained from New England Biolabs (Ipswich, MA). Primers used in this study are listed in Supplemental Table 10. For subcloning of PCR products pJET1.2 was used as an intermediate PCR cloning vector (ThermoScientific, Waltham, MA). GeneJET PCR purification, plasmid miniprep, and gel extraction kits (ThermoScientific, Waltham, MA) were routinely used. Ligation products were transformed by electroporation into E. coli EC100Dpir+ (Epicentre, Madison, WI).
Experimental evolution
Triplicate populations of the RifR strain Pseudomonas sp. H2 (RP4) were evolved for ca. 600 generations in serial batch cultures in the presence of Tc to select for the plasmid, and Rif to avoid contamination. In addition, triplicate plasmid-free populations were also evolved in the presence of Rif only. Each population was founded from a single colony that was inoculated into 5 ml TSB. Daily for a total of 60 days, 4.9 µl of culture was transferred to 5ml of fresh media (1024-fold dilution) to yield roughly 10 generations per day. Each of the populations was archived at −70 ℃ on day 0 and every 10 days in 30 % glycerol. After 60 days the populations were diluted and spread onto TSA, and three single colonies representing three unique clones from each population were randomly chosen and archived in glycerol at −70 ℃.
Comparison of plasmid persistence
Plasmid persistence in the evolving plasmid-containing populations was assayed every 100 generations as follows: 4.9 µl of each culture was transferred into 5ml TSB supplemented with Rif and maintained by serial batch transfer for 10 days. The growth conditions were thus the same as in the evolution experiment but without Tc selection for the plasmid. On a daily basis the fraction of TcR cells was determined as described previously (18) by spreading on TSA supplemented with either Rif only or both Rif and Tc to differentiate between plasmid-containing and plasmid-free cells.
Plasmid persistence for each the 9 evolved clones was measured for as described above, but starting from their respective archived glycerol stocks. Additionally, one of the first plasmid-free segregants that were obtained for each clone was archived at −70 ℃, similar to the original plasmid-containing clone. Persistence of the ancestral plasmid as well as other naive plasmids in these evolved plasmid-free segregrants (denoted with an ‘*’) and in clones from the plasmid-free evolved populations was assayed in the same way after the plasmids were introduced into the respective hosts by means of conjugation or mobilization.
Where necessary, the plasmid persistence profiles, defined as the time series obtained from the plasmid persistence assays, were compared and analyzed using the plasmid population dynamics models as described and implemented by us previously (7, 13, 18, 25, 45). The two models, defined as the segregation and selection (SS) or horizontal transfer (HT) models, describe the plasmid dynamics based on Maximum Likelihood Estimates (MLEs) of the segregation rate (λ), conjugation rate (γ), and cost (σ). The SS model employs only λ and σ, whereas the HT model employs all three parameters. As a first step in the analysis, the SS and HT models were compared to find the best fitting model based on the lowest negative log likelihood score for each plasmid persistence profile. Thereafter the different profiles were compared to each other to determine how similar or different they were based on the difference in the Bayesian Information Criteria (ΔBIC) score. In this method, the more negative the ΔBIC score the more different the profiles (see ref. 18 for more detailed information). To visualize the differences in plasmid persistence profiles a matrix of the ΔBIC scores was used to construct a complete linkage cluster dendrogram. To determine which of the resulting clusters or groups showed the highest plasmid persistence, the combined dynamics from each group was used to extrapolate the number of days till only 1% of the population retained the plasmid (T1%). Where necessary T1% was also extrapolated for individual clones based on their individual profiles and MLEs.
Estimation of plasmid cost and segregational loss frequencies
Plasmid cost and segregational loss frequencies were estimated by jointly analyzing data generated from competition and persistence experiments using the SS model. Competition experiments were done by mixing plasmid-carrying strains and their isogenic plasmid-free counterparts at a 1:1 ratio and growing them under conditions resembling the plasmid persistence assays. Specifically, the cultures were inoculated from glycerol stock into 5 ml TSB containing the appropriate antibiotics and grown for 16 h. Thereafter 1.5 ml of each culture was harvested by centrifugation to remove the spent media and antibiotics, and cell pellets were resuspended in 1.5 ml PBS and their optical density measured at 600 nm (OD600). The appropriate volumes that represented a similar cell count for each competitor were then mixed together in a total volume of 200 µl. Of each mixture, 4.9 µl was inoculated in triplicate into 5 ml TSB supplemented with Rif and grown for 24 h, after which the cultures were again diluted 1024-fold into 5 ml of fresh medium and grown for another 24 h. Dilution series of the original mixed culture and after each 24 h time period were spread onto TSA supplemented with Rif. From here, the fractions of plasmid-containing and -free cells within each population were determined by scoring the absence or presence of growth for 52 colonies replicated onto TSA supplemented with Rif and with or without Tc. To be able to leverage the information from the competition assay together with the information from the persistence assays to better estimate the plasmid cost, the joint likelihood function coming from these two experiments was maximized, using our R package “StabilityToolkit”. In an online appendix that is publicly available, we provide extensive instructions to download and install this package, and perform these calculations (see code availability).
We note that although our SS and HT models are useful to jointly estimate cost, segregation and conjugation, they scale the cost parameter σ slightly differently than the traditional cost s (10,11, 13–17). Despite the different scaling, in the Supplemental Methods we show that both formulations concur in what constitutes a plasmid cost and a plasmid benefit (positive and negative values of the cost parameters). A one to one transformation between σ and s is given by: (see Supplementary file).
Conjugation assays to determine the conjugation rate constant
Conjugation between plasmid-containing donors (D) and plasmid-free recipients (R) was measured under growth conditions similar to those used for measuring plasmid persistence, with one difference. To distinguish D and R, the plasmid-free recipient strain was isogenic to the plasmid-free ancestor H2 but resistant to nalidixic acid rather than Rif. D and R cultures were grown for 16 h, their densities adjusted based on their OD600 and 4.9 µl of each culture were inoculated together into tubes containing 5 ml TSB. To quantify the total initial (N0) cell densities, a subsample of the mixture was immediately diluted and spread onto D-, R- and Transconjugant (T)-selective TSA and incubated at 30℃ for 24 h. The cultures were incubated at 30℃ with shaking for 24 h before being diluted and spread again onto the appropriate agar media to quantify total (N), D, R and T cell densities. The cell densities were then used to calculate the transfer rate constant λ (in ml/(cfu × t)) as follows and as described by Simonsen et al. (46):
where ψ is the maximum growth rate for the combined donor and recipient cultures, which is calculated from two optical density (OD600) determinations, say a and b, taken during the exponential phase, as follows (see ref. 46):
DNA sequencing and analysis
Genomic DNA (gDNA) was extracted from populations using a GenElute™ Bacterial Genomic DNA Kit (Sigma-Aldrich) according to the manufacturer’s instructions. The quality and integrity of the gDNA was assessed on a 1% agarose gel and the concentrations were determined fluorometrically using Quant-iT™ PicoGreen® dsDNA Reagent (ThermoFisher Scientific) on a SpectraMax® Paradigm® Multi-Mode Microtiter Plate Reader (Molecular Devices). To identify mutations in evolved clones compared to ancestral clones the gDNA was sequenced using a whole-genome shotgun approach with paired 300-bp reads generated on MiSeq (Illumina) at the IBEST Genomics Resources Core (GRC) at the University of Idaho (USA). Sequencing adapters and low quality bases were trimmed using custom scripts after which the reads were mapped to the Pseudomonas sp. nov. H2 and RP4 genomes available in NCBI under accession numbers JRPO00000000 and L27758.1, respectively, using BreSeq version 0.26.1 (47). The genome and sequencing data were visualized using Circos version 0.69 (48).
To determine SNP frequency within heterogeneous populations amplicon libraries of the target regions were constructed using a dual barcoded two-step PCR amplification protocol for sequencing on an Illumina MiSeq sequencing platform. Briefly, target sequences were amplified using sequence-specific primers containing universal CS1 and CS2 adapter sequences (Supplemental Table 10). The PCR reactions consisted of PCR buffer (New England Biolabs® Inc), 3 mM MgCl2 (ThermoFisher Scientific), 0.24 mg.ml−1 BSA (ThermoFisher Scientific), 200 µM dNTPs (ThermoFisher Scientific), 50 nM each of the appropriate forward and reverse CS-tagged primers, 0.025 U.µl−1 Taq DNA polymerase (New England Biolabs® Inc) and 2 ng.µl−1 gDNA in a final volume of 50 µl. The BioRad thermocycler parameters were as follows; 95 ℃ for 2 min, 20 cycles of 95, 51 and 68 ℃ each for 1 min and finally 68 ℃ for 10 min. The resulting PCR products were assessed on a 1% agarose gel whereafter it was diluted 15-fold and used as template in a second PCR reaction to attach the appropriate barcodes. These PCR reactions consisted of PCR buffer New England Biolabs® Inc), 4.5 mM MgCl2 (ThermoFisher Scientific), 0.24 mg.ml−1 BSA (ThermoFisher Scientific), 200 µM dNTPs (ThermoFisher Scientific), 75 nM barcoded primer, 0.05 U.µl−1 Taq DNA polymerase (New England Biolabs® Inc) and 1:300 PCR product 1 in a final volume of 50 µl. The BioRad thermocycler parameters were as follows; 95 ℃ for 1 min, 10 cycles of 95 ℃ for 30 sec, 60 ℃ for 30 sec and 68 ℃ for 1 min and finally 68 ℃ for 5 min. The quality and yield of the final amplicon libraries were assessed on an Agilent Bioanalyzer (Agilent Technologies) using the Agilent DNA 1000 kit as per manufacturer’s instruction. The resulting libraries were sequenced using on an Illumina MiSeq sequencing platform using MiSeq Reagent kit v3 at the IBEST GRC. Sequencing adapters and low quality bases were trimmed, the read numbers normalized using custom scripts and the SNP frequency determined using the Geneious R8 software package.
Gene expression analysis
Stationary phase bacterial cultures were diluted to an OD600 of approximately 0.005 in fresh TB, supplemented with Rif and Tet as required, and grown at 30°C. When the cultures reached an optical density of approximately 0.5 and 1.0, 1mL of cells were combined with 200 µl stop solution (95% ethanol, 5% acid phenol chloroform). The cells were then harvested by centrifugation at 4°C (13,000 RPM, 2 min) and stored at −20°C until RNA extraction. RNA was extracted using GeneJET RNA Purification Kit (ThermoFisher Scientific) according to manufactures instructions. Residual genomic DNA contamination was removed by DNAse I (ThermoFisher) treatment at 37°C for 1h. The amount of RNA was quantitated using a NanoDrop (NanoDrop2000). For cDNA construction, 1 µg total RNA was added to the iScript Reverse Transcription Supermix kit (BIO-RAD) following manufacturers instructions. Quantitative Real-Time PCR was performed on ca. 150ng cDNA for uvrD and 3ng for 16S rRNA, using the iTaq Universal SYBR Green Supermix (BIO-RAD) on a BioRad CFX-connect qPCR machine. The uvrD and 16S rRNA cDNA concentrations were quantified using the UvrD_helicase-Fwd/-Rev and 16S rRNA primer sets. The samples were normalized using the 16S rRNA levels and the uvrD gene expression relative to the plasmid-free ancestor was determined using the ΔΔCt method. The statistical significance of differences in gene expression relative to the ancestor was determined using Tukey's multiple comparisons test.
Allelic exchange
Allelic exchange experiments were performed based on a modified protocol from Hmelo et al. (49). Briefly, the ancestral and evolved alleles were cloned into pPS04 and introduced into H2 by mobilization from E. coli S17.1pir+. Recombinants were selected on LB agar supplemented with kanamycin (50 µg.ml−1). For each allele, 46 merodiploid recombinants were replicated onto LB agar supplemented with kanamycin (50 µg.ml−1). Insertion of the vector into the H2 genome was confirmed by PCR using primers specific to the pPS04 aph-1 gene and by verifying vector absence using a plasmid miniprep kit (ThermoScientific, Waltham, MA). Thereafter, a single clone for each allele was inoculated into 5 ml ‘no salt LB’ (NSLB) supplemented with 15% (w/v) sucrose and incubated for ca. 16h at 30 ℃ to select and enrich for double cross-over mutants. The stationary phase cultures were diluted and spread onto NSLB agar supplemented with 15% (w/v) sucrose and ca. 1,000 colonies were screened for the absence of the aph-1 gene by replication onto NSLB agar supplemented with 15% (w/v) sucrose and with or without kanamycin. Absence of the pPS04 aph-1 gene was also confirmed by PCR. To verify the correct genotype for each allele PCR products were generated in each case from 24 clones using the H2_Xpd/Rad3_Seq and H2_UvrD_Seq primer pairs and sequenced by means of Sanger sequencing in both the forward and reverse directions (Elim Biopharm). In each case, a clone that retained the original allele was also saved and used to control for the effect of allelic exchange on the host.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases grant R01 AI084918 of the National Institutes of Health (NIH). The genome resequencing was done by the IBEST Genomics Research Core, and made possible thanks to an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences (NIGMS) of the NIH under grant number P30 GM103324. KB was in part supported by a University of Idaho Department of Biological Sciences Undergraduate Research Grants and by an NIGMS INBRE award, grant number P20 GM103408. HQ was supported by a National Science Foundation REU Site award, 1460696. We thank the lab of Dr. C. Marx for providing us with vector pPS04, and Dr. H. Merrikh for useful suggestions.
Footnotes
Data availability
All sequencing data is available under NCBI BioProject accession number PRJNA261945. All persistence and competition assay data are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.v25vk.
Code availability
The StabilityToolkit package and instructions are available at https://github.com/jmponciano/StabilityToolkit/blob/master/RunningStabToolsPack.zip
AUTHOR CONTRIBUTIONS
W.L. and E.M.T conceived the project and wrote the manuscript, W.L., K.B., H.Q., K.D., M.T and J.M. performed the experiments, W.L. and J.M.P. the statistical analyses, W.L. and S.H. the bioinformatic analysis, and H.M. facilitated part of the work and helped with data interpretation.
COMPETING FINANCIAL INTERESTS
The authors have no competing financial interests.
References
- 1.Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat. Rev. Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 2.Schwarz S, Johnson AP. Transferable resistance to colistin: a new but old threat. J Antimcrob. Chemother. 2016;71:2066–2070. doi: 10.1093/jac/dkw274. [DOI] [PubMed] [Google Scholar]
- 3.Baker KS, et al. The Murray collection of Enterobacteriacae: A unique resource. Genome Med. 2015;7:97–114. doi: 10.1186/s13073-015-0222-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas CM. The horizontal gene pool. Bacterial plasmids and gene spread. Harwood Academic Publishers; Amsterdam: 2000. [Google Scholar]
- 5.Datta N, Hughes VM. Plasmids of the same Inc groups in Enterobacteria before and after the medical use of antibiotics. Nature. 1983;306:616–617. doi: 10.1038/306616a0. [DOI] [PubMed] [Google Scholar]
- 6.Popowska M, Krawczyk-Balska A. Broad-host-range IncP-1 plasmids and their resistance potential. Front. Microbiol. 2013;4:1–8. doi: 10.3389/fmicb.2013.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Gelder L, Ponciano JM, Joyce P, Top EM. Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiol. 2007;153:452–463. doi: 10.1099/mic.0.2006/001784-0. [DOI] [PubMed] [Google Scholar]
- 8.Ebersbach G, Gerdes K. Plasmid segregation mechanisms. Annu. Rev. Genet. 2005;39:453–479. doi: 10.1146/annurev.genet.38.072902.091252. [DOI] [PubMed] [Google Scholar]
- 9.Stewart FM, Levin BR. The population biology of bacterial plasmids: a priori conditions for the existence of conjugationally transmitted factors. Genetics. 1977;87:209–228. doi: 10.1093/genetics/87.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison E, Guymer D, Spiers AJ, Paterson S, Brockhurst MA. Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr. Biol. 2015;25:2034–2039. doi: 10.1016/j.cub.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 11.San Millan A, Toll-Riera M, Qi Q, MacLean RC. Interactions between horizontally acquired genes create a fitness cost in Pseudomonas aeruginosa. Nat. Commun. 2015;6:6845. doi: 10.1038/ncomms7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shintani M, et al. Response of the Pseudomonas host chromosomal transcriptome to carriage of the IncP-7 plasmid pCAR1. Environ. Microbiol. 2010;12:1413–1426. doi: 10.1111/j.1462-2920.2009.02110.x. [DOI] [PubMed] [Google Scholar]
- 13.De Gelder L, Williams JJ, Ponciano JM, Sota M, Top EM. Adaptive plasmid evolution results in host-range expansion of a broad-host-range plasmid. Genetics. 2008;178:2179–2190. doi: 10.1534/genetics.107.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sota M, et al. Shifts in the host range of a promiscuous plasmid through parallel evolution of its replication initiation protein. ISME J. 2010;4:1568–1580. doi: 10.1038/ismej.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouma JE, Lenski RE. Evolution of a bacteria/plasmid association. Nature. 1988;335:351–352. doi: 10.1038/335351a0. [DOI] [PubMed] [Google Scholar]
- 16.San Millan A, et al. Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat. Commun. 2014;5:5208. doi: 10.1038/ncomms6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dahlberg C, Chao L. Amelioration of the cost of conjugative plasmid carriage in Eschericha coli K12. Genetics. 2003;165:1641–1649. doi: 10.1093/genetics/165.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loftie-Eaton W, et al. Evolutionary paths that expand plasmid host-range: Implications for spread of antibiotic resistance. Mol. Biol. Evol. 2016;33:885–897. doi: 10.1093/molbev/msv339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maestro B, Sanz JM, Díaz-Orejas R, Fernández-Tresguerres E. Modulation of pPS10 host range by plasmid-encoded RepA initiator protein. J. Bacteriol. 2003;185:1367–1375. doi: 10.1128/JB.185.4.1367-1375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yano, et al. Evolved plasmid-host interactions reduce plasmid interference cost. Mol. Microbiol. 2016;101:743–756. doi: 10.1111/mmi.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heuer H, Fox RE, Top EM. Frequent conjugative transfer accelerates adaptation of a broad-host-range plasmid to an unfavorable Pseudomonas putida host. FEMS Microbiol. Ecol. 2007;59:738–748. doi: 10.1111/j.1574-6941.2006.00223.x. [DOI] [PubMed] [Google Scholar]
- 22.Pansegrau W, et al. Complete nucleotide sequence of Birmingham IncPalpha plasmids: Compilation and analysis. J. Mol. Biol. 1994;239:626–663. doi: 10.1006/jmbi.1994.1404. [DOI] [PubMed] [Google Scholar]
- 23.Saunders JR, Grinsted J. Properties of RP4, an R factor which originated in Pseudomonas aeruginosa S8. J. Bacteriol. 1972;112:690–696. doi: 10.1128/jb.112.2.690-696.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baquero F, Coque TM, de la Cruz F. Ecology and evolution as targets: the need for novel eco-evo drugs and strategies to fight antibiotic resistance. Antimicrob. Agents Chemother. 2001;55:3649–3660. doi: 10.1128/AAC.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ponciano JM, De Gelder L, Top EM, Joyce P. The population biology of bacterial plasmids: a hidden Markov model approach. Genetics. 2007;176:957–968. doi: 10.1534/genetics.106.061937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenski RE, Simpson SC, Nguyen TT. Genetic analysis of a plasmid-encoded, host genotype-specific enhancement of bacterial fitness. J. Bacteriol. 1994;176:3140–3147. doi: 10.1128/jb.176.11.3140-3147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heuer H, Ebers J, Weinert N, Smalla K. Variation in permissiveness for broad-host-range plasmids among genetically indistinguishable isolates of Dickeya sp. from a small field plot. FEMS Microbiol. Ecol. 2010;73:190–196. doi: 10.1111/j.1574-6941.2010.00880.x. [DOI] [PubMed] [Google Scholar]
- 28.Klümper U, et al. Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J. 2014;9:934–945. doi: 10.1038/ismej.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelley AL, Mezulis S, Tayes MC, Wass NM, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protocols. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yachdav G, et al. PredictProtein - An open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 2014;42:337–343. doi: 10.1093/nar/gku366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Byrd AK, Raney KD. Superfamily 2 helicases. Front. Biosci. 2013;17:2070–2088. doi: 10.2741/4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merrikh H, Zhang Y, Grossman AD, Wang JD. Replication-transcription conflicts in bacteria. Nat. Rev. Microbiol. 2013;10:449–458. doi: 10.1038/nrmicro2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merrikh CN, Brewer BJ, Merrikh H. The B. subtilis accessory helicase PcrA facilitates DNA replication through transcription units. PLOS Gen. 2015;11:e1005289. doi: 10.1371/journal.pgen.1005289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fairman-Williams ME, Guenther U, Jankowsky E. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baharoglu Z, Bikard D, Mazel D. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLOS Gen. 2010;6:e1001165. doi: 10.1371/journal.pgen.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingmer H, Miller C, Cohen SN. The RepA protein of plasmid pSC101 controls Escherichia coli cell division through the SOS response. Mol. Microbiol. 2001;42:519–526. doi: 10.1046/j.1365-2958.2001.02661.x. [DOI] [PubMed] [Google Scholar]
- 37.Boubakri H, de Septenville AL, Viguera E, Michel B. The helicases DinG, Rep and UvrD cooperate to promote replication across transcriptional units in vivo. EMBO J. 2010;29:145–157. doi: 10.1038/emboj.2009.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Epshtein V, et al. UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature. 2014;505:372–377. doi: 10.1038/nature12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guy CP, et al. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Molecular Cell. 2009;36:654–666. doi: 10.1016/j.molcel.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borukhou S, Nudler E. RNA polymerase holoenzyme: structure, function and biological implications. Curr. Opin. Microbiol. 2003;6:93–100. doi: 10.1016/s1369-5274(03)00036-5. [DOI] [PubMed] [Google Scholar]
- 41.Kuznedelov K, et al. A role for interaction of the RNA polymerase flap domain with σ subunit in promoter recognition. Science. 2002;295:855–857. doi: 10.1126/science.1066303. [DOI] [PubMed] [Google Scholar]
- 42.Brandis G, Wrande M, Liljas L, Hughes D. Fitness-compensatory mutations in rifampicin-resistant RNA polymerase. Mol. Microbiol. 2012;85:142–151. doi: 10.1111/j.1365-2958.2012.08099.x. [DOI] [PubMed] [Google Scholar]
- 43.Reynolds MG. Compensatory evolution in rifampin-resistant Escherichia coli. Genetics. 2000;156:1471–1481. doi: 10.1093/genetics/156.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbour Laboratory Press; 2001. [Google Scholar]
- 45.Joyce P, et al. Modeling the impact of periodic bottlenecks, unidirectional mutation, and observational error in experimental evolution. J. Math. Biol. 2005;50:645–662. doi: 10.1007/s00285-004-0300-8. [DOI] [PubMed] [Google Scholar]
- 46.Simonsen L, Gordon DM, Stewart FM, Levin BR. Estimating the rate of plasmid transfer: an end-point method. J. Gen. Microbiol. 1990;136:2319–2325. doi: 10.1099/00221287-136-11-2319. [DOI] [PubMed] [Google Scholar]
- 47.Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 2014;1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krzywinski MI, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hmelo LR, et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat. Protoc. 2015;10:1820–1841. doi: 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.