Abstract
The broad objective of this study was to increase our knowledge of Muleshoe virus and other hantaviruses associated with cricetid rodents in Texas. Anti-hantavirus antibody was found in 38 (3.2%) of 1171 neotomine rodents and 6 (1.8%) of 332 sigmodontine rodents from 10 Texas counties; hantaviral RNA was detected in 23 (71.9%) of 32 antibody-positive rodents. Analyses of nucleocapsid protein gene sequences indicated Muleshoe virus infection in four hispid cotton rats (Sigmodon hispidus) from northern Texas; Bayou virus, three Texas marsh oryzomys (Oryzomys texensis) from the Gulf Coast; Limestone Canyon virus, five brush mice (Peromyscus boylii) from western Texas; and Sin Nombre virus—five Texas mice (P. attwateri), one Lacey's white-ankled deer mouse (P. laceianus), four white-footed mice (P. leucopus), and one fulvous harvest mouse (Reithrodontomys fulvescens) from northern, central, or southern Texas. The results of this study together with the results of a previous study revealed that Muleshoe virus, perhaps in association with S. hispidus, is distributed across northern Texas. Finally, the results of Bayesian analyses of glycoprotein precursor (GPC) gene sequences and pairwise comparisons of complete GPC (amino acid) sequences strengthened support for the notion that Muleshoe virus is distinct from Black Creek Canal virus, Bayou virus, and all other species included in the Bunyaviridae, genus Hantavirus.
Keywords: : cotton rat, hantavirus, Muleshoe virus, rodent borne, Sigmodon hispidus
Introduction
The broad objective of this study was to increase our knowledge of the hantaviruses (Bunyaviridae, genus Hantavirus) associated with cricetid rodents (Cricetidae) in North America, more specifically neotomine rodents (Neotominae) and sigmodontine rodents (Sigmodontinae) in Texas (Musser and Carleton 2005). Previous studies revealed anti-hantavirus antibody in rodents captured in 18 of the 254 counties in Texas (Fig. 1). Specific knowledge of hantaviruses associated with cricetid rodents in Texas previously was limited to Bayou virus (BAYV) RNA from Texas marsh oryzomys (Oryzomys texensis, formerly recognized as O. palustris; Hanson et al. 2010) and hispid cotton rats (Sigmodon hispidus) captured in Brazoria and Jefferson counties (Torrez-Martinez et al. 1998, Houck et al. 2001); Sin Nombre virus (SNV) RNA—a white-footed mouse (Peromyscus leucopus) and deer mouse (P. maniculatus) from Deaf Smith County (Rawlings et al. 1996); SNV RNA—deer mice from Castro, Loving, and Moore counties (Monroe et al. 1999); El Moro Canyon virus (ELMCV) RNA—western harvest mice (Reithrodontomys megalotis) and deer mice from Deaf Smith County (Rawlings et al. 1996); and Muleshoe virus (MULV) RNA—hispid cotton rats from Bailey and Deaf Smith counties (Rawlings et al. 1996).
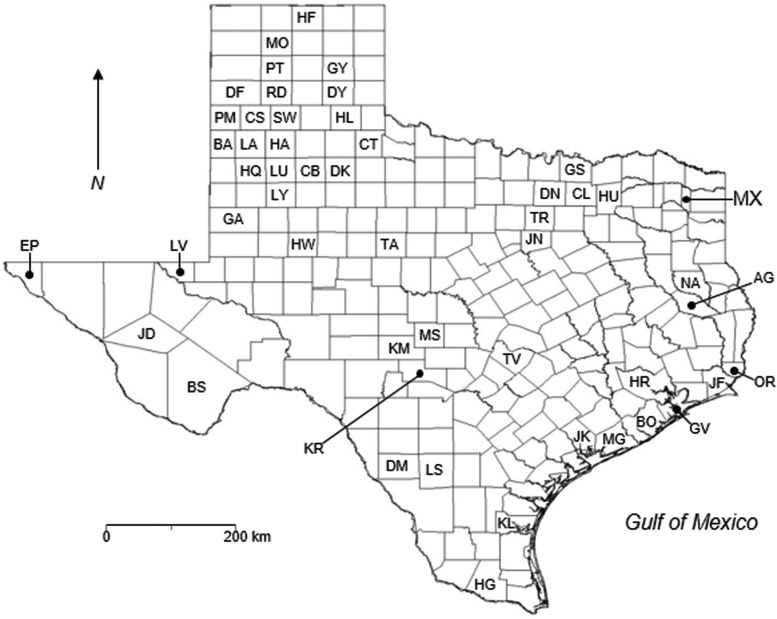
FIG. 1.
County map of Texas (696,241 km2). HF, Hansford; MO, Moore; PT, Potter; GY, Gray; DF, Deaf Smith; RD, Randall; DY, Donley; PM, Parmer; CS, Castro; SW, Swisher; HL, Hall; BA, Bailey; LA, Lamb; HA, Hale; CT, Cottle; HQ, Hockley; LU, Lubbock; CB, Crosby; DK, Dickens; GS, Grayson; LY, Lynn; DN, Denton; CL, Collin; HU, Hunt; MX, Morris; GA, Gaines; TR, Tarrant; HW, Howard; TA, Taylor; JN, Johnson; EP, El Paso; LV, Loving; NA, Nacogdoches; AG, Angelina; JD, Jeff Davis; BS, Brewster; KM, Kimble; MS, Mason; KR, Kerr; TV, Travis; HR, Harris; OR, Orange; JF, Jefferson; GV, Galveston; BO, Brazoria; JK, Jackson; MG, Matagorda; DM, Dimmit; LS, La Salle; KL, Kleberg; HG, Hidalgo. The eastern and western halves of the Chaparral Wildlife Management Area were located in western LS and eastern DM counties, respectively. The rodents that were positive for anti-hantavirus antibody in previous studies (Torrez-Martinez et al. 1998, Mantooth et al. 2001, Mauldin et al. 2013, Pitts et al. 2013) were from CT, LU, DK, GS, DN, CL, MX, JD, BS, KM, MS, KR, OR, JF, GV, BO, MG, and LS counties; the rodents that were positive for anti-hantavirus antibody in this study (Table 1) were from DY, HL, LU, LY, JN, JD, KR, and DM counties; and the 32 rodents tested for hantaviral RNA in this study were from CT, LU, LY, MX, JD, KR, GV, BO, MG, and DM counties. The 46 laboratory-confirmed cases of hantavirus pulmonary syndrome in Texas through April 2016 were reported from 29 counties: HF (n = 1), PT (n = 1), GY (n = 1), DF (n = 1), RD (n = 6), PM (n = 2), CS (n = 3), SW (n = 1), LA (n = 1), HA (n = 2), HQ (n = 1), LU (n = 2), CB (n = 2), HU (n = 1), GA (n = 3), TR (n = 1), HW (n = 1), TA (n = 1), EP (n = 2), AG (n = 1), TV (n = 1), HR (n = 2), OR (n = 1), JF (n = 3), BO (n = 1), JK (n = 1), MG (n = 1), KL (n = 1), and HG (n = 1) (Texas Department of State Health Services, unpublished data).
A specific aim of this study was to examine further the phylogenetic and taxonomical relationships of MULV to BAYV, Black Creek Canal virus (BCCV), and other members of the genus Hantavirus (Plyusnin et al. 2012). The genomes of hantaviruses comprise three negative-sense, single-stranded RNA molecules, designated S (small, 1.7–2.1 kb), M (medium, 3.6–3.7 kb), and L (large, 6.5–6.7 kb) (Plyusnin et al. 2012). These molecules encode the nucleocapsid (N) protein, glycoprotein precursor (GPC) to the envelope glycoproteins (Gn and Gc), and RNA-dependent RNA polymerase (L protein), respectively. Our knowledge of the genome of MULV previously was limited to the nucleotide sequences of a 397-nt fragment of the N protein gene of strain SH-Tx-311 (Deaf Smith County, GenBank accession no. U54576) and 1989-nt fragment of the S segment of strain SH-Tx-339 (Bailey County, U54575). The results of a maximum parsimony analysis of these nucleotide sequences and other hantaviral N protein gene sequences (Rawlings et al. 1996) suggested that MULV is closely related to BAYV and BCCV.
Materials and Methods
Blood samples from 1503 cricetid rodents captured in 10 Texas counties in January 2001–May 2012 were tested for anti-hantavirus antibody (Table 1). The rodents from Dimmit and La Salle counties were captured in January 2001–October 2002 at three sites on the Chaparral Wildlife Management Area during a prospective (mark-release-recapture) study on the ecology of Catarina virus (Arenaviridae, Milazzo et al. 2013). These sites were designated “Web I,” “Web II,” and “Web III”; Webs I and II were located in eastern Dimmit County; and Web III was located in western La Salle County. Samples of fresh-frozen lung from white-footed mouse TTU-M98174 (P. leucopus, Web II), 6 other cricetid rodents captured in 2001–2012, and 25 rodents captured in Texas in 1995–1999 (Table 2) were tested for hantaviral N protein gene RNA and hantavirus. The rodents from 1995 to 1999 were positive for anti-hantavirus antibody in a previous study (Mantooth et al. 2001).
Table 1.
Prevalence of Anti-Hantavirus Antibody in 1503 Cricetid Rodents Captured in Texas in 2001–2012, By County
| Speciesa,b | ||||||||
|---|---|---|---|---|---|---|---|---|
| County | Patt | Pboy | Plac | Pleu | Rmeg | Shis | Otherc | Totalb |
| Dimmit | 4/155 | 0/91 | 0/220 | 4/466 | ||||
| Donley | 0/16 | 8/27 | 0/3 | 0/7 | 0/32 | 8/85 | ||
| Hall | 3/23 | 0/1 | 0/10 | 3/34 | ||||
| Jeff Davis | 1/25 | 1/5 | 0/5 | 2/35 | ||||
| Johnson | 1/2 | 1/2 | ||||||
| Kerr | 14/99 | 0/15 | 0/2 | 0/1 | 14/117 | |||
| Kimble | 0/59 | 0/101 | 0/1 | 0/20 | 0/181 | |||
| La Salle | 0/139 | 0/40 | 0/135 | 0/314 | ||||
| Lubbock | 5/39 | 5/159 | 1/60d | 11/258 | ||||
| Lynn | 1/10 | 0/1 | 1/11 | |||||
| Total | 14/174 | 1/25 | 1/118 | 15/347 | 6/47 | 6/328 | 1/464 | 44/1503 |
Patt, Peromyscus attwateri; Pboy, P. boylii; Plac, P. laceianus (formerly recognized as Peromyscus pectoralis—Bradley et al. 2015); Pleu, P. leucopus; Rmeg, Reithrodontomys megalotis; Shis, Sigmodon hispidus; Other—Baiomys taylori (n = 11), Neotoma leucodon (n = 1), N. mexicana (n = 1), N. micropus (n = 306), Onychomys leucogaster (n = 27), P. maniculatus (n = 56), R. fulvescens (n = 57), R. montanus (n = 1), and S. ochrognathus (n = 4).
Number positive/number tested for anti-hantavirus IgG.
Dimmit—B. taylori (n = 2), N. micropus (n = 183), O. leucogaster (n = 8), R. fulvescens (n = 27); Donley—B. taylori (n = 1), N. micropus (n = 19), O. leucogaster (n = 2), P. maniculatus (n = 7), R. fulvescens (n = 2), R. montanensis (n = 1); Hall—B. taylori (n = 8), P. maniculatus (n = 1), R. fulvescens (n = 1); Jeff Davis—N. mexicana (n = 1), S. ochrognathus (n = 4); Kerr—N. leucodon (n = 1); La Salle—N. micropus (n = 104), O. leucogaster (n = 16), R. fulvescens (n = 15); Lubbock—P. maniculatus (n = 48), R. fulvescens (n = 12); and Lynn—O. leucogaster (n = 1).
One (2.1%) of the 48 deer mice (P. maniculatus) from Lubbock County was positive for anti-hantavirus antibody.
Table 2.
Prevalence of Anti-Hantavirus Antibody in 775 Cricetid Rodents Captured in Texas in 1998–2007, By Locality
| Speciesb,c | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Localitya | Otex | Patt | Pboy | Plac | Pleu | Rful | Rmeg | Shis | Otherd | Total |
| 5.5 km east of New Deal, Lubbock County (July 6, 2005) | ||||||||||
| 0/5 | 1/13 | 3/56 (0/1) | 0/26 | 4/100 (0/1) | ||||||
| 13.8 km northeast of Lubbock, Lubbock County (June 9, 2005) | ||||||||||
| 3/8 (0/2) | 2/35 (0/1) | 0/6 | 5/49 (0/3) | |||||||
| 8.9 km north-northeast of Tahoka, Lynn County (July 9, 2007) | ||||||||||
| 1/10 (1/1) | 0/1 | 1/11 (1/1) | ||||||||
| Matador Wildlife Management Area, Cottle County (February 7–9, 1998) | ||||||||||
| 2/18 (1/2) | 3/25 (3/3) | 2/5 (1/2) | 0/4 | 0/9 | 7/61 (5/7) | |||||
| White Oak Creek Wildlife Management Area, Morris County (July 10–14, 1999) | ||||||||||
| 0/1 | 3/18 (3/3) | 0/1 | 3/20 (3/3) | |||||||
| Mt. Livermore Preserve, Jeff Davis County (July 14–19, 1998) | ||||||||||
| 2/82 (2/2) | 0/1 | 0/3 | 0/8 | 2/94 (2/2) | ||||||
| Mt. Livermore Preserve, Jeff Davis County (July 21–23, 1999) | ||||||||||
| 9/114 (3/3) | 0/1 | 0/1 | 0/12 | 9/128 (3/3) | ||||||
| Kerr Wildlife Management Area, Kerr County (May 14–18, 1998) | ||||||||||
| 7/102 (4/5) | 1/7 (1/1) | 0/7 | 0/1 | 8/117 (5/6) | ||||||
| Virginia Point, Galveston County (May 24–28, 1998) | ||||||||||
| 5/32 (1/2) | 0/1 | 0/32 | 5/65 (1/2) | |||||||
| Peach Point Wildlife Management Area, Brazoria County (April 24, 1998) | ||||||||||
| 2/5 (1/1) | 0/8 | 0/1 | 2/14 (1/1) | |||||||
| Mad Island Wildlife Management Area, Matagorda County (April 25, 1998) | ||||||||||
| 1/7 (1/1) | 0/6 | 0/4 | 0/5 | 0/5 | 1/27 (1/1) | |||||
| Chaparral Wildlife Management Area-Web I, Dimmit County (March 13–15, 2001) | ||||||||||
| 1/20 (0/1) | 0/1 | 0/5 | 0/11 | 1/37 (0/1) | ||||||
| Chaparral Wildlife Management Area-Web II, Dimmit County (January 7–9, 2002) | ||||||||||
| 1/24 (1/1) | 0/4 | 0/3 | 0/21 | 1/52 (1/1) | ||||||
| Total | 8/44 (3/4) | 9/120 (5/7) | 11/196 (5/5) | 1/7 (1/1) | 5/84 (4/5) | 2/23 (1/2) | 4/24 (0/2) | 9/176 (4/6) | 0/101 | 49/775 (23/32) |
The antibody data from the rodents captured in 1998 or 1999 were published previously (Mantooth et al. 2001). One or more antibody-positive rodents from each locality were tested for hantaviral nucleocapsid (N) protein gene RNA.
New Deal—300 traps set on 1 night, 300 trap nights in tall grass alongside fields repopulated with native grasses, 33.3% trap success; near Lubbock—300 traps, 300 trap nights, tall grass alongside fields repopulated with native grasses, 16.3%; Tahoka—450 traps, 450 trap nights, tall grass alongside fields repopulated with native grasses, 2.4%; Matador Wildlife Management Area (WMA)—450 or 500 traps, 1450 trap nights, mesquite and juniper on grassland, 4.2%; White Oak Creek WMA—650 or 700 traps, 2700 trap nights, post oak on savannah, 0.7%; Mt. Livermore Preserve, 1998—500 traps, 2500 trap nights, montane pine-oak forest, 3.8%; Mt. Livermore Preserve, 1999—350, 400, or 700 traps, 1450 trap nights, montane pine-oak forest, 8.8%; Kerr WMA—200, 400, or 450 traps, 1050 trap nights, juniper grassland, 11.4%; Virginia Point—coastal marsh; Peach Point WMA—500 traps, 1000 trap nights, coastal marsh, 1.4%; Mad Island WMA—500 traps, 1000 trap nights, coastal marsh, 2.7%; Chaparral WMA, 2001—960 traps, 2880 trap nights, South Texas scrubland, 1.3%; Chaparral WMA, 2002—960 traps, 2880 trap nights, South Texas scrubland, 1.8%.
Otex, Oryzomys texensis, formerly recognized as O. palustris (Hanson et al. 2010); Patt, Peromyscus attwateri; Pboy, P. boylii; Plac, P. laceianus; Pleu, P. leucopus; Rful, Reithrodontomys fulvescens; Rmeg, R. megalotis; Shis, Sigmodon hispidus.
Number positive/number tested for anti-hantavirus antibody. (Number positive/number tested for nucleocapsid protein gene RNA.) All the rodents tested for nucleocapsid protein gene RNA were positive for anti-hantavirus antibody.
New Deal—P. maniculatus (n = 26); Lubbock—P. maniculatus (n = 6); Tahoka—Onychomys leucogaster (n = 1); Matador WMA—Neotoma micropus (n = 1), O. leucogaster (n = 2), P. maniculatus (n = 6); White Oak Creek WMA—P. gossypinus (n = 1); Mt. Livermore Preserve (1998)—N. mexicana (n = 5), O. arenicola (n = 1), S. ochrognathus (n = 2); Mt. Livermore Preserve (1999)—P. nasutus (n = 10), R. montanus (n = 1), S. ochrognathus (n = 1); Kerr WMA—N. leucodon (n = 1); Mad Island WMA—Baiomys taylori (n = 5); Chaparral WMA, Web I—N. micropus (n = 11); Chaparral WMA, Web II—N. micropus (n = 21).
Capture and processing of rodents
The rodents were collected and processed with approval from the Institutional Animal Care and Use Committee, Texas Tech University, and with permission from the Texas Parks and Wildlife Department and other property owners. Sherman live traps (H. B. Sherman Traps, Inc., Tallahassee, FL; model LFATDG) or Tomahawk traps (Tomahawk Live Traps, Mfg., Hazelhurst, WI; model 201) were set 1 h before sunset; baited with a mixture of rolled oats, cracked corn, and birdseed; and checked at dawn the following day. Each rodent was assigned a unique identification number, restrained with ketamine hydrochloride by intramuscular injection, and then examined and measured. The identification number, species identity, gender, total length (tip of nose to tip of tail), length of tail, and other information were recorded on a standardized form. Death was by intraperitoneal injection of a mixture of ketamine hydrochloride and xylazine hydrochloride, asphyxiation in an atmosphere of carbon dioxide, or exposure to a lethal dose of vaporized isoflurane. Voucher specimens from white-footed mouse TTU-M98174, 68 other rodents from the Chaparral Wildlife Management Area, and virtually all the rodents from the other localities in this study were deposited into the Natural Science Research Laboratory, Museum of Texas Tech University. The voucher specimens included skins, skulls, skeletons, samples of blood dried on Nobuto blood filter strips (Advantec MFS, Inc., Pleasanton, CA), and fresh-frozen samples of lung and other solid tissues.
Antibody assay
The blood samples were tested for anti-hantavirus immunoglobulin G (IgG), using an ELISA (Fulhorst et al. 1997). The test antigen was prepared from Vero E6 cells infected with Caño Delgadito virus (CADV) strain VHV-574 (Fulhorst et al. 1997). The comparison (control) antigen was prepared from uninfected Vero E6 cells in a manner quantitatively identical to that used to prepare the test antigen. Serial fourfold dilutions (from 1:80 through 1:5120) of each blood sample were tested against both antigens, IgG bound to antigen was detected using a mixture of goat anti-Rat IgG peroxidase conjugate and goat anti-Peromyscus leucopus IgG peroxidase conjugate in conjunction with the ABTS Microwell Peroxidase Substrate System (Kirkegaard and Perry Laboratories, Gaithersburg, MD), and the adjusted optical density (AOD) for a reaction was the optical density of the well coated with the test antigen less the optical density of the well coated with the comparison antigen. A sample was considered positive for anti-hantavirus IgG if the AOD at 1:80 was ≥0.200, the AOD at 1:320 was ≥0.200, and the sum of the AOD for the series of fourfold dilutions (from 1:80 through 1:5120) was ≥0.750.
Assays for nucleocapsid protein gene RNA
Samples of lung from white-footed mouse TTU-M98174, hispid cotton rat TTU-M80670, and 30 other antibody-positive rodents were tested for hantaviral N protein gene RNA (Milazzo et al. 2012). Insofar as possible, the 32 rodents represented the species diversity and geographical distribution of the antibody-positive rodents in this study and a previous study (Mantooth et al. 2001). First-strand cDNA was synthesized from total RNA using SuperScript II RNase H− Reverse Transcriptase (Invitrogen Life Technologies, Inc., Carlsbad, CA) with oligonucleotide 5′-GGTGGTTGTGGTAGTAGTAGACTCC-3′ (Morzunov et al. 1995) or HTS90 (Milazzo et al. 2010). The PCR assays used the MasterTaq® Kit (5 PRIME, Inc., Gaithersburg, MD); HTS90 and HTS12 (Milazzo et al. 2010) were used to prime the first-round assays; and HTS11 (5′-CGGGCAGCTGTGTCTGCATTGGA-3′) with HTS12 or HTS15 (Milazzo et al. 2010) with HTS12 were used to prime the second-round assays. Amplicons (including primer sequences) from the second-round assays were expected to be 443- or 592-bp long.
Virus assay
Samples of fresh-frozen lung from the rodents that were positive for N protein gene RNA were assayed for hantavirus by cultivation in monolayers of Vero E6 cells (Milazzo et al. 2010). The Vero E6 cells were inoculated with crude 10% w/v homogenates of lung, maintained under a fluid overlay, and blindly passaged twice at 12- or 13-day intervals. Cells harvested on Day 36 or 37 postinoculation were tested for hantaviral antigen, using an indirect fluorescent antibody test in which the primary antibody was a polyvalent anti-hantavirus hyperimmune mouse ascitic fluid.
Genetic characterization of hantaviruses HV B0530033 and HV F0260003
The nucleotide sequences of a 1907-nt fragment of the S genomic segment and 3615-nt fragment of the M genomic segment of HV B0530033 were determined from the RNA isolated from cotton rat TTU-M80670. The complete nucleotide sequences of the S and M segments of HV F0260003 were determined from RNA isolated from a monolayer of HV F0260003-infected Vero E6 cells (Milazzo et al. 2006). First-strand cDNA was synthesized with SuperScript II RNase H− Reverse Transcriptase; the PCR assays used the MasterTaq Kit; and the sequences of the oligonucleotides that were used to prime reverse transcription, the PCR assays, and cycle sequencing reactions are available from MNBC.
Genetic characterization of rodents
The complete nucleotide sequences of the cytochrome b (Cytb) genes of cotton rats TTU-M80670 and TTU-M119675 (Table 3) were determined from DNA isolated from liver. The PCR mixtures included GoTaq® DNA Polymerase (Promega Corp., Madison, WI) and either MVZ05 (Smith and Patton 1993) with H15915 (Irwin et al. 1991) or P3′ (Tiemann-Boege et al. 2000) with MVZ14 (Smith and Patton 1993). The oligonucleotides used to prime the sequencing reactions were MVZ05, H15915, P3′, MVZ14, F1 (Whiting et al. 2003), 400R (Tiemann-Boege et al. 2000), 700H and 700L (Peppers and Bradley 2000), and 870R (Peppers et al. 2002).
Table 3.
Hantaviruses Associated with 23 of 32 Rodents Tested for Nucleocapsid Protein Gene RNA
| Virusa | Strain | Rodentb | Speciesc | Date captured | County | Localityd | GenBank acc. no. |
|---|---|---|---|---|---|---|---|
| BAYV | HV F0260003 | TTU-M82910 | Otex | 5/24/1998 | Galveston | VP | GQ200820 |
| BAYV | HV B0530005 | TTU-M78329 | Otex | 4/24/1998 | Brazoria | PPWMA | KX066103 |
| BAYV | HV B0530006 | TTU-M78032 | Otex | 4/25/1998 | Matagorda | MIWMA | KX066104 |
| LSCV | HV B0770017 | TTU-M122340 | Pboy | 7/18/1998 | Jeff Davis | MLP | KX066105 |
| LSCV | HV B0770018 | TTU-M122341 | Pboy | 7/19/1998 | Jeff Davis | MLP | KX066106 |
| LSCV | HV B0530023 | TTU-M81118 | Pboy | 7/23/1999 | Jeff Davis | MLP | KX066107 |
| LSCV | HV B0530025 | TTU-M81139 | Pboy | 7/23/1999 | Jeff Davis | MLP | KX066108 |
| LSCV | HV B0530029 | TTU-M81165 | Pboy | 7/23/1999 | Jeff Davis | MLP | KX066109 |
| SNV | HV B0530012 | TTU-M77766 | Plac | 5/14/1998 | Kerr | KWMA | KX066110 |
| SNV | HV B0770007 | TTU-M77788 | Patt | 5/15/1998 | Kerr | KWMA | KX066111 |
| SNV | HV B0530013 | TTU-M77797 | Patt | 5/15/1998 | Kerr | KWMA | KX066112 |
| SNV | HV B0530015 | TTU-M77801 | Patt | 5/15/1998 | Kerr | KWMA | KX066113 |
| SNV | HV B0770009 | TTU-M77805 | Patt | 5/15/1998 | Kerr | KWMA | KX066114 |
| MULV | HV Q0020014 | TTU-M119675 | Shis | 7/9/2007 | Lynn | Tahoka | KX066123 |
| MULV | HV B0530031 | TTU-M80666 | Shis | 7/10/1999 | Morris | WOCWMA | KX066115 |
| MULV | HV B0770020 | TTU-M80668 | Shis | 7/10/1999 | Morris | WOCWMA | KX066116 |
| MULV | HV B0530033 | TTU-M80670 | Shis | 7/10/1999 | Morris | WOCWMA | KX066124 |
| SNV | HV B0770021 | TTU-M78838 | Patt | 2/9/1998 | Cottle | MWMA | KX066117 |
| SNV | HV B0770014 | TTU-M78841 | Pleu | 2/9/1998 | Cottle | MWMA | KX066118 |
| SNV | HV B0530001 | TTU-M78827 | Pleu | 2/9/1998 | Cottle | MWMA | KX066119 |
| SNV | HV B0770015 | TTU-M78845 | Pleu | 2/9/1998 | Cottle | MWMA | KX066120 |
| SNV | HV B0770016 | TTU-M78816 | Rful | 2/9/1998 | Cottle | MWMA | KX066121 |
| SNV | HV Q0020010 | TTU-M98174 | Pleu | 1/7/2002 | Dimmit | CWMA | KX066122 |
The assays for nucleocapsid protein gene RNA were negative in nine rodents.
BAYV, Bayou virus; LSCV, Limestone Canyon virus; SNV, Sin Nombre virus; MULV, Muleshoe virus.
Museum of Texas Tech University. The nucleotide sequences of the cytochrome b genes of TTU-M119675 and TTU-M80670 were deposited into GenBank under accession nos. KX866981 and KX866980, respectively.
Otex, Oryzomys texensis; Pboy, Peromyscus boylii; Plac, P. laceianus; Patt, P. attwateri; Shis, Sigmodon hispidus; Pleu, P. leucopus; Rful, Reithrodontomys fulvescens.
VP, Virginia Point; PPWMA, Peach Point Wildlife Management Area (WMA); MIWMA, Mad Island WMA; MLP, Mt. Livermore Preserve; KWMA, Kerr WMA; WOCWMA, White Oak Creek WMA; MWMA, Matador WMA; CWMA, Chaparral WMA.
Sequencing reactions and analysis
The sequences of both strands of each purified amplicon were determined directly, using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). The products of the sequencing reactions were analyzed on an ABI PRISM® 3100-Avant™ or 3130-Avant Genetic Analyzer (Applied Biosystems).
Analysis of nucleotide sequences
The N protein gene sequence dataset comprised the GenBank sequences under accession nos. L33683, L33816, L36929, L39949, U09488, U11427, U18100, U32591, U54575, U54576, U54578, U54582, U54583, AB620100, AB620103, AB620106, AF005727, AF291702, AF307322, AY267347, DQ256126, DQ285046, DQ285566, EF534079, FJ532244, GQ200820, JN097459, JN097461, JN097462, JN097466–JN097468, JN097470, JN097476, JN097477, and KX066103–KX066124. GPC gene sequences were L33474, L33684, L36930, L39950, U26828, U36801, AB620101, AB620104, AB620107, AF005728, AF291703, AF307323, AY363179, DQ177347, DQ284451, DQ285047, EF534080, FJ608550, GQ244521, and KX066125. Cytb gene sequences were AF425192 (Sigmodon hirsutus), EU073180 (S. toltecus), and sequences from 25 hispid cotton rats (S. hispidus). The S. hispidus sequences were AF155414 (Oklahoma), AF155415 (Refugio County, TX), AF155420 (Florida), AF188198 (New Mexico), AF425199 (Cameron Co., TX), AF425200 (Dimmit Co., TX), AF425201 (McMullen Co., TX), AF425202 (Tennessee), AF425203 (Missouri), AF425204 (Louisiana), AF425205 (LA), AF425206 (FL), AF425207 (FL), AF425209 (Kansas), AF425210 (Freestone Co., TX), AF425211 (Kimble Co., TX), AF425212 (Cottle Co., TX), AF425214 (Nacogdoches Co., TX), AF425227 (Lamar Co., TX), AF435110 (Lubbock Co., TX), EU073177 (Tamaulipas), EU073178 (Nuevo León), EU293749 (Arizona), KX866980 (Morris Co., TX), and KX866981 (Lynn Co., TX). The sequences under accession nos. GX200820, GQ244521, KX066103–KX066125 (Table 3), KX866980, and KX866981 were determined in this study.
The amino acid sequences predicted from the N protein gene sequences (and GPC gene sequences) were aligned using Clustal W (Thompson et al. 1994) in the computer software package MEGA5 (Tamura et al. 2011). The nucleotide sequence alignments (codons preserved) were generated in MEGA5, guided by the amino acid sequence alignments. Nonidentities between amino acid sequences were equivalent to uncorrected p distances. The alignment of Cytb gene sequences was constructed manually.
The Bayesian analyses were done with MrBayes v3.1.2 (Huelsenbeck and Ronquist 2001) and programs in the computer software package PAUP*, Version 4.0a150 (Swofford 2002). The analyses used the general time reversible + proportion invariant + Γ (GTR+I+G) model of nucleotide substitution, with a site-specific gamma distribution. The options in MrBayes were 2 simultaneous runs of 4 Markov chains, 10 million generations, and sample frequency = every 1000th generation. The out-groups in the analyses of the N protein, GPC, and Cytb gene sequences were AF291702 (Andes virus), AF291703 (Andes virus), and AF425192 (S. hirsutus), respectively. The first 1000 trees from each dataset were discarded after review of the likelihood scores, convergence statistics, and potential scale reduction factors. A consensus tree (50% majority rule) was constructed from the remaining trees, probability values in support of the clades were calculated a posteriori, and clades with probability values ≥0.95 were considered supported by the data (Erixon et al. 2003).
Results
Anti-hantavirus antibody (IgG) was found in 44 (2.9%) of 1503 cricetid rodents captured in 10 counties in Texas, 2001–2012 (Table 1). The antibody-positive rodents were Peromyscus attwateri (n = 14), P. boylii (n = 1), P. laceianus (formerly recognized as P. pectoralis; Bradley et al. 2015, n = 1), P. leucopus (n = 15), P. maniculatus (n = 1), Reithrodontomys megalotis (n = 6), and Sigmodon hispidus (n = 6).
Hantaviral N protein gene RNA was detected in 23 (71.9%) of 32 antibody-positive rodents, 21 of 25 captured in 1995–1999 and 2 of 7 captured in 2001–2012 (Table 3). Hantavirus HV F0260003 was isolated from a Texas marsh oryzomys (O. texensis) captured in Galveston County in 1998. More than 75% of cells in a sample harvested from the virus-positive culture on Day 36 PI were stained with fluorescein, the fluorophore used in the indirect fluorescent antibody test for hantaviral antigen. The assays for hantavirus in the 22 other N protein gene RNA-positive rodents were negative.
The complete sequences of the N protein genes of hantaviruses HV B0530033 (accession no. KX066124) and HV F0260003 (GQ200820) were 1284-nt long. Similarly, the lengths of the complete sequences of the GPC genes of HV B0530033 (KX066125) and HV F0260003 (GQ244521) were 3423-nt.
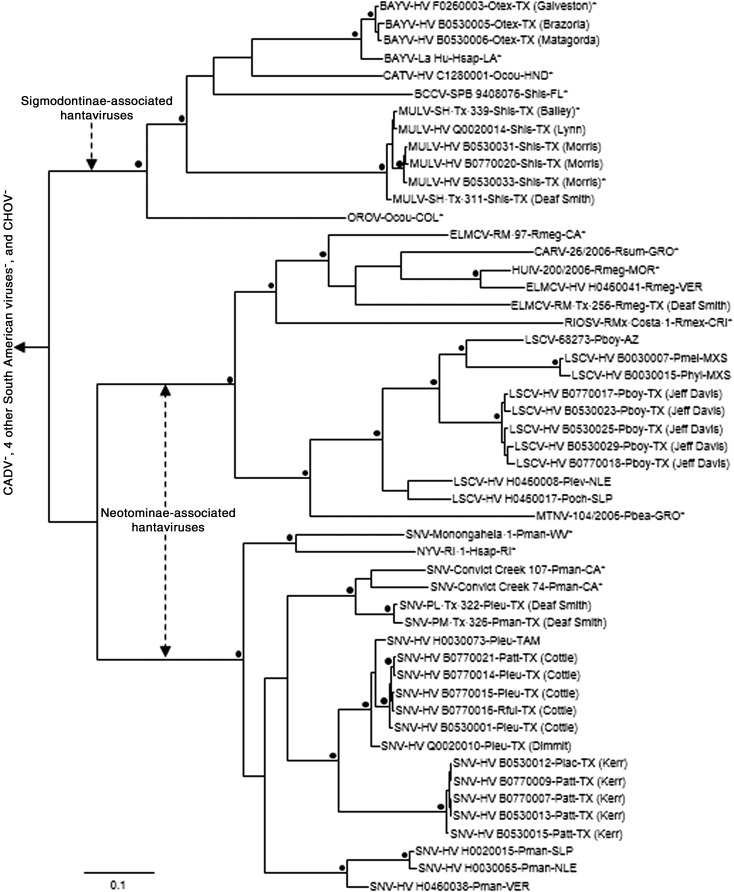
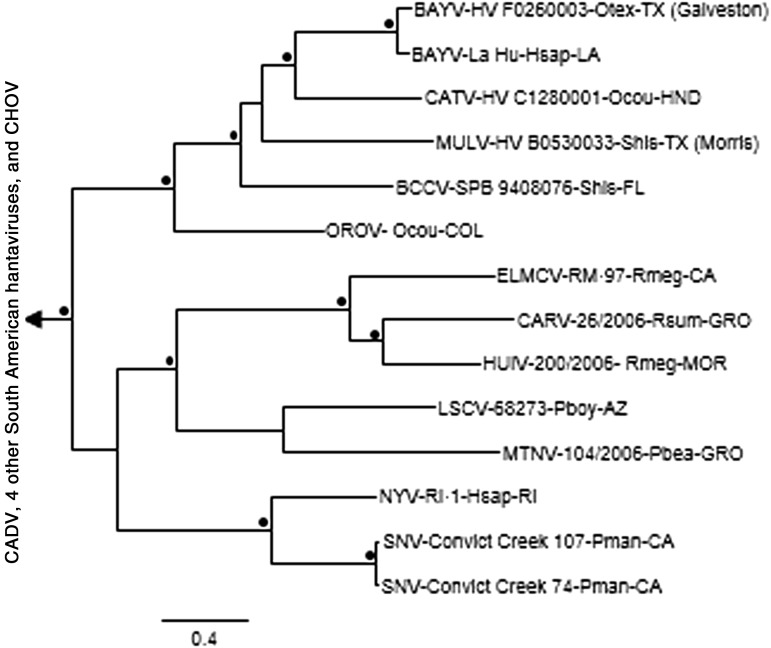
The results of the Bayesian analyses of N protein gene sequences (Fig. 2) indicated that HV F0260003 (Galveston County) is a strain of BAYV. The results of the N protein gene sequence analyses also indicated that BAYV was the hantavirus associated with the marsh oryzomys (O. texensis) from Brazoria and Matagorda counties; Limestone Canyon virus (LSCV)—brush mice (P. boylii), Jeff Davis County; MULV—hispid cotton rats (S. hispidus), Lynn and Morris counties; SNV—Texas mice (P. attwateri) and Lacey's white-ankled deer mouse (P. laceianus), Kerr County; SNV—white-footed mouse (P. leucopus), Dimmit County; and SNV—Texas mouse, white-footed mice, and fulvous harvest mouse (Reithrodontomys fulvescens), Cottle County (Table 3). The Bayesian analyses of N protein gene sequences (Fig. 2) and GPC gene sequences (Fig. 3) separated MULV and the 4 other hantaviruses associated with North American sigmodontine rodents (i.e., BAYV, BCCV, Catacamas virus [CATV], and Playa de Oro virus) from the hantaviruses associated with neotomine rodents (i.e., Carrizal virus, ELMCV, Huitzilac virus, LSCV, Montano virus, New York virus, Rio Segundo virus, and SNV). Monophyly of MULV, BAYV, BCCV, and CATV in the analyses of N protein gene sequences and GPC gene sequences was supported by analyses done a posteriori (clade probability values, 1.00).
FIG. 2.
Phylogenetic relationships among Muleshoe virus and other hantaviruses based on Bayesian analyses of nucleocapsid (N) protein gene sequences. BAYV, Bayou virus; CATV, Catacamas virus; BCCV, Black Creek Canal virus; MULV, Muleshoe virus; OROV, Playa de Oro virus; ELMCV, El Moro Canyon virus; CARV, Carrizal virus; HUIV, Huitzilac virus; RIOSV, Rio Segundo virus; LSCV, Limestone Canyon virus; MNTV, Montano virus; SNV, Sin Nombre virus; NYV, New York virus; CADV, Caño Delgadito virus; CHOV, Choclo virus. Otex, Oryzomys texensis; Hsap, Homo sapiens; Ocou, O. couesi; Shis, Sigmodon hispidus; Rmeg, Reithrodontomys megalotis; Rsum, R. sumichrasti; Rmex, R. mexicanus; Pboy, Peromyscus boylii; Pmel, P. melanotis; Phyl, P. hylocetes; Plev, P. levipes; Poch, P. ochraventer; Pbea, P. beatae; Pman, P. maniculatus; Pleu, P. leucopus; Patt, P. attwateri; Rful, fulvescens; Plac, P. laceianus. TX, Texas; LA, Louisiana; HND, Honduras; COL, Colima; CA, California; GRO, Guerrero; MOR, Morelos; VER, Veracruz; CRI, Costa Rica; AZ, Arizona; MXS, state of Mexico; NLE, Nuevo León; SLP, San Luis Potosí; WV, West Virginia; RI, Rhode Island; TAM, Tamaulipas. NM, New Mexico. The 4 “other” South American taxa were Andes virus, Laguna Negra virus, Maporal virus, and Rio Mamoré virus. An asterisk (*) denotes that the sequence was the complete N protein gene sequence. The other sequences in the N protein gene sequence dataset were 377-nt (LSCV strain HV H0460017), 397-nt (n = 26), 545-nt (n = 7), or 1194-nt (LSCV strain 68273). The length of the scale bar is equivalent to 0.1 substitution per site, a black dot (•) indicates that the clade probability values were ≥95.0, and Andes virus strain Chile-9717869 was the designated outgroup.
FIG. 3.
Phylogenetic relationships among Muleshoe virus and other hantaviruses based on Bayesian analyses of glycoprotein precursor gene sequences. The labels are the same as those in Figure 2. The LSCV and OROV sequences were 3328-nt and 1537-nt, respectively; the 12 other North American sequences were complete and ranged from 3420- to 3426-nt. The length of the scale bar is equivalent to 0.4 substitution per site, a black dot (•) indicates that the clade probability values were ≥95.0, and Andes virus strain Chile-9717869 was the designated outgroup.
Nonidentities between the amino acid sequence of the N protein of MULV strain HV B0530033 and the amino acid sequences of the N proteins of the four other hantaviruses associated with North American sigmodontine rodents ranged from 7.2% to 10.5% (Table 4). Similarly, nonidentities between the amino acid sequence of the GPC of HV B0530033 and the amino acid sequences of the GPC of the other hantaviruses associated with North American sigmodontine rodents ranged from 10.2% to 12.3% (Table 4).
Table 4.
Nonidentities Among the Predicted Amino Acid Sequences of the Nucleocapsid Proteins and Among the Predicted Amino Acid Sequences of the Glycoprotein Precursors of Hantaviruses Naturally Associated with Neotomine or Sigmodontine Rodents
| Nucleocapsid protein (% sequence nonidentity)b | ||||||
|---|---|---|---|---|---|---|
| Virusa | MULV | BAYV | BCCV | CATV | OROV | Other |
| MULV | — | 7.2–7.5 | 10.0 | 7.7 | 10.5 | ≥16.1 |
| BAYV | 11.0–11.3 | — | 7.5–7.7 | 4.9–5.1 | 7.2 | ≥11.7 |
| BCCV | 12.3 | 11.2–11.5 | — | 8.4 | 10.3 | ≥14.0 |
| CATV | 11.0 | 6.4–6.7 | 11.1 | — | 7.9 | ≥12.4 |
| OROV | 10.2 | 8.4–9.0 | 9.8 | 8.4 | — | ≥13.6 |
| Other | ≥20.8 | ≥19.7 | ≥19.9 | ≥19.9 | ≥12.1 | — |
| Glycoprotein precursor (% sequence nonidentity)a | ||||||
MULV, Muleshoe virus (strain HV B0530033); BAYV, Bayou virus (HV F0260003 and La Hu); BCCV, Black Creek Canal virus (SPB 9408076); CATV, Catacamas virus (HV C1280001); OROV, Playa de Oro virus; Other—Carrizal virus (26/2006), El Moro Canyon virus (RM ·97), Huitzilac virus (200/2006), Limestone Canyon virus (68273), Montano virus (104/2006), New York virus (NY-1), Sin Nombre virus (Convict Creek 74 and Convict Creek 107). The pairwise comparisons of N protein sequences also included Sin Nombre virus strain Monongahela-1 and Rio Segundo virus strain RMx·Costa·1.
Numbers above and below the diagonal are nonidentities among the amino acid sequences of the nucleocapsid proteins and glycoprotein precursors, respectively.
The Bayesian analyses of Cytb gene sequences separated the 25 hispid cotton rats into two groups (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/vbz): Group I—TTU-M80670 (Morris County); TTU-M119675 (Lynn County); the nine other rats captured in Texas in counties west of Morris County (Fig. 1); and the rats from Arizona, Kansas, New Mexico, Oklahoma, and Mexico (Nuevo León and Tamaulipas) and Group II—the rat from Nacogdoches County, Texas; the rat from Miami-Dade County, Florida; the two other rats from Florida; and the rats from Louisiana and Tennessee. Monophyly of the 18 rats in group I and monophyly of the 7 rats in group II were supported by analyses done a posteriori (clade probability values, 0.99). We note that MULV RNA was found in TTU-M80670 and TTU-M119675 in this study (Table 3) and that the rat from Miami-Dade County was captured alongside BCCV-infected hispid cotton rats (CFF, unpublished data).
Discussion
Specific rodents (usually one or two closely related species) are principal hosts of the hantaviruses for which natural host relationships have been well characterized. For example, the hispid cotton rat (S. hispidus) is the principal host of BCCV in southern Florida (Glass et al. 1998); the brush mouse (P. boylii), LSCV in Arizona (Sanchez et al. 2001); and the deer mouse (P. maniculatus), SNV in the western United States (Childs et al. 1994).
This study together with a previous study (Rawlings et al. 1996) revealed that MULV is distributed across northern Texas—from Morris County almost 700 km westward to Bailey and Deaf Smith counties. The results of the analyses of Cytb gene sequences herein support the notion that the hispid cotton rat associated with MULV is phylogenetically distinct from the hispid cotton rat that serves as the principal host of BCCV in southern Florida (Rawlings et al. 1996).
Presently, our knowledge of the natural host relationships of MULV is limited to virus-specific RNA from 4 (14.3%) of 28 cotton rats captured in Lynn or Morris county (Table 2) and 2 (3.8%) of 53 cotton rats captured in Bailey or Deaf Smith county (Rawlings et al. 1996). Arguably, the MULV RNA in the six infected cotton rats was a consequence of contemporary interspecific virus transmission rather than intraspecific (cotton rat-to-cotton rat) virus transmission. Clearly, further work is needed to determine whether S. hispidus in northern Texas is the principal host of MULV.
The results of the Bayesian analyses of N protein gene sequences (Fig. 2) and GPC gene sequences (Fig. 3) affirmed that MULV is closely related to BCCV but failed to determine whether MULV is most closely related to BCCV, BAYV, or CATV. The Ninth Report of the International Committee on Taxonomy of Viruses (Plyusnin et al. 2012) indicated that strains of different hantaviral species should exhibit at least 7% amino acid sequence nonidentity in (i) comparisons of complete N protein sequences and (ii) comparisons of complete GPC sequences. The results of the pairwise comparisons of complete N protein sequences and complete GPC sequences in this study (Table 4) strengthen support for the decision by the Committee to acknowledge that Muleshoe virus is distinct from Black Creek Canal virus, Bayou virus, and all other species included in the Bunyaviridae, genus Hantavirus.
This study showed that LSCV was enzootic in Texas in 1998–1999 and that BAYV is associated with O. texensis in Galveston and Matagorda counties, as well as Brazoria and Jefferson counties (Torrez-Martinez et al. 1998, Houck et al. 2001). In addition, this study along with previous studies (Rawlings et al. 1996, Monroe at al. 1999) revealed that SNV in association with various Peromyscus spp. is geographically widely distributed in Texas.
Hantavirus pulmonary syndrome (HPS, also known as hantavirus cardiopulmonary syndrome or HCPS) was first described as a distinct clinical entity in human medicine in 1993 (MacNeil et al. 2011). Through April 2016, 46 HPS cases were reported to the Texas Department of State Health Services from 29 counties in Texas (Fig. 1). The etiologic agent was BAYV in 5 cases, SNV in 29 cases, and indeterminate in 12 cases.
The geographic distribution of S. hispidus in Texas (Rawlings et al. 1996, Schmidly and Bradley 2016) includes 28 of the 29 counties that reported HPS cases to the Department of State Health Services. Hypothetically, MULV was the etiologic agent in 1 or more of the 12 cases in which the identity of the hantavirus was not determined.
A previous study (Fulhorst et al. 1997) demonstrated that IgG to BCCV and selected other hantaviruses can be highly reactive with CADV in ELISA. Even so, the ELISA in this study may have failed to detect IgG to hantaviruses that are antigenically more distantly related to CADV than BCCV or the hantaviruses found in rodents in this study (Table 3).
Supplementary Material
Acknowledgments
Nicole Evert (Zoonosis Control Branch, Texas Department of State Health Services, Austin, TX) provided the information on HPS cases. Taylor Soniat (Department of Biological Sciences, Texas Tech University, Lubbock, TX) determined the nucleotide sequences of the Cytb genes of cotton rats TTU-M80670 and TTU-M119675. This research was financially supported by National Institutes of Health grant AI-41435, “Ecology of emerging arenaviruses in the southwestern United States.”
Author Disclosure Statement
No competing financial interests exist.
References
- Bradley RD, Schmidly DJ, Amman BR, Platt RN II, et al. . Molecular and morphometric data reveal multiple species in Peromyscus pectoralis. J Mammal 2015; 96:446–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs JE, Ksiazek TG, Spiropoulou CF, Krebs JW, et al. . Serologic and genetic identification of Peromyscus maniculatus as the primary rodent reservoir for a new hantavirus in the southwestern United States. J Infect Dis 1994; 169:1271–1280 [DOI] [PubMed] [Google Scholar]
- Erixon P, Svennblad B, Britton T, Oxelman B. Reliability of Bayesian posterior probabilities and bootstrap frequencies in phylogenetics. Syst Biol 2003; 52:665–673 [DOI] [PubMed] [Google Scholar]
- Fulhorst CF, Monroe MC, Salas RA, Duno G, et al. . Isolation, characterization and geographic distribution of Caño Delgadito virus, a newly discovered South American hantavirus (family Bunyaviridae). Virus Res 1997; 51:159–171 [DOI] [PubMed] [Google Scholar]
- Glass GE, Livingstone W, Mills JN, Hlady WG, et al. . Black Creek Canal virus infection in Sigmodon hispidus in southern Florida. Am J Trop Med Hyg 1998; 59:699–703 [DOI] [PubMed] [Google Scholar]
- Hanson JD, Indorf JL, Swier VJ, Bradley RD. Molecular divergence in the Oryzomys palustris complex: Evidence for multiple species. J Mammal 2010; 91:336–347 [Google Scholar]
- Houck MA, Qin H, Roberts HR. Hantavirus transmission: Potential role of ectoparasites. Vector-Borne Zoonotic Dis 2001; 1:75–79 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist FR. MrBayes: Bayesian inference for phylogeny. Biometrics 2001; 17:754–756 [DOI] [PubMed] [Google Scholar]
- Irwin DM, Kocher TD, Wilson AC. Evolution of the cytochrome b gene of mammals. J Mol Evol 1991; 32:128–144 [DOI] [PubMed] [Google Scholar]
- MacNeil A, Ksiazek TG, Rollin PE. Hantavirus pulmonary syndrome, United States, 1993–2009. Emerg Infect Dis 2011; 17:1195–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantooth SJ, Milazzo ML, Bradley RD, Hice CL, et al. . Geographical distribution of rodent-associated hantaviruses in Texas. J Vector Ecol 2001; 26:7–14 [PubMed] [Google Scholar]
- Mauldin MR, Keith MS, Milazzo ML, Hanson JD. Assessment of hantavirus and arenavirus antibody prevalence and associated rodent species in Dickens County, Texas. Texas J Sci 2013; 64:157–171 [Google Scholar]
- Milazzo ML, Cajimat MNB, Hanson JD, Bradley RD, et al. . Catacamas virus, a hantaviral species naturally associated with Oryzomys couesi (Coues' oryzomys) in Honduras. Am J Trop Med Hyg 2006; 75:1003–1010 [PMC free article] [PubMed] [Google Scholar]
- Milazzo ML, Duno G, Utrera A, Richter MH, et al. . Natural host relationships of hantaviruses native to western Venezuela. Vector-Borne Zoonotic Dis 2010; 10:605–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milazzo ML, Cajimat MNB, Romo HE, Estrada-Franco J, et al. . Geographic distribution of hantaviruses associated with neotomine and sigmodontine rodents, Mexico. Emerg Infect Dis 2012; 18:571–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milazzo ML, Amman BR, Cajimat MNB, Méndez-Harclerode FM, et al. . Ecology of Catarina virus (family Arenaviridae) in southern Texas, 2001–2004. Vector-Borne Zoonotic Dis 2013; 13:50–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe MC, Morzunov SP, Johnson AM, Bowen MD, et al. . Genetic diversity and distribution of Peromyscus-borne hantaviruses in North America. Emerg Infect Dis 1999; 5:75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morzunov SP, Feldmann H, Spiropoulou CF, Semenova VA, et al. . A newly recognized virus associated with a fatal case of hantavirus pulmonary syndrome in Louisiana. J Virol 1995; 69:1980–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser GG, Carleton MD. Superfamily Muroidea. In: Wilson DE, Reeder DM, eds. Mammal Species of the World. A Taxonomic and Geographic Reference. Third edition. Baltimore: Johns Hopkins University Press, 2005:894–1522 [Google Scholar]
- Peppers LL, Bradley RD. Cryptic species in Sigmodon hispidus: Evidence from DNA sequences. J Mammal 2000; 81:332–343 [Google Scholar]
- Peppers LL, Carroll DS, Bradley RD. Molecular systematics of the genus Sigmodon: Evidence from the mitochondrial cytochrome b gene. J Mammal 2002; 83:396–407 [Google Scholar]
- Pitts RM, Mauldin MR, Thompson CW, Choate JR. Evidence of hantavirus exposure in rodents from North Texas. West North Am Nat 2013; 73:386–391 [Google Scholar]
- Plyusnin A, Beaty BJ, Elliott RM, Goldbach R, et al. . Family Bunyaviridae. In: King AMQ, Lefkowitz EJ, Adams MJ, Carstens EB, eds. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses (ICTV). San Diego: Elsevier Academic Press, 2012: 725–741 [Google Scholar]
- Rawlings JA, Torrez-Martinez N, Neill SU, Moore GM, et al. . Cocirculation of multiple hantaviruses in Texas, with characterization of the small (S) genome of a previously undescribed virus of cotton rats (Sigmodon hispidus). Am J Trop Med Hyg 1996; 55:672–679 [DOI] [PubMed] [Google Scholar]
- Sanchez AJ, Abbott KD, Nichol ST. Genetic identification and characterization of Limestone Canyon virus, a unique Peromyscus-borne hantavirus. Virology 2001; 286:345–353 [DOI] [PubMed] [Google Scholar]
- Schmidly DJ, Bradley RD. The Mammals of Texas. Seventh edition. Austin: University of Texas Press, 2016:694 [Google Scholar]
- Smith MF, Patton JL. The diversification of South American rodents: Evidence from mitochondrial sequence data for the akodontine tribe. Bio J Linn Soc 1993; 50:149–177 [Google Scholar]
- Swofford DL. PAUP: Phylogenetic Analysis Using Parsimony (* and other methods), version 4.0a150. Sunderland, MA: Sinauer Associates, Inc., Publishers, 2002 [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, et al. . MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W (1.7): Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choices. Nucleic Acids Res 1994; 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemann-Boege I, Kilpatrick CW, Schmidly DJ, Bradley RD. Molecular phylogenetics of the Peromyscus boylii species group (Rodentia: Muridae) based on mitochondrial cytochrome b sequences. Mol Phylogenet Evol 2000; 16:366–378 [DOI] [PubMed] [Google Scholar]
- Torrez-Martinez N, Bharadwaj M, Goade D, Delury J, et al. . Bayou virus-associated hantavirus pulmonary syndrome in eastern Texas: Identification of the rice rat, Oryzomys palustris, as reservoir host. Emerg Infect Dis 1998; 4:105–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiting AS, Bauer AM, Sites JW., Jr. Phylogenetic relationships and limb loss in sub-Saharan African scincine lizards (Squamata: Scincidae). Mol Phylogenet Evol 2003; 29:583–598 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.