Abstract
Objective: Dysfunctional remodeling of the extracellular matrix contributes to the formation of TLR-dependent feed forward loops that drive chronic inflammation. We have previously shown that two Type III domains of Fibronectin, FnEDA and FnIII-1c, cooperate to induce the synergistic release of interleukin 8 (IL-8) from dermal fibroblasts. We now identify steps in the TLR4 pathway where synergy can be demonstrated as well as additional kinases functioning in fibronectin activation of TLR4 signaling. We also evaluate the ligand and cell-type specificity of this synergistic response.
Approach: FnEDA, FnIII-1c, and lipopolysaccharide (LPS)-induced genes in fibroblasts were analyzed by a quantitative reverse transcription-polymerase chain reaction (qPCR) and protein was measured by an enzyme-linked immunosorbent assay (ELISA). Kinases functioning in gene expression were identified by using specific inhibitors. Activated TLR4-dependent effector molecules were identified by cell fractionation and Western blot and quantified by image analysis.
Results: The addition of FnEDA and FnIII-1c to dermal fibroblasts resulted in a synergistic increase in the expression of IL-8, tumor necrosis factor alpha (TNF-α), and vascular cell adhesion molecule (VCAM-1). Synergy between these domains was detected at the level of nuclear factor kappa-light chain enhancer of activated B cells (NF-κB) and inhibitor of kappa B kinase (IKK) activation. Induction of IL-8 by fibronectin ligands was partially attenuated in the presence of inhibitors to either epidermal growth factor receptor or Src kinases. FnIII-1c also synergized with LPS to induce IL-8 in dermal fibroblasts, whereas the combined effect of FnEDA and LPS on IL-8 synthesis was additive. In contrast, synergistic responses to these ligands were not observed in THP-1 monocytic cells.
Innovation: The data suggest that chronic inflammation may be driven by matrix- and pathogen-derived TLR4 ligands that work in synergy to promote an exuberant innate response.
Conclusion: The data suggest that the molecular mechanism underlying synergistic responses to TLR4 ligands lies upstream of IKK activation, likely in the molecular composition of the TLR4 receptor complex that assembles in response to each ligand. In addition, synergistic responses to TLR4 activation may be both cell-type and ligand specific.
Keywords: : fibronectin, TLR4, IL-8, inflammation, LPS

Paula J. McKeown-Longo, PhD
Introduction
Prolonged inflammation in response to injury is a contributory factor to the development of chronic wounds. Extracellular matrix (ECM)-derived products of tissue damage, known as damage-associated molecular pattern (DAMP) molecules, promote inflammation by activating Toll-like receptors (TLRs) to induce expression of inflammatory cytokines.1 After injury, dysregulation of the ECM remodeling process results in increased proteolytic activity and the continual generation of matrix fragments that function as DAMPs to promote persistent inflammation.2,3 In contrast to DAMPs that serve as endogenous agonists to TLRs, pathogen-associated molecular pattern (PAMP) molecules act as exogenous TLR agonists to signal the response to pathogens.4 The best-characterized PAMP is the bacterial cell wall component lipopolysaccharide (LPS), which is the canonical TLR4 ligand.5
After injury, stromal fibroblasts synthesize an alternatively spliced isoform of fibronectin, which includes an additional Type III domain, extra domain A (EDA).6,7 EDA+fibronectin is not expressed in normal adult tissue but is upregulated during wound healing and highly expressed in exudates from chronic wounds.8 EDA+fibronectin is increased in the skin of patients with scleroderma,9 psoriasis,4,10 and keloid scars.11 Several reports have identified the EDA+fibronectin as a DAMP molecule that functions as a TLR4 agonist to induce the synthesis of inflammatory cytokines in various cell types, including dermal fibroblasts.9,12–16 EDA+fibronectin has also been shown to promote fibro-inflammatory responses in several mouse models.9,17,18 The EDA domain of fibronectin can activate TLR4 signaling as either the individual domain, proteolytic fragment or in the context of the intact molecule.19,20
Being responsible for matrix synthesis, assembly, and turnover, stromal fibroblasts are in constant dialogue with the fibronectin matrix in its polymerized and fragmented forms. Changes in tissue mechanical forces have been shown to cause the structurally labile fibronectin type III (FnIII) domains to unfold, revealing otherwise hidden bioactive sites.21 We have identified the unfolded III-1 domain of fibronectin (III-1c) as a second domain within fibronectin that activates TLR4.13,15 This domain is conformationally labile and believed to unfold to support fibronectin polymerization.22,23 This unfolded domain can also be released from the matrix through proteolysis as it is a substrate for MMP2.24 We have recently shown that the EDA and III-1c domains of fibronectin can synergistically regulate TLR4-dependent release of the inflammatory cytokine, interleukin 8 (IL-8), in human dermal fibroblasts13; however, the molecular basis of this synergy is not known. In the current study, we demonstrate that the synergy between the two FnIII domains can be observed downstream of TLR4 activation at the level of inhibitor of kappa B kinase (IKK) phosphorylation, nuclear factor kappa-light chain enhancer of activated B cells (NF-κB) translocation to the nucleus, and IL-8, TNF-α and VCAM-1 mRNA production in human dermal fibroblasts. In addition, we show that synergistic induction of cytokines by TLR4 is both cell-type and ligand specific.
Clinical Problem Addressed
Chronic inflammation is a hallmark of poor wound healing. Identification of the biochemical and cellular mediators leading to overly robust and prolonged inflammatory responses will identify novel targets to control the exuberant inflammation driving the development of chronic wounds.
Materials and Methods
Reagents
Unless otherwise indicated, reagents were purchased from Sigma Chemical Co. (St. Louis, MO). Rabbit polyclonal antibodies to focal adhesion kinase (FAK) and Lamin A/C (H-110) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibodies to phospho-IKKα/β (Ser176/180) and NF-κB were obtained from Cell Signaling Technology (Danvers, MA). The RNeasy Mini KIT, RT2 SYBR Green qPCR Master Mix, RT2 First Strand Kit, and qPCR Primer Assays (IL-8, VCAM1, and TNF) were obtained from SA Biosciences (Frederick, MD). His-tagged recombinant fibronectin modules, FnEDA, FnIII-1c, and FnIII-10n were prepared as previously described.13 A peptide representing an unfolded stable intermediate of the III-10 domain of fibronectin (FnIII-10n) was utilized as a negative control in these experiments.25
Cell culture, treatment, and lysis
Human dermal fibroblasts (A1-F) were grown and maintained in Dulbecco's modified Eagle's medium (DMEM; Life Technologies, Grand Island, NY) containing 10% fetal bovine serum (FBS; HyClone Laboratories, Logan, UT) in an 8% CO2 humidified atmosphere at 37°C. THP-1 cells, purchased from ATCC, were maintained in Roswell Park Memorial Institute (RPMI) medium-1640 with 10% FBS, 0.05 mM 2-mercaptoethanol in a 5% CO2 atmosphere at 37°C. Before use in experiments, fibroblast monolayers were serum starved overnight in DMEM supplemented with 0.1% bovine serum albumin (BSA) and THP-1 monocytes rinsed with RPMI supplemented with 0.1% BSA. Specific treatment amounts and durations are provided in the figure legends.
Cells in 48-well culture plates were treated with fibronectin modules for 4 h, after which cell conditioned medium was collected and analyzed for IL-8 protein expression by using a human enzyme-linked immunosorbent assay (ELISA) kit (BD Biosciences, San Diego, CA), as directed by the manufacturer. For analysis of whole cell lysates, cells were placed on ice after treatment and rinsed once with phosphate-buffered saline (PBS) before lysis with cytoplasmic lysis buffer (20 mM Tris, pH 7.4, 0.1 M NaCl, 40 mM NaF, 2 mM EGTA, 0.5 mM PMSF, 1% Triton X-100, and 0.5% NP-40) supplemented with Complete Protease Inhibitor (Roche Applied Science, Indianapolis, IN).
For analysis of nuclear extracts, cells were placed on ice post-treatment and then washed once with PBS and once with low-salt buffer (10 mM HEPES, pH 7.9, 1 mM Na3VO4, 5 mM NaF, 5 mM KCl, 1.5 mM MgCl2, and 0.5 mM PMSF). Cells were then lysed in nuclear isolation buffer (low-salt buffer supplemented with 0.5% NP-40, and Complete Protease Inhibitor) and passed through a 23-gauge needle three times before centrifugation at 3,000 g and 4°C for 5 min. Nuclear pellets were washed with nuclear isolation buffer, and nuclear proteins were extracted with a high-salt buffer (nuclear isolation buffer supplemented with 0.5 M NaCl). Resuspended nuclear extracts were centrifuged at 16,000 g and 4°C for 5 min, and supernatant was collected as nuclear extract.
Immunoblot analysis
Samples were combined with 4 × Reducing Sample Buffer (0.25 M Tris, pH 6.8, 8% SDS, 40% glycerol, 20% β-mercaptoethanol, and 0.01% bromophenol blue) and boiled for 5 min before separation by electrophoresis on SDS-polyacrylamide gels. Proteins were transferred onto nitrocellulose membranes (GE Healthcare); then, they were blocked in TBST (Tris-HCl, pH 7.4, 150 mM NaCl, and 0.1% Tween 20) supplemented with 5% w/v BSA. Nitrocellulose membranes were incubated overnight with primary antibodies at 4°C. Secondary antibodies conjugated with horseradish peroxidase (HRP) were used to detect proteins with an enhanced chemiluminescence reagent (GE Healthcare). Nitrocellulose membranes were stripped in stripping buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, and 1% β-mercaptoethanol) for 10 min at 60°C to remove previously bound antibodies.
Real-time RT-PCR
qPCR was performed in quadruplicate with specific primer pairs in MicroAmp Optical 96-well reaction plates with Optical Caps by using the MyiQ cycler system from Bio-Rad Laboratories (Hercules, CA). In short, the RNeasy Mini Kit and the RT2 First Strand Kit were used to isolate and convert total RNA into first-strand cDNA in accordance with the manufacturer's protocol. Data were normalized to β-actin mRNA levels as a housekeeping gene and shown as fold change by using the ΔΔCt method.
Statistical analysis
Data are presented as the mean ± standard error of the mean of at least three independent experiments. Analysis was performed with Sigma Plot version 11.0 (Systat Software, Chicago, IL), with p < 0.05 considered significant. Synergy was determined by comparing the combined effect of TLR4 ligands against the predicted additive effect.
Results
FnIII-1c and FnEDA synergistically regulate cytokine release upstream of NF-κB activation
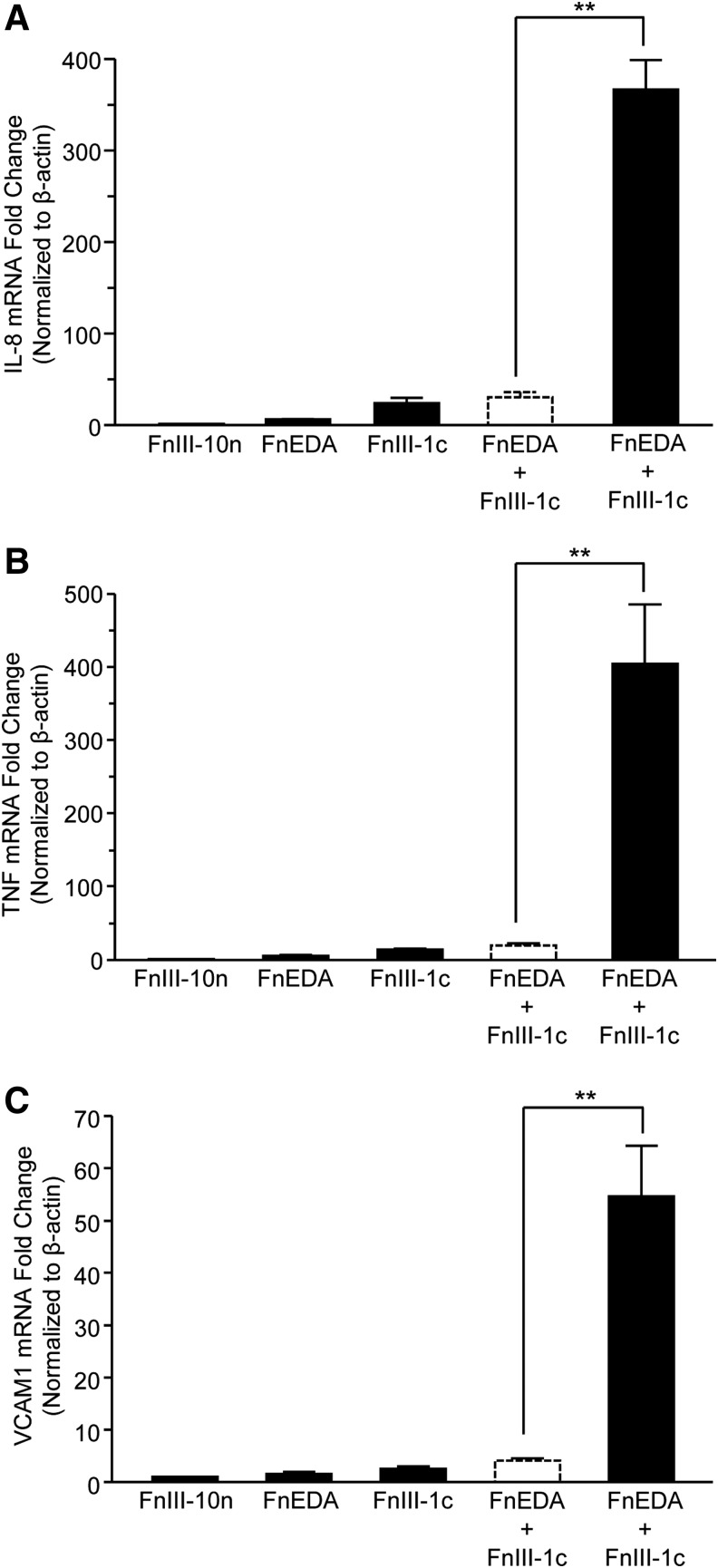
Previously, we have demonstrated that two Type III domains of fibronectin (FnIII-1c and FnEDA) synergistically induce the TLR4-dependent expression of inflammatory cytokines in human dermal fibroblasts.13,15 In an effort to understand the mechanistic basis for this synergy, we examined three steps of the TLR4-NF-κB pathway upstream of IL-8 protein secretion: IL-8 mRNA levels, NF-κB translocation to the nucleus, and activation of the upstream kinase, IKK. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis indicated that individually FnEDA and FnIII-1c induced a 6- and 24-fold increase in IL-8 mRNA, respectively; however, when cells were incubated with both domains, IL-8 mRNA was induced by 366-fold (Fig. 1A). These findings indicate that the combination of the two domains results in a synergistic increase (i.e., significantly greater than the predicted additive increase shown in the dashed line bars) in mRNA production. In addition to IL-8, we have identified several pro-inflammatory genes that are upregulated in human dermal fibroblasts after FnIII-1c and FnEDA treatment, including tumor necrosis factor-alpha (TNF-α)13,15 and vascular cell adhesion molecule-1 (VCAM-1) (unpublished observations). A synergistic increase in both TNF-α and VCAM-1 mRNA was also seen when cells were incubated with FnIII-1c and FnEDA. Analysis by qPCR showed that stimulation with FnEDA and FnIII-1c resulted in a 5.6- and 14.2-fold increase in TNF-α mRNA levels, respectively, when added individually to cells. TNF-α induction was increased to more than 400-fold when cells were incubated with both FnIII domains (Fig. 1B). Similarly, FnEDA and FnIII-1c individually induced a 1.6- and 2.6-fold increase, respectively in VCAM-1 gene expression whereas both fibronectin domains induced a 54-fold change when combined (Fig. 1C). These data indicate that the synergistic induction seen between the two FnIII domains is not limited to IL-8 but includes additional pro-inflammatory genes.
Figure 1.
FnIII-1c and FnEDA modules stimulate a synergistic increase in the expression of fibro-inflammatory genes. Confluent monolayers of human dermal fibroblasts were serum starved overnight; then, they were treated with 2 μM each of FnIII domains, alone and in combination. Data are presented as fold increase over the control module, FnIII-10n, which was set at 1. The theoretical expected additive effect of combining FnEDA and FnIII-1c is represented by the open dashed line bar. After a 2 h incubation period, mRNA was isolated and analyzed via qRT-PCR by using primers to human IL-8 (A), TNF-α (B), and human VCAM-1 (C). Statistical analysis was performed by using a Student's t-test. **p < 0.01. Data are presented as the mean ± S.E.M. (n = 3). EDA, extra domain A; IL-8, interleukin 8; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; SEM, standard error of the mean.
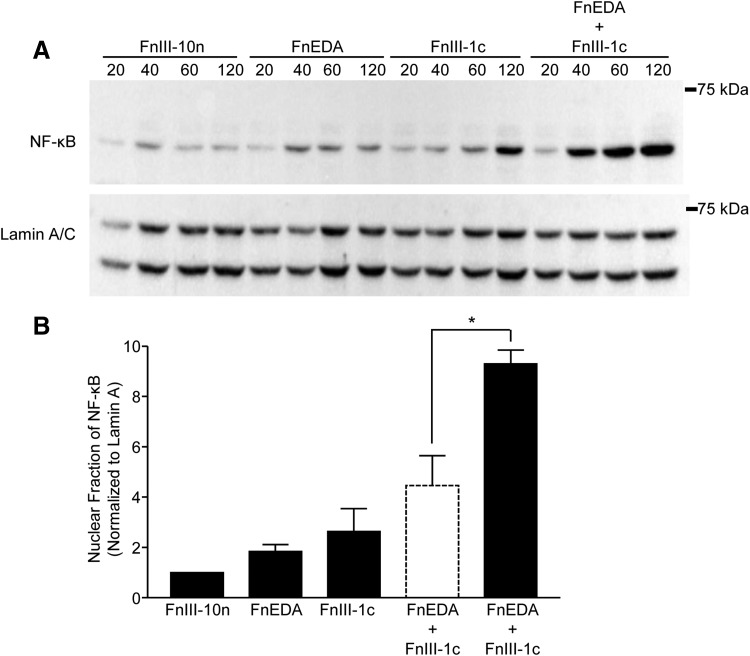
Induction of IL-8 and other inflammatory genes is dependent on the translocation of the transcription factor, NF-κB, from the cytoplasm to the nucleus, where the active subunit of NF-κB, p65 functions to initiate transcription. To assess the impact of FnIII domains on activation of NF-κB, cells were incubated with FnIII domains, either individually or in combination, and nuclear lysates were analyzed for NF-κB-p65 by Western blot (Fig. 2A). The data for the 60-min time point are quantified in Fig. 2B and show that FnEDA and FnIII-1c individually stimulated a 1.8- and 2.6-fold increase in NF-κB-p65 translocation to the nucleus, respectively. When added together, the two domains stimulated a 9.3-fold increase in the nuclear localization of NF-κB-p65, which is significantly greater than the expected additive effect of 4.5-fold (dashed line bar). A similar synergy could also be seen at the 120 min time point (data not shown).
Figure 2.
FnIII-1c and FnEDA synergistically regulate NF-κB translocation to the nucleus. Monolayers of human dermal fibroblasts were serum starved overnight before treatment with FnIII domains, either individually (2 μM) or in combination, for the indicated amount of time. Nuclear fractions were collected, and NF-κB was visualized by Western blot (A). Blots were quantified by using densitometry and values for NF-κB were normalized to Lamin A/C, which served as a loading control. Resulting values were normalized to the control (FnIII-10n) at each time point. The theoretical expected additive effect of combining FnEDA and FnIII-1c is represented by the open dashed line bar. Data are presented as fold change at 60 min (B). Statistical analysis was performed by using a Student's t-test. *p < 0.05. Data are presented as the mean ± S.E.M. (n = 3). NF-κB, nuclear factor kappa-light chain enhancer of activated B cells.
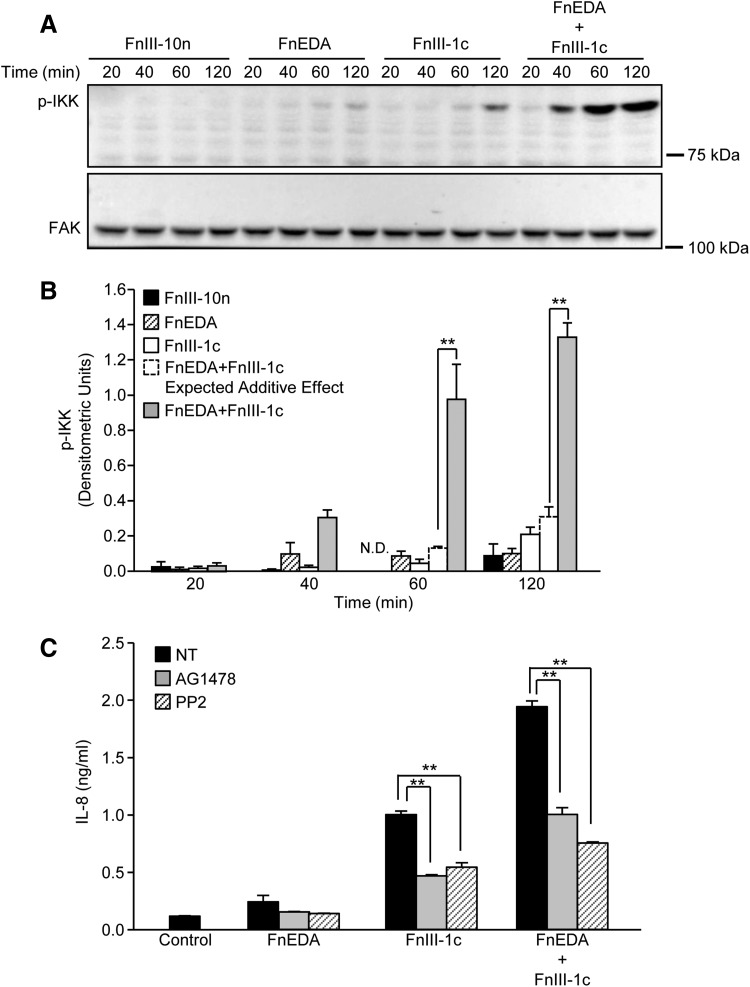
Activation of the TLR4-NF-κB signaling pathway initially results in the homodimerization of TLR4 receptors, ultimately leading to the assembly of a scaffold of associated kinases, which regulate the phosphorylation of IKK. Once activated, IKK phosphorylates the NF-κB inhibitor, IκB, thereby releasing the transcriptionally active p65 subunit to enter the nucleus. To determine whether the phosphorylation of IKK reflected the synergism observed at the level of NF-κB translocation, IL-8 transcription, and protein expression, fibroblast lysates were collected after the addition of FnIII-1c and FnEDA, alone and in combination. Cells incubated with the control domain (FnIII-10n) showed no activation of IKK over the 120 min time course (Fig. 3A). The FnEDA and FnIII-1c domains showed modest increases in IKK phosphorylation; however, there was a marked increase in IKK phosphorylation when both domains were incubated with cells. Quantification of data obtained by image analysis of Western blots representing three different experiments demonstrated that synergy between the two modules is evident at the 60 and 120 min time points (Fig. 3B). Densitometric units representing p-IKK indicate that incubation of dermal fibroblasts with FnEDA and FnIII-1c for 60 min resulted in densitometric units of 0.086 and 0.045, respectively; however, incubation with both FnEDA and FnIII-1c for 60 min resulted in densitometric units of 0.98, consistent with a synergistic increase in IKK phosphorylation. A similar result was seen after 120 min where the individual densitometric units, 0.10 and 0.21, of FnEDA and FnIII-1c increased to 1.33 when the domains were added simultaneously. Together, these data indicate that the synergy between the two modules is established upstream of the phosphorylation of IKK, perhaps at the level of TLR4 receptor complex assembly. TLR complexes are dynamic structures consisting of various ancillary molecules and co-receptors that influence TLR activation and downstream signaling. The molecular components of the TLR4 receptor complex formed in response to FnIII domains are not well understood. Based on recent findings linking TLR4 to epidermal growth factor receptor (EGFR)/Src signaling,26–28 we evaluated whether either EGFR or Src kinases contributed to IL-8 release initiated by FnEDA and FnIII-1c. As shown in Fig. 3C, IL-8 induction in response to FnEDA and FnIII-1c, individually or combined, was attenuated (∼50%) by inhibitors for EGFR (AG1478) and Src kinases (PP2). The data suggest that FnIII domain/TLR4-dependent cytokine release was partially dependent on EGFR and Src signaling and that in response to fibronectin DAMPs, TLR4 receptors on dermal fibroblasts form functional complexes with EGFR and Src.
Figure 3.
FnIII-1c and FnEDA synergistically regulate IKK activation. Monolayers of human dermal fibroblasts were serum starved overnight; then, they were treated with the designated FnIII domains (2 μM), alone and in combination. At the indicated time, cells were lysed and phospho-IKK (pIKK) was assessed by Western blot. FAK served as a loading control (A). Blots were quantified by densitometry, and values for p-IKK were normalized to FAK. The theoretical expected additive effect of combining FnEDA and FnIII-1c is represented by the open dashed line bar at the 60 and 120 min time points. Graph represents the mean ± S.E.M. of three individual experiments (B). Statistical analysis was performed by using a Student's t-test. **p < 0.01. Fibroblasts were preincubated for 1 h with inhibitors to EGFR kinase, AG1478 (10 μM), Src kinase, PP2 (10 μM), or NT before incubation with the designated FnIII domains (4 μM), individually or in combination. Control cells received just buffer. After an additional 4 h, IL-8 in the conditioned medium was determined by ELISA (C). Statistical analysis was performed by using a Student's t-test. **p < 0.01. Data represent the mean ± S.E.M. of triplicate samples from one representative experiment. EGFR, epidermal growth factor receptor; ELISA, enzyme-linked immunosorbent assay; FAK, focal adhesion kinase; IKK, inhibitor of kappa B kinase; NT, not treated.
Fibronectin matrix-derived DAMPs selectively synergize with LPS to regulate IL-8 expression
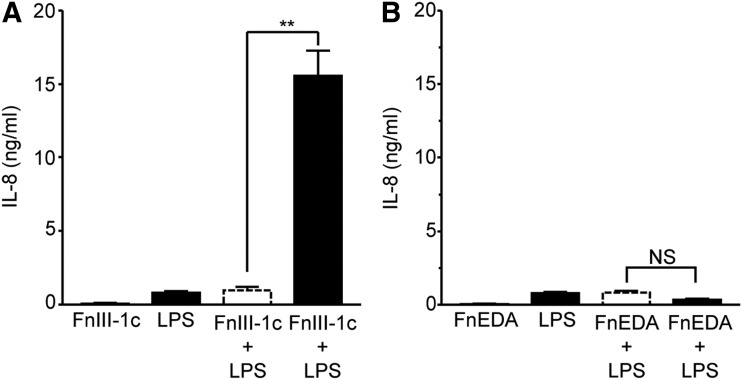
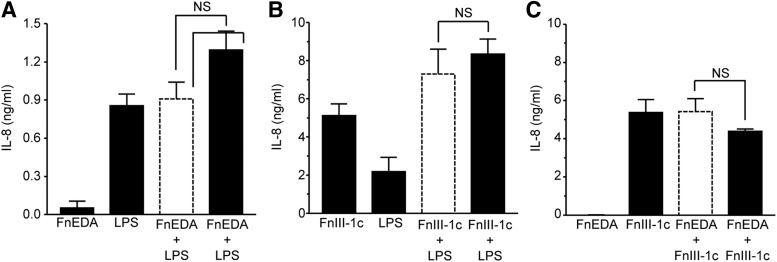
TLRs function as surveillance for both exogenous pathogens and endogenous products of tissue damage. TLR4 recognizes gram-negative bacteria and is activated in response to LPS. To determine whether fibronectin modules synergize with LPS to induce expression of cytokines, human dermal fibroblasts were treated with LPS, FnIII-1c, and FnEDA, individually and in combination. Individually, FnIII-1c and LPS induced <1 ng/mL of IL-8; however, when combined, they stimulated the release of more than 15 ng/mL of IL-8, indicating synergy between FnIII-1c and LPS (Fig. 4A). This synergy was not observed between FnEDA and LPS (Fig. 4B).
Figure 4.
FnIII-1c and LPS synergistically regulate IL-8 expression in dermal fibroblasts. Monolayers of human dermal fibroblasts (A, B) were serum starved overnight before treatment with 2 μM of the designated FnIII domains and 2 μg/mL LPS, alone or in combination. The theoretical expected additive effect of combining FnEDA and FnIII-1c is represented by the open dashed line bar. After 4 h, conditioned medium was collected and IL-8 was determined by ELISA. Statistical analysis was performed by using a Student's t-test. **p < 0.01. NS, not significant. Data are presented as the mean ± S.E.M. (n = 3). LPS, lipopolysaccharide.
To determine whether the synergy seen between TLR4 ligands in fibroblasts was also observed in immune cells, similar experiments were performed by using THP-1 monocytic cells. As shown in Fig. 5A, incubation of monocytic cells with either FnEDA or LPS elicited an IL-8 response of 0.1 and 0.9 ng/mL, respectively. When treated with both LPS and FnEDA, the cells released ∼1.2 ng of IL-8, indicating that their combined effect was additive and not synergistic. A similar lack of synergy was seen between FnIII-1c and LPS (Fig. 5B) or between FnIII-1c and FnEDA (Fig. 5C). The data suggest that although both DAMPs and PAMPs can synergize to induce a rapid, robust innate immune response in the resident stromal cells within a tissue, this synergy is both ligand- and cell-type specific.
Figure 5.
Fibronectin modules and LPS do not synergistically regulate IL-8 expression in human monocytes. THP-1 monocytes were rinsed with serum-starving medium before treatment with 2 μM of the designated FnIII domains and 2 μg/mL LPS, alone or in combination. Conditioned medium was collected after 4 h; then, it was analyzed by ELISA for IL-8. The theoretical expected additive effect of combining FnEDA and FnIII-1c is represented by the open dashed line bar. Data represent the mean ± S.E.M. (n = 3, A/B; n = 5, C). Statistical analysis was performed by using a Student's t-test. NS, not significant.
Discussion
Remodeling of the fibronectin matrix is characterized by the synthesis of EDA+fibronectin and by the unfolding of Type III domains. In previous studies, we have shown that FnEDA and the unfolded FnIII-1c domain stimulate the release of pro-inflammatory cytokines from dermal fibroblasts via the TLR4-NF-κB pathway.13 When combined, these domains promote a synergistic increase in the expression of the pro-inflammatory cytokine, IL-8. We now show that in addition to IL-8, the fibronectin domains also synergistically regulate the expression of the pro-inflammatory genes, TNF-α and VCAM-1, in human dermal fibroblasts. Examination of the TLR4 signaling pathway after FnIII-1c and FnEDA stimulation showed synergistic regulation at the level of IL-8 message transcription, NF-κB translocation to the nucleus, and phosphorylation of IKK, suggesting that the molecular basis for this phenomenon lies further upstream in the signaling pathway, possibly at the formation of the myddosome or at the assembly of the TLR4 receptor complex.
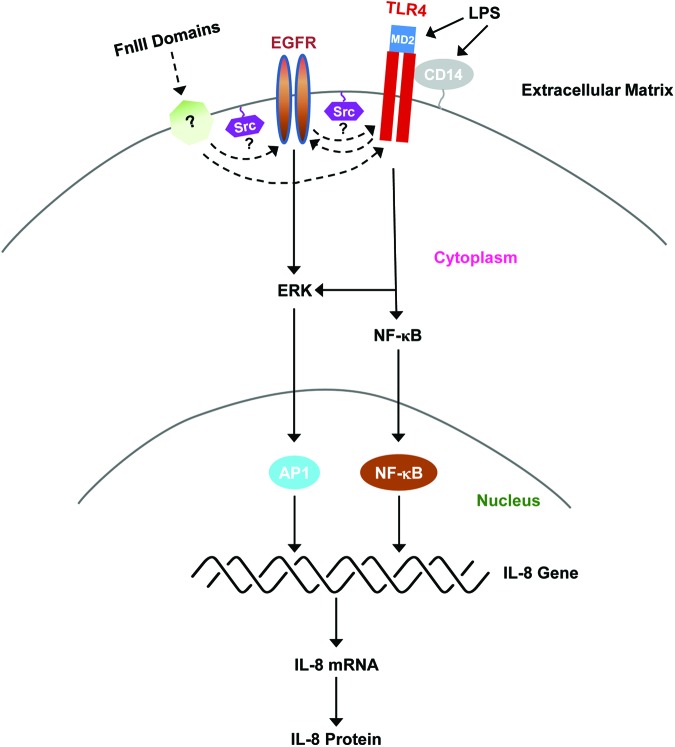
The myddosome is a signaling platform that assembles on the cytoplasmic tail of the TLR receptor and consists of the adaptor proteins MyD88 and TIRAP, which initiate signaling through the binding of IL-1 receptor-associated kinases (IRAKs), leading to the activation of IKK and NF-κB.29,30 TLR4 activation occurs in lipid rafts and depends on the activity of ancillary molecules and co-receptors that together can form physical or functional complexes.31 TLR complexes can be dynamic and assemble in a ligand-specific manner, resulting in the activation of distinct downstream effectors.32 As shown in Fig. 6, the activation of TLR4 by LPS is dependent on CD14 and MD2, which function to bind LPS and induce TLR4 dimerization and downstream signaling. Whether FnIII-1c or FnEDA binds directly to TLR4 or to TLR4 co-receptors is not known. Recent studies have now identified Src and EGFR as additional raft-associated proteins that are involved in transmitting LPS/TLR4-dependent signals, although the interrelationship among these proteins is not well understood.28,33,34 Our studies show that inhibitors of EGFR and Src kinases attenuated IL-8 release in response to the matrix-derived DAMPs, FnEDA and FnIII-1c. The inhibition, although significant, was only partial (∼50%), suggesting that some of the signaling downstream of TLR4 was independent of EGFR and Src activity.
Figure 6.
Regulation of IL-8 expression by LPS and FnIII domains. The model depicts the activation of TLR4 signaling in response to LPS and FnIII domains. LPS binds to both CD14, a GPI-linked protein, and MD2, a TLR4-associated protein. The interaction of LPS with these molecules triggers TLR4 dimerization and activation of downstream signaling to NF-κB and mitogen-active protein kinase pathways such as ERK. Activation of the NF-κB and AP1 transcription factors then induces the expression of IL-8 mRNA and protein synthesis. EGFR and Src kinases have been demonstrated to regulate the TLR4-dependent release of cytokines by LPS. The current study has now implicated Src and EGFR in cytokine release in response to FnIII domains. The specific roles of Src kinases and EGFR in TLR4 activation and signaling are not well understood. Possible interrelationships are depicted by dotted lines. The molecule labeled with a question mark (?) represents possible binding partners for FnIII domains that might participate in TLR4 activation. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/wound
Our data indicate that individual fibronectin Type III domains can synergize with each other and with LPS to regulate IL-8 expression in human dermal fibroblasts. How this synergy may reveal itself in vivo is not well understood. After tissue injury, fibroblasts synthesize fibronectin fibers that are rich in EDA-fibronectin, which promotes TLR4-dependent inflammatory responses.6–8,11 Several studies have now demonstrated that changes in the balance of tissue mechanical forces lead to the unfolding of fibronectin Type III domains.21,35,36 Our data suggest a model whereby placing EDA-rich fibers under mechanical strain as a result of increased tissue stiffness or cellular contractile force would unmask the pro-inflammatory activity present in the III-1 domain, thus synergistically enhancing the release of cytokines by fibroblasts that are adherent to the matrix.
Our study also demonstrates that the FnIII-1c domain and LPS synergize to enhance IL-8 release, indicating a previously undescribed synergy between tissue damage and pathogen invasion in the regulation of the innate immune response. Interestingly, increased levels of circulating LPS have been noted in patients with hepatic fibrosis.37 In addition, increased TLR4 signaling and fibronectin matrix deposition by hepatic stellate cells have been implicated in the progression of liver fibrosis.38 Activated stellate cells are highly contractile, providing a potential mechanism for Type III domain unfolding to contribute to a synergistic release of cytokines in response to circulating LPS. Alternatively, activated stellate cells could stimulate the release of FnIII-1c activity from matrix fibers through the increased synthesis of MMP2.24,39 These data suggest that in the presence of pathogens the contractile or proteolytic unveiling of TLR4 binding sites within fibronectin's III-1 domain would result in an exuberant fibro-inflammatory response. In contrast, incubation of fibroblasts with FnEDA and LPS resulted in an additive increase in IL-8 expression, demonstrating that only selective ligands synergize to induce inflammatory gene expression.
Interestingly, synergy between FnIII-1c and LPS or between FnIII-1c and FnEDA was not seen in THP-1 monocytic cells, suggesting that synergistic responses to TLR4 agonists are cell-type specific. The molecular components of the active TLR4 complex regulating the response to fibronectin DAMPs are not known. Therefore, it is possible that the cell type and ligand specificity of the synergistic response initiated by selected TLR4 ligands is dependent on these components. Identification of the molecular mechanisms underlying synergy could provide novel target(s) that could be exploited to either enhance immunity or dampen the inflammatory response. Furthermore, an understanding of the biochemical basis for the synergistic interplay among TLR4 ligands represents an opportunity to selectively regulate the immune response to either pathogens or tissue damage.
Key Findings.
• Synergism in the TLR4 signaling pathway activated in skin fibroblasts by DAMPs (FnEDA and FnIII-1c) was demonstrated at the level of IKK phosphorylation and in the nuclear translocation of NF-κB.
• Inhibitors of Src and EGFR kinases partially blocked cytokine release in response to Fn DAMPs, suggesting that these kinases are functionally linked to TLR4 activation.
• Synergism in the release of cytokines from skin fibroblasts was also seen between FnIII-1c and pathogen-derived LPS but not between FnEDA and LPS, indicating ligand specificity to the synergistic response.
• Combination of Fn DAMPs and LPS did not yield synergistic cytokine release in monocytic cells, indicating a cell-type response to TLR4 ligands.
Innovation
The data indicate that TLR4 ligands can work in synergy to drive inflammation and that this synergy occurs during the initial steps after TLR4 activation. The data also indicate that the synergy is selective to specific TLR4 ligands and cell types. Identification of the molecular steps that are unique to the synergistic activation of NF-κB will provide a framework for the identification of therapeutic targets to treat the exuberant inflammatory responses that are associated with poor wound healing.
Abbreviations and Acronyms
- A1-F
human dermal fibroblasts
- BSA
bovine serum albumin
- CD14
cluster of differentiation 14
- cDNA
complementary deoxyribonucleic acid
- DAMP
damage-associated molecular pattern
- DMEM
Dulbecco's modified eagle medium
- ECM
extracellular matrix
- EDA
extra domain A
- EGFR
epidermal growth factor receptor
- EGTA
ethylene glycol-bis(β-aminoethyl ether)-N,N,N',N'-tetraacetic acid
- ELISA
enzyme-linked immunosorbent assay
- FAK
focal adhesion kinase
- FBS
fetal bovine serum
- Fn
fibronectin
- HEPES
hydroxyethylpiperazineethane sulfonic acid
- HRP
horseradish peroxidase
- IκB
inhibitor of kappa B
- IKK
inhibitor of kappa B kinase
- IL-8
interleukin 8
- IRAK
interleukin-1 receptor-associated kinases
- KCl
potassium chloride
- LPS
lipopolysaccharide
- MD2
message digest algorithm 2
- MMP2
matrix metallopeptidase-2
- mRNA
message ribonucleic acid
- MyD
myeloid differentiation primary response gene
- NaCl
sodium chloride
- NaF
sodium fluoride
- NF-κB
nuclear factor kappa-light chain enhancer of activated B cells
- NT
not treated
- PAMP
pathogen-associated molecular pattern
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- p-IKK
phospho-inhibitor of kappa B
- PMSF
phenylmethane sulfonly fluoride
- qPCR
quantitative polymerase chain reaction
- qRT-PCR
quantitative reverse transcription-polymerase chain reaction
- RNA
ribonucleic acid
- RPMI
Roswell Park Memorial Institute medium
- SDS
sodium dodecyl sulfate
- S.E.M.
standard error of the mean
- TBST
tris-buffered saline Tween 20
- THP-1
human leukemia monocytic cell line
- TLR
toll-like receptor
- TNF-α
tumor necrosis factor alpha
- Tris
Tris(hydroxymethyl)aminomethane
- VCAM-1
vascular cell adhesion molecule
Acknowledgments and Funding Sources
This work was supported by grants from the National Institutes of Health R01-CA058636 and R21-AR067956 to P.J.M.-L.
Author Disclosure and Ghostwriting
There are no competing financial interests for any author. The content of this article was expressly written by the authors listed. No ghostwriters were used to write this article.
About the Authors
Lauren M. Valenty, completed the requirements for an MS degree in Biomedical Sciences at the Albany Medical Center (AMC) and is currently a student in the Physician Assistant Program at AMC. Christine M. Longo, received an MD from Ross University, completed an internship in General Surgery at AMC, and is currently a postdoctoral fellow in the Department of Regenerative and Cancer Cell Biology (RCCB) at AMC. Carol Horzempa, received a BS from the State University of New York at Geneseo and is manager of lab operations in RCCB at AMC. Anthony Ambesi, received a PhD from the Medical University of South Carolina and completed a postdoctoral fellowship at Yale University. He is an assistant professor in RCCB at AMC. Paula J. McKeown-Longo, received a PhD from the University of Connecticut and completed a postdoctoral fellowship at the University of Wisconsin. She is professor and co-chair of RCCB at AMC.
References
- 1.Schaefer L. Complexity of danger: the diverse nature of damage-associated molecular patterns. J Biol Chem 2014;289:35237–35245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eming SA, Koch M, Krieger A, et al. . Differential proteomic analysis distinguishes tissue repair biomarker signatures in wound exudates obtained from normal healing and chronic wounds. J Proteome Res 2010;9:4758–4766 [DOI] [PubMed] [Google Scholar]
- 3.McCarty SM, Percival SL. Proteases and delayed wound healing. Adv Wound Care (New Rochelle) 2013;2:438–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gubán B, Vas K, Balog Z, et al. . Abnormal regulation of fibronectin production by fibroblasts in psoriasis. Br J Dermatol 2016;174:533–541 [DOI] [PubMed] [Google Scholar]
- 5.Beutler B. TLR4 as the mammalian endotoxin sensor. Curr Top Microbiol Immunol 2002;270:109–120 [DOI] [PubMed] [Google Scholar]
- 6.Muro AF, Chauhan AK, Gajovic S, et al. . Regulated splicing of the fibronectin EDA exon is essential for proper skin wound healing and normal lifespan. J Cell Biol 2003;162:149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ffrench-Constant C, van de Water L, Dvorak HF, Hynes RO. Reappearance of an embryonic pattern of fibronectin splicing during wound healing in the adult rat. J Cell Biol 1989;109:903–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stanley CM, Wang Y, Pal S, et al. . Fibronectin fragmentation is a feature of peridontal disease sites and diabetic foot and leg wounds and modifies cell behavior. J Peritontol 2008;79:861–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhattacharyya S, Tamaki Z, Wang W, et al. . FibronectinEDA promotes chronic cutaneous fibrosis through toll-like receptor signaling. Sci Transl Med 2014;6:232ra50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McFadden JP, Basketter DA, Dearman RJ, Kimber IR. Extra domain A-positive fibronectin-positive feedback loops and their association with cutaneous inflammatory disease. Clin Dermatol 2011;29:257–265 [DOI] [PubMed] [Google Scholar]
- 11.Andrews JP, Marttala J, Macarak E, Rosenbloom J, Uitto J. Keloid pathogenesis: potential role of cellular fibronectin with the EDA domain. J Invest Dermatol 2015;135:1921–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gondokaryono SP, Ushio H, Niyonsaba F, et al. . The extra domain A of fibronectin stimulates murine mast cells via toll-like receptor 4. J Leukocyte Biol 2007;82:657–665 [DOI] [PubMed] [Google Scholar]
- 13.Kelsh R, You R, Horzempa C, Zheng M, McKeown-Longo PJ. Regulation of the innate immune response by fibronectin: synergism between the III-1 and EDA domains. PLoS One 2014;9:e102974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng M, Jones DM, Horzempa C, Prasad A, McKeown-Longo PM. The first type III domain of fibronectin is associated with the expression of cytokines within the lung tumor microenvironment. J Cancer 2011;2:478–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.You R, Zheng M, McKeown-Longo PJ. The first type III repeat in fibronectin activates an inflammatory pathway in dermal fibroblasts. J Biol Chem 2010;285:36255–36259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lasarte JJ, Casares N, Gorraiz M, et al. . The extra domain A from fibronectin targets antigens to TLR4-expressing cells and induces cytotoxic T cell responses in vivo. J Immunol 2007;178:748–756 [DOI] [PubMed] [Google Scholar]
- 17.Muro AF, Moretti FA, Moore BB, et al. . An essential role for fibronectin extra type III domain A in pulmonary fibrosis. Am J Resp Crit Care Med 2008;177:638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Booth AJ, Wood SC, Cornett AM, et al. . Recipient-derived EDA fibronectin promotes cardiac allograft fibrosis. J Pathol 2012;226:609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Julier Z, Martino MM, de Titta A, Jeanbart L, Hubbelll JA. The TLR4 agonist fibronectin extra domain A is cryptic, exposed by elastase-2; use in a fibrin matrix cancer vaccine. Sci Rep 2015;5:8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mogami H, Kishore AH, Shi H, Keller PW, Akgul Y, Word RA. Fetal fibronectin signaling induces matrix metalloproteases and cyclooxygenase-2 (COX-2) in amnion cells and preterm birth in mice. J Biol Chem 2013;288:1953–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klotzsch E, Smith ML, Kubow KE, et al. . Fibronectin forms the most extensible biological fibers displaying switchable force-exposed cryptic binding sites. Proc Natl Acad Sci U S A 2009;106:18267–18272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hocking DC, Sottile J, McKeown-Longo PJ. Fibronectin's III-1 module contains a conformation-dependent binding site for the amino-terminal region of fibronectin. J Biol Chem 1994;269:19183–19187 [PubMed] [Google Scholar]
- 23.Früh SM, Schoen I, Ries J, Vogel V. Molecular architecture of native fibronectin fibrils. Nat Commun 2015;6:7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doucet A, Overall CM. Broad coverage identification of multiple proteolytic cleavage site sequences in complex high molecular weight proteins using quantitative proteomics as a complement to edman sequencing. Mol Cell Proteomics 2011;10:M110..003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gee EP, Ingber DE, Stultz CM. Fibronectin unfolding revisited: modeling cell traction-mediated unfolding of the tenth type-III repeat. PLoS One 2008;3:e2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang Q, Zou C, Zhong P, et al. . EGFR mediates hyperlipidemia-induced renal injury via regulating inflammation and oxidative stress: the detrimental role and mechanism of EGFR activation. Oncotarget 2016;7:24361–24373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W, Fang Q, Zhong P, et al. . EGFR inhibition blocks palmitic acid-induced inflammation in cardiomyocytes and prevents hyperlipidemia-induced cardiac injury in mice. Sci Rep 2016;6:24580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trussoni CE, Tabibian JH, Splinter PL, O'Hara SP. Lipopolysaccharide (LPS)-induced biliary epithelial cell NRas activation requires epidermal growth factor receptor (EGFR). PLoS One 2015;10:e0125793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Motshwene PG, Moncrieffe MC, Grossman JG, et al. . An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J Biol Chem 2009;284:25404–25411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin S-C, Lo Y-C, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature 2010;465:885–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plociennikowska A, Hromada-Judycka , Borzecka K, Kwiatrkowska K. Co-operation of TLR4 and raft proteins in LPS-induced pro-inflammatory signaling. Cell Mol Life Sci 2015;72:557–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piccinini AM, Zulliani-Alvarez L, Lim JMP, Midwood KS. Distinct microenvironment cues stimulate divergent TLR4-mediated signaling pathways in macrophages. Sci Signal 2016;9:ra86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McElroy SJ, Hobbs S, Kallen M, et al. . Transactivation of EGFR by LPS induces COX-2 expression in enterocytes. PLoS One 2012;7:e38373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho R-L, Yang C-C, Lee I-T, et al. . Lipopolysaccharide induces ICAM-1 expression via a c-Src/NADPH oxidase/ROS-dependent NF-kB pathway in human pulmonary alveolar epithelial cells. Am J Physiol Lung Cell Mol Physiol 2016;310:L639–L657 [DOI] [PubMed] [Google Scholar]
- 35.Chandler EM, Saunders MP, Yoon CJ, Gourdon D, Fishbach C. Adipose progenitor cells increase fibronectin matrix strain and unfolding in breast tumors. Phys Biol 2011;8:015008. [DOI] [PubMed] [Google Scholar]
- 36.Wang K, Seo BR, Fischbach C, Gourdon D. Fibronectin mechanobiology regulates tumorigenesis. Cell Mol Bioengin 2016;9:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riordan SM, Skinner N, Nagee A, et al. . Peripheral blood mononuclear cell expression of Toll-like receptors and relation to cytokine levels in cirrhosis. Hepatol 2003;37:1154–1164 [DOI] [PubMed] [Google Scholar]
- 38.Seki E, De Minicis S, Österreicher CH, et al. . TLR4 enhances TGF-β signaling and hepatic fibrosis. Nat Med 2007;13:1324–1332 [DOI] [PubMed] [Google Scholar]
- 39.Friedman SL. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J Biol Chem 2000;275:2247–2250 [DOI] [PubMed] [Google Scholar]