Abstract
The present study aimed to investigate the role of homeobox B4 (HOXB4) in breast cancer. Analysis of The Cancer Genome Atlas data revealed that HOXB4 expression was positively associated with expression of the StAR-related lipid transfer domain protein 13 (STARD13), and the overall survival of patients with breast cancer. Immunohistochemistry and quantitative polymerase chain reaction assays demonstrated that HOXB4 expression was downregulated in breast cancer tissues compared with adjacent normal tissues and was additionally positively associated with STARD13 expression. HOXB4 promoted STARD13 expression in breast cancer cells. Chromatin immunoprecipitation and luciferase reporter assays confirmed that HOXB4 directly bound to the STARD13 promoter. Additionally, HOXB4 inhibited breast cancer cell migration and the epithelial-mesenchymal transition through the STARD13/Ras homolog (Rho) family member A/Rho associated protein kinase signaling pathway. HOXB4 overexpression enhanced the sensitivity of breast cancer cells to doxorubicin and reversed resistance in doxorubicin-resistant cells. Overall, the results indicated that HOXB4 inhibited breast cancer cell migration and enhanced the sensitivity of breast cancer cells to doxorubicin by targeting STARD13.
Keywords: breast cancer, cell migration, doxorubicin, HOXB4, STARD13
Introduction
Breast cancer is the leading cause of cancer-related death in women worldwide (1). Although chemotherapy has greatly improved the survival of patients with breast cancer, drug resistance often occurs (2). Therefore, it is important to identify new targets for preventing breast cancer metastasis and drug resistance.
HOXB4, a member of HOX family, is preferentially expressed in most hematopoietic cell types and elicits the selective expansion of more primitive populations (3). Recently, HOXB4 was shown to control stem cell amplification via its unique proline-rich region (4). However, the roles of HOXB4 in cancers are unclear.
StAR-related lipid transfer domain protein 13 (STARD13) acts as a tumor-suppressor in various types of cancers (5,6), its expression level is downregulated in metastatic breast cancer, potentially mediating a competitive endogenous RNA (ceRNA) network to inhibit breast cancer metastasis (7). Additionally, STARD13 can act as a ceRNA for Fas to promote apoptosis of hepatocellular carcinoma cells (8). Although STARD13 has been identified as a potential target for miR-125b in breast (6) and gastric cancer (9), the mechanisms regulating the transcription of STARD13 in breast cancer are unclear.
Here, we found that HOXB4 expression was positively correlated with STARD13 expression in breast cancer tissues and cells with different metastatic potentials. Additionally, we showed that HOXB4 could bind directly to the STARD13 promoter, thus inducing STARD13 expression in breast cancer. HOXB4 could also inhibit breast cancer cell migration and the EMT through the STARD13-RhoA-ROCK signaling pathway. Finally, we showed that forced expression of HOXB4 enhanced the sensitivity of breast cancer cells to doxorubicin and reversed resistance in doxorubicin-resistant cells. Therefore, STARD13 could be a downstream effector of HOBX4 in breast cancer, and HOXB4 could be targeted as a potential inhibitor of breast cancer metastasis.
Materials and methods
The cancer genome atlas (TCGA) data and patient samples
Twenty primary breast tumors with lymph node metastasis, twenty seven metastasis-free primary breast tumors, and adjacent normal tissues were obtained from the Affiliated Hospital of Jining Medical University from May 2015 to June 2016. Approval from the Institute Research Ethics Committee was obtained for the use of these clinical materials for research purposes. The R2: Genomics Analysis and Visualization Platform (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi) was used to download and analyze the HOXB4 and STARD13 mRNA expression profiling data.
Cell culture
293T, MCF-10A (normal breast epithelial cell lines), MCF-7 (relative low metastatic breast cancer cell lines), MDA-MB-435 (relative median metastatic breast cancer cell lines) and MDA-MB-231 (relative high metastatic breast cancer cell lines) cells were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China). Doxorubicin resistant MCF-7 cells (MCF-7-ADR) were purchased from KeyGen Biotech (Nanjing, China). MCF-10A, MCF-7 and MCF-7-ADR cells were cultured in Dulbecco's Modified Eagle Medium (Gibco, Grand Island, NY, USA), MDA-MB-435 cells were cultured in 1640 medium (Gibco,), MDA-MB-231 cells were cultured in L-15 medium (Gibco) with 10% fetal bovine serum, 80 U/ml penicillin and 0.08 mg/ml streptomycin at 37°C under humidified air with 5% CO2. Y-27632 (B1293), a ROCK inhibitor, was purchased from ApexBio (Hsinchu City, Taiwan).
Construction of stable cell lines
Lentivirus short hairpin(sh)RNA against human HOXB4 and a scramble non-targeting shRNA (sc-38692) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA), and inserted into pLKO.1. HOXB4 coding sequences were amplified by PCR and cloned into the SpeI and Xbal sites of pLVX-IRES-ZsGreen1, referred to as Lenti-HOXB4-CDS. The primers were described in Table I. As described previously (8). Quantitative real-time PCR (qRT-PCR) and western blot analyses were used for verification. Cells infected with Lenti-HOXB4-CDS were selected by fluorescent cell sorting.
Table I.
qRT-PCR Primer sequences and the sequence(s) for PCR.
| Gene | Sequences (5′ to 3′) |
|---|---|
| HOXB4-CDS forward (Spel) | ACTAGTATGGCTATGAGTTCTTTTTTGATCA |
| HOXB4-CDS reverse (Xbal) | TCTAGACTAGAGCGCGCGGGGGCCTCCATTG |
| GAPDH forward | CGGAGTCAACGGATTTGGTCGTAT |
| GAPDH reverse | AGCCTTCTCCATGGTGGTGAAGAC |
| HOXB4-qRT-PCR forward | ACACACCCAAACAAGGACACAGCA |
| HOXB4-qRT-PCR reverse | ACACACACGGAGAGAGGGAGAAAG |
| STARD13-qRT-PCR forward | ACAGGAGGGATTCTGGTGTAGGGG |
| STARD13-qRT-PCR reverse | AGGGAAGTTTTCATTCATTTGGCG |
| STARD13promoter (for ChIP) forward | GAGGAAAAGCAATACACGCACAAA |
| STARD13promoter (for ChIP) forward | TCAGGACAGGACCAAGAACAAGGT |
| Vimentin-qRT-PCR forward | AGGAACCAATGAGTCCCTGGAACG |
| Vimentin-qRT-PCR reverse | CTGCAGAAAGGCACTTGAAAGCTG |
| E-cadherin-qRT-PCR forward | CTCACATTTCCCAACTCCTCTCCT |
| E-cadherin-qRT-PCR reverse | ACCTTCAGCCATCCTGTTTCTCTT |
Adenovirus vectors construction
The adenovirus vectors containing the STARD13 coding area (Ad-STARD13-CDS) or STARD13 shRNA (Ad-STARD13-shRNA) were constructed by Hanbio (Shanghai, China). The constructs were verified by DNA sequencing.
Promoter activity assay
The STARD13 promoter sequences were introduced into the pGL3 vector (Promega, Madison, WI, USA) (pGL3-STARD13) for the STARD13 promoter transcriptional activity assays. The β-gal vector was used as an internal control. 293T cells were co-transfected with pGL3-STARD13 (0.2 µg) and β-gal (0.2 µg) for 24 h using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) in 24 wells plate, then further infected with Lenti-HOXB4-CDS.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed to assess in vivo DNA-protein interactions at the STRAD13 promoter using the EZ-CHIP™ Kit (EMDMillipore, Darmstadt, Germany). The recovered DNA was used as the template to amplify the STARD13 promoter. The primers for detecting the STARD13 promoter containing the putative HOXB4 binding sites were shown in Table I.
Cell migration assays
The detailed procedure was referred to the previous study (6).
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
Cells (3×103/well) were plated in 96-well plates and treated with the IC50 of doxorubicin after adhesion. Cell viability was assessed after 24, 48, and 72 h of culturing. MTT (KeyGen Biotech) was added into the medium at 0.25 mg/ml and the absorbance was measured at 570 nm using a microplate reader.
Cell adhesion assays
Cell adhesion was assessed as described previously (10).
Apoptosis assay
Apoptosis was evaluated using flow cytometry with AnnexinV-FITC and propidium iodide (PI) staining (Vazyme Biotech, Nanjing, China). Cells with and without HOXB4 overexpressionwere harvested and washed with ice-cold PBS. The cells were then stained with Annexin V-FITC and PI (BD Biosciences, Franklin Lakes, NY, USA) following the manufacturer's protocol. Flow cytometry utilized an instrument from BD Biosciences.
Quantitative real-time PCR (qRT-PCR)
Total RNA was prepared from cells using TransZol Up (Transgen Biotech, Beijing, China) according to the manufacturer's protocols. Total RNA was reverse transcribed into cDNA using EasyScript Reverse Transcriptase (M-MLV, RNaseH-) (Transgen Biotech) following the standard protocols. mRNA expression levels were determined according to the TransScript Probe qPCR SuperMix protocols (Transgen Biotech) and performed on an ABI Prism 7500 Detection System (Applied Biosystems, Foster City, CA, USA). The primers for qRT-PCR were listed in Table I. The expression of each transcript was calculated using the 2−ΔΔct method.
Western blotting
The detailed western blotting procedure was described previously (11). Antibodies against Bcl-2 (sc-509), Bax (sc-4239) and STARD13 (sc-67843) were purchased from Santa Cruz Biotechnology. Antibodies against HOXB4 (ab76093), E-cadherin (ab40772), vimentin (ab8978) and β-actin (ab8227) were purchased from Abcam (Cambridge, UK), Blots were washed and incubated with a peroxidase-conjugated secondary antibody. Chemiluminescence was detected using Super Signal West Pico (Thermo Fisher Scientific, Waltham, MA, USA) followed by exposure with Tanon 5200.
Immunohistochemistry
Immunohistochemistry procedures were described previously (7).
Statistical analysis
All data are presented as means ± SD from three independent experiments. Differences between the groups were analyzed with One-way ANOVA and then Tukey's test, and *P<0.05 or less was considered significant.
Results
HOXB4 expression was positively correlated with STARD13 expression in patients with breast cancer
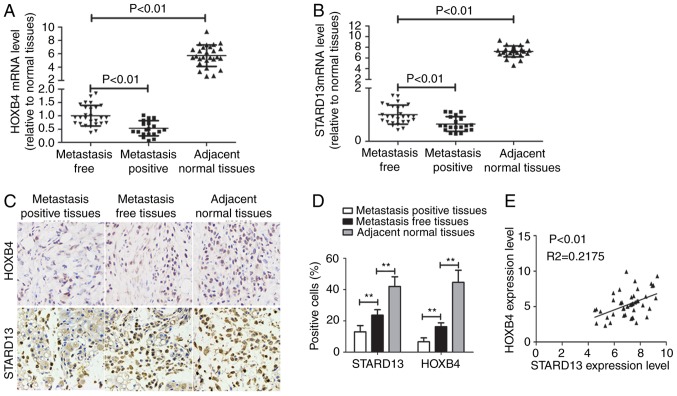
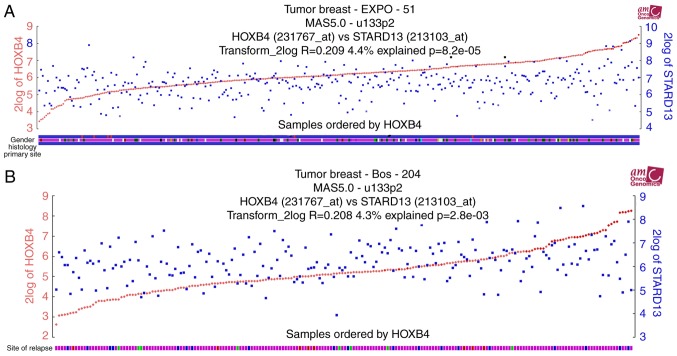
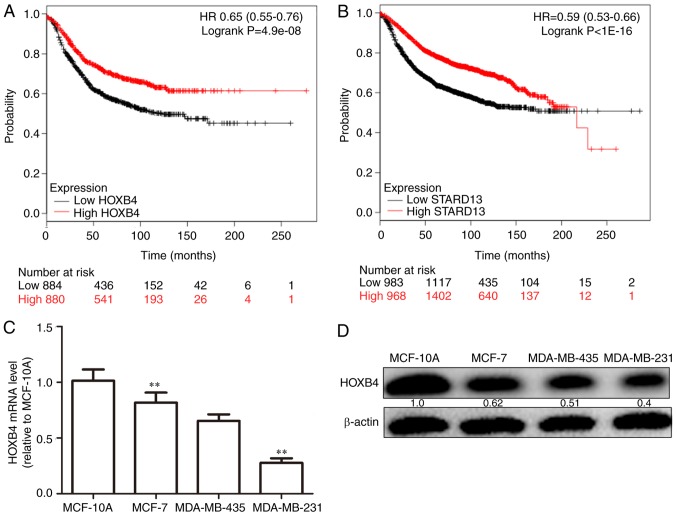
Immunohistochemistry and qRT-PCR assays were used to examine the expression levels of HOXB4 and STARD13 in breast cancer and adjacent normal tissues. HOXB4 and STARD13 expression were both decreased (Fig. 1A-D) and positively correlated in breast cancer tissues (Fig. 1E). Importantly, HOXB4 and STARD13 expression levels were downregulated in breast cancer tissues with lymph node metastasis compared with primary tissues. TCGA data analysis was further performed to determine expression profiles in patients with breast cancer. Notably, HOXB4 and STARD13 expression levels were positively correlated in two different clinical samples (Fig. 2A and B). Additionally, higher HOXB4 and STARD13 expression was correlated with longer survival in patients with breast cancer (Fig. 3A and B). Next, as shown in Fig. 3C and D, HOXB4 mRNA and protein levels were higher in normal MCF-10A breast epithelial cells than in breast cancer cells. In addition, HOXB4 expression was negatively correlated with cell metastatic ability (MCF-7 cells, showing low metastatic ability, vs., MDA-MB-435 cells, showing moderate metastatic ability, and MDA-MB-231 cells, showing high metastatic ability), consistent with STARD13 expression patterns reported in a previous study (7), and this result was similar with the results obtained from clinical samples.
Figure 1.
HOXB4 and STARD13 expression levels are downregulated in breast cancer tissues and cells. (A and B) HOXB4 and STARD13 mRNA levels were detected in breast cancer with metastasis (n=20) or not (n=27) and normal tissues (n=27) by qRT-PCR. (C and D) HOXB4 and STARD13 protein levels were examined in breast cancer and normal tissues by immunohistochemistry analysis. (E) qRT-PCR results showed that HOXB4 and STARD13 expression levels were positively correlated. Data are presented as means ± SD; n=3, *P<0.05, **P<0.01 vs. the control.
Figure 2.
HOXB4 and STARD13 expression levels are positively correlated. (A and B) TCGA data set was used to test the correlation between STARD13 and HOXB4 expression levels in two different clinical samples.
Figure 3.
HOXB4 and STARD13 expression levels are positively correlated with patients survival. (A and B) TCGA data were used to test the correlations between survival and HOXB4/STARD13-expression levels. (C and D) HOXB4 mRNA and protein levels were determined in normal breast epithelial cells (MCF-10A) and breast cancer cell lines (MCF-7, MDA-MB-435 and MDA-MB-231). Data are presented as means ± SD; n=3, *P<0.05, **P<0.01 vs. the control.
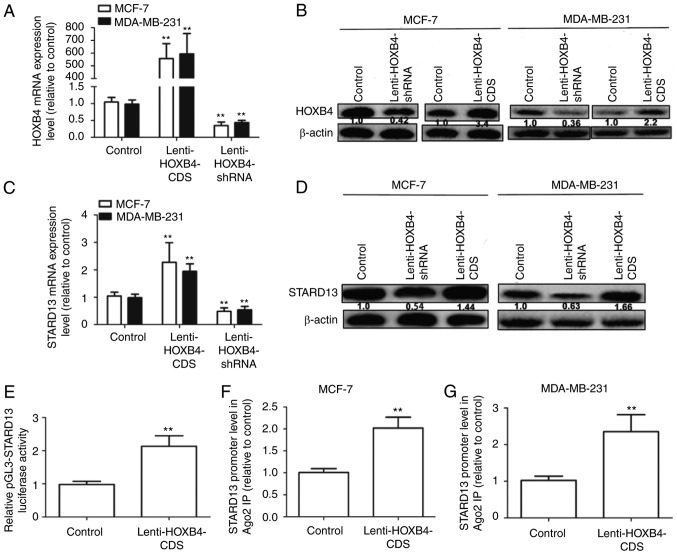
HOXB4 promoted STARD13 expression by directly binding to the STARD13 promoter
MCF-7 cells (showing low metastatic ability) and MDA-MB-231 cells (showing high metastatic ability) were used for further studies. We constructed MCF-7 and MDA-MB-231 cells with stable expression of HOXB4 or HOXB4 shRNA. As shown in Fig. 4A and B, infection with Lenti-HOXB4-CDS or Lenti-HOXB4-shRNA markedly upregulated or downregulated HOXB4 expression respectively, in MCF-7 and MDA-MB-231 cells. STARD13 expression levels were significantly upregulated in cells overexpressing HOXB4 (Fig. 4C and D), but were downregulated in cells with HOXB4 knockdown. The Genomatix Software Suite (https://www.genomatix.de/cgi-bin) was used to predict which transcription factors could bind to the STARD13 promoter. As expected, HOXB4 had five potential binding sites in the STARD13 promoter. We then assessed whether HOXB4 could enhance STARD13 promoter activity. Infection with Lenti-HOXB4-CDS could promote pGL3-STARD13 activity in 293T cells (Fig. 4E). ChIP analysis was performed to confirm whether HOXB4 could bind to the STARD13 promoter directly. The HOXB4 binding complex was pulled down with HOXB4 antibodies in MCF-7 and MDA-MB-231 cells, regardless of HOXB4 overexpression. We then examined the bound STARD13 promoter sequence containing HOXB4 binding sites by qRT-PCR. The STARD13 promoter sequence containing the HOXB4 binding sites was increased in cells with HOXB4 overexpression (Fig. 4F and G). These results suggest that HOXB4 could directly bind to the STARD13 promoter in breast cancer cells.
Figure 4.
HOXB4 promotes STARD13 expression by directly binding to the STARD13 promoter. (A and B) The infection efficiencies of Lenti-HOXB4-CDS and Lenti-HOXB4-shRNA were detected by qRT-PCR and western blot analyses in MCF-7 and MDA-MB-231 cells. (C and D) STARD13 mRNA and protein levels were determined in cells with stable expression of HOXB4-CDS or knockdown of HOXB4. (E) The promoter activity of STARD13 was detected by luciferase reporter assay in 293T cells. (F and G) qRT-PCR was used to measure the enrichment of STARD13 promoter sequences in the immunoprecipitated complexes using promoter-specific primer sets in MCF-7 and MDA-MB-231 cells with stable expression of HOXB4. Data are presented as means ± SDs; n=3, **P<0.01 vs. the control.
HOXB4 inhibited breast cancer cell migration, adhesion ability and the EMT in a STARD13-dependent manner
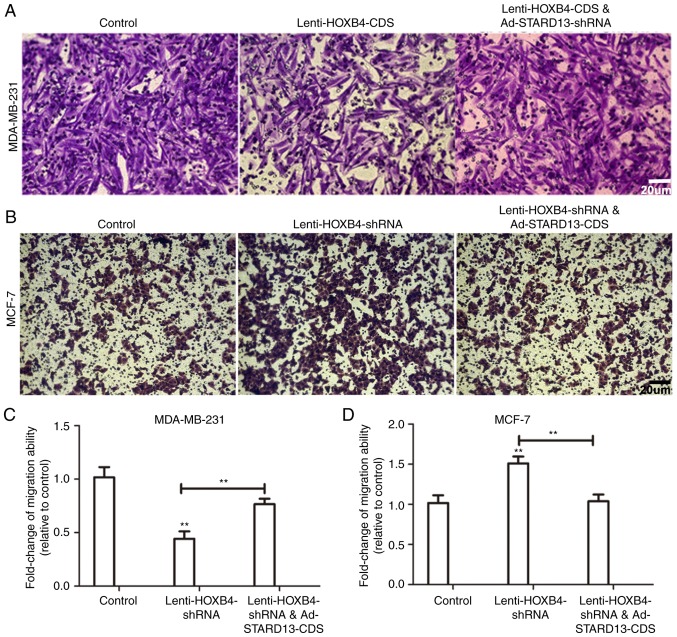
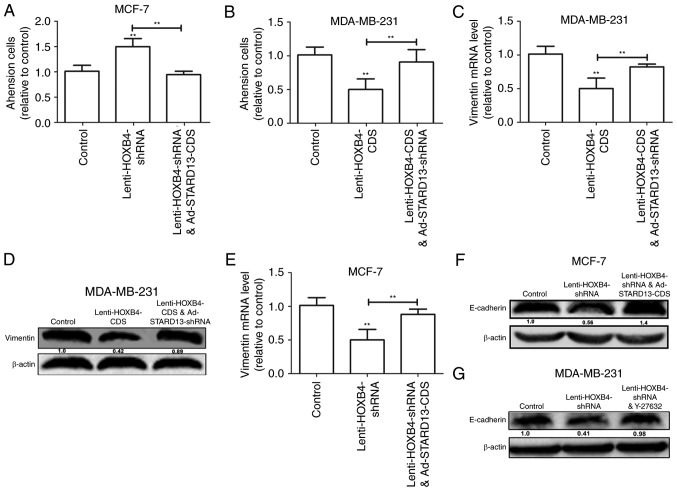
Because STARD13 is involved in breast cancer metastasis and the EMT (6,7), we speculated that HOXB4 could hold similar roles in a STARD13-dependent manner. We induced HOXB4 overexpression in MDA-MB-231 cells and HOXB4 knockdown in MCF-7 cells (Fig. 5). Decreased migration ability was observed in Leni-HOXB4-CDS-infected MDA-MB-231 cells, and this ability was attenuated when cells were co-infected with Ad-STARD13-shRNA (Fig. 5A and C). Similarly, cell migration was increased in Lenti-HOXB4-shRNA-infected MCF-7 cells, and this effect was reversed with Ad-STARD13-CDS co-infection (Fig. 5B and D). Adhesion of tumor cells to the extra-cellular matrix is thought to be the initial step in tumor invasion (12). HOXB4 knockdown-induced downregulation of cell adhesion was reversed by overexpression of STARD13 in MCF-7 cells (Fig. 6A). In contrast, Overexpression of HOXB4 decreased the adhesion ability of MDA-MB-231 cells, which was attenuated by STARD13 knockdown (Fig. 6B). Moreover, forced expression of HOXB4 inhibited the expression of vimentin in MDA-MB-231 cells (Fig. 6C and D). This effect was also attenuated by Ad-STARD13-shRNA infection. In contrast, knockdown of HOXB4 decreased the expression levels of E-cadherin in MCF-7 cells, and Ad-STARD13-CDS infection reversed this effect (Fig. 6E and F). Because STARD13 can inhibit the activity of RhoA (13). Treatment of HOXB4-knockdown MCF-7 cells with Y-27632 (an inhibitor of ROCK) completely blocked the HOXB4 knockdown-dependent downregulation of E-cadherin (Fig. 5G). These results demonstrate that ectopic expression of HOXB4 could inhibit breast cancer cell migration and the EMT process in a STARD13-dependent manner.
Figure 5.
HOXB4 inhibites breast cancer cell migration in a STARD13-dependent manner. (A-D) Transwell migration assays were used to detect the migration ability of MCF-7 and MDA-MB-231 cells. Overexpression of HOXB4 in MDA-MB-231 cells or knockdown of HOXB4 in MCF-7 cells inhibited or promoted migration, respectively, and this effect was attenuated or reversed by co-infection with Ad-STARD13-shRNA or Ad-STARD13-CDS. Data are presented as means ± SDs; n=3, **P<0.01 vs. the control.
Figure 6.
HOXB4 inhibites breast cancer adhesion ability and the EMT in a STARD13-dependent manner. (A and B) Cell adhesion assay of MDA-MB-231 cells with HOXB4 overexpression and MCF-7 cells with HOXB4 knockdown. (C-F) qRT-PCR and western blotting were used to detect the expression levels of EMT markers (Vimentin and E-cadherin) in MDA-MB-231 cells co-treated with Lenti-HOXB4-CDS and Ad-STARD13-shRNA and MCF-7 cells co-treated with Lenti-HOXB4-shRNA and Ad-STARD13-CDS. (G) Western blotting was used to detect E-cadherin protein levels in MCF-7 cells co-treated with Lenti-HOXB4-shRNA and Y-27632. Data are presented as means ± SD. n=3, **P<0.01.
Overexpression of HOXB4 enhanced doxorubicin sensitivity in breast cancer cells
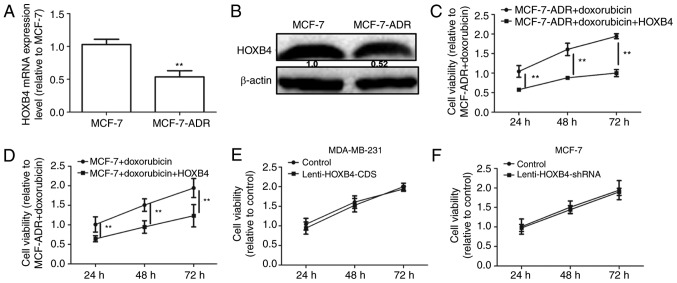
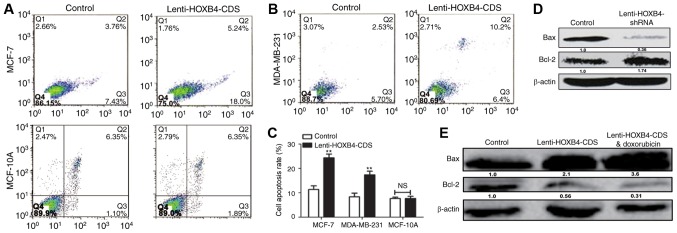
A previous study reported that the EMT could lead to chemoresistance in cancer cells (14). HOXB4 expression levels were detected in MCF-7 and MCF-7-ADR cells. As shown in Fig. 7A and B, HOXB4 expression levels were downregulated in MCF-7-ADR compared to MCF-7 cells. We then analyzed HOXB4 overexpression in MCF-7-ADR cells with Lenti-HOXB4-CDS infection. The MTT assay showed that forced expression of HOXB4 attenuated doxorubicin resistance in MCF-7-ADR cells (Fig. 7C). Additionally, overexpression of HOXB4 enhanced doxorubicin sensitivity in MCF-7 cells (Fig. 7D). However, the proliferation ability was not altered in MDA-MB-231 cells with or without HOXB4 overexpression and in MCF-7 cells with or without HOXB4 knockdown (Fig. 7E and F). Furthermore, increased apoptosis was observed in HOXB4 overexpressing cells (Fig. 8A-C), while ectopic expression of HOXB4 had no effect on MCF-10A cell apoptosis. MDA-MB-231 cells with HOXB4 overexpression exhibited increased expression of Bax and decreased expression of Bcl-2, which was enhanced by additive doxorubicin treatment (Fig. 8D). Conversely, HOXB4 knockdown inhibited Bax expression but increased Bcl-2 expression in MCF-7 cells (Fig. 8E). Hence, our results indicate that forced expression of HOXB4 could increase doxorubicin sensitivity and attenuate doxorubicin resistance in MCF-7 cells.
Figure 7.
Overexpression of HOXB4 enhanced doxorubicin-mediated downregulation of cell viability in breast cancer cells. (A and B) HOXB4 mRNA and protein levels were detected in MCF-7 and MCF-7-ADR cells. (C) Cell viability was examined using the MTT assays in MCF-7-ADR cells treated with doxorubicin with or without Ad-HOXB4-CDS. (D) Cell viability of MCF-7 cells treated with doxorubicin with or without Ad-HOXB4-CDS. (E and F) Cell viability in MCF-7 and MDA-MB-231 cells with HOXB4 knockdown or overexpression, respectively. **P<0.01 vs. the control.
Figure 8.
Overexpression of HOXB4 enhanced doxorubicin-mediated downregulation of cell apoptosis in breast cancer cells. (A-C) Apoptosis assays was assessed in MCF-7, MCF-10Aand MDA-MB-231 cells with or without HOXB4 overexpression. (D) Bcl-2 and Bax protein levels were determined by western blotting in MCF-7 cells treated with Lenti-HOXB4-CDS with or without doxorubicin. (E) Bcl-2 and Bax protein levels were determined by western blotting in MCF-7 cells with or without HOXB4 knockdown. Data are presented as means ± SD. n=3, *P<0.05, **P<0.01.
Discussion
Upregulation of HOXB4 promotes is strongly associated with the overall survival of patients with acute myeloid leukemia (15). However, the roles of HOXB4 in somatic tumors are unclear.
In the present study, we investigated the effects of HOXB4 on breast cancer metastasis, the EMT process and doxorubicin sensitivity. Bioinformatics analysis was used to predict the potential targets of HOXB4. STARD13 attracted our interest because it has suppressive roles in breast cancer. Further analysis of clinical samples and TCGA data, as well as ChIP assays, confirmed our prediction that HOXB4 could bind to the STARD13 promoter directly. Our results obtained from gain and loss of function approaches indicated that HOXB4 was negatively correlated with the migratory abilities of breast cancer cells in vitro. For MCF-7 cells with low metastatic ability and high expression of HOXB4, knockdown of HOXB4 permitted these cells to gain higher metastatic potential. In MDA-MB-231 cells with low expression of HOXB4, cell migration ability was significantly inhibited with HOXB4 overexpression. Importantly, the inhibitory effects of HOXB4 on cell migration was dependent on STARD13 expression.
The EMT occurs aberrantly during tumor progression and initiates the metastasis cascade and drug resistance (7,16). Here, we found that enforced expression of HOXB4 strongly suppressed the EMT process in breast cancer cells. To demonstrate the mechanisms through which HOXB4 regulates the EMT, we used an inhibitor of ROCK. We showed that HOXB4 regulated E-cadherin expression through the STARD13-RhoA-ROCK signaling pathway. Interestingly, the proliferation ability of cells was unchanged as HOXB4 expression was altered, indicating that HOXB4 may affect the EMT process in breast cancer and thereby modulate cell migration ability. Finally, our results showed that overexpression of HOXB4 could promote apoptosis, enhance doxorubicin sensitivity and attenuate doxorubicin resistance in MCF-7 cells. These results suggested that HOXB4 modulated doxorubicin sensitivity, possibly through regulating the EMT process or cell apoptosis, but not through modulating cell proliferation. In contrast to our results, a previous study reported that HOXB4 knockdown reversed multidrug resistance of human myelogenous leukemia K562/ADM cells (17). This difference may be due to the different types of tumors, suggesting that HOXB4 may have various roles depending on the tumors. Further studies are needed to elucidate the roles of HOXB4 in other tumors.
To the best of our knowledge, this is the first study to demonstrate the potential role of HOXB4 in breast cancer. These results provided strong evidence in support of the inhibitory activities of HOXB4 in the EMT, migration process and doxorubicin resistance in breast cancer r. It is possible that these characteristics may be exploited for application in gene therapy of cancer.
References
- 1.Giordano SB, Gradishar W. Breast cancer: Updates and advances in 2016. Curr Opin Obstet Gynecol. 2017;29:12–17. doi: 10.1097/GCO.0000000000000343. [DOI] [PubMed] [Google Scholar]
- 2.Lou PJ, Lai PS, Shieh MJ, Macrobert AJ, Berg K, Bown SG. Reversal of doxorubicin resistance in breast cancer cells by photochemical internalization. Int J Cancer. 2006;119:2692–2698. doi: 10.1002/ijc.22098. [DOI] [PubMed] [Google Scholar]
- 3.Helgason CD, Sauvageau G, Lawrence HJ, Largman C, Humphries RK. Overexpression of HOXB4 enhances the hematopoietic potential of embryonic stem cells differentiated in vitro. Blood. 1996;87:2740–2749. [PubMed] [Google Scholar]
- 4.Cusan M, Vegi NM, Mulaw MA, Bamezai S, Kaiser LM, Deshpande AJ, Greif PA, Quintanilla-Fend L, Göllner S, Müller-Tidow C, et al. Controlled stem cell amplification by HOXB4 depends on its unique proline-rich region near the N-terminus. Blood. 2017;129:319–323. doi: 10.1182/blood-2016-04-706978. [DOI] [PubMed] [Google Scholar]
- 5.Petzold KM, Naumann H, Spagnoli FM. Rho signalling restriction by the RhoGAP Stard13 integrates growth and morphogenesis in the pancreas. Development. 2013;140:126–135. doi: 10.1242/dev.082701. [DOI] [PubMed] [Google Scholar]
- 6.Tang F, Zhang R, He Y, Zou M, Guo L, Xi T. MicroRNA-125b induces metastasis by targeting STARD13 in MCF-7 and MDA-MB-231 breast cancer cells. PLoS One. 2012;7:e35435. doi: 10.1371/journal.pone.0035435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Zheng L, Zhang F, Hu J, Chou J, Liu Y, Xing Y, Xi T. STARD13-correlated ceRNA network inhibits EMT and metastasis of breast cancer. Oncotarget. 2016;7:23197–23211. doi: 10.18632/oncotarget.8099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H, Wang F, Hu Y. STARD13 promotes hepatocellular carcinoma apoptosis by acting as a ceRNA for Fas. Biotechnol Lett. 2017;39:207–217. doi: 10.1007/s10529-016-2253-6. [DOI] [PubMed] [Google Scholar]
- 9.Chang S, He S, Qiu G, Lu J, Wang J, Liu J, Fan L, Zhao W, Che X. MicroRNA-125b promotes invasion and metastasis of gastric cancer by targeting STARD13 and NEU1. Tumour Biol. 2016;37:12141–12151. doi: 10.1007/s13277-016-5094-y. [DOI] [PubMed] [Google Scholar]
- 10.Yang J, Li T, Gao C, Lv X, Liu K, Song H, Xing Y, Xi T. FOXO1 3′UTR functions as a ceRNA in repressing the metastases of breast cancer cells via regulating miRNA activity. FEBS Lett. 2014;588:3218–3224. doi: 10.1016/j.febslet.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Zheng L, Li X, Meng X, Chou J, Hu J, Zhang F, Zhang Z, Xing Y, Liu Y, Xi T. Competing endogenous RNA networks of CYP4Z1 and pseudogene CYP4Z2P confer tamoxifen resistance in breast cancer. Mol Cell Endocrinol. 2016;427:133–142. doi: 10.1016/j.mce.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Wang B, Zheng L, Chou J, Li C, Zhang Y, Meng X, Xi T. CYP4Z1 3′UTR represses migration of human breast cancer cells. Biochem Biophys Res Commun. 2016;478:900–907. doi: 10.1016/j.bbrc.2016.08.048. [DOI] [PubMed] [Google Scholar]
- 13.Nagaraja GM, Kandpal RP. Chromosome 13q12 encoded Rho GTPase activating protein suppresses growth of breast carcinoma cells, and yeast two-hybrid screen shows its interaction with several proteins. Biochem Biophys Res Commun. 2004;313:654–665. doi: 10.1016/j.bbrc.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Xu T, Zhang J, Chen W, Pan S, Zhi X, Wen L, Zhou Y, Chen BW, Qiu J, Zhang Y, et al. ARK5 promotes doxorubicin resistance in hepatocellular carcinoma via epithelial-mesenchymal transition. Cancer Lett. 2016;377:140–148. doi: 10.1016/j.canlet.2016.04.026. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Zhao CT, Cui BL, Wu SL, Liu XD, Su Z, Yang J, Wang W, Cui ZG, Zhao HG. Expression of HOXB4, PRDM16 and HOXA9 in patients with acute myeloid leukemia and its clinical significance. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2016;24:326–331. doi: 10.7534/j.issn.1009-2137.2016.02.004. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 16.Sun W, Tang L. MDM2 increases drug resistance in cancer cells by inducing EMT independent of p53. Curr Med Chem. 2016;23:4529–4539. doi: 10.2174/0929867323666160926150820. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Jia XH, Chen JR, Yi YJ, Wang JY, Li YJ, Xie SY. HOXB4 knockdown reverses multidrug resistance of human myelogenous leukemia K562/ADM cells by downregulating P-gp, MRP1 and BCRP expression via PI3K/Akt signaling pathway. Int J Oncol. 2016;49:2529–2537. doi: 10.3892/ijo.2016.3738. [DOI] [PubMed] [Google Scholar]