Abstract
It has previously been demonstrated that the long non-coding RNA (lncRNA) nuclear enriched abundant transcript (NEAT)-1 is increased in multiple cancers and may be associated with cancer development. However, the function and mechanism of NEAT1 in non-small cell lung cancer (NSCLC) has not yet been fully elucidated. In the present study, the expression of NEAT1 in NSCLC was detected using quantitative polymerase chain reaction and association with survival was estimated. The effect of NEAT1 on proliferation was detected by growth curve and cell cycle analysis. Bioinformatics analysis was used to identify miRNAs that interact with NEAT1. Following this, a series of molecular biological techniques were used to verify the mechanism of NEAT1. The results indicated that NEAT1 was highly expressed in NSCLC, and high NEAT1 expression was associated with a shorter overall survival. NEAT1 promoted NSCLC cell growth and affected the cell cycle process in vitro. Furthermore, NEAT1 was observed to bind hsa-miR-377-3p, functioning as a competing endogenous RNA, which resulted in de-repression of its target gene E2F transcription factor 3 (E2F3). E2F3, as an oncogene, may promote NSCLC progression. These results suggested that NEAT1 may promote the development of NSCLC through the miR-377-3p-E2F3 pathway.
Keywords: NEAT1, miR-377-3p, E2F3, non-small cell lung cancer, proliferation
Introduction
Lung cancer is the main cause of cancer-related death worldwide. Non-small cell lung carcinoma (NSCLC), accounting for approximately 80% of lung cancer, is the most common histopathological type. Surgery is the only treatment method for early stages of NSCLC, but the 5-year overall survival rate is only 40% (1). Although remarkable progress has been made in the past decades, the molecular mechanisms of NSCLC carcinogenesis are still not clear.
Long non-coding RNAs (lncRNAs), which are non-coding RNAs with more than 200 nucleotides in length, are frequently cell-type specific and contribute to the progression of multiple human tumors (2,3). Moreover, lncRNAs also have the potential function as diagnostic or prognostic markers of cancer (4–10). Nuclear enriched abundant transcript 1 (NEAT1) is a nuclear-restricted lncRNA that has two isoforms: NEAT1-1 and NEAT1-2 (11). It was recently revealed as an essential architectural component of paraspeckle structure (12,13). NEAT1 plays a role in several biological processes, including stress response and cellular differentiation through paraspeckles pathway (14). NEAT1 is also a marker for poor survival of breast cancer patients (15). In prostate cancer, NEAT1 was proved to promote oncogenic growth by altering gene promoters and then inducing transcription (16). But until now, the specific mechanism of NEAT1 is still not clear in NSCLC.
The aim of this study is to explore the function and mechanism of NEAT1 in NSCLC. We first found the increased expression of NEAT1 in NSCLC and its correlation with poor survival. Then, we found that NEAT1 (represented by NEAT1-2) harbors three conserved miR-377-3p cognate sites, and E2F3 harbors two conserved miR-377-3p cognate sites using Biological information technology. At last, our results showed that NEAT1 might be a crucial oncogenic regulator involved in NSCLC development by functioning as a ceRNA, which binding to miR-377-3p and then further activated the E2F3 pathway.
Materials and methods
Tissue collection
All the samples (20 paired NSCLC tissues and adjacent normal tissues) were obtained from patients suffering from NSCLC who operated at the second affiliated hospital of Harbin Medical University between 2011.5 and 2012.8. All these patients did not recieve any local or systemic treatment before operation. All collected tissue samples were immediately treated by liquid nitrogen, then stored at −80°C before RNA extraction. All patients signed their informed consent. The study has got approval from the Research Ethics Committee of Harbin Medical University, and thus meets the standards of the Declaration of Helsinki in its revised version of 1975 and its amendments of 1983, 1989, and 1996.
Cell lines and culture conditions
Normal human embryonic lung fibroblasts cell line (HELF) and several NSCLC cancer cell lines, including A549, SPC-A1, H1299, 95D, SK-MES-1, and NCI-H520, were used in this study. These cells were cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) or Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS) at 37°C in humidified air with 5% CO2.
Transfection of cell lines
The siRNA (small interfering RNA) sequences (GenePharma, Shanghai, China) were shown in Table I. Synthetic sequence-scrambled siRNA was used as negative controls (NC). Human E2F3 gene was ligated into pGCMV vector (GenePharma). The empty vector was used as NC. Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used to complete transfection in A549 and H1299 cells according to the manufacturer's protocol.
Table I.
Sequence of NEAT1 siRNA.
| NEAT1 siRNA | Sequence |
|---|---|
| siRNA1 | 5′-GCCAUCAGCUUUGAAUAAAUU-3′ |
| siRNA2 | 5′-GGUGUUAUCAAGUGAAUUAUU-3′ |
| siRNA3 | 5′-GCCUUGUAAAUGCCUAUAUUU-3′ |
Cell proliferation ability analysis
Different groups of cells were seeded into 6 wells plate at 1×105 cells per well. The cell number was counted for three consecutive days and then growth curve was drawn according to the cell number at 24, 48 and 72 h. 1×106 cells were prepared for cell cycle analysis. First, the cells were fixed with 70% cold ethanol. After 12 h at 4°C, the cells were washed with PBS and stained with propidium iodide (PI). Finally, flow cytometry (BD Biosciences, San Jose, CA, USA) was conducted to analyze cell cycle.
Luciferase reporter assays
The wild-type and mutant 3′-UTR of human E2F3 or lncRNA NEAT1 were inserted into the pmiR-RB-REPORT TM (Guangzhou RiboBio Co., Ltd., Guangzhou, China) through XhoI and NotI sites. Co-transfected wild-type/mutant reporter plasmid and miR-Ribo TM negative control into A549 and H1299 cells by Lipofectamine 2000 (Invitrogen), and then measured luciferase activities using the Dual-Luciferase Reporter Assay system 48 h post-transfection according to the manufacturer's instructions (Promega Corp., Madison, WI, USA).
RNA isolation and quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA in cells and tissues was extracted using Trizol reagent (Invitrogen). RNA was reversed into cDNA through reverse transcription kits (Guangzhou RiboBio Co., Ltd.). Then the levels of target mRNAs were detected using qRT-PCR by SYBR® Premix Ex Taq™II (Takara Bio, Inc., Otsu, Japan) in the ABI PRISM® 7300 real-time PCR system (Applied Biosystems, Foster City, CA, USA). GAPDH and U6 were used as endogenous controls. The primer sequences involved in this study were shown in Table II.
Table II.
Sequence of primers.
| Gene | Primer sequence |
|---|---|
| GAPDH | Forward 5′-TGTGGGCATCAATGGATTTGG-3′ |
| Reverse 5′-ACACCATGTATTCCGGGTCAAT-3′ | |
| NEAT1_2 | Forward 5′-CAGTTAGTTTATCAGTTCTCCCATCCA-3′ |
| Reverse 5′-GTTGTTGTCGTCACCTTTCAACTCT-3′ | |
| E2F3 | Forward 5′-AGAAAGCGGTCATCAGTACCT-3′ |
| Reverse 5′-TGGACTTCGTAGTGCAGCTCT-3′ | |
| miR-377-3p | RT: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACacaa-3′ |
| U6 | RT: 5′-AAAATATGGAACGCTTCACGAATTTG-3′ |
| miR-377-3p | Forward 5′-GGGAGGCAGTGTATTGTTA-3′ |
| Reverse 5′-GTCGTATCCAGTGCAGGGTCCGAGGT-3′ |
Western blot analysis
Total protein was obtained from the A549 and H1299 cells after 48 h of transfection using cell lysis reagent. BCA protein assay reagent kit (Thermo Fisher Scientific, Inc.) was used to detected the protein concentrations in supernatant. The protein was separated in 10% SDS-polyacrylamide gels, transferred to PVDF membranes (Hertfordshire, UK), incubated in 5% milk powder to block non-specific reaction, and then treated with antibodies overnight at 4°C, including anti-E2F3 (1:1,000 dilutions; Affinity), anti-cyclin D1 (1:1,000 dilutions; Affinity), anti-cyclinD2 (1:1,000 dilutions; Affinity), anti-CDK4 (1:1,000 dilutions; Affinity), anti-p21 (1:1,000 dilutions; Affinity), anti-p57 (1:1,000 dilutions; Affinity) and anti-GADPH (1:5,000 dilutions; Affinity). At last, the membranes were incubated with second antibody (conjugated to horseradish peroxidase, 1:12,000 dilutions) for 2 h and exposed to ECL Advance reagent (GE Healthcare Biosciences, Buckinghamshire, UK) for 2 min. The band was visualized with Fluor S Multimager and Quantity One 4.1 (Bio-Rad Laboratories, Hercules, CA, USA).
Statistical analysis
In the present study, all the data are presented as means ± SD. A two-tailed paired t-test was used to reflect the difference between groups. P<0.05 indicated that the difference was statistically significant.
Results
NEAT1 is up-regulated in NSCLC tissues and correlates with poor prognosis
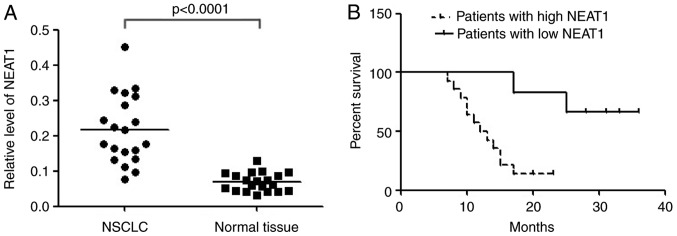
In order to validate the differential expression, NEAT1 was detected in 20 paired NSCLC tissues and adjacent normal tissues with qRT-PCR. The result showed that NEAT1 expression was significantly up-regulated in cancerous tissues than normal counterparts (Fig. 1A). Compared with patients whose cancer tissue expressed less NEAT1 (≤2 folds of increase, n=6), patients whose cancer tissue expressed more NEAT1 (>2 folds of increase, n=14) had shorter overall survival according to Kaplan-Meier survival analysis (Fig. 1B). The results demonstrated that high levels of NEAT1 were associated with NSCLC poor prognosis. Upregulation of NEAT1 might play important roles in NSCLC.
Figure 1.
NEAT1 was up-regulated in NSCLC tissues and correlates with poor prognosis. (A) Relative expression of NEAT1 in NSCLC tissues (n=20) and in paired adjacent normal tissues (n=20). NEAT1 expression was examined by qPCR and normalized to GAPDH expression. (shown as 2-ΔCT). (B) The Kaplan-Meier survival analysis indicated that NSCLC patients with high NEAT1 (n=14) have a worse overall survival compared to the low expression subgroup (n=6). P<0.001.
NEAT1 promotes NSCLC cells growth in vitro
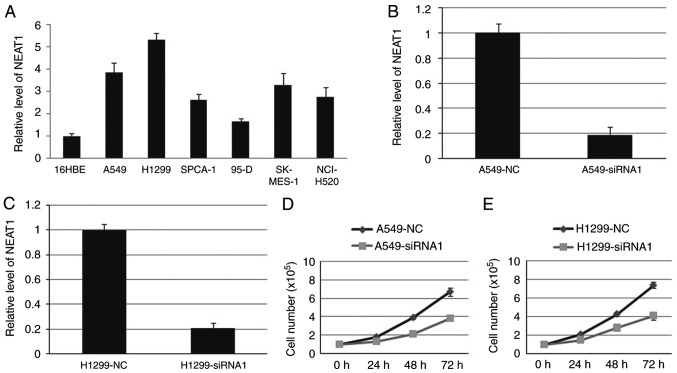
We first detected NEAT1 expression in NSCLC cell lines, including A549, H1299, SPCA-1, 95-D, SK-MES-1 and NCI-H520 cell line. The result showed that the expression level of NEAT1 in NSCLC cell lines was higher than that in 16HBE cell line (Fig. 2A). Then, endogenous NEAT1 expression was down-regulated by approximately 80% through small interfering RNAs (siRNAs) in A549 and H1299 cells (Fig. 2B and C). Compared with blank cells, the proliferation ability was decreased in A549-siRNA1 and H1299-siRNA1 cells (Fig. 2D and E). Cell cycle analysis indicated that deletion of NEAT1 induced cell cycle arrest in G1 phase.
Figure 2.
NEAT1 promotes NSCLC cells growth in vitro. The expression of NEAT1 was higher in NSCLC cell lines (A). The NETA1 was significantly inhibited by siRNA1in both A549 (B) and H1299 (C). Cell growth ability was decreased when NEAT1 was inhibited in both A549 (D) and H1299 (E). P<0.05.
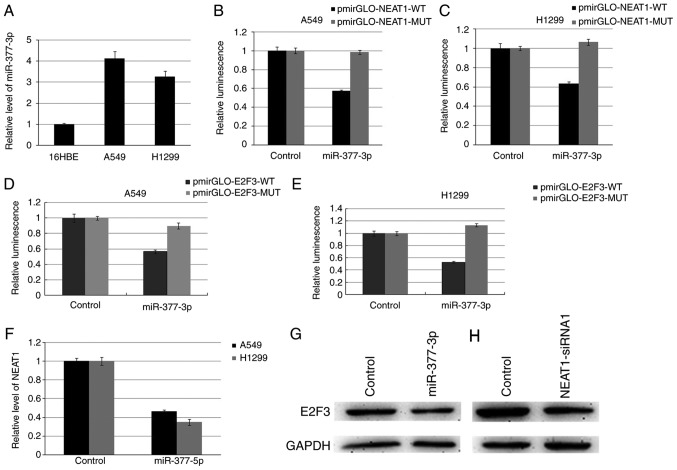
NEAT1 promotes E2F3 expression in part through competitive binding miRNA-377-3p
In view of the potential function of lncRNA as a ceRNA or a molecular sponge of miRNAs, we screened the miRNAs interacting with NEAT1 using the bioinformatics, starBase2.0. Among these miRNAs, miR-377-3p (MIMAT0000730) was selected for further study. We first verified miR-377-3p expression in A549 and H1299 cells, and it is significantly higher than 16HBE cell (Fig. 3A). Dual-luciferase reporter assay indicated that miR-377-3p could decrease the luciferase activity of pmirGLO-NEAT1-WT (wild-type), however, it had no affect on the pmirGLO-NEAT1-MUT (Fig. 3B and C). Targetscan, miRanda and PicTar were used to predict the possible targets of miR-377-3p, and found that E2F3 involving in cell cycle regulation harbors two conserved miR-377-3p cognate sites. Luciferase reporter assays indicated that luciferase activity was declined more than 50% after co-transfected with miR-377-3p and the reporter vector with E2F3-3′-WT (Fig. 3D and E). Up-regulating miR-377-3p in A549 and H1299 cells suppressed the expression of NEAT1 and E2F3 (Fig. 3F and G). Inhibition of NEAT1 resulted in down-regulation of E2F3 protein level (Fig. 3H).
Figure 3.
NEAT1 promotes E2F3 expression in part through competitive binding miRNA-377-3p. The relative level of miR-377-3p in A549 and H1299 (A). The luciferase report assay demonstrated that miR-377-3p could reduce the intensity of fluorescence in A549 (B) and H1299 (C) transfected with the NEAT1-WT vector. Luciferase reporter assays indicated that luciferase activity generated by the reporter vector with E2F3-3′-WT decreased more than 50% after co-transfected with miR-377-3p in both A549 (D) and H1299 (E). Overexpression of miR-377-3p inhibited NEAT1 expression (F). Overexpression of miR-377-3p inhibited E2F expression in A549 (G). Inhibiting NEAT1 resulted in a decrease of E2F3 in A549 (H). P<0.05.
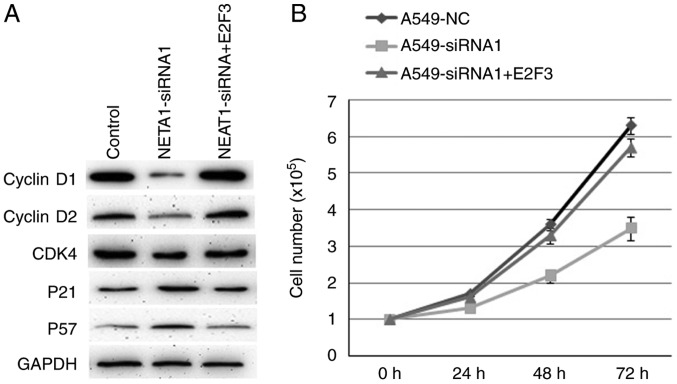
E2F3 promotes NSCLC proliferation through regulating cell cycle in NSCLC
Inhibiting NEAT1 in A549 cells suppressed the protein expression of cyclinD1, cyclinD2 and CDK4, whereas increased the protein levels of p21 and p57 (Fig. 4A). Consistently, over-expression of E2F3 in A549-siRNA1 cells reversed the expression of these proteins mentioned above (Fig. 4A). Over-expression of E2F3 also reversed the cell proliferation ability inhibited by decreased NEAT1 (Fig. 4B). So, E2F3 mediated the cell proliferation induced by NEAT1 by regulating cell cycle associated proteins expression.
Figure 4.
E2F3 promotes NSCLC proliferation through regulating the cell cycle in NSCLC. The expression of cell cycle-associated proteins in A549 were detected by western blot (A). E2F3 can rescue the cell growth inhibition induced by decreased NEAT1 (B). P<0.05.
Discussion
Recent studies have demonstrated lncRNAs play an important role in human cancers (17–20). Our research indicated that NEAT1 was up-regulated in NSCLC. NEAT1 acted as an oncogene to promote the proliferation ability of NSCLC cells, which is largely due to its ceRNAs function to suppress miR-377-3p and subsequently derepress E2F3, and then activate the E2F3 signaling pathway. This study is important to illustrate the relationship between NEAT1 and NSCLC development.
This study indicated that NEAT1 was significantly up-regulated in NSCLC tissues compared to adjacent normal lung tissues. Particularly, our study also demonstrated an association between increased NEAT1 expression levels and NSCLC poor prognosis. This result indicated that NEAT1 expression may be a helpful biomarker for doctor to recognize patients with poor prognosis. It was in accordance with Pan and his colleagues' findings (21). In addition, NEAT1 significantly improved NSCLC cell proliferative ability. In summary, the results of this research suggested that NEAT1 may act as an oncogene as well as contribute significantly to NSCLC development and progression.
It has been reported that paraspeckles pathway is one of the molecular mechanisms by which NEAT1 plays roles in controlling several biological processes (12,13). Recently, NEAT1 was found to repress the transcription of some genes by protein sequestration into paraspeckles (22). Here, we discovered that NEAT1 might function by another potential molecular mechanism in NSCLC progression-acting as ‘molecular sponges’ to regulate microRNAs. Many studies have proved that lncRNAs can work as ceRNAs to regulate microRNAs, such as GAS5 (23). For another instance, CCAT1 can promote the development of gallbladder cancer by acting as miR-218-5p sponges (24). In the present study, we found that NEAT1, acting as a ceRNA, bound miR-377-3p and restricted its function.
As we have known, miRNA can regulate the expression of multiple target genes generally. It was reported that miR-377 can inhibit human clear cell renal cell carcinoma by targeting ETS1 (25), and miR-377 can also suppress cell proliferation and invasion in hepatocellular carcinoma by inhibiting TIAM1 expression (26). Additionally, Wen et al revealed miR-377 regulates mesenchymal stem cell-induced angiogenesis in ischemic hearts by targeting VEGF (27). But among the target genes of miR-377, E2F3 transcription factor caught our attention. The G1/S transition of the cell cycle is regulated in part by E2Fs transcription factors and is a tightly orchestrated process that culminates in DNA synthesis (28). E2Fs regulate expression of genes involved in many cellular processes, including apoptosis, mitosis, differentiation, and development with E2F1-3 as transcriptional activators and E2F4-8 as transcriptional repressors of these genes (29). E2F3 plays an essential role in normal proliferation (30). Furthermore, many cancers have been found to be associated with aberrant E2F expression, including lung cancer, bladder cancer and prostate cancer (31,32). In our study, we proved that E2F3 can increase the protein expression of cyclinD1, cyclinD2 and CDK4, whereas suppressing the protein levels of p21 and p57.
In summary, our results suggested lncRNA NEAT1 promoted the growth of NSLC by competitively binding to miR-377 and then removing the suppressive effect of miR-377 on E2F3. NEAT1 may also be seen as a factor that prompt the traces of NSCLC. Our research has provided a better understanding of the NEAT1 function in NSCLC, which might facilitate the development of lncRNA-directed diagnostics and therapeutics against this disease.
Acknowledgements
This work was supported by the Heilongjiang Provincial Health Bureau (No. 2016-176).
References
- 1.Laskin JJ, Sandler AB. State of the art in therapy for non-small cell lung cancer. Cancer Invest. 2005;23:427–442. doi: 10.1081/CNV-67172. [DOI] [PubMed] [Google Scholar]
- 2.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 4.Zhou M, Sun Y, Sun Y, Xu W, Zhang Z, Zhao H, Zhong Z, Sun J. Comprehensive analysis of lncRNA expression profiles reveals a novel lncRNA signature to discriminate nonequivalent outcomes in patients with ovarian cancer. Oncotarget. 2016;7:32433–32448. doi: 10.18632/oncotarget.8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou M, Guo M, He D, Wang X, Cui Y, Yang H, Hao D, Sun J. A potential signature of eight long non-coding RNAs predicts survival in patients with non-small cell lung cancer. J Transl Med. 2015;13:231. doi: 10.1186/s12967-015-0556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou M, Zhao H, Wang Z, Cheng L, Yang L, Shi H, Yang H, Sun J. Identification and validation of potential prognostic lncRNA biomarkers for predicting survival in patients with multiple myeloma. J Exp Clin Cancer Res. 2015;34:102. doi: 10.1186/s13046-015-0219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou M, Xu W, Yue X, Zhao H, Wang Z, Shi H, Cheng L, Sun J. Relapse-related long non-coding RNA signature to improve prognosis prediction of lung adenocarcinoma. Oncotarget. 2016;7:29720–29738. doi: 10.18632/oncotarget.8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou M, Wang X, Shi H, Cheng L, Wang Z, Zhao H, Yang L, Sun J. Characterization of long non-coding RNA-associated ceRNA network to reveal potential prognostic lncRNA biomarkers in human ovarian cancer. Oncotarget. 2016;7:12598–12611. doi: 10.18632/oncotarget.7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou M, Zhao H, Xu W, Bao S, Cheng L, Sun J. Discovery and validation of immune-associated long non-coding RNA biomarkers associated with clinically molecular subtype and prognosis in diffuse large B cell lymphoma. Mol Cancer. 2017;16:16. doi: 10.1186/s12943-017-0580-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan SX, Yang F, Yang Y, Tao QF, Zhang J, Huang G, Yang Y, Wang RY, Yang S, Huo XS, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology. 2012;56:2231–2241. doi: 10.1002/hep.25895. [DOI] [PubMed] [Google Scholar]
- 11.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci USA. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naganuma T, Hirose T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013;10:456–461. doi: 10.4161/rna.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choudhry H, Albukhari A, Morotti M, Haider S, Moralli D, Smythies J, Schödel J, Green CM, Camps C, Buffa F, et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2α dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2015;34:4546. doi: 10.1038/onc.2014.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nie FQ, Zhu Q, Xu TP, Zou YF, Xie M, Sun M, Xia R, Lu KH. Long non-coding RNA MVIH indicates a poor prognosis for non-small cell lung cancer and promotes cell proliferation and invasion. Tumour Biol. 2014;35:7587–7594. doi: 10.1007/s13277-014-2009-7. [DOI] [PubMed] [Google Scholar]
- 18.Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Zhou M, Zhong L, Xu W, Sun Y, Zhang Z, Zhao H, Yang L, Sun J. Discovery of potential prognostic long non-coding RNA biomarkers for predicting the risk of tumor recurrence of breast cancer patients. Sci Rep. 2016;6:31038. doi: 10.1038/srep31038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan LJ, Zhong TF, Tang RX, Li P, Dang YW, Huang SN, Chen G. Upregulation and clinicopathological significance of long non-coding NEAT1 RNA in NSCLC tissues. Asian Pac J Cancer Prev. 2015;16:2851–2855. doi: 10.7314/APJCP.2015.16.7.2851. [DOI] [PubMed] [Google Scholar]
- 22.Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, Yokoi T, Nakagawa S, Bénard M, Fox AH, Pierron G. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell. 2014;25:169–183. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou M, Diao Z, Yue X, Chen Y, Zhao H, Cheng L, Sun J. Construction and analysis of dysregulated lncRNA-associated ceRNA network identified novel lncRNA biomarkers for early diagnosis of human pancreatic cancer. Oncotarget. 2016;7:56383–56394. doi: 10.18632/oncotarget.10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY, Gong W, Quan ZW. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e1583. doi: 10.1038/cddis.2014.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang R, Ma Y, Yu D, Zhao J, Ma P. miR-377 functions as a tumor suppressor in human clear cell renal cell carcinoma by targeting ETS1. Biomed Pharmacother. 2015;70:64–71. doi: 10.1016/j.biopha.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Chen G, Lu L, Liu C, Shan L, Yuan D. MicroRNA-377 suppresses cell proliferation and invasion by inhibiting TIAM1 expression in hepatocellular carcinoma. PLoS One. 2015;10:e0117714. doi: 10.1371/journal.pone.0117714. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Wen Z, Huang W, Feng Y, Cai W, Wang Y, Wang X, Liang J, Wani M, Chen J, Zhu P, et al. MicroRNA-377 regulates mesenchymal stem cell-induced angiogenesis in ischemic hearts by targeting VEGF. PLoS One. 2014;9:e104666. doi: 10.1371/journal.pone.0104666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: Unraveling the biology. Trends Biochem Sci. 2004;29:409–417. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and pRB families in vivo: Distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14:804–816. [PMC free article] [PubMed] [Google Scholar]
- 30.Humbert PO, Verona R, Trimarchi JM, Rogers C, Dandapani S, Lees JA. E2f3 is critical for normal cellular proliferation. Genes Dev. 2000;14:690–703. [PMC free article] [PubMed] [Google Scholar]
- 31.Foster CS, Falconer A, Dodson AR, Norman AR, Dennis N, Fletcher A, Southgate C, Dowe A, Dearnaley D, Jhavar S, et al. Transcription factor E2F3 overexpressed in prostate cancer independently predicts clinical outcome. Oncogene. 2004;23:5871–5879. doi: 10.1038/sj.onc.1207800. [DOI] [PubMed] [Google Scholar]
- 32.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/S1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]