Abstract
The gut microbiome is a novel frontier in xenobiotic metabolism. Polybrominated diphenyl ethers (PBDEs), especially BDE-47 (2, 2′, 4, 4′-tetrabromodiphenyl ether) and BDE-99 (2, 2′, 4, 4′,5-pentabromodiphenyl ether), are among the most abundant and persistent environmental contaminants that produce a variety of toxicities. Little is known about how the gut microbiome affects the hepatic metabolism of PBDEs and the PBDE-mediated regulation of drug-processing genes (DPGs) in vivo. The goal of this study was to determine the role of gut microbiome in modulating the hepatic biotransformation of PBDEs. Nine-week-old male C57BL/6J conventional (CV) or germ-free (GF) mice were treated with vehicle, BDE-47 or BDE-99 (100 μmol/kg) for 4 days. Following BDE-47 treatment, GF mice had higher levels of 5-OH-BDE-47 but lower levels of four other metabolites in liver than CV mice; whereas following BDE-99 treatment GF mice had lower levels of four minor metabolites in liver than CV mice. RNA sequencing demonstrated that the hepatic expression of DPGs was regulated by both PBDEs and enterotypes. Under basal conditions, the lack of gut microbiome upregulated the Cyp2c subfamily but downregulated the Cyp3a subfamily. Following PBDE exposure, certain DPGs were differentially regulated by PBDEs in a gut microbiome–dependent manner. Interestingly, the lack of gut microbiome augmented PBDE-mediated upregulation of many DPGs, such as Cyp1a2 and Cyp3a11 in mouse liver, which was further confirmed by targeted metabolomics. The lack of gut microbiome also augmented the Cyp3a enzyme activity in liver. In conclusion, our study has unveiled a novel interaction between gut microbiome and the hepatic biotransformation of PBDEs.
Introduction
The liver is a major organ for the metabolism and excretion of xenobiotics. Many genes encoding drug-metabolizing enzymes or transporters [together called drug-processing genes (DPGs)] are highly expressed in liver. Drug-metabolizing enzymes include phase-I enzymes that perform oxidation, reduction, and hydrolysis reactions; and phase-II enzymes that catalyze various conjugation reactions (Aleksunes and Klaassen, 2012; Li et al., 2016). Uptake transporters bring various chemicals into hepatocytes for biotransformation, whereas efflux transporters export chemicals out of hepatocytes for elimination into urine or bile (Klaassen and Aleksunes, 2010). The expression of DPGs is regulated by xenobiotic-sensing transcription factors such as the aryl hydrocarbon receptor, constitutive androstane receptor [(CAR)/Nr1i3], and pregnane X receptor [(PXR)/Nr1i2] (Klaassen and Slitt, 2005; Cui et al., 2009; Aleksunes and Klaassen, 2012; Cui and Klaassen, 2016; Li et al., 2016; Park et al., 2016). Activation of these transcription factors by drugs or other xenobiotics alters the expression of DPGs, which may influence the pharmacokinetics of coadministered drugs or other chemicals, resulting in drug-drug interactions or drug-food reactions.
Polybrominated diphenyl ethers (PBDEs) are widely used as flame retardants and are persistently present in the environment due to their lipophilic and bio-accumulative properties. Accumulating evidence has shown the adverse effects of PBDEs on laboratory animals, including thyroid hormone disorders (Zhou et al., 2002), neurotoxicity (Madia et al., 2004), hepatic oxidative stress (Fernie et al., 2005), and carcinogenesis (National Toxicology Program, 1986). Among all 209 known PBDE congeners, two bromodiphenyl ethers (BDEs), i.e., 2,2′,4,4′-tetrabromodiphenyl ether (BDE-47) and 2,2′,4,4′,5-pentabromodiphenyl ether (BDE-99), are the most predominant congeners detected in humans (Sjödin et al., 2001; Schecter et al., 2003; Hites, 2004). BDE-47 and BDE-99 are also activators of CAR and PXR in both rodents and human hepatocytes, leading to induced expression of genes important in xenobiotic metabolism and disposition (Pacyniak et al., 2007; Sueyoshi et al., 2014). Studies incubating BDE-47 and BDE-99 with rat or human microsomes have demonstrated that PBDEs are first biotransformed to hydroxylated PDBE (OH-BDE) metabolites by cytochrome P450 (P450) enzymes (Hamers et al., 2008; Lupton et al., 2009; Stapleton et al., 2009; Dong et al., 2010; Erratico et al., 2010, 2011; Feo et al., 2013). OH-BDEs have been identified in blood samples of women and children who were environmentally exposed to PBDEs (Qiu et al., 2009), and in blood, bile, urine, and feces of rodents after exposure to BDE-47, BDE-99, or PBDE mixtures (Orn and Klasson-Wehler, 1998; Hakk et al., 2002; Malmberg et al., 2005; Chen et al., 2006; Marsh et al., 2006; Staskal et al., 2006). The OH-BDEs are potentially more potent and toxic than their parent compounds in disrupting Ca2+ homeostasis, altering neuronal viability, and competing with thyroxine for binding to human transthyretin (Meerts et al., 2000; Cantón et al., 2005; Hamers et al., 2006; Dingemans et al., 2011). The OH-BDEs are then further metabolized by phase-II enzymes such as UDP-glucuronosyltransferases (Ugts) and sulfotransferases (Sults) before elimination (Erratico et al., 2015).
The gut microbiome is recognized as an additional regulator of DPGs in liver. Profound differences have been reported in the expression patterns of DPGs between conventional (CV) and germ-free (GF) mice (Björkholm et al., 2009; Toda et al., 2009; Selwyn et al., 2015a,b; 2016). Alterations in the gut microbiome by either diet or other environmental factors have been suggested to contribute to interindividual variations in drug response and altered expression of DPGs. This in turn may also change the composition of gut microbiota and ultimately exaggerate the adverse effects of the initial insults (Klaassen and Cui, 2015; Nayak and Turnbaugh, 2016). Therefore, classification of the human gut microbiome into distinct enterotypes, defined by their bacterial composition, has been proposed to serve as a new research paradigm to better understand the interplay between microbial variation and human diseases (Arumugam et al., 2011; Knights et al., 2014; Nayak and Turnbaugh, 2016).
Despite the known importance of hepatic DPGs and gut microbiome in xenobiotic biotransformation, very little is known regarding the potential contribution of the liver-gut-bacteria axis on hepatic PBDE metabolism and disposition in vivo. The goals of the present study were to investigate: 1) how gut microbiome modifies the PBDE-mediated regulation of the hepatic DPGs, and 2) how the lack of gut microbiome modifies the hepatic hydroxylation of PBDEs. To accomplish these goals, we used GF mouse models, PBDE-targeted metabolomics, RNA sequencing (RNA-Seq), and liquid chromatography–tandem mass spectroscopy (LC-MS/MS)–based targeted proteomics to strategically investigate the novel interactions between gut microbiome and host DPGs and their effect on the metabolism of the major PBDE congeners BDE-47 and BDE-99 in liver.
Materials and Methods
Chemicals.
BDE-47 was purchased from Chem Service, Inc. (West Chester, PA). BDE-99 was purchased from AccuStandard, Inc. (New Haven, CT). Phosphate-buffered saline (10×, pH 7.4) was purchased from Thermo Fisher Scientific (Grand Island, NY). Synthetic heavy labeled peptides (Supplemental Table 1) were obtained from Thermo Fisher Scientific (Rockford, IL). Ammonium bicarbonate (98% purity) and Pierce Trypsin protease (mass spectrometry grade) were purchased from Thermo Fisher Scientific (Rockford, IL). Chloroform, ethyl ether, Optima mass spectrometry–grade acetonitrile, methanol, and formic acid were purchased from Fischer Scientific (Fair Lawn, NJ). All other chemicals and reagents, unless indicated otherwise, were purchased from Sigma-Aldrich (St. Louis, MO).
Animals.
Eight-week-old male C57BL/6J CV mice were purchased from the Jackson Laboratory (Bar Harbor, ME), and were acclimated to the animal facility at the University of Washington for 1 week prior to experiments. The initial breeding colony of GF mice in C57BL/6 background was established with mice purchased from the National Gnotobiotic Rodent Resource Center (University of North Carolina, Chapel Hill, NC). All mice were housed according to the Association for Assessment and Accreditation of Laboratory Animal Care International guidelines (https://aaalac.org/resources/theguide.cfm), and the studies were approved by the Institutional Animal Care and Use Committee at the University of Washington. The CV and GF mice were exposed to the same diet [laboratory autoclaved rodent diet (LabDiet #5010)] (LabDiet, St. Louis, MO), water (nonacidified autoclaved water), and bedding (autoclaved Enrich-N’Pure) (Andersons, Maumee, OH). All chemical solutions were sterilized using the Steriflip Vacuum-Driven Filtration System with a 0.22 μm Millipore Express Plus Membrane (EMD Millipore, Temecula, CA). All gavage needles and syringes were sterilized by autoclave. At 9 weeks of age, CV mice (n = 5 per group) were randomly allocated for the treatment of vehicle (corn oil, 10 ml/kg), BDE-47 (10 and 100 μmol/kg), or BDE-99 (10 and 100 μmol/kg) via oral gavage once daily for 4 days. Livers were collected 24 hours after the last dosing on the fifth day and immediately frozen in liquid nitrogen. Based on the highest induction of Cyp2b10 and Cyp3a11 mRNAs in CV mice, age-matched GF mice (n = 3 or 4 per group) were treated with vehicle (corn oil, 10 ml/kg), BDE-47 (100 μmol/kg), or BDE-99 (100 μmol/kg) following the same procedure (as shown in Fig. 1). Due to the potential variations caused by the estrous cycle in adult female mice, only male livers were used for this study. All tissues were immediately frozen in liquid nitrogen and stored at −80°C until further analysis.
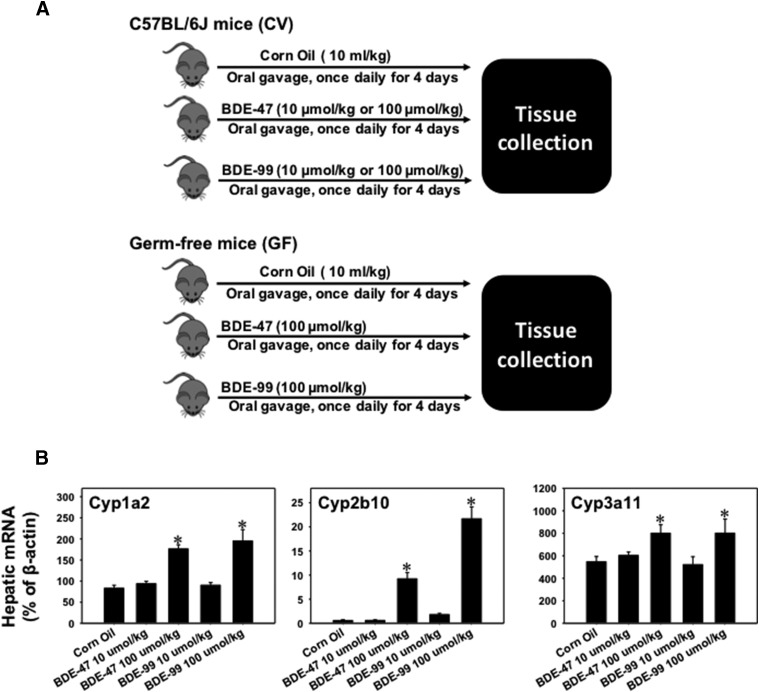
Fig. 1.
(A) Diagram illustrating the experimental design and dosing regimen of mice. Briefly, 9-week-old C57BL/6J CV mice were treated with vehicle (corn oil, 10 ml/kg, oral gavage), BDE-47 (10 or 100 μmol/kg, oral gavage), or BDE-99 (10 or 100 μmol/kg, oral gavage) once daily for 4 days (n = 5 per group). Tissues were collected 24 hours after the final dose. Age-matched GF mice were treated with vehicle (corn oil, 10 ml/kg, oral gavage), BDE-47 (100 μmol/kg, oral gavage), or BDE-99 (100 μmol/kg, oral gavage) following the same procedure (n = 3 or 4 per group). (B) RT-qPCR of mRNAs of the prototypical aryl hydrocarbon receptor–target gene Cyp1a2, the prototypical CAR-target gene Cyp2b10, and the prototypical PXR-target gene Cyp3a11 in liver of CV mice treated with corn oil, BDE-47 (10 or 100 μmol/kg), or BDE-99 (10 or 100 μmol/kg). Data are expressed as percentage of the housekeeping gene β-actin (n = 5 per group). Asterisks (*) indicate statistically significant differences between corn oil–treated and PBDE-treated groups in liver (P < 0.05).
Serum Alanine Aminotransferase (ALT) Quantification and H&E Staining.
Serum samples were analyzed by standard enzymatic-colorimetric assays using ALT kits according to the manufacturer’s protocol (Pointe Scientific, Inc. Canton, MI). H&E staining of liver sections collected from CV and GF mice treated with corn oil, BDE-47, or BDE-99 was performed at the Histology and Imaging Core Facility at the University of Washington. Representative slides are shown (×100).
Total RNA Isolation.
Total RNA was isolated from frozen tissues using RNA-Bee reagent (Tel-Test lnc., Friendswood, TX) according to the manufacturer’s protocol. RNA concentrations were quantified using NanoDrop 1000 Spectrophotometer (Thermo Scientific, Waltham, MA) at 260 nm. The integrity of total RNA samples was evaluated by formaldehyde-agarose gel electrophoresis with visualization of 18S and 28S rRNA bands under UV light, and by an Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA). Samples with RNA integrity numbers above 8.0 were used for RNA-Seq.
Quantification of Gene Expression by Quantitative Reverse Transcription Polymerase Chain Reaction (RT-PCR).
Total RNA was transcribed to cDNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). The cDNAs were amplified by PCR using SsoAdvanced Universal SYBR Green Supermix in a Bio-Rad CFX384 Real-Time PCR Detection System (Bio-Rad, Hercules, CA). The PCR primers were synthesized by Integrated DNA Technologies (Coralville, IA). The primers for β-actin were GGCCAACCGTGAAAAGATGA (forward) and CAGCCTGGATGGCTACGTACA (reverse), the primers for Cyp1a2 were GACATGGCCTAACGTGCAG (forward) and GGTCAGAAAGCCGTGGTTG (reverse), the primers for Cyp2b10 were AAGGAGAAGTCCAACCAGCA (forward) and CTCTGCAACATGGGGGTACT (reverse), and the primers for Cyp3a11 were ACAAACAAGCAGGGATGGAC (forward) and GGTAGAGGAGCACCAAGCTG (reverse). The delta delta Cq (ddCq) values were calculated for each target gene and normalized to the expression of the housekeeping gene β-actin.
cDNA Library Preparation and RNA-Seq.
The cDNA libraries were constructed from total RNA samples using an Illumina TruSeq Stranded mRNA kit (Illumina, San Diego, CA). Briefly, 1.25 μg of total RNA was used as the RNA input. The mRNAs were selected from the total RNAs by purifying the poly-A containing molecules using poly-T primers. The RNA fragmentation, first and second strand cDNA syntheses, end repair, adaptor ligation, and PCR amplification were performed according to the manufacturer’s protocol. The cDNA libraries were validated for integrity and quantity using an Agilent 2100 Bioanalyzer (Agilent Technologies) before sequencing. The cDNA libraries were sequenced on an Illumina HiSeq2000 sequencer using a 50 base pair, paired-end multiplexing strategy with five samples per lane at Dr. Nickerson’s Sequencing Facility at the Genome Sciences Department, University of Washington.
RNA-Seq Data Analysis.
FASTQ files were demultiplexed and concatenated per sample for downstream analysis. Quality control of all FASTQ files was performed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). RNA-Seq reads from the FASTQ files were mapped to the reference genome [National Center for Biotechnology Information (NCBI GRCm38/mm10)] using HISAT version 0.1.6 beta (Kim et al., 2015). Sequencing alignment/map files were converted to binary format using SAMtools version 1.2 (Li et al., 2009). The output files in binary alignment/map format were analyzed by Cufflinks version 2.2.1 to estimate the transcript abundance (Trapnell et al., 2010). The mRNA abundance was expressed as fragments per kilobase of transcript per million mapped reads (FPKM). Differential expression analysis was performed using Cuffdiff (Trapnell et al., 2010). The differentially expressed genes were defined as false discovery rate–adjusted P value, <0.05, in at least one of the chemical-treated groups compared with the vehicle-treated group of the same enterotypes of mice, as well as genes that were differentially expressed by the lack of gut microbiome under the same treatment. The RNA-Seq generated an average of 70 million reads per sample, and more than 80% of the reads were mapped to the mouse reference genome (NCBI GRCm38/mm10). The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through Gene Expression Omnibus accession number GSE101650.
Extraction and Quantification of PBDE Hydroxylated Metabolites.
The extraction and measurement of OH-BDEs was modified from the methods described by Hovander et al. (2000). Briefly, liver samples were first homogenized with four volumes of deionized water and then spiked with the surrogate, 4-OH-PCB-159. Next, the homogenates were denatured by adding 6 M HCl and 2-propanol. After 5 minutes at room temperature, the mixture was extracted with hexane/methyl tert-butyl ether (1:1 v/v). The organic extract was volume reduced and the phenolic fraction (containing the OH-BDEs) was separated from neutral compounds by adding a solution of potassium hydroxide (0.5 M in 50% ethanol). The aqueous layer, which contained phenolic compounds, was acidified with 2 M HCl and then extracted with hexane/methyl tert-butyl ether mix (9:1 v/v). The hexane/ether extract, which now contained the phenolic compounds, was dried over ∼2 g of sodium sulfate for 1 hour, and then volume reduced to 2 ml and mixed with 200 µl of diazomethane for derivatization. After overnight incubation at room temperature, the derivatized extract was further cleaned using an acid silica gel column. The columns were prepared with 2 g of acid silica, 1 g of neutral silica, and a top layer of sodium sulfate. Prior to sample loading, the column was exhaustively rinsed with dichloromethane and hexane. After loading, the samples were eluted from the column with 40 ml of dichloromethane. Afterward, the dichloromethane extract was evaporated, solvent exchanged with hexane, and then subsequently evaporated to approximately 100 μl in volume and spiked with an internal standard (BDE-166) for analysis. Quantification of OH-BDEs in the hexane extract was by gas chromatography mass spectroscopy (GCMS) in the negative chemical ionization mode using an Agilent 7890B/5977A GCMS system (Agilent Technologies Inc.). Calibration curves were made using authentic OH-BDE standards.
LC/MS-MS Protein Quantification.
The procedure was similar to that previously published with few modifications (Selwyn et al., 2015a). Briefly, the membrane proteins of the mouse livers were isolated using the Mem-PER plus membrane protein extraction kit (Pierce Biotechnology, Rockford, IL). The total membrane protein concentration was quantified using a bicinchoninic acid kit, followed by digestion using an in-solution trypsin digestion kit (Pierce Biotechnology). Eighty microliters of the tissue extract (2 mg/ml total protein) was mixed with 10 µl dithiothreitol (DTT) (250 mM), 20 µl of 0.2 mg/ml bovine serum albumin, 10 µl of 10 mg/ml human serum albumin, and 40 µl ammonium bicarbonate buffer (100 mM, pH 7.8), and then incubated at 95°C for 5 minutes (denaturation and reduction). Subsequently, 20 µl iodoacetamide (500 mM) was added and the sample was incubated for 30 minutes at ambient temperature in the dark (alkylation). Ice-cold methanol (0.5 ml), chloroform (0.1 ml), and water (0.4 ml) were added to each sample. After centrifugation at 16,000g for 5 minutes at 4°C, the upper and lower layers were removed and the pellet was washed with ice-cold methanol (0.5 ml) and centrifuged at 8000g for 5 minutes at 4°C. The pellet was resuspended with 40 µl 50 mM ammonium bicarbonate buffer. Trypsin (20 µl) was added at a 1:80 trypsin:protein ratio (w/w) and the samples were incubated for 18 hours at 37°C. Trypsin digestion was stopped by adding 10 µl of chilled quenching solvent (80% acetonitrile with 0.5% formic acid) and 30 µl heavy peptide internal standard, the samples were centrifuged for 5 minutes at 4000g and 4°C, and then the supernatant was collected in LC-MS/MS vials. The stable isotope-labeled heavy peptides were used as internal standards (Thermo Fisher Scientific, Rockford, IL). LC-MS/MS consisted of an Acquity LC (Waters Technologies, Milford, MA) coupled to a Triple Quadrupole 6500 mass spectrometry system (AB Sciex, Framingham, MA). One to three surrogate peptides per protein were designed for the quantification of selected proteins (Supplemental Table 1) according to a previously published protocol (Vrana et al., 2017) (QPrOmics; www.qpromics.uw.edu/qpromics/assay/). The peptide separation was achieved on an Acquity ultra-performance liquid chromatography HSS T3 column (1.8 μm, 2.1 × 100 mm; Waters, Hertfordshire, United Kingdom). Mobile phase A (water with formic acid 0.1%; v/v) and mobile phase B (acetonitrile with formic acid 0.1%; v/v) were used with a flow rate of 0.3 ml/min in a gradient manner. Peak integration and quantification were performed using Analyst (version 1.6, Mass Spectrometry Toolkit version 3.3; Applied Biosystems, Framingham, MA). A robust strategy was used to ensure optimal reproducibility when quantifying these proteins, for example, ion suppression was addressed by using heavy peptide. Bovine serum albumin was used as an exogenous internal standard, which was added to each sample to correct for protein loss during processing and digestion efficiency. In total, three-step data normalization was used; first, average light peak areas for specific peptide daughter fragments were divided by corresponding average heavy peak areas. This ratio was further divided by the bovine serum albumin light/heavy area ratio. Data were further normalized to average quality control values (pooled representative sample). The protein abundance data are expressed as the mean (normalized to total protein) of the three biologic replicates with S.E.
Preparation of Crude Membrane and Microsomal Fractions (for Enzyme Activities).
For isolation of crude membranes, livers were homogenized in Sucrose-Tris buffer (0.25 M sucrose, 10 mM Tris-HCl, pH 7.4) containing protease inhibitors (1:100; Sigma-Aldrich), and centrifuged at 100,000g for 60 minutes at 4°C. The resulting pellet constituted the crude membrane fraction and was resuspended in Sucrose-Tris buffer. For isolation of microsomes, homogenates were first centrifuged at 10,000g for 20 minutes at 4°C. The supernatant was then centrifuged at 100,000g for 60 minutes. Sucrose-Tris buffer was used to resuspend the microsomal pellet. Protein concentration was determined using the Qubit Protein Assay Kit (Thermo Fisher Scientific, Grand Island, NY).
Enzyme Activity of Cyp1a, Cyp2b, and Cyp3a in Mouse Liver Microsomes.
The Cyp1a, Cyp2b, and Cyp3a enzyme activities in liver microsomes from CV and GF mice were determined using a P450-Glo Screening System (Promega Corporation, Madison, WI) as described previously. Briefly, a luminogenic P450-Glo substrate (100 μM luciferin 6′-methyl ether (luciferin-ME) for Cyp1a, 3 μM luciferin-2B6 for Cyp2b, or 3 μM luciferin isopropyl acetal (luciferin-IPA) for Cyp3a) was incubated at 37°C with 10 μg of liver microsomal protein, control membrane, or positive P450 enzyme for 10 minutes. The reactions were initiated by adding the NADPH-regeneration system and incubated for 10 minutes. At the end of the incubation, an equal volume of the luciferin detection reagent (50 μl) was added at room temperature, and luminescence was quantified 20 minutes later using a Glomax 96 Microplate Luminometer (Promega Corporation). The magnitude of the light signal was dependent on and directly proportional to the amount of luciferin product generated by the P450 reaction.
Clustering Analysis.
A hierarchical clustering dendrogram (Ward’s minimum variance method, distance scale) of the differentially expressed DPGs was generated based on their standardized mean FPKM values, using JMP Genomics software (SAS Institute, Inc., Cary, NC).
Statistical Analysis.
RNA-Seq data are expressed as mean FPKM ± S.E. Genes with mean FPKM values per treatment group lower than one in all groups were defined as not expressed in liver. Asterisks (*) represent significant differences between corn oil–treated and PBDE-treated groups of the same enterotypes of mice. Pound signs (#) represent significant differences between CV and GF mice under the same treatment. The mean FPKM and fold change of differentially regulated DPGs by gut microbiome or by PBDEs are summarized in Supplemental Tables 2 and 3.
Results
Dose-Response of PBDE Exposure on Hepatic Gene Expression.
To determine at which dose PBDEs produce more changes in DPG expression in livers of CV mice, CV mice were treated with vehicle and a low dose (10 μmol/kg) or a high dose (100 μmol/kg) of BDE-47 or BDE-99 as described in Fig. 1A (upper panel). The mRNAs of phase-I drug-metabolizing enzymes Cyp1a2 (prototypical aryl hydrocarbon receptor–target gene), Cyp2b10 (prototypical CAR-target gene), and Cyp3a11 (prototypical PXR-target gene) were quantified in livers of CV mice using RT-qPCR. As shown in Fig. 1B, Cyp1a2, Cyp2b10, and Cyp3a11 mRNAs were all upregulated by BDE-47 and BDE-99 only at the high dose. Therefore, the high doses of PBDEs were selected for the GF mice treatment to compare the effect of the lack of gut microbiota on the regulation of DPGs in liver (Fig. 1A, lower panel).
Serum ALT and Liver Histology after PBDE Exposure.
To ensure that the doses used in this study did not produce any off-target effects due to liver injuries, serum ALT and liver histology were performed as shown in Supplemental Fig. 1. Serum ALT levels were within the normal range (between 4 and 36 IU/l at 37°C) in all treatment groups of CV and GF mice (Supplemental Fig. 1A). Consistent with the serum ALT data, H&E staining showed no apparent hepatic injuries in livers of CV and GF mice treated with corn oil, BDE-47, or BDE-99 (Supplemental Fig. 1B).
Hydroxylated Metabolites of PBDEs in Livers of CV and GF Mice.
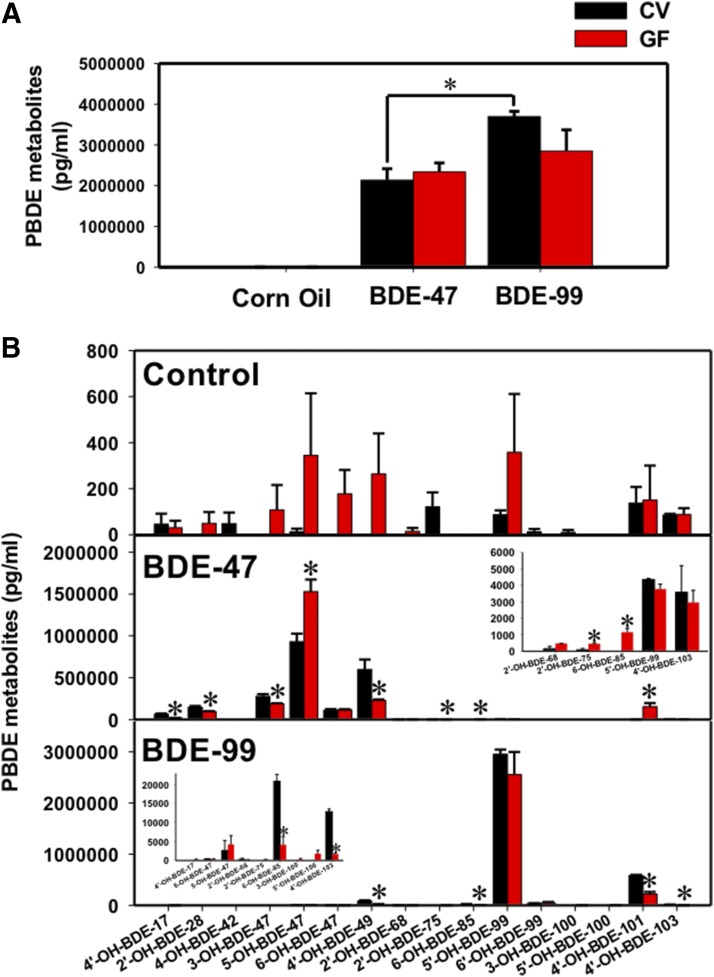
Sixteen hydroxylated metabolites of BDE-47 and BDE-99 were measured by PBDE-targeted metabolomics (GCMS negative chemical ionization) in livers of CV and GF mice treated with corn oil, BDE-47 (100 μmol/kg), or BDE-99 (100 μmol/kg) (Fig. 2). Total hepatic levels of PBDE hydroxylated metabolites were similar between CV and GF mice treated with the same PBDE congener (Fig. 2A). However, at equal molar dose, the total hydroxylated metabolites of BDE-99 were higher than those of BDE-47 in CV mice, whereas this difference was not observed in GF mice. This suggests that the bioavailability of BDE-99 is higher than BDE-47, which may be due to a combination of the host and microbiota effects, including different substrate specificities of the host transporters or P450s, or different microbial biotransformation pathways in gut microbiome that influence the absorption of the two PBDE congeners. Alternatively, this may also suggest that BDE-99 is more readily hydroxylated than BDE-47 due to substrate specificities and/or levels of preferred P450s.
Fig. 2.
Concentrations of hydroxylated PBDE metabolites (picograms per milliliters, mean ± S.E.) in livers of CV and GF mice treated with corn oil, BDE-47 (100 μmol/kg), or BDE-99 (100 μmol/kg). (A) Total hepatic levels of hydroxylated PBDE metabolites in CV and GF mice treated with corn oil, BDE-47 (100 μmol/kg), or BDE-99 (100 μmol/kg). Asterisks (*) represent statistically significant differences between comparison groups. (B) Concentrations of individual hydroxylated metabolites of BDE-47 and BDE-99 in livers of CV and GF mice treated with corn oil, BDE-47 (100 μmol/kg), or BDE-99 (100 μmol/kg). Asterisks (*) represent statistically significant differences between CV and GF mice.
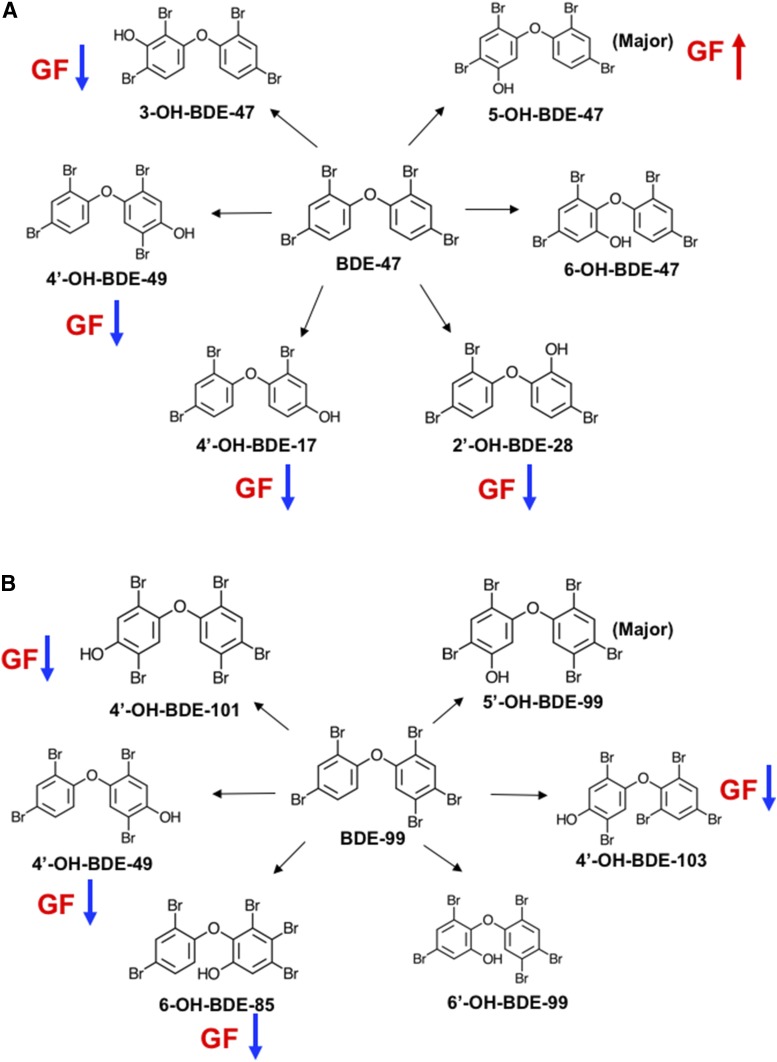
Regarding individual hydroxylated metabolites of PBDEs (Fig. 2B), low levels of PBDE hydroxylated metabolites were detected in livers of CV mice treated with corn oil. These PBDE metabolites are likely a result of a background exposure to PBDEs from diet, bedding, and other environmental factors within the animal facility. Control GF mice appeared to have higher levels of many PBDE metabolites, although statistical significance was not achieved (Fig. 2B, top panel). Following BDE-47 treatment, 12 out of 16 PBDE hydroxylated metabolites were detected in livers of CV and GF mice (Fig. 2B, middle panel). The major hydroxylated metabolite of BDE-47 was 5-OH-BDE-47 in livers of both CV and GF mice, with a higher level observed in GF conditions. Four minor BDE-47 metabolites were decreased in livers of GF mice compared with livers of BDE-47-treated CV mice, including 4′-OH-BDE-17, 2′-OH-BDE-28, 3-OH-BDE-47, and 4′-OH-BDE-49. There was no difference for 6-OH-BDE-47 between BDE-47-treated CV and GF mice. Unexpectedly, although mice were treated exclusively with BDE-47, which contains four bromine atoms, there were low levels of other tetraBDEs (2′-OH-BDE-68 and 2′-OH-BDE-75) and also pentaBDEs (6-OH-BDE-85, 5′-OH-BDE-99, 4′-OH-BDE-101, and 4′-OH-BDE-103) detected in livers of BDE-47-treated CV and/or GF mice. Among these, 2′-OH-BDE-75, 6-OH-BDE-85, and 4′-OH-BDE-101 were increased in livers of GF mice compared with livers of BDE-47-treated CV mice. These hydroxylated tetraBDE and pentaBDE metabolites may come from the diet or other environmental sources. Following BDE-99 treatment, 13 out of 16 hydroxylated PBDE metabolites were detected in livers of CV and GF mice (Fig. 2B, bottom panel). The major hydroxylated metabolite of BDE-99 was 5′-OH-BDE-99 in livers of both CV and GF mice, and it remained unchanged between CV and GF conditions. Four minor BDE-99 hydroxylated metabolites were decreased in livers of GF mice compared with livers of BDE-99-treated CV mice, including 4′-OH-BDE-49, 6-OH-BDE-85, 4′-OH-BDE-101, and 4′-OH-BDE-103. There was no difference for 6′-OH-BDE-99 between BDE-99-treated CV and GF mice. In addition, very low amounts of tetraBDEs (4′-OH-BDE-17, 5-OH-BDE-47, 6-OH-BDE-47, 2′-OH-BDE-68, and 2′-OH-BDE-75) and pentaBDEs (3-OH-BDE-100 and 5′-OH-BDE-100) were detected in livers of BDE-99-treated CV and GF mice. Chemical structures of individual BDE-47 and BDE-99 metabolites, especially the major metabolite and the metabolites that were altered in GF condition, were plotted using ChemDoodle (iChemLabs, LLC., Chesterfield, VA) and are shown in Fig. 3. In summary, the lack of gut microbiota modifies the bioavailability and the hepatic metabolism of PBDEs. Divergent regulatory patterns were observed regarding individual hydroxylated PBDE metabolites between CV and GF conditions, indicating the involvement of multiple P450s that are divergently regulated by gut microbiota in PBDE hepatic oxidative metabolism.
Fig. 3.
Chemical structures (plotted by ChemDoodle) of hydroxylated metabolites of BDE-47 (A) and BDE-99 (B) in livers of CV and GF mice treated with corn oil, BDE-47 (100 μmol/kg), or BDE-99 (100 μmol/kg). The major hydroxylated metabolite of BDE-47 or BDE-99 is labeled. Compared with CV mice, the upregulated (red arrow) or downregulated (blue arrow) individual metabolite in GF conditions are highlighted.
To determine the role of the gut microbiome on hepatic transcriptome that is likely to be involved in PBDE metabolism and disposition, the expression of various phase-I (Cyp1, 2, and 3 family members, which are critically involved in xenobiotic metabolism) and phase-II drug-metabolizing enzymes [Ugts, Sults, and glutathione S-transferases (Gsts)], as well as transporters [solute carrier organic anion (Slco) and ATP-binding cassette subfamily c members, (Abcc)] was examined. Only DPGs that were differentially expressed by PBDEs or by the lack of gut microbiome are shown in Figs. 4–7. DPGs were plotted separately to better visualize the fold changes, and plotted together to compare the absolute abundance within the same family/subfamily.
Fig. 4.
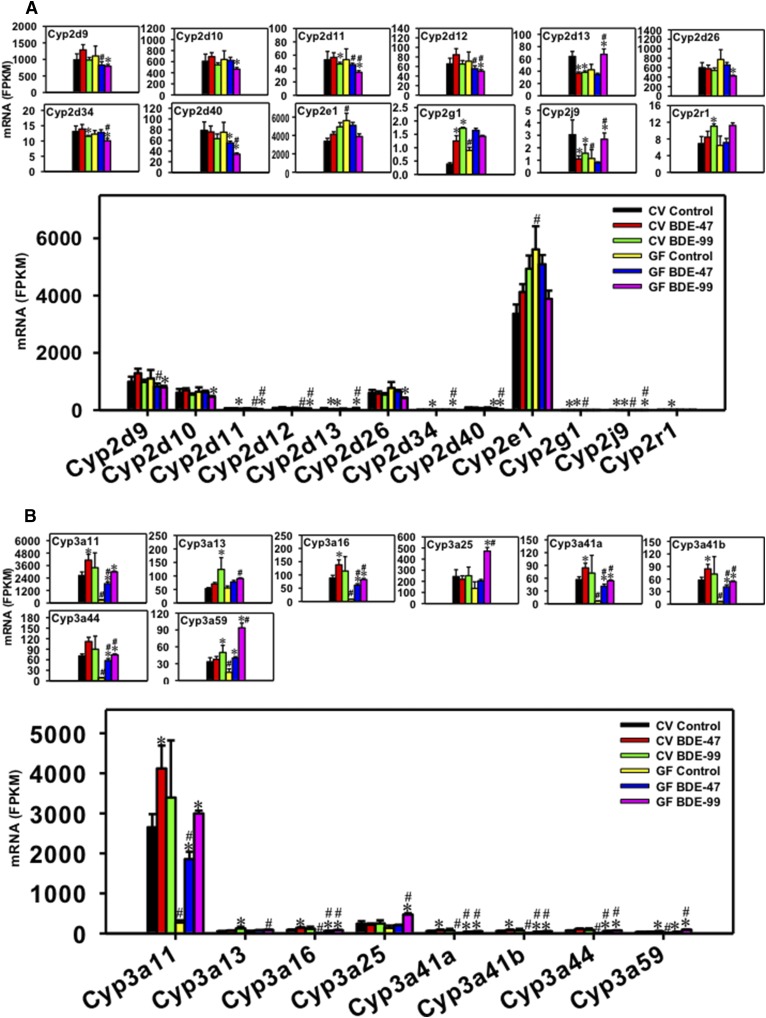
Regulation of phase-I Cyp1 (A), Cyp2a and Cyp2b (B), and Cyp2c (C) drug-metabolizing enzymes by BDE-47 or BDE-99 in livers of CV and GF mice. Only Cyps that were differentially expressed by PBDEs or by lack of gut microbiota are shown. Genes in the same family are grouped together to quantitatively compare the mRNA abundance, and are also presented individually to better visualize the mRNA fold changes by BDE-47 or BDE-99 in CV and GF mice. Asterisks (*) represent statistically significant differences between corn oil–treated and PBDE-treated groups of the same enterotypes of mice (false discovery rate–adjusted P value, <0.05). Pound signs (#) represent statistically significant differences between CV and GF mice under the same treatment.
Fig. 7.
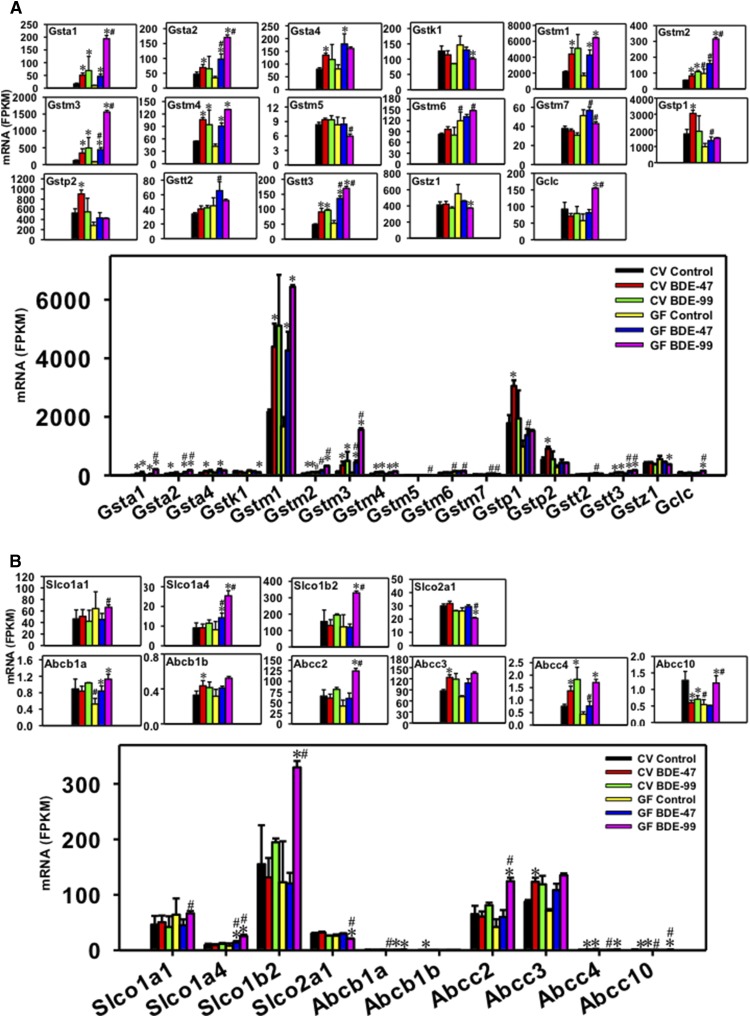
Regulation of phase-II Gsts (A), as well as transporters in the Slco and ATP-binding cassette families (B) by BDE-47 or BDE-99 in livers of CV and GF mice. Only DPGs differentially expressed by PBDEs or by lack of gut microbiota are shown. Genes in the same family are grouped together to quantitatively compare the mRNA abundance, and are also presented individually to better visualize the mRNA fold changes by BDE-47 or BDE-99 in CV and GF mice. Asterisks (*) represent statistically significant differences between corn oil–treated and PBDE-treated groups of the same enterotypes of mice (false discovery rate–adjusted P value, <0.05). Pound signs (#) represent statistically significant differences between CV and GF mice under the same treatment.
Regulation of Hepatic Cyp1 and Cyp2a-c Genes by BDE-47 and BDE-99 in CV and GF Mice.
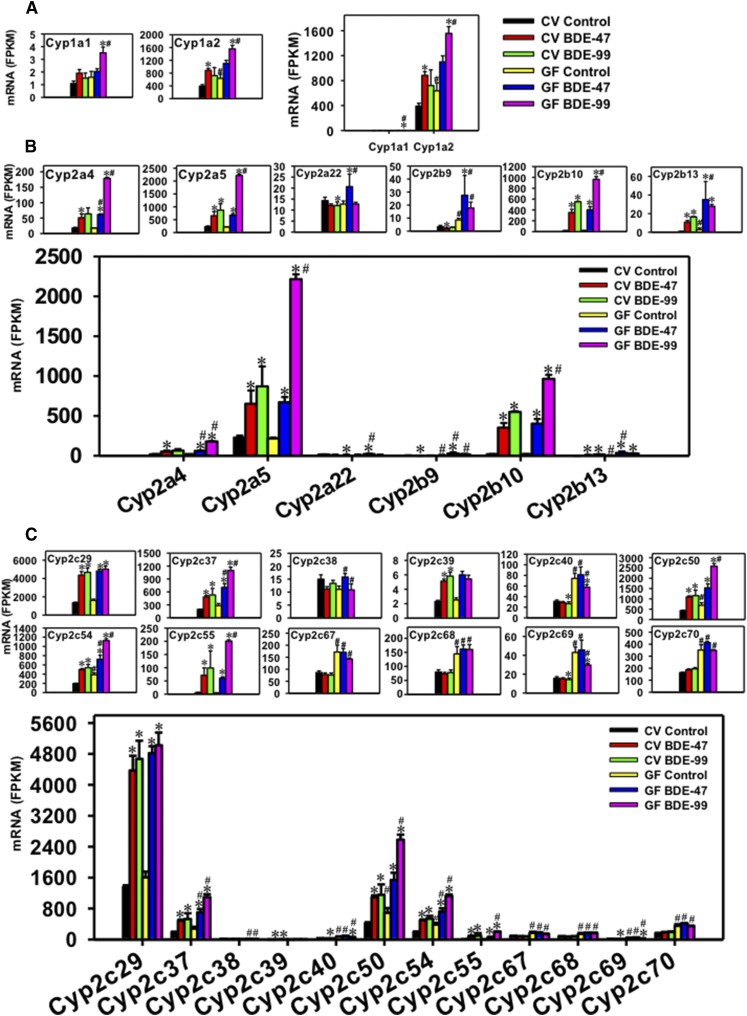
For the Cyp1 family, both Cyp1a1 and Cyp1a2 were differentially regulated by PBDEs (Fig. 4A), whereas Cyp1b1 remained unchanged (data not shown). Cyp1a2 was the predominant Cyp1 isoform that was much more abundantly expressed in the liver compared with Cyp1a1. Under basal conditions, Cyp1a1 mRNA remained unchanged between CV and GF mice, whereas the lack of gut microbiome led to a moderate increase in the constitutive expression of Cyp1a2 mRNA (1.62-fold). In livers of CV mice, BDE-47 did not alter Cyp1a1 mRNA expression, but upregulated Cyp1a2 mRNA (2.26-fold); whereas in livers of GF mice, BDE-47 did not alter the mRNAs of Cyp1a1 or Cyp1a2. In livers of CV mice, BDE-99 did not alter the mRNAs of Cyp1a1 or 1a2; however, it markedly increased the mRNAs of both Cyp1a1 (2.22-fold) and Cyp1a2 (2.45-fold) in livers of GF mice, suggesting that the lack of gut microbiome augmented BDE-99-mediated upregulation of Cyp1a1 and Cyp1a2 mRNAs in mouse liver.
For the Cyp2a subfamily, Cyp2a4, 2a5, and 2a22 were differentially regulated by PBDEs in liver, among which Cyp2a5 mRNA was the highest expressed isoform in all treatment groups (Fig. 4B). Lack of gut microbiome had no effect on the basal expression of these Cyp2a isoforms. In livers of CV mice, BDE-47 markedly increased the mRNAs of Cyp2a4 (2.87-fold) and Cyp2a5 (2.86-fold); whereas in livers of GF mice, BDE-47 also increased the mRNAs of Cyp2a4 (3.47-fold) and Cyp2a5 (3.10-fold), with higher fold increase in Cyp2a4 mRNA compared with BDE-47-treated CV mice. Cyp2a22 mRNA was not altered by BDE-47 in livers of CV mice, but was upregulated by BDE-47 in livers of GF mice compared with the GF control group (1.62-fold), suggesting that the lack of gut microbiome augmented BDE-47-mediated upregulation of Cyp2a22 in mouse liver. In livers of CV mice, BDE-99 increased the Cyp2a5 mRNA (3.82-fold) and tended to increase Cyp2a4 mRNA (3.58-fold, although a statistical significance was not achieved), but slightly decreased the mRNA of Cyp2a22 (15.75%). In livers of GF mice, the mRNA fold increases were much greater for BDE-99-mediated upregulation of Cyp2a4 (10.09-fold) and Cyp2a5 (10.23-fold), whereas Cyp2a22 mRNA remained unchanged. In summary, BDE-99 had a more prominent effect than BDE-47 on the upregulation of Cyp2a4 and Cyp2a5 mRNAs in GF conditions; lack of gut microbiome augmented BDE-47-mediated upregulation of Cyp2a4 and Cyp2a22; and also augmented BDE-99-mediated upregulation of Cyp2a4 and Cyp2a5 in mouse liver.
For the Cyp2b subfamily, Cyp2b9, 2b10, and 2b13 were differentially regulated by PBDEs in liver and all of them were lowly expressed under basal conditions in CV mice. The lack of gut microbiome resulted in a constitutive increase in the basal mRNA expression of Cyp2b9 (2.59-fold) and Cyp2b13 (3.69-fold) (Fig. 4B). In livers of CV mice, BDE-47 upregulated the mRNAs of both Cyp2b10 (18.84-fold) and Cyp2b13 (14.38-fold), and such upregulation was also observed in livers of GF mice (22.41-fold for Cyp2b10 and 12.71-fold for Cyp2b13). The BDE-47-mediated upregulation of Cyp2b13 was greater in livers of GF mice compared with BDE-47-treated CV mice. Interestingly, BDE-47 decreased Cyp2b9 mRNA in CV conditions (45.20%) but this pattern was reversed in GF conditions as evidenced by a 3.22-fold increase in Cyp2b9 mRNA in livers of BDE-47-treated GF mice. In livers of CV mice, BDE-99 markedly increased the mRNAs of Cyp2b10 (29.40-fold) and Cyp2b13 (22.22-fold), and such upregulation was also observed in livers of GF mice (53.56-fold for Cyp2b10 and 10.08-fold for Cyp2b13). The BDE-99-mediated upregulation of Cyp2b10 was greater in GF mice compared with BDE-99-treated CV mice. BDE-99 did not alter the Cyp2b9 mRNA in CV or GF conditions compared with the control group of the same enterotype. In summary, the lack of gut microbiome increased the basal expression of Cyp2b9 and 2b13 mRNAs, and reversed the BDE-47-mediated regulatory pattern of Cyp2b9 mRNA; BDE-99 appeared to have a more prominent effect than BDE-47 on the mRNA expression of Cyp2b10, which is the highest expressed Cyp2b isoform under treatment conditions; the lack of gut microbiome augmented BDE-47-mediated upregulation of Cyp2b9 and Cyp2b13, and also augmented BDE-99-mediated upregulation of Cyp2b10 in mouse liver.
For the Cyp2c subfamily, 12 Cyp2c members (Cyp2c29, Cyp2c37, Cyp2c38, Cyp2c39, Cyp2c40, Cyp2c50, Cyp2c54, Cyp2c55, Cyp2c67, Cyp2c68, Cyp2c69, and Cyp2c70) were differentially regulated by PBDEs in liver, among which Cyp2c29 mRNA was the highest expressed isoform under basal and treatment conditions, followed by Cyp2c50, and then Cyp2c37 and 2c54 at similar levels (Fig. 4C). Under basal conditions, the lack of gut microbiome led to a constitutive increase in the basal mRNA expression of Cyp2c40 (2.38-fold), Cyp2c50 (1.61-fold), Cyp2c54 (1.96-fold), Cyp2c67 (1.99-fold), Cyp2c68 (1.81-fold), Cyp2c69 (2.77-fold), and Cyp2c70 (2.15-fold); whereas other Cyp2c isoforms remained unchanged (Fig. 4C). In livers of CV mice, BDE-47 upregulated the mRNAs of most Cyp2c isoforms, including Cyp2c29 (3.22-fold), Cyp2c37 (2.50-fold), Cyp2c39 (2.21-fold), Cyp2c50 (2.56-fold), Cyp2c54 (2.52-fold), and Cyp2c55 (13.45-fold). The BDE-47-mediated upregulation was also observed in livers of GF mice (3.00-fold for Cyp2c29, 2.51-fold for Cyp2c37, 2.23-fold for Cyp2c50, 1.87-fold for Cyp2c54, and 15.88-fold for Cyp2c55). Compared with BDE-47-treated CV mice, BDE-47-treated GF mice had higher hepatic levels of Cyp2c37 and 2c54, indicating a augmentation effect of BDE-47 due to lack of gut microbiota. In livers of CV mice, BDE-99 increased the mRNAs of Cyp2c29 (3.44-fold), Cyp2c37 (2.73-fold), Cyp2c39 (2.51-fold), Cyp2c50 (2.70-fold), Cyp2c54 (2.73-fold), and Cyp2c55 (18.68-fold). The BDE-99-mediated upregulation was also observed in livers of GF mice (3.13-fold for Cyp2c29, 3.91-fold for Cyp2c37, 3.74-fold for Cyp2c50, 2.91-fold for Cyp2c54, and 52.83-fold for Cyp2c55). BDE-99 slightly downregulated the mRNAs of Cyp2c40 (14.74%) and Cyp2c69 (8.90%) in livers of CV mice, and such downregulation was also observed in livers of GF mice (22.37% for Cyp2c40 and 31.11% for Cyp2c69). The lack of gut microbiome augmented the BDE-99-mediated mRNA increase in Cyp2c37, 2c40, 2c50, 2c54, and 2c55 compared with BDE-99-treated CV mice. In summary, the lack of gut microbiome increased the basal expression of many Cyp2c isoforms; BDE-99 appeared to have a more prominent effect than BDE-47 on the mRNA expression of Cyp2c37, 2c50, 2c54, and 2c55; and the lack of gut microbiome augmented PBDE-mediated upregulation of many Cyp2c isoforms in mouse livers.
Regulation of Hepatic Cyp2d, Cyp2e1, Cyp2g1, Cyp2j9, and Cyp2r1 Genes by BDE-47 and BDE-99 in CV and GF Mice.
For the Cyp2d subfamily, eight Cyp2d family members (Cyp2d9, Cyp2d10, Cyp2d11, Cyp2d12, Cyp2d13, Cyp2d26, Cyp2d34, and Cyp2d40) were differentially regulated by PBDEs in liver (Fig. 5A). In general, among these the Cyp2d isoforms Cyp2d9, 2d10, and 2d26 were the most abundantly expressed. The lack of gut microbiome had no effect on the basal expression of these Cyp2d isoforms. In livers of CV mice, BDE-47 had minimal effect on the mRNA expression of Cyp2d9, 2d10, 2d11, 2d12, 2d26, 2d34, and 2d40, but markedly decreased Cyp2d13 mRNA (42.66%); whereas in livers of GF mice, BDE-47 decreased Cyp2d40 mRNA (26.47%). Compared with BDE-47-treated CV mice, BDE-47-treated GF mice had lower hepatic expression of Cyp2d9, 2d11, and 2d12. In livers of CV mice, BDE-99 downregulated the mRNAs of Cyp2d11 (12.08%), Cyp2d13 (40.34%), and Cyp2d34 (12.12%). BDE-99 also downregulated Cyp2d11 (35.58%) and Cyp2d34 (17.86%) in livers of GF mice, but reversed the downregulation pattern for Cyp2d13, as evidenced by a 1.59-fold mRNA increase in GF conditions. In addition, in livers of GF mice, BDE-99 decreased the mRNAs of Cyp2d9 (28.10%), Cyp2d10 (27.41%), Cyp2d12 (29.47%), Cyp2d26 (45.16%), and Cyp2d40 (54.63%). The lack of gut microbiome augmented BDE-99-mediated downregulation of Cyp2d11, 2c12, 2d34, and 2d40, but also augmented the BDE-99-mediated upregulation of Cyp2d13. In summary, the lack of gut microbiome had no effect on the basal expression of Cyp2d isoforms; whereas PBDEs tended to downregulate certain Cyp2d isoforms in either CV or GF conditions— except for Cyp2d13, which was upregulated in livers of BDE-99-treated GF mice. In general, BDE-99 had a more prominent effect than BDE-47 on the regulation of most Cyp2d isoforms.
Fig. 5.
Regulation of phase-I Cyp2d, Cyp2e1, Cyp2g1, Cyp2j9, Cyp2r1 (A), and Cyp3a (B) drug-metabolizing enzymes by BDE-47 or BDE-99 in livers of CV and GF mice. Only Cyps that were differentially expressed by PBDEs or by lack of gut microbiota are shown. Genes in the same family are grouped together to quantitatively compare the mRNA abundance, and are also presented individually to better visualize the mRNA fold changes by BDE-47 or BDE-99 in CV and GF mice. Asterisks (*) represent statistically significant differences between corn oil–treated and PBDE-treated groups of the same enterotypes of mice (false discovery rate–adjusted P value, <0.05). Pound signs (#) represent statistically significant differences between CV and GF mice under the same treatment.
For the Cyp2e1, Cyp2g1, Cyp2j9, and Cyp2r1 genes, under basal conditions, the lack of gut microbiome increased the mRNAs of Cyp2e1 (1.67-fold) and Cyp2g1 (2.33-fold), but decreased Cyp2j9 mRNA (62.55%). In livers of CV mice, BDE-47 markedly increased Cyp2g1 mRNA (3.25-fold) and decreased Cyp2j9 mRNA (64.05%), but had no effect on Cyp2e1 and Cyp2r1. In livers of GF mice, BDE-47 had minimal effect on the mRNA expression of Cyp2e1, Cyp2g1, Cyp2j9, and Cyp2r1. No difference was observed for their mRNA abundances between BDE-47-treated CV and GF mice. In livers of CV mice, BDE-99 increased the mRNAs of Cyp2g1 (4.49-fold) and Cyp2r1 (1.61-fold), and tended to increase their mRNAs in GF mice, although a statistical significance was not achieved. Interestingly, BDE-99 decreased Cyp2j9 mRNA in CV conditions (48.87%), but this pattern was reversed in GF conditions as evidenced by an increase in its mRNA (2.36-fold).
Regulation of Hepatic Cyp3a Genes by BDE-47 and BDE-99 in CV and GF Mice.
For the Cyp3a subfamily, eight Cyp3a family members (Cyp3a11, Cyp3a13, Cyp3a16, Cyp3a25, Cyp3a41a, Cyp3a41b, Cyp3a44, and Cyp3a59) were differentially regulated by PBDEs in liver, among which Cyp3a11 mRNA was the highest expressed isoform (Fig. 5B). Under basal conditions, the lack of gut microbiome led to a dramatic decrease of most Cyp3a isoforms, including Cyp3a11 (89.69%), Cyp3a16 (91.13%), Cyp3a41a (89.82%), Cyp3a41b (89.96%), Cyp3a44 (87.75%), and Cyp3a59 (56.16%). In livers of CV mice, BDE-47 increased the mRNAs of Cyp3a11 (1.55-fold), Cyp3a16 (1.57-fold), Cyp3a41a (1.48-fold), and Cyp3a41b (1.46-fold), and such upregulation was further increased in GF conditions (6.78-fold for Cyp3a11, 7.87-fold for Cyp3a16, 7.12-fold for Cyp3a41a, and 7.12-fold for Cyp3a41b). In livers of GF mice, BDE-47 also increased the mRNAs of Cyp3a44 (6.63-fold) and Cyp3a59 (2.74-fold), indicating the augmentation effect of GF conditions in BDE-47-mediated transcriptional regulation. In livers of CV mice, BDE-99 increased the mRNAs of Cyp3a13 (2.33-fold) and Cyp3a59 (1.50-fold), whereas the lack of gut microbiota augmented the BDE-99-mediated mRNA increase of Cyp3a11 (10.96-fold), Cyp3a16 (10.54-fold), Cyp3a25 (3.48-fold), Cyp3a41a (9.34-fold), Cyp3a41b (9.34-fold), Cyp3a44 (8.53-fold), and Cyp3a59 (6.42-fold). In summary, the lack of gut microbiome resulted in a constitutive decrease in the basal mRNA expression of most Cyp3a isoforms, and also augmented PBDE-mediated transcriptional increase of many Cyp3a isoforms in mouse liver.
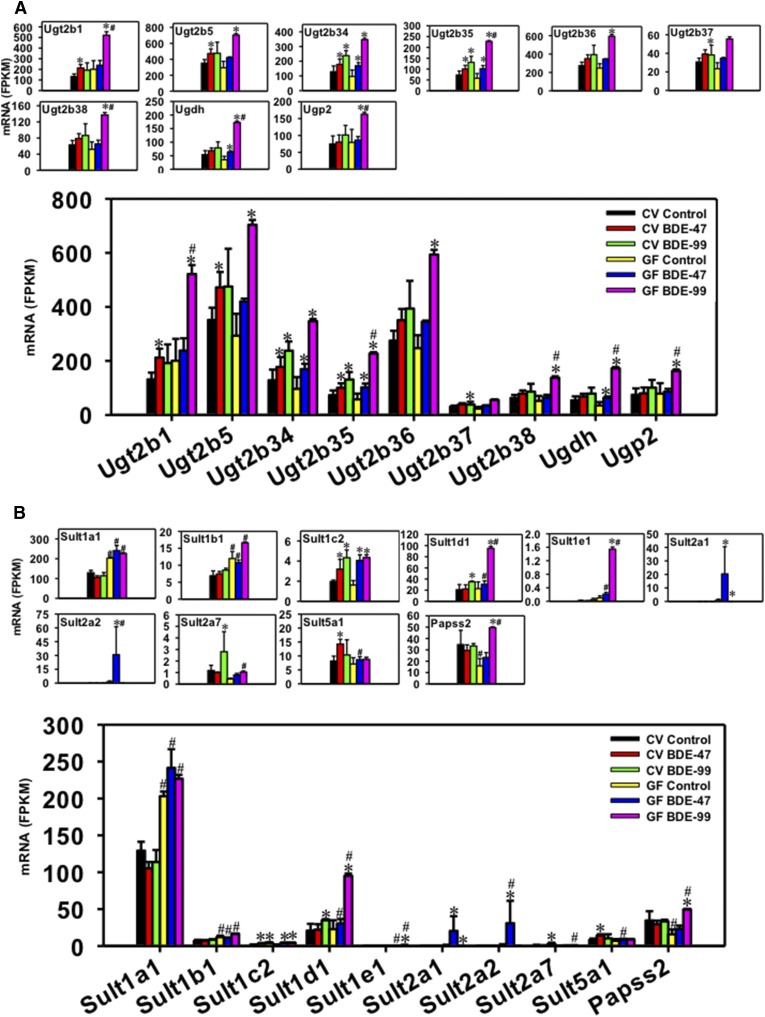
Regulation of Hepatic Ugts by BDE-47 and BDE-99 in CV and GF Mice.
For the Ugt family, Ugt2b1, Ugt2b5, Ugt2b34, Ugt2b35, Ugt2b36, Ugt2b37, Ugt2b38, UDP-glucose 6-dehydrogenase (Ugdh), and UDP-glucose pyrophosphorylase 2 (Ugp2) were differentially regulated by PBDEs in liver (Fig. 6A). Ugdh and Ugp2 are enzymes involved in the synthesis of cosubstrates for phase-II glucuronidation reaction. Ugp2 catalyzes the synthesis from glucose-1-phosphate to UDP-glucose, and Ugdh converts UDP-glucose to UDP-glucuronic acid, which is the cosubstrate for UDP-glucuronidation. Lack of gut microbiome had minimal effect on the basal expression of these genes. In livers of CV mice, BDE-47 increased the mRNAs of Ugt2b1 (1.61-fold), Ugt2b5 (1.34-fold), Ugt2b34 (1.38-fold), and Ugt2b35 (1.37-fold), and upregulation of Ugt2b34 and Ugt2b35 was also observed in GF mice (1.76-fold for Ugt2b34 and 1.77-fold for Ugt2b35). BDE-47 also increased Ugdh mRNA in livers of GF mice (1.82-fold). In livers of CV mice, BDE-99 increased the mRNAs of Ugt2b34 (1.85-fold), Ugt2b35 (1.79-fold), and Ugt2b37 (1.25-fold); whereas in livers of GF mice, BDE-99 markedly increased the mRNAs of Ugt2b1 (2.60-fold), Ugt2b5 (2.40-fold), Ugt2b34 (3.60-fold), Ugt2b35 (3.94-fold), Ugt2b36 (2.40-fold), Ugt2b38 (2.63-fold), Ugdh (4.91-fold), and Ugp2 (2.05-fold), and tended to increase Ugt2b37 (statistical significance was not achieved). Under BDE-99 treatment conditions the lack of gut microbiome led to a greater fold increase of the mRNAs of Ugt2b1, 2b35, and 2b38, as well as Ugdh and Ugp2, suggesting that it augmented BDE-99-mediated upregulation of these genes in mouse liver.
Fig. 6.
Regulation of phase-II Ugts (A) and Sults (B) by BDE-47 or BDE-99 in livers of CV and GF mice. Only DPGs differentially expressed by PBDEs or by lack of gut microbiota are shown. Genes in the same family are grouped together to quantitatively compare the mRNA abundance, and are also presented individually to better visualize the mRNA fold changes by BDE-47 or BDE-99 in CV and GF mice. Asterisks (*) represent statistically significant differences between corn oil–treated and PBDE-treated groups of the same enterotypes of mice (false discovery rate–adjusted P value, <0.05). Pound signs (#) represent statistically significant differences between CV and GF mice under the same treatment.
Regulation of Hepatic Sults by BDE-47 and BDE-99 in CV and GF Mice.
For the Sult family, Sult1a1, Sult1b1, Sult1c2, Sult1d1, Sult1e1, Sult2a1, Sult2a2, Sult2a7, Sult5a1, and 3′-phosphoadenosine 5′-phosphosulfate synthase 2 (Papss2) were differentially regulated by PBDEs in liver, among which Sult1a1 mRNA was the highest expressed isoform (Fig. 6B). Papss2 catalyzes the synthesis of 5′- phosphoadenosine-5′-phosphosulfate, the sulfate donor for all sulfation reactions. Under basal conditions, the lack of gut microbiome increased the mRNAs of Sult1a1 (1.57-fold) and Sult1b1 (1.74-fold), but decreased Papss2 mRNA (54.13%). In livers of CV mice, BDE-47 increased the mRNAs of Sult1c2 (1.62-fold) and Sult5a1 (1.74-fold); whereas in livers of GF mice, BDE-47 increased the mRNAs of Sult1c2 (2.52-fold), Sult2a1 (24.13-fold), and Sult2a2 (25.39-fold). Compared with BDE-47-treated CV mice, BDE-47-treated GF mice had higher levels of Sult1a1, 1b1, 1d1, 1e1, and 2a2. In livers of CV mice, BDE-99 increased the mRNAs of Sult1c2 (2.20-fold), Sult1d1 (1.68-fold), and Sult2a7 (2.38-fold); whereas in livers of GF mice, BDE-99 increased the mRNAs of Sult1c2, Sult1d1, Sult1e1, and Papss2 (2.70-fold for Sult1c2, 4.17-fold for Sult1d1, 15.48-fold for Sult1e1, and 3.12-fold for Papss2), but decreased the mRNA of Sult2a1 (99.64%). Compared with BDE-99-treated CV mice, BDE-99-treated GF mice had higher levels of Sult1a1, 1b1, 1d1, 1e1, and Papss2. In summary, the lack of gut microbiome increased the basal expression of Sult1a1 and Sult1b1, but decreased Papss2; the lack of gut microbiome augmented BDE-47-mediated upregulation of Sult2a2, and augmented BDE-99-mediated upregulation of Sult1d1, Sult1e1, and Papss2 in mouse livers.
Regulation of Hepatic Gsts by BDE-47 and BDE-99 in CV and GF Mice.
For the Gst family, Gsta1, Gsta2, Gsta4, Gstk1, Gstm1-m7, Gstp1, Gstp2, Gstt2, Gstt3, Gstz1, and Gclc were differentially regulated by PBDEs in liver, among which Gstm1 was the highest expressed isoform (Fig. 7A). Under basal conditions, the lack of gut microbiome increased the basal mRNAs of Gstm2 (1.76-fold) and Gstm6 (1.45-fold). In livers of CV mice, BDE-47 markedly increased the mRNAs of Gsta1 (3.04-fold), Gsta2 (1.48-fold), Gsta4 (1.69-fold), Gstm1 (2.03-fold), Gstm2 (1.49-fold), Gstm3 (2.86-fold), Gstm4 (1.96-fold), Gstp1 (1.71-fold), Gstp2 (1.72-fold), and Gstt3 (1.86-fold). In livers of GF mice, BDE-47 also increased the mRNAs of many Gst isoforms (4.55-fold for Gsta1, 2.82-fold for Gsta2, 2.20-fold for Gsta4, 2.54-fold for Gstm1, 5.24-fold for Gstm3, 2.16-fold for Gstm4, and 2.56-fold for Gstt3). However, the BDE-47-mediated mRNA increase in Gstm2, Gstp1, and Gstp2 was gut microbiota dependent. Conversely, the BDE-47-mediated mRNA increase in Gsta2, m3, and t3 was augmented by lack of gut microbiota. In livers of CV mice, BDE-99 upregulated Gsta1 (4.14-fold), Gstm2 (1.97-fold), Gstm3 (4.12-fold), Gstm4 (1.74-fold), and Gstt3 (1.98-fold), and such upregulation was also observed in GF mice (19.15-fold for Gsta1, 3.25-fold for Gstm2, 18.41-fold for Gstm3, 3.12-fold for Gstm4, and 3.19-fold for Gstt3). In GF mice, BDE-99 also upregulated the mRNAs of Gsta2 (4.93-fold), Gstm1 (3.84-fold), and Gclc (2.70-fold), but downregulated the mRNAs of Gstk1 (31.12%) and Gstz1 (32.33%). The lack of gut microbiota augmented the BDE-99-mediated mRNA increase in Gsta1, Gsta2, Gstm2, Gstm3, Gstt3, and Gclc, and the mRNA decrease in Gstk1 and Gstz1. In summary, the lack of gut microbiome increased the basal expression of Gstm2 and Gstm6; lack of gut microbiome augmented BDE-47-mediated upregulation of Gsta2, Gstm3, and Gstt3, and augmented BDE-99-mediated upregulation of Gsta1, Gsta2, Gstm2, Gstm3, Gstt3, and Gclc.
Regulation of Hepatic Uptake Transporters by BDE-47 and BDE-99 in CV and GF Mice.
For the Slco uptake transporters, Slco1a1, Slco1a4, Slco1b2, and Slco2a1 were differentially regulated in liver, among which Slco1b2 was the highest expressed isoform (Fig. 7B). The lack of gut microbiome had no effect on the basal mRNAs of these Slco isoforms. In livers of CV mice, BDE-47 had minimal effect on the mRNAs of Slco1a1, 1a4, 1b2, and 2a1; whereas in livers of GF mice, BDE-47 increased Slco1a4 mRNA (1.74-fold). BDE-99 also did not affect the mRNAs of these Slco transporters in liver of CV mice; however, in livers of GF mice, BDE-99 increased the mRNAs of Slco1a4 (3.11-fold) and Slco1b2 (2.69-fold), but slightly decreased Slco2a1 (20.72%). In summary, lack of gut microbiota augmented PBDE-mediated (and especially BDE-99-mediated) transcriptional changes of these hepatic uptake transporters.
Regulation of Hepatic Efflux Transporters by BDE-47 and BDE-99 in CV and GF Mice.
For the ATP-binding cassette efflux transporters, Abcb1a, Abcb1b, Abcc2, Abcc3, Abcc4, and Abcc10 were differentially regulated in liver, among which Abcc3 was the highest expressed isoform (Fig. 7B). Under basal conditions, the lack of gut microbiome decreased the constitutive mRNA expression of Abcb1a (41.23%) and Abcc10 (57.60%). BDE-47 increased the mRNAs of Abcb1b (1.32-fold), Abcc3 (1.42-fold), and Abcc4 (1.82-fold), but decreased Abcc10 mRNA (53.27%) in liver, all in a gut microbiome–dependent manner. In livers of CV mice, BDE-99 increased Abcc4 mRNA (2.43-fold), and such upregulation was also observed in GF mice (3.91-fold). Conversely, BDE-99 decreased Abcc10 mRNA in CV conditions (44.81%) and this pattern was reversed in GF conditions as evidenced by an increase in its mRNA (2.21-fold) by BDE-99 treatment. The lack of gut microbiota also augmented the BDE-99-mediated increase in the mRNAs of Abcc2 (2.97-fold) only in livers of GF mice. In summary, the lack of gut microbiome decreased the basal mRNAs of Abcb1a and Abcc10; the BDE-47-mediated upregulation of Abcb1b, Abcc3, and Abcc4 depended on the presence of gut microbiota, whereas the lack of gut microbiome augmented BDE-99-mediated upregulation of Abcc2, and reversed the BDE-99-mediated regulatory pattern on the expression of Abcc10 in mouse liver.
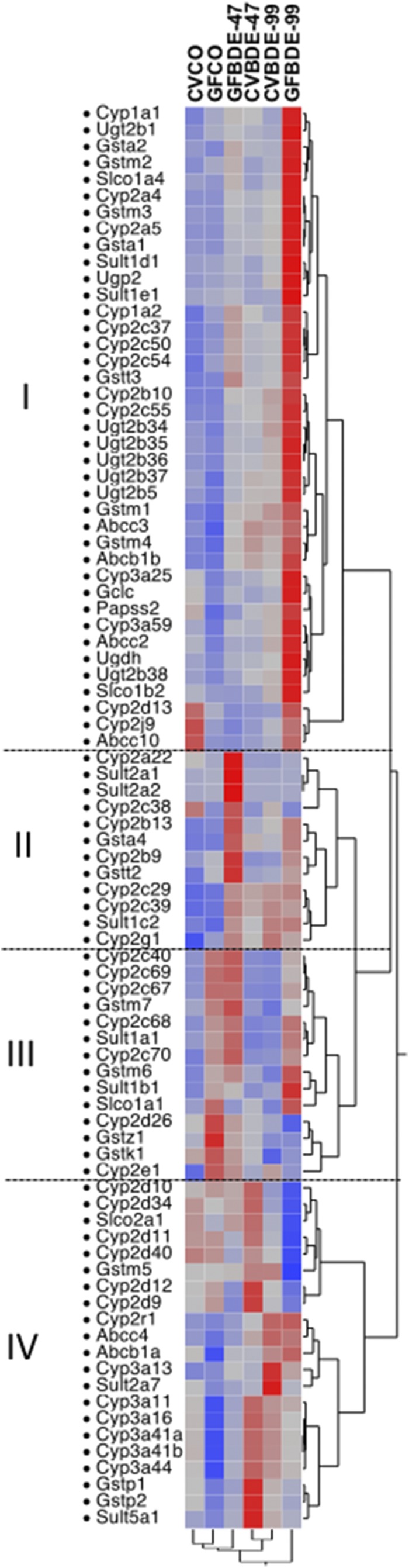
As shown in Fig. 8, a two-way hierarchical clustering dendrogram of the mean FPKM values revealed four distinct patterns of the 86 DPGs that were differentially expressed and summarized in Figs. 1–7. PBDEs appeared to have a more prominent effect than enterotypes in modulating the expression of these genes; as evidenced by a greater separation between vehicle-treated versus PBDE-treated groups and CV control versus GF control groups, BDE-99 appeared to have a more prominent effect than BDE-47 in the differential expression of these DPGs. Genes in pattern I (39 DPGs) were sensitized to BDE-99-mediated differential regulation due to the lack of gut microbiota. Genes in pattern II (12 DPGs) were sensitized to BDE-47-mediated differential regulation due to lack of gut microbiota. Genes in pattern III (14 DPGs) were affected more by enterotypes because they were in general upregulated in livers of GF mice under both basal and PBDE-treated conditions. Conversely, Genes in pattern IV (21 DPGs) were in general downregulated in livers of GF mice. For example, many Cyp3a and Cyp2d subfamily members were downregulated in GF mice under both control and PBDE-treated conditions (Fig. 8). In summary, both PBDEs (to a greater extent) and enterotypes (to a lesser extent) regulate the expression of DPGs in liver. The gut microbiome-dependent effect and GF augmentation effect are summarized in Table 1, which is complimentary to Fig. 8 since it displays the differentially regulated DPGs per category (namely, phase-I, phase-II, and transporters) and also highlights three distinct regulatory patterns: 1) genes that were regulated by gut microbiota under basal conditions (category A); 2) genes that were regulated by PBDEs in a gut microbiome-dependent manner (category B); and 3) genes that were regulated by PBDEs in the absence of gut microbiome (category C). Information between Fig. 8 and Table 1 can be cross-referenced, for example, genes in pattern I (Fig. 8) were upregulated to a greater extent by BDE-99 due to lack of gut microbiome. The majority of these genes also belong to category C in Table 1 (augmentation effect due to lack of gut microbiome), including Cyp1a1, Cyp1a2, Cyp2a4, Cyp2a5, Cyp2b10, Cyp2cs, Ugt2bs, Slco1a4, Slco1b2, Abcc10, etc. Similarly, genes in pattern II (Fig. 8) were upregulated to a greater extent by the BDE-47 effect due to lack of gut microbiome, and they are also shown in category C of Table 1, including Cyp2a22, Sult2a2, Cyp2b13, Cyp2b9, etc. Genes in pattern III (Fig. 8) were affected more by enterotype and were generally upregulated in livers of GF mice. These genes are shown in category A of Table 1, such as Cyp2c40, Cyp2c69, Cyp2c67, Cyp2c68, Cyp2e1, Gstm6, and Sult1a1. Genes in pattern IV (Fig. 8) were in general downregulated in livers of GF mice, such as Gstp1, Gstp2, and Sult5a1, and these genes also belong to category B in Table 1 (gut microbiome-dependent effect).
Fig. 8.
Two-way hierarchical clustering dendrogram of critical DPGs differentially regulated by PBDEs or by the lack of gut microbiome in livers of CV and GF mice. Red color represents upregulation and blue color represents downregulation.
TABLE 1.
DPGs differentially regulated by the lack of gut microbiome and by PBDEs in livers of CV and GF mice treated with corn oil, BDE-47 (100 μmol/kg), or BDE-99 (100 μmol/kg). CV, conventional mice; GF, germ-free mice; CO, corn oil.
| Category | Treatment | Phase I | Phase II | Transporter |
|---|---|---|---|---|
| A. Genes differentially regulated by gut microbiome (CVCO vs. GFCO) | Corn oil | Cyp1a2, Cyp2b9, Cyp2b13, Cyp2c (2c40, 2c50, 2c54, 2c67, 2c68, 2c69, 2c70), Cyp2e1, Cyp2g1, Cyp2j9, Cyp3a (3a11, 3a16, 3a41a, 3a41b, 3a44, 3a59) | Sult1a1, Sult1b1, Papss2, Gstm2, Gstm6 | Abcb1a, Abcc10 |
| B. Genes differentially regulated by PBDEs only in CV mice (gut microbiome–dependent effect) | BDE-47 | Cyp1a2, Cyp2c39, Cyp2d13, Cyp2g1, Cyp2j9 | Ugt2b1, Ugt2b5, Sult5a1, Gstm2, Gstp1, Gstp2 | Abcb1b, Abcc3, Abcc4, Abcc10 |
| BDE-99 | Cyp2a22, Cyp2c39, Cyp2g1, Cyp2r1, Cyp3a13 | Ugt2b37, Sult2a7 | ||
| C. Genes differentially regulated by PBDEs preferably in GF mice (augmentation effect due to the lack of gut microbiome) | BDE-47 | Cyp2a4, Cyp2a22, Cyp2b9, Cyp2b13, Cyp2c37, Cyp2c54, Cyp3a11, Cyp3a16, Cyp3a41a, Cyp3a41b, Cyp3a44 | Sult2a2, Gsta2, Gstm3, Gstt3 | Slco1a4 |
| BDE-99 | Cyp1a1, Cyp1a2, Cyp2a4, Cyp2a5, Cyp2b10, Cyp2c (2c37, 2c40, 2c50, 2c54, 2c55, 2c69), Cyp2d (2d11, 2d12, 2d13, 2d34, 2d40), Cyp2j9, Cyp3a (3a16, 3a25, 3a41a, 3a41b, 3a44, 3a59) | Ugt2b1, Ugt2b35, Ugt2b38, Ugdh, Ugp2, Sult1d1, Sult1e1, Papss2, Gsta1, Gsta2, Gstm2, Gstm3, Gstt3, Gclc | Slco1a4, Slco1b2, Slco2a1, Abcc2, Abcc10 |
LC-MS/MS Proteomic Quantification of Cyp1a2 and Cyp3a11 in CV and GF Mouse Livers.
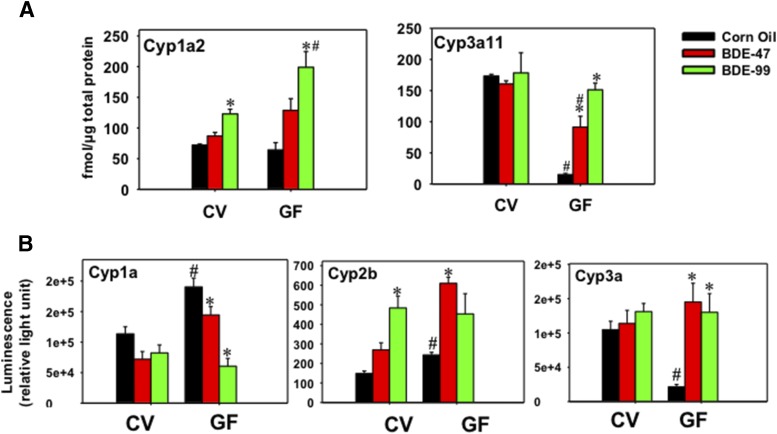
The hepatic Cyp1a2 and Cyp3a11 proteins in CV and GF mice were further quantified by a LC-MS/MS proteomic approach as described in Materials and Methods. These two proteins were quantified based on either their importance in drug processing or availability of surrogate peptides. As shown in Fig. 9A, Cyp1a2 protein was not altered by BDE-47; however, it was increased by BDE-99 in both CV and GF mice, with a greater effect observed in GF conditions (1.70-fold in CV mice and 3.09-fold in GF mice). Compared with BDE-99-treated CV mice, the lack of gut microbiome augmented BDE-99-mediated upregulation of Cyp1a2 protein expression in GF mouse livers (1.62-fold). Consistent with the literature (Selwyn et al., 2015b), the constitutive level of Cyp3a11 protein was downregulated by the lack of gut microbiome (decreased by 91.27%). Interestingly, in livers of GF mice, both BDE-47 and BDE-99 upregulated or restored Cyp3a11 protein level, with a greater effect observed for BDE-99 (6.05-fold by BDE-47 and 10.00-fold by BDE-99). However, the increased Cyp3a11 protein in GF mice under BDE-47 treatment was still lower than its vehicle control level in CV mice (43.10% lower). As for BDE-99 effect, there was no significant difference between CV and GF mice. These observations suggested that at the given dose, BDE-99 is more effective than BDE-47 in upregulating Cyp3a11 protein expression under GF conditions (Fig. 9A).
Fig. 9.
(A) Ultra-performance liquid chromatography mass spectroscopy quantification of Cyp1a2 and Cyp3a11 protein levels in livers of CV and GF mice treated with corn oil, BDE-47 (100 μmol/kg), or BDE-99 (100 μmol/kg). (B) Enzyme activities of Cyp1a, Cyp2b, and Cyp3a in livers of CV or GF mice using the Promega P450-Glo CYP1A, CYP2B, and CYP3A assays. Asterisks (*) represent statistically significant differences between corn oil–treated and PBDE-treated groups of the same enterotypes of mice (P < 0.05). Pound signs (#) represent statistically significant differences between CV and GF mice under the same treatment (P < 0.05).
Enzyme Activities of Cyp1a, Cyp2b, and Cyp3a in CV and GF Mouse Livers.
Because of the importance of Cyp1, Cyp2, and Cyp3 enzymes in xenobiotic metabolism and their dramatic changes induced by PBDE in CV and GF mice, the enzyme activities of Cyp1a, Cyp2b, and Cyp3a were quantified in CV and GF mice treated with corn oil, BDE-47, or BDE-99 (Fig. 9B). Under control conditions, Cyp1a enzyme activity increased 1.68-fold in GF mice, and this was consistent with the basal mRNA increase of Cyp1a2 (Fig. 4A). Although Cyp1a2 mRNA was upregulated by BDE-47 in livers of CV mice (Fig. 4A), Cyp1a2 protein and Cyp1a activity were not altered by BDE-47 in livers of CV mice. The discrepancies between Cyp1a2 mRNA, protein, and activity by BDE-47 could be due to post-transcriptional modification such as the production of microRNAs, which are known to simultaneously inhibit mRNA translation into protein. In livers of CV mice, Cyp1a2 mRNA also tended to be upregulated by BDE-99, followed by the increased expression of Cyp1a2 protein (Fig. 9A). However, Cyp1a activity was not altered by BDE-99 in livers of CV mice, suggesting the presence of post-translational modification that regulates protein degradation. In livers of GF mice, Cyp1a2 mRNA and protein were both upregulated by BDE-99. However, Cyp1a activity was decreased by both BDE-47 (24.13%) and BDE-99 (68.18%) in livers of GF mice, suggesting that there may be extensive post-translational downregulation of Cyp1a gene expression to prevent the induction of Cyp1a activity in GF conditions.
Under control conditions, Cyp2b enzyme activity increased 1.64-fold in GF mice, and this correlates with the basal mRNA increase in Cyp2b9 and 2b13, but not Cyp2b10 (Fig. 4B). In livers of CV mice, Cyp2b10 mRNA was upregulated by both BDE-47 and BDE-99 (Fig. 4B). Expectedly, Cyp2b enzyme activity was increased by BDE-99 (3.26-fold), and tended to be increased by BDE-47 (although a statistical significance was not achieved) (Fig. 9B). In livers of GF mice, Cyp2b enzyme activity was increased by BDE-47 (2.51-fold), and this correlates with a greater fold increase in the mRNAs of Cyp2b9 and Cyp2b13 in BDE-47-treated GF mice, but not with the Cyp2b10 data that were equally upregulated by BDE-47 between CV and GF conditions (Fig. 4B). Therefore, surprisingly, the major Cyp2b isoform Cyp2b10 did not appear to contribute to the PBDE-mediated increase in Cyp2b enzyme activities, and the minor Cyp2b isoforms Cyp2b9 and 2b13 may be more important in contributing to the overall increase in the Cyp2b enzyme activities following PBDE exposure.
Regarding Cyp3a enzyme activities, in livers of CV mice, Cyp3a11 mRNA was moderately upregulated by BDE-47 (Fig. 5B); however, its protein and enzyme activity was not altered by PBDEs, suggesting the involvement of post-transcriptional modification mechanisms that may suppress the expression and activity of Cyp3a11. In livers of GF mice, consistent with the mRNA and proteomic data (Figs. 5B and 9A), Cyp3a activity was markedly decreased by the lack of gut microbiome in the control group (79.57%). Following PBDE exposure, consistent with mRNA and proteomics data, the lack of gut microbiota markedly augmented the PBDE-mediated increase in Cyp3a enzyme activities (BDE-47: 6.78-fold increase; BDE-99: 6.08-fold increase) (Fig. 9B).
Discussion
The present study has unveiled a novel interaction between the gut microbiome and host DPGs on the metabolism of BDE-47 and BDE-99 in CV and GF mice. A systematic comparison of the hepatic DPG expression between CV and GF mice has revealed three notable findings. 1) Under basal condition, the lack of gut microbiome displays divergent regulatory patterns on DPG expression, with a marked upregulation of Cyp2c genes (e.g., Cyp2c40, Cyp2c50, Cyp2c54, Cyp2c67, Cyp2c68, Cyp2c69, and Cyp2c70), and a marked downregulation of Cyp3a genes (e.g., Cyp3a11, Cyp3a16, Cyp3a41a, Cyp3a41b, Cyp3a44, and Cyp3a59). 2) The lack of gut microbiome abolishes PBDE-mediated up- or downregulation of certain DPGs as observed in CV mice, suggesting that the expression of these DPGs is gut microbiome dependent. For example, the upregulation of Ugt2b1, Ugt2b5, Sult5a1, Gstp1, Gstp2, Abcb1b, Abcc3, and Abcc4 by BDE-47 is only observed with the presence of gut microbiome in livers of CV mice but not in GF mice. Similarly, the downregulation of Abcc10 by BDE-47 is only observed in CV mice but not in GF mice. 3) The lack of gut microbiome potentiates PBDE-mediated alterations on some DPG expression in mouse liver. For example, many Cyp2 family members (such as Cyp2a5, Cyp2b10, Cyp2c37, Cyp2c50, Cyp2c54, and Cyp2c55) are upregulated by BDE-99 in livers of both CV and GF mice, with a much greater fold change observed in GF conditions (Table 1).
Intestinal microbiota can regulate certain host DPGs (Selwyn et al., 2015b). In this study, the lack of gut microbiome upregulates Cyp1a2, Cyp2b9, Cyp2c40, Cyp2c50, Cyp2c54, Cyp2c67, Cyp2c68, Cyp2c69, Sult1a1, and Sult1b1, but downregulates Cyp3a11, Cyp3a16, Cyp3a44, and Cyp3a59 in livers of GF mice, both of which are consistent with previous reports in adult male GF mice by RNA-Seq (Selwyn et al., 2015a,b, 2016). The observations of upregulated Cyp2b9 and Cyp2b13 and the downregulated Cyp3a subfamily by the lack of gut microbiome are also consistent with findings from two other groups (Björkholm et al., 2009; Toda et al., 2009). Although a previous study has shown decreased Cyp1a2, Cyp2b9, and Cyp2b13 mRNAs in GF mice compared with CV mice (Toda et al., 2009), in the present study these genes appeared to be upregulated in livers of GF mice. Toda et al. (2009) also showed decreased mRNAs of Sult1d1 and Abcc3 in GF mice; however, in the present study the mRNA levels of Sult1d1 and Abcc3 remained the same in GF and CV mice. These discrepancies are likely due to differences in the strain of mice (IQI GF mice versus C57BL/6J GF mice) and experimental conditions. Additionally, the observations of upregulated Cyp2g1 and downregulated Cyp2j9, Cyp3a41b, Papss2, and Abcc10 by the lack of gut microbiome has not been reported previously.
The present study has added new knowledge to the existing paradigm regarding the role of gut microbiome in xenobiotic metabolism. It is known that PBDEs can modulate the expression of the prototypical target genes of CAR (mouse Cyp2b10 and human CYP2B6) and PXR (mouse Cyp3a11 and human CYP3A4) in mouse liver and human hepatocyptes (Erratico et al., 2012; Sueyoshi et al., 2014). The present study provides the first evidence showing that the lack of gut microbiome can modify PBDE-mediated alterations in DPG expression in mouse liver. For example, the lack of gut microbiome abolishes BDE-47-mediated upregulation of Cyp1a2, Ugt2b1, Ugt2b5, Sult5a1, Gstp1, Gstp2, Abcc3, and Abcc4, and downregulation of Cyp2d13 and Abcc10 in GF mice. Similarly, the lack of gut microbiome also abolishes BDE-99-mediated upregulation of Cyp2a22, Cyp2c39, Cyp2g1, Cyp2r1, Cyp3a13, and Sult2a7 in GF mice. Interestingly, the lack of gut microbiome can also potentiate PBDE-mediated alterations, as evidenced by a greater fold change observed in GF conditions. Many DPGs that were minimally or moderately regulated by PBDEs in the livers of CV mice were markedly altered by PBDEs with the absence of gut microbiome in GF mice. These genes include Cyp2a22 and Cyp3a11 by BDE-47; Cyp3a59, Ugt2b35, Ugdh, Sult1d1, and Sult1e1 by BDE-99; as well as Cyp3a16, Cyp3a41a, Cyp3a41b, Cyp3a44, Gsta2, Gstt3, and Slco1a4 by both BDE-47 and BDE-99. The augmentation effect of the lack of gut microbiome seemed to be more prominent for BDE-99. For example, the lack of gut microbiome potentiates BDE-99-mediated upregulation of over half of the DPGs as shown in this study, including the phase I gene family (Cyp1 to Cyp3), phase II gene families (Ugts, Sults, and Gsts), as well as transporters (Slco1a4, Slco1b2, Abcb1a, Abcc2, and Abcc10).
The present study has provided critical information on the in vivo hepatic metabolism of BDE-47 and BDE-99 in CV and GF mice, with a primary focus on the hydroxylated PBDE metabolites. Accumulated evidence has demonstrated that PBDEs are first biotransformed to hydroxylated metabolites in mammals (Hamers et al., 2008; Lupton et al., 2009; Stapleton et al., 2009; Dong et al., 2010; Erratico et al., 2010). Almost 20 OH-BDE metabolites have been identified in rat blood after exposure to a mixture of tetra- to decaBDEs. In the present study, 16 hydroxylated metabolites of PBDEs were measured by PBDE-targeted metabolomics (GCMS negative chemical ionization) in livers of CV and GF mice treated with BDE-47 or BDE-99. Twelve out of 16 measured OH-BDEs were detected in livers of CV and GF mice treated with BDE-47. The major hydroxylated metabolite of BDE-47 was 5-OH-BDE-47 in livers of both CV and GF mice, with a higher level observed in GF conditions. The findings of six hydroxylated BDE-47 metabolites in the present study, namely, 5-OH-BDE-47, 4′-OH-BDE-49, 3-OH-BDE-47, 6-OH-BDE-47, 4′-OH-BDE-17, and 2′-OH-BDE-28, are consistent with previous in vitro studies that incubated BDE-47 with phenobarbital-induced rat liver microsomes or recombinant human P450s (Hamers et al., 2008; Erratico et al., 2013; Feo et al., 2013). Following BDE-99 treatment, 13 out of 16 measured OH-BDEs were detected in livers of CV and GF mice. The major hydroxylated metabolite of BDE-99 was 5′-OH-BDE-99 in livers of both CV and GF mice. The findings of hydroxylated BDE-99 metabolites, namely, 5′-OH-BDE-99, 6′-OH-BDE-99, and 4′-OH-BDE-101, are consistent with previous studies that incubated BDE-99 with rat liver microsomes (Stapleton et al., 2009; Erratico et al., 2011). In human blood, 5-OH-BDE-47 and 5′-OH-BDE-99 are also reported as being the two most abundant metabolites of BDE-47 and BDE-99, respectively. The mean concentrations of BDE-47 and BDE-99 are 60 ± 29 ng/g lipid and 19 ± 8.6 ng/g lipid in human fetal and maternal blood samples from the United States (Qiu et al., 2009). However, the present study did not measure 4-OH-BDE-90 and 2,4,5-Tribromophenol (TBP) as observed in the previous report, due to the lack of corresponding standards. Compared with prior in vitro and in vivo studies, the present study is the first study to introduce gut microbiome as key determinants of the biotransformation of BDE-47 and BDE-99.
The present study has suggested the involvement of multiple DPGs in the biotransformation of BDE-47 and BDE-99 with the absence of gut microbiome. Evidence from human or rat microsomes and recombinant human P450 enzymes has demonstrated that BDE-47 and BDE-99 are initially oxidized to hydroxylated metabolites (OH-BDEs) by various P450s, particularly by CYP2A2 and CYP3A1 in rats and CYP2B6 in humans (Erratico et al., 2011; Feo et al., 2013). The lack of gut microbiome increased the major metabolite of BDE-47 in livers of GF mice, but decreased the other four minor metabolites, suggesting the involvement of multiple Cyps in the biotransformation of BDE-47. For example, the lack of gut microbiome upregulated the expression of Cyp2b9, Cyp2b13, and many Cyp2c subfamily members, which could contribute to the increased formation of the major BDE-47 metabolite; whereas the lack of gut microbiome significantly downregulated Cyp3a enzymes (such as Cyp3a11, Cyp3a16, Cyp3a41a, Cyp3a41b, Cyp3a44, and Cyp3a59) in livers of GF mice, which may be responsible for the decreased four minor BDE-47 metabolites. Conversely, the lack of gut microbiome not only affects the biotransformation of PBDEs through the baseline expression levels of these enzymes, it also modulates how these PBDEs and their metabolites in turn affect the expression of these and other drug-metabolizing enzymes.
The P450-Glo assay (Promega Corporation) used in the present study provides a highly sensitive and selective substrate for each specific P450 enzyme (CYP1 to 3) in liver microsomes (Cali et al., 2006, 2012; Hrycay and Bandiera, 2009). Although it was initially designed for human samples, it has been widely used in mouse studies as shown in prior publications (Sekiya and Suzuki, 2011; Lee et al., 2013; Selwyn et al., 2015a). For example, the CYP1A1 activity was measured in mouse with C57BL/6 background using the P450-Glo CYP1A1 assay (containing luciferin 6′-chloroethyl ether [luciferin-CEE] substrate) (Lee et al., 2013). The CYP1A2 activity was measured in mouse embryonic fibroblasts, defined transcription factor–induced hepatocyte-like cells, and mouse primary hepatocytes using P450-Glo CYP1A2 (containing luciferin 6′-methyl ether substrate) (Lim et al., 2016). The Cyp3a activity was measured in GF mice and mouse hepatocytes using the P450-Glo CYP3A4 assay (containing luciferin isopropyl acetal) (Sekiya and Suzuki, 2011; Selwyn et al., 2015a). The recombinant human P450 isoforms have shown that CYP2B6 was the predominant isoform that could metabolize Luciferin-2B6 to luciferin, and CYP3A4 was the predominant isoform that could metabolize luciferin isopropyl acetal to luciferin. According to the NCBI Homologene Database, mouse Cyp2b and Cyp3a are sequence homologs of the corresponding human P450s within the same families, and the human P450s are not sequence homologs of the mouse P450s of other families such as Cyp1a, Cyp2a, or Cyp2c. Therefore, the substrates used in this study are most likely substrates for the mouse P450s of the same family as humans. However, it is possible that the CYP2B substrates can be metabolized by multiple mouse Cyp2b isoforms other than Cyp2b10.
Because there lacks a systematical comparison of PBDE exposure in humans for various groups such as newborns, children, the elderly, adults, and occupational workers who are at a high risk of PBDE exposure, the low dose used in the present study was based on the exposure to PBDEs in newborns because these data are readily available (Frederiksen et al., 2009). This human-relevant dose is one of the examples that represent one of the most vulnerable populations. In the present study, the mice were dosed with 5 and 50 mg/kg of PBDEs; these doses are calculated based on the potential exposure for infants, who are more vulnerable to adverse drug reactions than adults. The dose estimations are based on the fact that BDE-47 and BDE-99 levels in human breast milk are approximately 27.8 and 5.7 ng/g liquid weight (Frederiksen et al., 2009). Assuming newborns weigh 5 kg on average, and consume 960 g/d, the 1-day human exposure levels of BDE-47 and BDE-99 in newborns are approximately 5.337 mg/kg (10.2 μmol/kg) and 1.094 mg/kg (1.8 μmol/kg), respectively. Therefore, the low dose is actually within the range of acute-to-subchronic exposure in the newborn vulnerable populations, whereas the high dose represents the dose of choice across species. Results from the National Health and Nutrition Examination Survey have shown that the adult intake dose of total PBDEs was estimated to be 7.7 ng/kg body weight per day, and children’s estimated intakes were higher at 49.3 ng/kg per day for ages 1–5, 14.4 ng/kg per day for ages 6–11, and 9.1 ng/kg per day for ages 12–19 (Lorber, 2008). Therefore, the acute low doses of PBDEs used in the present study (four consecutive days) are similar to newborn subchronic exposure in humans (approximately 3 months), whereas the acute high doses of PBDEs considered an extrapolation of 10 across different species. These doses are similar to doses that have been used in adult mice to induce P450s (Pacyniak et al., 2007).
In conclusion, we report a novel interaction between the gut microbiome and host hepatic DPGs in vivo. Most notably, the lack of gut microbiome augmented BDE-99-mediated upregulation of many DPGs and also impacted the hydroxylation of PBDEs. Using GF mice as a research model, the present study is the first to characterize the effect of the gut microbiome on the host metabolism of environmental PBDEs. However, there are some limitations to this study. Future studies are needed to systematically determine the distribution of PBDEs and quantify the blood and lipid metabolome profiles of PBDEs. Future studies are also needed to determine members of the gut microbiota that have the most significant effect on host DPG expression involved in PBDE metabolism.
Acknowledgments
The authors thank the Gnotobiotic Animal Core facility (University of Washington) for technical support in the germ-free mice experiments; Dr. Debbie Nickerson’s laboratory for sequencer access and support for the RNA-Seq experiments; Brian High and the IT team from the Department of Environmental and Occupational Health Sciences (University of Washington) for providing server access and advice to improve the RNA-Seq bioinformatics pipeline; Matt Karasu from the School of Pharmacy (University of Washington) for the proteomics data analysis; the members of Dr. Cui’s laboratory for help in animal work and tissue collection; and the Department of Comparative Medicine (University of Washington) for processing the H&E staining.
Abbreviations
- ALT
alanine aminotransferase
- BDE
bromodiphenyl ether
- BDE-47
2,2′,4,4′-tetrabromodiphenyl ether
- BDE-99
2,2′,4,4′,5-pentabromodiphenyl ether
- CAR
constitutive androstane receptor
- CV
conventional
- DPG
drug-processing gene
- FPKM
fragments per kilobase of transcript per million mapped reads
- GCMS
gas chromatography mass spectroscopy
- GF
germ free
- Gst
glutathione S-transferase
- LC-MS/MS
liquid chromatography–tandem mass spectroscopy
- NCBI
National Center for Biotechnology Information
- OH-BDE
hydroxylated polybrominated diphenyl ether
- Papss2
3′-phosphoadenosine 5′-phosphosulfate synthase 2
- P450
cytochrome P450
- PBDE
polybrominated diphenyl ether
- PCR
polymerase chain reaction
- PXR
pregnane X receptor
- RNA-Seq
RNA sequencing
- Slco
solute carrier organic anion
- Sult
sulfotransferase
- Ugdh
UDP-glucose 6-dehydrogenase
- Ugp2
UDP-glucose pyrophosphorylase 2
- Ugt
UDP glucuronosyltransferase
Authorship Contributions
Participated in research design: Li, Bammler, Cui.
Conducted experiments: Li, Lee, Cade, Kuo, Schultz, Bhatt, Cui.
Performed data analysis: Li, Bhatt, Prasad, Cui.
Wrote or contributed to the writing to the manuscript: Li, Cade, Schultz, Bhatt, Prasad, Bammler, Cui.
Footnotes
This work was supported by the National Institutes of Health [Grants GM111381, ES019487, and ES025708], and the University of Washington Center for Exposures, Diseases, Genomics, and Environment [Grant P30 ES0007033], as well as the Murphy Endowment.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Aleksunes LM, Klaassen CD. (2012) Coordinated regulation of hepatic phase I and II drug-metabolizing genes and transporters using AhR-, CAR-, PXR-, PPARα-, and Nrf2-null mice. Drug Metab Dispos 40:1366–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. MetaHIT Consortium (2011) Enterotypes of the human gut microbiome. Nature 473:174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkholm B, Bok CM, Lundin A, Rafter J, Hibberd ML, Pettersson S. (2009) Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS One 4:e6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cali JJ, Ma D, Sobol M, Simpson DJ, Frackman S, Good TD, Daily WJ, Liu D. (2006) Luminogenic cytochrome P450 assays. Expert Opin Drug Metab Toxicol 2:629–645. [DOI] [PubMed] [Google Scholar]
- Cali JJ, Ma D, Wood MG, Meisenheimer PL, Klaubert DH. (2012) Bioluminescent assays for ADME evaluation: dialing in CYP selectivity with luminogenic substrates. Expert Opin Drug Metab Toxicol 8:1115–1130. [DOI] [PubMed] [Google Scholar]
- Cantón RF, Sanderson JT, Letcher RJ, Bergman A, van den Berg M. (2005) Inhibition and induction of aromatase (CYP19) activity by brominated flame retardants in H295R human adrenocortical carcinoma cells. Toxicol Sci 88:447–455. [DOI] [PubMed] [Google Scholar]
- Chen LJ, Lebetkin EH, Sanders JM, Burka LT. (2006) Metabolism and disposition of 2,2′,4,4′,5-pentabromodiphenyl ether (BDE99) following a single or repeated administration to rats or mice. Xenobiotica 36:515–534. [DOI] [PubMed] [Google Scholar]
- Cui JY, Klaassen CD. (2016) RNA-Seq reveals common and unique PXR- and CAR-target gene signatures in the mouse liver transcriptome. Biochim Biophys Acta 1859:1198–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui YJ, Cheng X, Weaver YM, Klaassen CD. (2009) Tissue distribution, gender-divergent expression, ontogeny, and chemical induction of multidrug resistance transporter genes (Mdr1a, Mdr1b, Mdr2) in mice. Drug Metab Dispos 37:203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingemans MM, van den Berg M, Westerink RH. (2011) Neurotoxicity of brominated flame retardants: (in)direct effects of parent and hydroxylated polybrominated diphenyl ethers on the (developing) nervous system. Environ Health Perspect 119:900–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Li Z, Man X, Zhou J, Lu H, Wang S. (2010) Identification of the metabolites of polybrominated diphenyl ether 99 and its related cytochrome P450s. J Biomed Res 24:223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erratico C, Zheng X, Ryden A, Marsh G, Maho W, Covaci A. (2015) Human hydroxylated metabolites of BDE-47 and BDE-99 are glucuronidated and sulfated in vitro. Toxicol Lett 236:98–109. [DOI] [PubMed] [Google Scholar]
- Erratico CA, Moffatt SC, Bandiera SM. (2011) Comparative oxidative metabolism of BDE-47 and BDE-99 by rat hepatic microsomes. Toxicol Sci 123:37–47. [DOI] [PubMed] [Google Scholar]
- Erratico CA, Szeitz A, Bandiera SM. (2010) Validation of a novel in vitro assay using ultra performance liquid chromatography-mass spectrometry (UPLC/MS) to detect and quantify hydroxylated metabolites of BDE-99 in rat liver microsomes. J Chromatogr B Analyt Technol Biomed Life Sci 878:1562–1568. [DOI] [PubMed] [Google Scholar]
- Erratico CA, Szeitz A, Bandiera SM. (2012) Oxidative metabolism of BDE-99 by human liver microsomes: predominant role of CYP2B6. Toxicol Sci 129:280–292. [DOI] [PubMed] [Google Scholar]
- Erratico CA, Szeitz A, Bandiera SM. (2013) Biotransformation of 2,2′,4,4′-tetrabromodiphenyl ether (BDE-47) by human liver microsomes: identification of cytochrome P450 2B6 as the major enzyme involved. Chem Res Toxicol 26:721–731. [DOI] [PubMed] [Google Scholar]
- Feo ML, Gross MS, McGarrigle BP, Eljarrat E, Barceló D, Aga DS, Olson JR. (2013) Biotransformation of BDE-47 to potentially toxic metabolites is predominantly mediated by human CYP2B6. Environ Health Perspect 121:440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie KJ, Shutt JL, Mayne G, Hoffman D, Letcher RJ, Drouillard KG, Ritchie IJ. (2005) Exposure to polybrominated diphenyl ethers (PBDEs): changes in thyroid, vitamin A, glutathione homeostasis, and oxidative stress in American kestrels (Falco sparverius). Toxicol Sci 88:375–383. [DOI] [PubMed] [Google Scholar]
- Frederiksen M, Vorkamp K, Thomsen M, Knudsen LE. (2009) Human internal and external exposure to PBDEs—a review of levels and sources. Int J Hyg Environ Health 212:109–134. [DOI] [PubMed] [Google Scholar]
- Hakk H, Larsen G, Klasson-Wehler E. (2002) Tissue disposition, excretion and metabolism of 2,2′,4,4′,5-pentabromodiphenyl ether (BDE-99) in the male Sprague-Dawley rat. Xenobiotica 32:369–382. [DOI] [PubMed] [Google Scholar]
- Hamers T, Kamstra JH, Sonneveld E, Murk AJ, Kester MH, Andersson PL, Legler J, Brouwer A. (2006) In vitro profiling of the endocrine-disrupting potency of brominated flame retardants. Toxicol Sci 92:157–173. [DOI] [PubMed] [Google Scholar]
- Hamers T, Kamstra JH, Sonneveld E, Murk AJ, Visser TJ, Van Velzen MJ, Brouwer A, Bergman A. (2008) Biotransformation of brominated flame retardants into potentially endocrine-disrupting metabolites, with special attention to 2,2′,4,4′-tetrabromodiphenyl ether (BDE-47). Mol Nutr Food Res 52:284–298. [DOI] [PubMed] [Google Scholar]
- Hites RA. (2004) Polybrominated diphenyl ethers in the environment and in people: a meta-analysis of concentrations. Environ Sci Technol 38:945–956. [DOI] [PubMed] [Google Scholar]
- Hovander L, Athanasiadou M, Asplund L, Jensen S, Wehler EK. (2000) Extraction and cleanup methods for analysis of phenolic and neutral organohalogens in plasma. J Anal Toxicol 24:696–703. [DOI] [PubMed] [Google Scholar]
- Hrycay EG, Bandiera SM. (2009) Expression, function and regulation of mouse cytochrome P450 enzymes: comparison with human P450 enzymes. Curr Drug Metab 10:1151–1183. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen CD, Aleksunes LM. (2010) Xenobiotic, bile acid, and cholesterol transporters: function and regulation. Pharmacol Rev 62:1–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen CD, Cui JY. (2015) Review: mechanisms of how the intestinal microbiota alters the effects of drugs and bile acids. Drug Metab Dispos 43:1505–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen CD, Slitt AL. (2005) Regulation of hepatic transporters by xenobiotic receptors. Curr Drug Metab 6:309–328. [DOI] [PubMed] [Google Scholar]
- Knights D, Ward TL, McKinlay CE, Miller H, Gonzalez A, McDonald D, Knight R. (2014) Rethinking “enterotypes”. Cell Host Microbe 16:433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Ding X, Riddick DS. (2013) The role of cytochrome P450-dependent metabolism in the regulation of mouse hepatic growth hormone signaling components and target genes by 3-methylcholanthrene. Drug Metab Dispos 41:457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Renaud HJ, Klaassen CD, Cui JY. (2016) Age-specific regulation of drug-processing genes in mouse liver by ligands of xenobiotic-sensing transcription factors. Drug Metab Dispos 44:1038–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KT, Lee SC, Gao Y, Kim KP, Song G, An SY, Adachi K, Jang YJ, Kim J, Oh KJ, et al. (2016) Small molecules facilitate single factor-mediated hepatic reprogramming. Cell Rep 15:814–829. [DOI] [PubMed] [Google Scholar]
- Lorber M. (2008) Exposure of Americans to polybrominated diphenyl ethers. J Expo Sci Environ Epidemiol 18:2–19. [DOI] [PubMed] [Google Scholar]
- Lupton SJ, McGarrigle BP, Olson JR, Wood TD, Aga DS. (2009) Human liver microsome-mediated metabolism of brominated diphenyl ethers 47, 99, and 153 and identification of their major metabolites. Chem Res Toxicol 22:1802–1809. [DOI] [PubMed] [Google Scholar]
- Madia F, Giordano G, Fattori V, Vitalone A, Branchi I, Capone F, Costa LG. (2004) Differential in vitro neurotoxicity of the flame retardant PBDE-99 and of the PCB Aroclor 1254 in human astrocytoma cells. Toxicol Lett 154:11–21. [DOI] [PubMed] [Google Scholar]
- Malmberg T, Athanasiadou M, Marsh G, Brandt I, Bergman A. (2005) Identification of hydroxylated polybrominated diphenyl ether metabolites in blood plasma from polybrominated diphenyl ether exposed rats. Environ Sci Technol 39:5342–5348. [DOI] [PubMed] [Google Scholar]
- Marsh G, Athanasiadou M, Athanassiadis I, Sandholm A. (2006) Identification of hydroxylated metabolites in 2,2′,4,4′-tetrabromodiphenyl ether exposed rats. Chemosphere 63:690–697. [DOI] [PubMed] [Google Scholar]
- Meerts IA, van Zanden JJ, Luijks EA, van Leeuwen-Bol I, Marsh G, Jakobsson E, Bergman A, Brouwer A. (2000) Potent competitive interactions of some brominated flame retardants and related compounds with human transthyretin in vitro. Toxicol Sci 56:95–104. [DOI] [PubMed] [Google Scholar]
- National Toxicology Program (1986) NTP toxicology and carcinogenesis studies of decabromodiphenyl oxide (CAS No. 1163-19-5) in F344/N rats and B6C3F1 mice (feed studies). Natl Toxicol Program Tech Rep Ser 309:1–242. [PubMed] [Google Scholar]
- Nayak RR, Turnbaugh PJ. (2016) Mirror, mirror on the wall: which microbiomes will help heal them all? BMC Med 14:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orn U, Klasson-Wehler E. (1998) Metabolism of 2,2′,4,4′-tetrabromodiphenyl ether in rat and mouse. Xenobiotica 28:199–211. [PubMed] [Google Scholar]
- Pacyniak EK, Cheng X, Cunningham ML, Crofton K, Klaassen CD, Guo GL. (2007) The flame retardants, polybrominated diphenyl ethers, are pregnane X receptor activators. Toxicol Sci 97:94–102. [DOI] [PubMed] [Google Scholar]
- Park S, Cheng SL, Cui JY. (2016) Characterizing drug-metabolizing enzymes and transporters that are bona fide CAR-target genes in mouse intestine. Acta Pharm Sin B 6:475–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Bigsby RM, Hites RA. (2009) Hydroxylated metabolites of polybrominated diphenyl ethers in human blood samples from the United States. Environ Health Perspect 117:93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schecter A, Pavuk M, Päpke O, Ryan JJ, Birnbaum L, Rosen R. (2003) Polybrominated diphenyl ethers (PBDEs) in U.S. mothers’ milk. Environ Health Perspect 111:1723–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya S, Suzuki A. (2011) Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475:390–393. [DOI] [PubMed] [Google Scholar]
- Selwyn FP, Cheng SL, Bammler TK, Prasad B, Vrana M, Klaassen C, Cui JY. (2015a) Developmental regulation of drug-processing genes in livers of germ-free mice. Toxicol Sci 147:84–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn FP, Cheng SL, Klaassen CD, Cui JY. (2016) Regulation of hepatic drug-metabolizing enzymes in germ-free mice by conventionalization and probiotics. Drug Metab Dispos 44:262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn FP, Cui JY, Klaassen CD. (2015b) RNA-seq quantification of hepatic drug processing genes in germ-free mice. Drug Metab Dispos 43:1572–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjödin A, Patterson DG, Jr, Bergman A. (2001) Brominated flame retardants in serum from U.S. blood donors. Environ Sci Technol 35:3830–3833. [DOI] [PubMed] [Google Scholar]
- Stapleton HM, Kelly SM, Pei R, Letcher RJ, Gunsch C. (2009) Metabolism of polybrominated diphenyl ethers (PBDEs) by human hepatocytes in vitro. Environ Health Perspect 117:197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskal DF, Hakk H, Bauer D, Diliberto JJ, Birnbaum LS. (2006) Toxicokinetics of polybrominated diphenyl ether congeners 47, 99, 100, and 153 in mice. Toxicol Sci 94:28–37. [DOI] [PubMed] [Google Scholar]
- Sueyoshi T, Li L, Wang H, Moore R, Kodavanti PR, Lehmler HJ, Negishi M, Birnbaum LS. (2014) Flame retardant BDE-47 effectively activates nuclear receptor CAR in human primary hepatocytes. Toxicol Sci 137:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Saito N, Ikarashi N, Ito K, Yamamoto M, Ishige A, Watanabe K, Sugiyama K. (2009) Intestinal flora induces the expression of Cyp3a in the mouse liver. Xenobiotica 39:323–334. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrana M, Whittington D, Nautiyal V, Prasad B. (2017) Database of optimized proteomic quantitative methods for human drug disposition-related proteins for applications in physiologically based pharmacokinetic modeling. CPT Pharmacometrics Syst Pharmacol 6:267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Taylor MM, DeVito MJ, Crofton KM. (2002) Developmental exposure to brominated diphenyl ethers results in thyroid hormone disruption. Toxicol Sci 66:105–116. [DOI] [PubMed] [Google Scholar]