Abstract
Receptor of activated C Kinase 1 (RACK1) is an essential scaffold and anchoring protein, which serves an important role in multiple tumorigenesis signaling pathways. The present study aimed to investigate the expression of RACK1 in gastric cancer (GC), and its association with the occurrence and development of GC. In addition, the effect and mechanism of RACK1 overexpression on the growth, and proliferation of GC cells was examined. Firstly, the protein expression of RACK1 was detected in 70 cases of GC tissues and 30 cases of noncancerous tissues using immunohistochemical staining, and the association between clinical and pathological features of GC was analyzed. Secondly, the mRNA and protein expression of RACK1 was determined in the poorly-differentiated human gastric cancer cell line HGC27 and gastric epithelial cell line GES-1. The growth of HGC27 cells following the upregulation of RACK1 was detected using MTT method. Subsequently, the interaction and co-location between RACK1, and WEE1 homolog (S. pombe) (WEE1) in HGC27 cells was confirmed using co-immunoprecipitation and indirect immunofluorescence. The expression level of RACK1 in GC was significantly lower compared with that in pericarcinous tissues (P<0.05). The protein level of RACK1 expression correlated with tumor node metastasis stage, tumor differentiation and lymph node metastasis. The mRNA and protein levels of RACK1 in HGC27 cells were significantly reduced, and overexpressed RACK1 downregulated WEE1 protein expression, thus inhibiting the growth of HGC27 cells. Co-immunoprecipitation and immunofluorescence confirmed that RACK1, and WEE1 interacted and co-located in the cytoplasm of HGC27 cells. Therefore, the abnormal expression of RACK1 in GC tissues was identified to be involved in the occurrence and development of GC. Overexpression of RACK1 was able to inhibit the growth of HGC27 cells. The current study suggests that low expression of RACK1 is an important indicator of poor prognosis of GC. RACK1 and WEE1 interact to regulate the growth of HGC27 cells.
Keywords: RACK1, WEE1, gastric cancer, HGC27
Introduction
Gastric cancer (GC) is one of the most common malignancies of the digestive tract and the second leading cause of cancer-related death worldwide (1,2). The incidence of GC in China is in the second place of malignant tumors, only to lung cancer. Especially in the rural areas of China, the annual incidence of GC is about 36.2/10 million, which located in the first place of a variety of malignant tumors (3,4). The wall of the stomach is composed of four layers, an outer fibrous membrane called the serosa, a three-ply layer of muscle, a submucous layer, and a mucous layer called the gastric mucosa. serosa invasion means subserosa (5,6). GC usually occurs in the mucosal layer of the stomach wall, and it can be removed with surgery. However, in cases of metastasis to other organs, surgical methods are not suitable for the treatment of GC metastasis (7). The metastasis of GC include the following forms: i) direct invasion, invasion of the lower end of the esophagus, duodenum, omentum, colon, liver, spleen, pancreas and other adjacent organs according to the different growth sites of GC; ii) hematogenous metastasis, the common metastasis organs are liver, lung, pancreas, bone and so on, and hepatic metastasis is commonly seen in the blood route metastasis of GC iii) peritoneal metastasis iv) lymph node metastasis. Lymph node metastasis is the main route of metastasis of GC. The rate of advanced GC lymph node metastasis is ~70%. There also will occur lymph node metastasis in early GC. The lymph node metastasis rate of GC is positively related to the depth of tumor invasion. There were 16 groups of regional lymph nodes draining the stomach, which can be divided into 3 stations according to their distance from the stomach. GC is metastasis from primary site to lymph node through lymphatic network to the first station, then, the cancer cells with vascular innervating the stomach, transfer to the second station disposition along the blood vessels surrounding lymph nodes, and the lymph node metastasis to the distant third station, can be regarded as a distant metastasis (5,6,8–11). Therefore, it is of great importance in the early diagnosis and treatment of GC. In the research field of molecular biology, it is important to search the suitable molecular markers for GC, and to provide theoretical basis for clinical treatment.
Receptor of activated protein kinase C (RACK1), a 36-kilodalton (kDa) protein with a propeller-like structure of seven WD40 (Trp-Asp) motifs, was originally identified on the basis of its ability to bind the activated form of protein kinase C (PKC), because it has homology with G protein beta subunit, also known as guanine nucleotide binding protein (G protein) beta polypeptide 2-like 1 (GNB2L1), and it is highly conserved in eukaryotes (12–14). RACK1 is a cellular shuttle protein, which can be located in cytoplasm, mitochondria, endoplasmic reticulum and nucleus. As a scaffold protein, it provides a platform for the interaction of a variety of proteins, thus integrating inputs from distinct signaling pathways. RACK1 interacts with PKC, phosphodiesterase4D5 (PDE4D5), tyrosine kinases/phosphatases, and signal transducers and activators of transcription 3 (STAT3) to regulate a multitude of cellular actions (15–17). For example, RACK1 interacts with activated PKC to regulate its intracellular localization (18). RACK1 regulates the stability of JNK or HIF1α protein as an anchored protein (19). RACK1 interacts with ribosomal proteins to regulate the translation of intracellular proteins (20). RACK1 combines with signal molecules from different transduction pathway and plays a key role in a variety of mammalian animal development (21). Therefore, RACK1 is a multifunctional scaffold protein, involving in regulating various biological processes, including signal transduction, immune response, cell growth, migration, differentiation, angiogenesis, tumor growth, neuronal response, apoptosis, chromatin remodeling and normal function of clock (22–23). In recent years, RACK1 is considered to be an important protein in regulating multiple signaling pathways and many biological functions of tumor such as proliferation, apoptosis, migration, especially its role in tumor invasion and metastasis. RACK1 promotes the invasion and metastasis of tumor and many kinds of cell function by activating PKC (24). RACK1 combines with PKC to regulate ribosome translation and promotes the expression of invasion and metastasis of related factors (25).
RACK1 is highly expressed in breast cancer, colon cancer, pancreatic ductal adenocarcinoma, melanoma, esophageal squamous cell carcinoma, lung cancer and oral squamous cell carcinoma and other tumors, and is considered to be a good marker (26). For example, RACK1 is an even superior predictor of breast cancer prognosis compared with commonly used diagnostic biomarkers (27). The high expression level of RACK1 is closely related to late clinical status, and silencing of RACK1 inhibits the tumorigenicity of epithelial ovarian cancer in vitro and in vivo (28). The high expression of RACK1 is correlated to the pathological stage and tumor size of lung adenocarcinoma, and is also a potential marker for clinical diagnosis (29). The expression of RACK1 in oral squamous cell carcinoma was significantly increased, and the expression level was negatively correlated with the prognosis of patients (30). In GC research, RACK1 suppresses the gastric tumorigenesis by negatively regulating Wnt signaling pathway through stabilizing the β-catenin destruction complex and act as a tumor suppressor in GC cells (31). Downregulation of RACK1 resulted in enhance of GC cell metastasis, via promoting the autocrine of interleukin (IL)-8 in vitro and in vivo (32). RACK1 inhibits GC progression through the NF-κB pathway (33). However, it is not clear whether RACK1 plays a tumor-suppressive role in GC cells through unknown mechanisms. Recent studies have indicated that RACK1 plays an important role in cell cycle progression, and it has attracted much attention. Genetic analysis of yeast (pombe S.) showed that RACK1/Cpc2 regulates cell cycle progression, and negatively regulates WEE1 homolog (S. pombe) (WEE1) protein levels and thus regulates mitosis (34). However, how RACK1 and WEE1 interact to regulate the occurrence and development of GC is still under investigation.
In the present study, the expression level of RACK1 is decreased in GC and was correlated to TNM stage, tumor differentiation and lymph node metastasis. In GC cells HGC27, the mRNA and protein levels of RACK1 were significantly reduced, and overexpression of RACK1 downregulated WEE1 protein expression, thus inhibits the growth and proliferation of HGC27 cells. Mechanistically, RACK1 and WEE1 interacted in HGC27 cells and co-located in the cytoplasm of HGC27 cells. Our results suggest that the abnormal expression of RACK1 in the tissues of GC was involved in the occurrence and development of GC. RACK1 and WEE1 interact to regulate the growth and proliferation of GC cells.
Materials and methods
Patient samples
All 70 tumors were diagnosed as GC and selected to ensure a broad range of clinical behavior (Table I). GC tissue specimens were obtained after written informed consent from patients undergoing GC surgery at the First Affiliated Hospital of Jinzhou Medical University (Jinzhou, China) during 2012–2013. All patients had not received chemotherapy and radiotherapy before operation. The study was approved by the Regional Ethics Committee of Jinzhou Medical University. Another 30 cases of normal GC adjacent to the edge of the cancer tissue were selected as the control. Samples of tumor and pericarcinous tissues were cut from the surgical specimens immediately fixed in buffered formalin for 48 h, embedded in paraffin, and sectioned before immunohistochemical staining. All biopsies were examined and classified by two histopathologist (Jing Y and Miao G) according to the World Health Organization (WHO) criteria.
Table I.
Clinicopathological features of 70 cases of GC.
| Clinical characteristic | n (%) |
|---|---|
| Sex | |
| Male | 43 (61.43) |
| Female | 27 (38.57) |
| Age (years) | |
| ≥60 | 26 (37.14) |
| <60 | 44 (62.86) |
| Tumor size (cm) | |
| ≤5 | 28 (40) |
| >5 | 42 (60) |
| Infiltrate depth | |
| Mucous membrane | 0 (0) |
| Submucosa | 3 (4.29) |
| Muscular layer | 9 (12.86) |
| Fibrous membrane | 47 (67.14) |
| Outside | 11 (15.71) |
| Lymph node metastasis | |
| ≤6 | 44 (62.85) |
| 7–14 | 18 (25.71) |
| ≥15 | 8 (11.43) |
| Distant metastasis | |
| Yes | 33 (47.14) |
| No | 37 (52.86) |
| Differentiation level | |
| High-Middle | 30 (42.86) |
| Low | 40 (57.14) |
| TNM stage | |
| I–II | 41 (58.57) |
| III–IV | 29 (41.43) |
T, tumor size; N, lymph node; M, metastasis.
Immunohistochemical staining
Ten-micrometer-thick consecutive sections were cut and mounted on glass slides. After deparaffinizing, rehydrating, antigen retrieval, and blocking endogenous peroxidases, the sections were washed three times in 0.01 mol/l phosphate-buffered saline (PBS) (8 mmol/l Na2HPO4, 2 mmol/l NaH2PO4, and 150 mmol/l NaCl) for 5 min each and blocked for 1 h in PBS supplemented with 0.3% Triton X-100 and 5% normal goat serum, followed by incubation of mouse monoclonal anti-human RACK1 antibody (610177; 1:200 dilution; BD Biosciences, San Jose, USA) at 4°C overnight. After brief washes in PBS, sections were exposed for 2 h to Polink-2 plus® Polymer HRP Detection System (PV-9002; ZSGB-BIO, Beijing, China) followed by development with 0.003% H2O2 and 0.03% 3,3′-diaminobenzidine in 0.05 mol/l Tris-HCl (pH 7.6). All sections were counterstained with hematoxylin.
The immunohistochemical evaluation was performed according to Xie lab (30) and slightly modified. The German semiquantitative scoring system was used, considering the staining intensity and area extent. Generally, each specimen was assigned a score according to the percentage of stained cells (0, <5%; 1, 5–25%; 2, 26–50%; 3, 51–75%; 4, 76–100%) and the intensity of the staining (0, no staining; 1, weak staining; 2, moderate staining and 3, strong staining). The final immunoreactive score was determined by multiplying the intensity score by the extent of score of stained cells. As a result, 9 grades were scored as 0, 1, 2, 3, 4, 6, 8, 9, and 12. When evaluating the protein expression of RACK1, we defined a score of 0–9 as low and 12 as high, respectively.
Construction of pcDNA3.1A-flag-RACK1 plasmid
Total RNA was extracted from the human embryonic kidney (HEK) 293 cells using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the procedure supplied by the manufacturer. Extracted RNA (1 µg) was used for cDNA synthesis using the PrimeScript® RT reagent kit (Takara, Dalian, China). The reaction system was prepared in a total volume of 20 µl containing 12.5 µl RNA primer mix, 4 µl 5xRT reaction buffer, 2 µl dNTPs, 1 µl RevertAid reverse transcriptase, 0.5 µl RiboLock RNase inhibitor and ddH2O up to 20 µl. A pair of primers was designed based on the RACK1 mRNA sequence (Genebank ID: NM_006098.4): kpnI (Takara) tailed forward (5′-ggcggGGTACCatgactgagcagatgacccttcg-3′) and XbaI (Takara) tailed reverse (5′-ggc ggT CTA GAT TAC TTG TCA TCG TCG TCC TTG TAG TCg cgt gtg cca atg gtc acc-3′) primers (restriction sites are underlined). The length of the amplification segment was 3,765 bp. The PCR mixture was mixed in a total volume of 50 µl containing1 µl cDNA, 1 µl each primer (20 µmol/l), 5 µl10x EasyPfu Buffer (Mg2+), 0.5 µl EasyPfu DNA Polymerase, 4 µl dNTP mix (2.5 mmol/l) and ddH2O up to 50 µl. The PCR program was started at 94°C for 7 min, followed by 35 cycles at 94°C for 45 sec, 55°C for 30 sec, 72°C for 3 min and completed with a final extension at 72°C for 10 min. The final PCR products were separated by electrophoresis using 1% polyacrylamide gels, and the target fragment was purified and recovered using agarose gel extraction kit (Axygen, Hangzhou, China). Double restriction enzyme digestion was applied to the purified target fragments and eukaryotic expression vector pcDNA3.1A-myc-plus(+), respectively. The enzyme reaction contained 3 µl target gene fragment or vector pcDNA3.1A-myc-plus(+), 5 µl 10x Fast Digest buffer, 3 µl KpnI, 3 µl XbaI and ddH2O up to 50 µl. Under the guidance of the T4 DNA ligase system instructions (Takara), the purified target fragment of the RACK1 was directionally ligated into pcDNA3.1A-myc-plus (+) vector in a 20 µl reaction system containing 15 µl target fragment, 2 µl pcDNA3.1A-myc-plus(+), 1 µl T4 DNA ligase, 2 µl 10xT4 buffer. The reactants were mixed at 16°C for 2 h, then the ligation was transformed into competent E. coli DH5a cells and inoculated into Luria-Bertani culture media containing 100 µg/ml ampicillin. After amplification by shaking the culture overnight at 37°C, the target plasmids were extracted from the bacterial liquid according to the instructions for the EndoFree Maxi Plasmid kit (QIAGEN, Duesseldorf, Germany). The resulting recombinant eukaryotic expression vector was named pcDNA3.1A-flag-RACK1. The recombinant plasmids was digested with KpnI and XbaI, and then evaluated by agarose gel electrophoresis. The recombinant plasmids was further sequenced to confirm its sequence by Beijing dingguochangsheng Biotechnology Co., Ltd. (Beijing, China).
Cell culture and transfection
The gastric epithelial cell line GES-1 (31) and GC cell line HGC27 were used in this study. The GES-1 cells and HGC27 cells were maintained and cultured in Dulbecco's modified Eagle medium (Invitrogen) and RPMI-1640 (Invitrogen) medium supplemented with 10% fetal bovine serum (FBS; PAA Laboratories, Pasching, Austria), 100 U/ml penicillin and 50 µg/ml streptomycin (Biochrom KG, Berlin, Germany) at 37°C in a humidified 5% CO2 incubator, respectively. The cells were transiently transfected with the plasmids pcDNA3.1A-flag-RACK1or pcDNA3.1A using Lipofectamine 2000 reagent (Invitrogen) following the manufacturer's instructions.
Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA of each sample was extracted using TRIzol reagent (Invitrogen) following the manufacturer's protocol. Equal amounts of RNAs (1 µg) were used as templates in each reaction (50 µl total volume) with the one-step RNA PCR kit (TakaRa, Kyoto, Japan). The nucleotide sequences of the sense and antisense primers used for RACK1 (Genebank ID: NM_006098.4) amplification were 5′-GGGGTCACTCCCACTTTGTT-3′ and 5′-AATCTGCCGGTTGTCAGAGG-3′, respectively (263 bp). The primers for β-actin (Genebank ID: NM_001101.1) amplification were 5′-TGACGGGGTCACCCACACTGTGCCCATCT-3′ and 5′-CTAGAAGCATTTGCGGTGGACGATGGAGGG-3′ (223 bp). The RT-PCR for RACK1 and β-actin included one round of reverse transcription at 50°C for 30 min, and 30 cycles of PCR amplification with 94°C for 45 sec, 55°C for 30 sec and 72°C for 1 min. The RT-PCR products were analyzed on 1% agarose gels and viewed under ultraviolet light (Bio-Rad, Hercules, CA).
Protein isolation and immunoblotting
Total protein lysates were prepared in 50 mM Tris-HCl (pH 7.5) containing 150 mM NaCl and 0.1% NP-40, supplied with protease inhibitor cocktail (Roche Molecular Biochemical, Indianapolis, USA). Cell debris was removed by centrifugation for 15 min at 13,000 g at 4°C. Protein concentration was measured with Pierce™ Bicinchoninic acid (BCA) Protein Assay Kit (Fisher Scientific). Equal amounts of total protein samples were separated on 10% sodiumdodecyl sulphate (SDS)-polyacrylamide gels (PAGE) (Life Technologies, Grand Island, NY, USA) with electrophoresis, and separated proteins were transferred onto 0.45 µm polyvinylidene fluoride (PVDF) membranes (Bio-Rad, Hercules, CA, USA) blocked with Tris-Buffered Saline and Tween-20 (TBST; 20 mM Tris-HCl pH 7.5, 150 mM NaCl and 1% Tween-20) containing 5% fat free dry milk for 2 h and incubated for 16 h with anti-RACK1 antibody (dilution, 1:1,000), rabbit polyclonal anti-human WEE1 antibody (ab203236; Abcam, CA, USA) (dilution, 1:1,000) and mouse anti-β ACTIN monoclonal antibody (TA-09; ZSGB-BIO, Beijing, China) (dilution, 1:1,000) in TBST. After primary antibody incubation, membranes were washed three times in TBST, followed by incubation with secondary antibodies cross-linked with horseradish peroxidase (HRP) (dilution, 1:5,000). Immunoreactive proteins were visualized with an enhanced chemiluminescence (ECL) detection system (Beyotime, Jiangsu, China). The relative expression of the target protein was calculated as the gray value ratio of target protein content to β-ACTIN content (target protein/β-ACTIN) using Image J software analysis.
Cell viability analysis
The cell viability measurements were carried out using 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay. The total cell number was quantified at 24 h intervals up to 96 h. Approximately 5×103 of HGC27 cells were seeded into 96-well plates, washed twice with PBS and 20 µl MTT (5 mg/ml with PBS, pH 7.4) was added to each well. Then, the cells were incubated at 37°C for 4 h and 150 µl dimethylsulfoxide (DMSO; Sigma-Aldrich, St. Louis, USA) was added to dissolve the formazan crystals. After shaking the plate for 10 min, cell viability was obtained by measuring the absorbance at 490 nm wavelength with enzyme-labeling instrument (Bio-Tek ELX800, Winooski, VT, USA), this assay was done six times. The proliferation rate was calculated according to the following formula: cell viability rate (%)=average absorbance of experimental group/average absorbance of blank control group ×100%.
Immunoprecipitation and immunoblotting
HGC27 cells were collected after transfection for 48 h, washed with PBS (pH 7.4), and lysed in modified RIPA buffer (50 mM Tris (pH 7.8), 150 mM NaCl, 5 mM EDTA, 15 mM MgCl2, 1% NP-40, 0.5% sodium deoxycholate, 1 mM DTT, and 20 mM N-ethylmaleimide) supplemented with complete protease inhibitor cocktail. Lysates were cleared by centrifugation at 12,000 g for 15 min at 4°C and quantified protein according to BCA kit. The same amount of protein precipitation was taken and added 20 µl lysate and 5 µl 5xSDS sample buffer, boiled for 5 min and preserved at −20°C. The precipitation was added 20 µl precold mixed suspension of protein A/G agarose beads (Santa Cruz Biotechnology, Santa Cruz, USA) and 1 µg IgG and the mixtures were centrifuged at 12,000 g for 30 min at 4°C. The supernatant was incubated on ice for 2 h with l-2 µg RACK1 antibody, then 50 µl protein A/G agarose immunoprecipitation reagent was added to each lysate and incubated with rotation for overnight at 4°C. The beads were retrieved by centrifugation and washed five times with RIPA buffer and once with PBS. Protein bound to the beads were eluted by boiling in 2xSDS sample buffer, separated by SDS-PAGE, transferred protein to PVDF membrane and blocked as described above. WEE1 antibody were incubated overnight at 4°C and washed 3 times in TBST followed by incubation with HRP conjugated goat anti rabbit IgG (dilution ratio 1:5,000) for 2 h. The signals were detected by ECL detection system.
Immunofluorescence analysis
HGC27 cells seeded on 6-well chamber slides were fixed in 4% paraformaldehyde, permeabilized in 0.2% Triton X-100 for 5 min and blocked in 1% BSA for 1 h. Protein levels were detected using RACK1 and WEE1 antibodies overnight at 4°C, The cells were washed with PBS for 5 min three times followed by incubation for 45 min at 37°C with Cy3-conjugated or FITC-conjugated secondary antibodies (Amersham Biosciences). The coverslips were washed with PBS, stained nucleus with DAPI (Sigma-Aldrich, St. Louis, USA) for 5 min at room temperature, and mounted in PBS containing 50% glycerol, and viewed on a Leica laser scanning confocal microscope equipped with a Photometrics Cool SnapES N&B camera driven by MetaMorph software (Universal Imaging Corporation, Downingtown, USA).
Statistical analysis
All data were representative of at least three independent experiments with similar results. Data were presented as mean ± SD. Graphpad prism 5 software was used for all statistical analysis. A student's t test was used to determine significant differences (two-tailed, P<0.05). Pearson's correlation coefficients were used to determine whether two prognosis-related factors were correlated to each other over all cases.
Results
RACK1 proteins were lowly expressed in GC patients
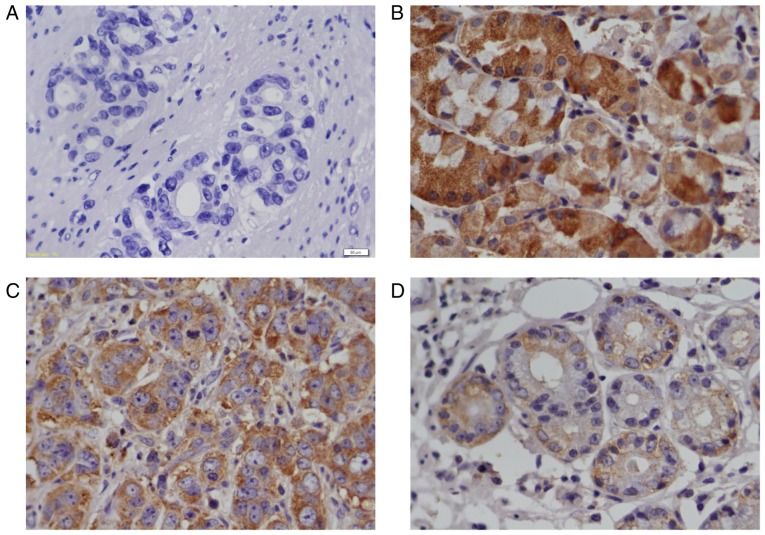
Fig. 1 presents the results of the immunostaining assay on RACK1 protein expression in GC tissues and adjacent gastric tissues. The immunohistochemical staining showed a significant decrease of RACK1 protein in the GC tissues (Fig. 1C and D, Table II) compared with pericarcinous tissues (Fig. 1B), and the expression of RACK1 in high and middle differentiation of GC tissues (Fig. 1C) are higher than that in poorly differentiated GC tissues (Fig. 1D). The positive expression of RACK1 protein was located in the cytoplasm of GC tissues and adjacent gastric tissues (Fig. 1B-D).
Figure 1.
Immunostaining showing the protein expression of RACK1 in GC tissues and adjacent gastric tissues (A). negative control, omitting antibody against RACK1 in high and middle differentiation of GC tissues (B). The high protein expression of RACK1 in adjacent gastric tissues (C). The high protein expression of RACK1 in high and middle differentiation of GC tissues (D). The low protein expression of RACK1 in low differentiated GC tissues. Pictures were taken with a Lecia 6000 microscope at magnification, ×400. Scale bar, 50 µm.
Table II.
The protein expression of RACK1 in GC tissues (n=70) and pericarcinous tissues (n=30).
| RACK1 expression | |||||
|---|---|---|---|---|---|
| Histological type | n | High (stage III–IV) | Low (stage I–II) | Ratio (%) | P-value |
| Pericarcinous tissues | 30 | 26 | 4 | 86.67 | 0.032 |
| GC tissues | 70 | 24 | 46 | 34.29 | |
The low expression of RACK1 correlated with pathological parameters in GC patients
The protein expression of RACK1 in stage I–II of GC tissues was higher than that in stage III–IV of GC tissues (P<0.01), and the protein expression of RACK1 in the high-middle differentiated GC tissues was higher than that in the low differentiated GC tissues (P<0.01), and the decreased expression of RACK1 was associated withlymph node metastasis (P<0.05). The protein expression level of RACK1 in GC was related to TNM stage, tumor differentiation, and lymph node metastasis, while it has no correlation with age, sex, tumor size and depth of tumor invasion (Table III).
Table III.
The relationship between expression of RACK1 in GC tissues and clinical pathological parameters.
| RACK1 expression | ||||
|---|---|---|---|---|
| Clinical features | n | Low | High | P-value |
| Sex | ||||
| Male | 43 | 31 | 12 | 0.156 |
| Female | 27 | 15 | 12 | |
| Age | ||||
| ≥60 | 26 | 15 | 11 | 0.227 |
| <60 | 44 | 31 | 13 | |
| Tumor Size | ||||
| ≤5 | 28 | 18 | 10 | 0.837 |
| >5 | 42 | 28 | 14 | |
| Differentiation level | ||||
| High-Middle | 30 | 12 | 18 | <0.001 |
| Low | 40 | 34 | 6 | |
| Infiltrate depth | ||||
| Submucosa | 3 | 2 | 1 | 0.923 |
| Muscular layer | 9 | 5 | 4 | |
| Fibrous membrane | 47 | 32 | 15 | |
| Outside | 11 | 7 | 4 | |
| Lymph node metastasis | ||||
| ≤6 | 44 | 28 | 16 | 0.028 |
| 7–14 | 18 | 12 | 6 | |
| ≥15 | 8 | 5 | 3 | |
| TNM stage | ||||
| StageI–II | 41 | 20 | 21 | <0.001 |
| Stage III–IV | 29 | 26 | 3 | |
T, tumor size; N, lymph node; M, metastasis.
Downregulation of RACK1 expression in GC cell lines
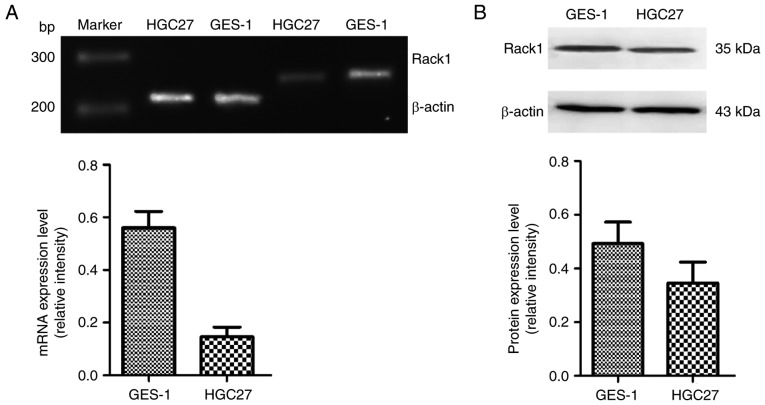
Agarose gel electrophoresis analysis showed that the RACK1 mRNA expression in normal gastric mucosal cells GES-1 was higher than that in GC cells HGC27 (Fig. 2A). The protein expression of RACK1 in HGC27 cells was lower than that in GES-1 cells (Fig. 2B).
Figure 2.
The mRNA and protein expression level of RACK1 in GES-1 cells and HGC27cells (A). The mRNA expression of RACK1 in HGC27 cells and GES-1 cells. The up-pannel is an analysis of agarose gel electrophoresis, while the down-pannel is relative mRNA expression level using β-actin as a loading control (B). The protein expression of RACK1 in HGC27 cells and GES-1 cells. The up-pannel is an analysis of western blotting, while the down-pannel is relative protein expression level using β-ACTIN as a loading control.
Overexpression of RACK1 inhibited tumor growth in vivo
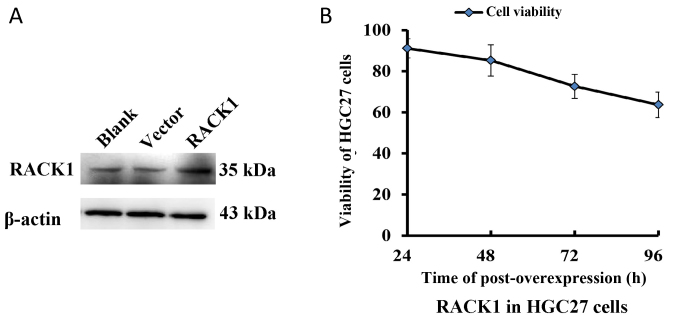
The protein expression of RACK1 was significantly increased in the HGC27 cells transfected with pcDNA3.1-RACK1 compared with the cells transfected with or without pcDNA3.1 vector (Fig. 3A). The survival rate of HGC27 cells transfected with pcDNA3.1-RACK1 was significantly reduced at 72 h and 96 h (P<0.01, Fig. 3B), which showed that the overexpression of RACK1 could significantly inhibit the growth of HGC27 cells.
Figure 3.
The viability of HGC27 cells transfected with pcDNA3.1-flag-RACK1 (A). The protein expression of RACK1 increased after HGC27 cells transfected with pcDNA3.1-flag-RACK1. Blank, the HGC27 cells transfected without pcDNA3.1 or pcDNA3.1-flag-RACK1. Vector, the HGC27 cells transfected with pcDNA3.1. RACK1, the HGC27 cells transfected with pcDNA3.1-flag-RACK1 (B). The survival rate of HGC27 cells transfected with pcDNA3.1-RACK1 is reduced. The cell survival rate was detected at 24, 48, 72 and 96 h, by MTT after transfection of pcDNA3.1-RACK1 and pcDNA3.1 to HGC27 cells, respectively.
The interaction between RACK1 and WEE1 in HCG27 cells
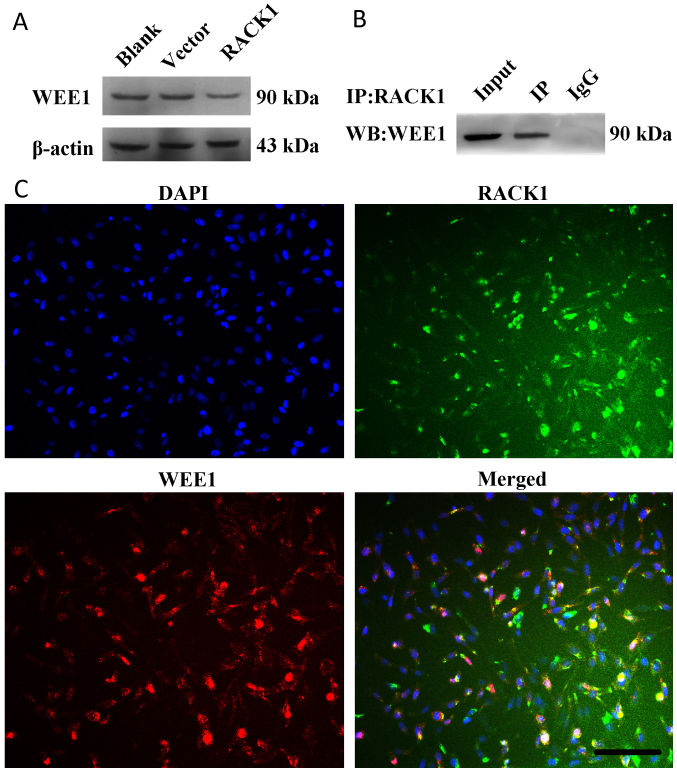
The protein expression of WEE1 was significantly decreased in HGC27 cells transfected with pcDNA3.1-RACK1 compared with cells with or without pcDNA3.1 vector, and there was no significant difference between two control groups (Fig. 4A). In order to explore possible mechanism(s) underlying RACK1 regulation, interactions between RACK1 and WEE1 using HGC27 cells via immunoprecipitation and immunofluorescence analyses were performed. Co-immunoprecipitation was investigated in HGC27 cells incubated with RACK1 antibody and detected with WEE1 antibody. As shown in Fig. 4B, RACK1 was co-immunoprecipitated with WEE1. This interaction was further confirmed to detect endogenous RACK1 and WEE1 in HGC27 cells. Immunofluorescence analysis revealed that RACK1 co-localized with WEE1 mainly in the cytoplasm of HGC27 cells (Fig. 4C).
Figure 4.
The interaction between RACK1 and WEE1 in HGC27 cells (A). The protein expression of WEE1 in HGC27 cells. The protein expression of WEE1 was detected in HGC27 cells at 48 h after transfection of pcDNA3.1-RACK1 and pcDNA3.1 empty vector (B). The interaction between RACK1 and WEE1 in HGC27 cells was confirmed by co-immunoprecipitation. HGC27 cells were collected and lysed, protein was extracted, prewashed and incubated with RACK1 antibody, added the protein A+G beads. After denaturation, the PVDF membrane was detected with WEE1 antibody for Western blotting (C). The location of RACK1 and WEE1 in HGC27 cells was observed by indirect immunofluorescence assay. Scale bar, 200 µm.
Discussion
RACK1 has been identified as an anchoring or adaptor protein in multiple intracellular signal transduction pathways and showed heterogeneity in different tumors (32–36). It was found that RACK1 is overexpressed in several types of cancers such as breast, colon, melanomas and lung (36), suggesting that RACK1 is involved in the occurrence and development of tumor as an oncogene. On the other hand, recent studies have reported that RACK1 is expressed lowly in GC tissues and cells, suggesting that RACK1 plays a tumor suppressor role in the development of GC (32–35).
In order to verify the function of RACK1 in GC tissues and cells, in this study, firstly, we found that RACK1 was downregulated in GC tissues using immunohistochemical staining, research on clinicopathological characteristics of these patients indicated that RACK1 expression was significantly correlated with TNM stage, tumor differentiation and lymph node metastasis, suggesting that the expression level of RACK1 is negatively regulated the development and metastasis of GC. The study is consistent with results of Deng et al (32). Secondly, we detected that the mRNA and protein level of RACK1 in HGC27 cells was significantly lower than that of GES-1 cells, which is consistent with the result of GC cell line SGC 7901 (19). Then, upregulation of RACK1 inhibits the proliferation of HGC27 cells, which is consistent with the findings of Deng et al and Yong-Zheng et al (32,34), suggesting that RACK1 negatively regulates the process of GC cells.
WEE1 is a member of the serine/threonine protein kinase family involved in terminal phosphorylation and inactivation of cyclin dependent kinase 1 (CDK1) and is a key regulator of cell cycle progression (7). Some studies have found that WEE1 is highly expressed in malignant melanoma, breast cancer, osteosarcoma and glioma (37–40). Kim HY reported that high expression of WEE1 is associated with poor prognosis in male GC patients with lymph node metastasis, and WEE1 expression was detected in 12 GC cell lines, 7 strains with high WEE1 expression, 5 strains with little or no WEE1 expression, but there is no information about HGC27 cells (7). Normal cells repair damaged DNA during G1-arrest, however cancer cells often have deficient G1-arrest and largely depend on G2-arrest. Thus, cancer cells have increased DNA damage at the G2-checkpoint compared to normal cells. The molecular switch for the G2/M transition is held by WEE1 and is pushed forward by Cell division cycle 25 (CDC25) (7). To study the possible mechanism of overexpressed RACK1 inhibits the growth and proliferation of GC cells, we detected the down expression level of WEE1 in HGC27 cells. Therefore, the overexpression of RACK1 in HGC27 cells destroyed the balance of G2/M checkpoint and inhibited cell proliferation. To further study the functional relationship between RACK1 and WEE1 in GC cells, we found RACK1 interacted with WEE1 by immuneprecipitation and both were co-localized in the cytoplasm by immunofluorescence using HGC27 cells. Therefore, the interaction between RACK1 and WEE1 is one of the molecular mechanism in regulating the growth and proliferation of GC cells.
In summary, the abnormal expression of RACK1 is involved in the occurrence and development of GC, and negatively regulate the process of GC cells. The interaction of RACK1 and WEE1 is one of the molecular mechanisms in regulating development of GC. However, in this study, we verified the interaction and localization of RACK1 and WEE1 in HGC27 cells with antibody, the result may be affected by antibody quality, protein molecular weight, therefore, exogenous plasmids were transfected into the cells for further testing. Furthermore, how RACK1 and WEE1 interact to regulate the molecular mechanism(s) of GC is still under investigation.
Acknowledgments
The authors thank Professor Li Feng of China Medical University for gifting gastric epithelial cell line GES-1 and gastric cancer cell line HGC27. The authors also thank Dr. Luan Zhidong of Jinzhou Medical University for giving pcDNA3.1A-myc-plus(+) plasmid. The authors acknowledge grant supports received from the National Nature Science Foundation of China (grant nos. 31371173, 81270698, and 81401199) and the Project of Science and Technology Plan of Liaoning Province (grant no. 2015020697).
Glossary
Abbreviations
- RACK1
receptor of activated C Kinase 1
- GC
gastric cancer
- GNB2L1
guanine nucleotide binding protein beta polypeptide 2-like 1
- WEE1
WEE1 homolog (S. pombe)
References
- 1.McLean MH, El-Omar EM. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol. 2014;11:664–674. doi: 10.1038/nrgastro.2014.143. [DOI] [PubMed] [Google Scholar]
- 2.Badgwell B. Multimodality therapy of localized gastric adenocarcinoma. J Natl Compr Canc Netw. 2016;14:1321–1327. doi: 10.6004/jnccn.2016.0139. [DOI] [PubMed] [Google Scholar]
- 3.Fang M, Wu J, Lai X, Ai H, Tao Y, Zhu B, Huang L. CD44 and CD44v6 are correlated with gastric cancer progression and poor patient prognosis: Evidence from 42 studies. Cell Physiol Biochem. 2016;40:567–578. doi: 10.1159/000452570. [DOI] [PubMed] [Google Scholar]
- 4.Wang J, Fei X, Wu W, Chen X, Su L, Zhu Z, Zhou Y. SLC7A5 Functions as a Downstream Target Modulated by CRKL in Metastasis Process of Gastric Cancer SGC-7901 Cells. PLoS One. 2016;11:e0166147. doi: 10.1371/journal.pone.0166147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawrence W. Way, Gerard M. Doherty. Current surgical diagnosis & treatment. 11th. McGraw-Hill Companies, Inc.; New York: 2003. pp. 285–287. [Google Scholar]
- 6.Japanese Gastric Cancer Association: Japanese classification of gastric carcinoma: 3rd English edition. Gastric Cancer. 2001;14:101–112. doi: 10.1007/s10120-011-0041-5. [DOI] [PubMed] [Google Scholar]
- 7.Kim HY, Cho Y, Kang H, Yim YS, Kim SJ, Song J, Chun KH. Targeting the WEE1 kinase as a molecular targeted therapy for gastric cancer. Oncotarget. 2016;7:49902–49916. doi: 10.18632/oncotarget.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amin MB, Edge S, Greene F, Byrd DR, Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR, Sullivan DC, et al. AJCC Cancer Staging Manual. 8th. Springer; New York: 2017. pp. 93–99. [Google Scholar]
- 9.Roder JD, Böttcher K, Busch R, Wittekind C, Hermanek P, Siewert JR. Classification of regional lymph node metastasis from gastric carcinoma. German Gastric Cancer Study Group. Cancer. 1998;82:621–631. doi: 10.1002/(SICI)1097-0142(19980215)82:4<621::AID-CNCR2>3.3.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Lim DH, Kim HS, Park YS, Lee J, Park SH, Lim HY, Ji SH, Park MJ, Yi SY, An JY, et al. Metastatic lymph node in gastric cancer; is it a real distant metastasis? BMC Cancer. 2010;10:25. doi: 10.1186/1471-2407-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akagi T, Shiraishi N, Kitano S. Lymph Node Metastasis of Gastric Cancer. Cancers (Basel) 2011;3:2141–2159. doi: 10.3390/cancers3022141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li JJ, Xie D. RACK1, a versatile hub in cancer. Oncogene. 2015;34:1890–1898. doi: 10.1038/onc.2014.127. [DOI] [PubMed] [Google Scholar]
- 13.Gandin V, Senft D, Topisirovic I, Ronai ZA. RACK1 function in cell motility and protein synthesis. Genes Cancer. 2013;4:369–377. doi: 10.1177/1947601913486348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson TJ. RACK1 research-ships passing in the night? FEBS Lett. 2012;586:2787–2789. doi: 10.1016/j.febslet.2012.04.048. [DOI] [PubMed] [Google Scholar]
- 15.Robles MS, Boyault C, Knutti D, Padmanabhan K, Weitz CJ. Identification of RACK1 and protein kinase Calpha as integral components of the mammalian circadian clock. Science. 2010;327:463–466. doi: 10.1126/science.1180067. [DOI] [PubMed] [Google Scholar]
- 16.Bird RJ, Baillie GS, Yarwood SJ. Interaction with receptor for activated C-kinase 1 (RACK1) sensitizes the phosphodiesterase PDE4D5 towards hydrolysis of cAMP and activation by protein kinase C. Biochem J. 2010;432:207–216. doi: 10.1042/BJ20101010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang W, Zong CS, Hermanto U, Lopez-Bergami P, Ronai Z, Wang LH. RACK1 recruits STAT3 specifically to insulin and insulin-like growth factor 1 receptors for activation, which is important for regulating anchorage-independent growth. Mol Cell Biol. 2006;26:413–424. doi: 10.1128/MCB.26.2.413-424.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haberman Y, Alon LT, Eliyahu E, Shalgi R. Receptor for activated C kinase (RACK) and protein kinase C (PKC) in egg activation. Theriogenology. 2011;75:80–89. doi: 10.1016/j.theriogenology.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 19.Li M, Liu Y, Jin F, Sun X, Li Z, Liu Y, Fang P, Shi H, Jiang X. Endothelin-1 induces hypoxia inducible factor 1α expression in pulmonary artery smooth muscle cells. FEBS Lett. 2012;586:3888–3893. doi: 10.1016/j.febslet.2012.08.036. [DOI] [PubMed] [Google Scholar]
- 20.Liu M, Peng P, Wang J, Wang L, Duan F, Jia D, Ruan Y, Gu J. RACK1-mediated translation control promotes liver fibrogenesis. Biochem Biophys Res Commun. 2015;463:255–261. doi: 10.1016/j.bbrc.2015.05.040. [DOI] [PubMed] [Google Scholar]
- 21.Cheng D, Zhu X, Barchiesi F, Gillespie DG, Dubey RK, Jackson EK. Receptor for activated protein kinase C1 regulates cell proliferation by modulating calcium signaling. Hypertension. 2011;58:689–695. doi: 10.1161/HYPERTENSIONAHA.111.174508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su J, Xu J, Zhang S. RACK1, scaffolding a heterotrimeric G protein and a MAPK cascade. Trends Plant Sci. 2015;20:405–407. doi: 10.1016/j.tplants.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 23.Jia X, Zhang L, Mao X. S-propranolol protected H9C2 cells from ischemia/reperfusion-induced apoptosis via downregultion of RACK1 Gene. Int J Clin Exp Patho. 2015;l8:10335–10344. [PMC free article] [PubMed] [Google Scholar]
- 24.Grosso S, Volta V, Sala LA, Vietri M, Marchisio PC, Ron D, Biffo S. PKCbetaII modulates translation independently from mTOR and through RACK1. Biochem J. 2008;415:77–85. doi: 10.1042/BJ20080463. [DOI] [PubMed] [Google Scholar]
- 25.Ruan Y, Sun L, Hao Y, Wang L, Xu J, Zhang W, Xie J, Guo L, Zhou L, Yun X, et al. Ribosomal RACK1 promotes chemoresistance and growth in human hepatocellular carcinoma. J Clin Invest. 2012;122:2554–2566. doi: 10.1172/JCI58488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campagne C, Julé S, Alleaume C, Bernex F, Ezagal J, Château-Joubert S, Estrada M, Aubin-Houzelstein G, Panthier JJ, Egidy G. Canine melanoma diagnosis: RACK1 as a potential biological marker. Vet Pathol. 2013;50:1083–1090. doi: 10.1177/0300985813490754. [DOI] [PubMed] [Google Scholar]
- 27.Cao XX, Xu JD, Liu XL, Xu JW, Wang WJ, Li QQ, Chen Q, Xu ZD, Liu XP. RACK1: A superior independent predictor for poor clinical outcome in breast cancer. Int J Cancer. 2010;127:1172–1179. doi: 10.1002/ijc.25120. [DOI] [PubMed] [Google Scholar]
- 28.Lin Y, Cui M, Teng H, Wang F, Yu W, Xu T. Silencing the receptor of activated C-kinase 1 (RACK1) suppresses tumorigenicity in epithelial ovarian cancer in vitro in vivo. Int J Oncol. 2014;44:1252–1258. doi: 10.3892/ijo.2014.2274. [DOI] [PubMed] [Google Scholar]
- 29.Choi YY, Lee SY, Lee WK, Jeon HS, Lee EB, Lee HC, Choi JE, Kang HG, Lee EJ, Bae EY, et al. RACK1 is a candidate gene associated with the prognosis of patients with early stage non-small cell lung cancer. Oncotarget. 2015;6:4451–4466. doi: 10.18632/oncotarget.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Liu N, Ma D, Liu L, Jiang L, Zhou Y, Zeng X, Li J, Chen Q. Receptor for activated C kinase 1 (RACK1) promotes the progression of OSCC via the AKT/mTOR pathway. Int J Oncol. 2016;49:539–548. doi: 10.3892/ijo.2016.3562. [DOI] [PubMed] [Google Scholar]
- 31.Li X, Ke Q, Li Y, Liu F, Zhu G, Li F. DGCR6L, a novel PAK4 interaction protein, regulates PAK4-mediated migration of human gastric cancer cell via LIMK1. Int J Biochem Cell Biol. 2010;42:70–79. doi: 10.1016/j.biocel.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Deng YZ, Yao F, Li JJ, Mao ZF, Hu PT, Long LY, Li G, Ji XD, Shi S, Guan DX, et al. RACK1 suppresses gastric tumorigenesis by stabilizing the β-catenin destruction complex. Gastroenterology. 2012;142:812–823.e15. doi: 10.1053/j.gastro.2011.12.046. [DOI] [PubMed] [Google Scholar]
- 33.Chen L, Min L, Wang X, Zhao J, Chen H, Qin J, Chen W, Shen Z, Tang Z, Gan Q, et al. Loss of RACK1 promotes metastasis of gastric cancer by inducing a miR-302c/IL8 signaling loop. Cancer Res. 2015;75:3832–4381. doi: 10.1158/0008-5472.CAN-14-3690. [DOI] [PubMed] [Google Scholar]
- 34.Yong-Zheng X, Wan-Li M, Ji-Ming M, Xue-Qun R. Receptor for activated protein kinase C 1 suppresses gastric tumor progression through nuclear factor-kB pathway. Indian J Cancer. 2015;52(Suppl 3):E172–E175. doi: 10.4103/0019-509X.186572. [DOI] [PubMed] [Google Scholar]
- 35.Núñez A, Franco A, Soto T, Vicente J, Gacto M, Cansado J. Fission yeast receptor of activated C kinase (RACK1) ortholog Cpc2 regulates mitotic commitment through Wee1 kinase. J Biol Chem. 2010;285:41366–41373. doi: 10.1074/jbc.M110.173815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang F, Osawa T, Tsuchida R, Yuasa Y, Shibuya M. Downregulation of receptor for activated C-kinase 1 (RACK1) suppresses tumor growth by inhibiting tumor cell proliferation and tumor-associated angiogenesis. Cancer Sci. 2011;102:2007–2013. doi: 10.1111/j.1349-7006.2011.02065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mueller S, Hashizume R, Yang X, Kolkowitz I, Olow AK, Phillips J, Smirnov I, Tom MW, Prados MD, James CD, et al. Targeting Wee1 for the treatment of pediatric high-grade gliomas. Neuro Oncol. 2014;16:352–360. doi: 10.1093/neuonc/not220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magnussen GI, Holm R, Emilsen E, Rosnes AK, Slipicevic A, Flørenes VA. High expression of wee1 is associated with poor disease-free survival in malignant melanoma: Potential for targeted therapy. PLoS One. 2012;7:e38254. doi: 10.1371/journal.pone.0038254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kreahling JM, Foroutan P, Reed D, Martinez G, Razabdouski T, Bui MM, Raghavan M, Letson D, Gillies RJ, Altiok S. Wee1 inhibition by MK-1775 leads to tumor inhibition and enhances efficacy of gemcitabine in human sarcomas. PLoS One. 2013;8:e57523. doi: 10.1371/journal.pone.0057523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murrow LM, Garimella SV, Jones TL, Caplen NJ, Lipkowitz S. Identification of WEE1 as a potential molecular target in cancer cells by RNAi screening of the human tyrosine kinome. Breast Cancer Res Treat. 2010;122:347–357. doi: 10.1007/s10549-009-0571-2. [DOI] [PMC free article] [PubMed] [Google Scholar]