Abstract
Breast cancer remains the most prevalent cancer among women worldwide. The expression of estrogen receptor-α (ER-α) is an important marker for prognosis. ER-α status may be positive or negative in breast cancer cells, although the cause of negative or positive status is not yet fully characterized. In the present study, the expression of ER-α and miRNA-148a was assessed in two breast cancer cell lines, HCC1937 and MCF7. An association between ER-α and miRNA-148a expression was identified. It was then demonstrated that DNA methyltransferase 1 (DNMT1) is a target of miRNA-148a, which may suppress the expression of ER-α via DNA methylation. Finally, an miRNA-148a mimic or inhibitor was transfected into MCF7 cells; the miRNA-148a mimic increased ER-α expression whereas the miRNA-148a inhibitor decreased ER-α expression. In conclusion, it was identified that miRNA-148a regulates ER-α expression through DNMT1-mediated DNA methylation in breast cancer cells. This may represent a potential miRNA-based strategy to modulate the expression of ER-α and provide a novel perspective for investigating the role of miRNAs in treating breast cancer.
Keywords: breast cancer cells, miRNA-148a, estrogen receptor, DNA methyltransferase, DNA methylation
Introduction
Breast cancer is the most prevalent type of cancer among women worldwide (1). The majority of primary breast cancer cases are estrogen receptor (ER) positive; approximately one-third of patients with breast cancer are ER− at the time of diagnosis (2). ER status is predictive for the response to endocrine therapy of breast cancer, and is thus an important marker of prognosis. Therapies targeting the ER, including tamoxifen and aromatase inhibitors, have been successfully applied for treating breast cancer patients with tumors that express ER (3). However, a fraction of breast cancers that are initially ER+ lose the expression of ER during tumor progression, and thus the long-term effectiveness of endocrine therapies is limited (2). Methylation of ESR1, the gene encoding ER-α, is associated with ER status and implicated in the tumor resistance to hormone-based treatment in breast cancer (4).
MicroRNAs (miRNAs) are a group of highly conserved, non-coding RNAs that suppress gene expression post-transcriptionally by binding to the 3′-untranslated region (3′UTR) of target mRNAs. Dysregulation of miRNAs is associated with the initiation and progression of a number of types of human cancer, including breast cancer (5,6). miRNAs are increasingly regarded as essential regulators of gene expression in breast cancer, which may act as oncogenes (e.g., miR-21) or tumor suppressors (e.g., let-7) (7).
Previous studies have demonstrated that the absence of ER expression may result from the hypermethylation of CpG islands in the 5′ region of the ESR1 gene in a proportion of breast cancer cases (2). DNA methylation is catalyzed by DNA methyltransferases (DNMTs). It has been reported that the specific inhibition of DNMT1 expression is sufficient to induce ER-α re-expression in human breast cancer cell lines (8). DNMT1 expression has been demonstrated to be inversely correlated with miRNA-148a expression in breast cancer tissue, which may imply that there is a negative feedback regulatory loop between miRNA-148a and DNMT1 (9).
In the present study, it was demonstrated that ER and miRNA-148a expression in HCC1937 and MCF7 cells were associated, then further confirmed that miRNA-148a increases the expression of ER-α through targeting DNMT1.
Materials and methods
Cell culture and transfection
The human breast cancer cell lines MCF7 and HCC1937 were purchased from the Cell Resource Center, Shanghai Institute of Biochemistry and Cell Biology at the Chinese Academy of Sciences (Shanghai, China). Cells were maintained at 37°C in a humidified air atmosphere containing 5% CO2 in RPMI-1640 (MCF7) or Dulbecco's modified Eagle's medium (HCC1937), supplemented with 10% fetal bovine serum (all from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
siRNA against DNMT1 (stB0002746C-1-5), miRNA-148a mimic (miR10000516-1-5) and miRNA-148a inhibitor (miR20000243-1-5) were obtained from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). miRNA mimic/inhibitor control (miR01101-1-5) and siRNA control (siN05815122147-1-5) were also obtained from Guangzhou RiboBio Co., Ltd. Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used for transfection into MCF-7 cells according to the manufacturer's protocol. The transfected concentrations of siRNA, miRNA-148a mimic and inhibitor were 50, 50 and 100 nM, respectively.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using TRIzol reagent (Sangon Biotech Co., Ltd., Shanghai, China). ReverTra Ace qPCR RT kit (Toyobo Co., Ltd., Osaka, Japan) was used to generate cDNA from mRNA and SYBR Premix Ex Taq (Takara Bio, Inc., Otsu, Japan) was used to perform qPCR with the ABI 7500 Real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. A total of 1 µg cDNA was used for qPCR. The primer sequences were as follows: DNMT1 forward, 5′-GCACAAACTGACCTGCTTCA-3′ and reverse, 5′-GCCTTTTCACCTCCATCAAA-3′; ER-α forward, 5′-TTCGGCTCCAACGGCCTGGGGGGTTT-3′ and reverse, 5′-GGTACTGGCCAATCTTTCTCTGCCACCCT-3′; GAPDH forward, 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse, 5′-GAAGATGGTGATGGGATTTC-3′. The thermocycling conditions for qPCR were as follows: 95°C for 30 sec, 40 cycles of 95°C for 5 sec and 60°C for 30 sec. The relative expression level was determined using the 2−ΔΔCq method (10).
In order to amplify miRNA sequences, total RNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and the microRNA qRT-PCR syb kit (TaqMan™ MicroRNA Assays, Thermo Fisher Scientific, Inc.) was used for generating cDNA from microRNA and its subsequent qPCR detection, according to the manufacturer's protocols. Primers for miRNA-148a and U6 were purchased from Invitrogen (Thermo Fisher Scientific, Inc.). The miRNA-148a primers were 5′-ATGCTCAGTGCACTACAGAA-3′ (forward) and 5′-GTGCAGGGTCCGAGGT-3′ (reverse). The internal standard was U6 and its primer was 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse).
Western blot analysis
Cultured cells were harvested and lysed in radioimmunoprecipitation assay buffer supplemented with complete protease inhibitor cocktail tablets (Roche Diagnostics GmbH, Mannheim, Germany). Cell debris was removed by centrifugation at 13,000 × g for 20 min at 4°C. The concentration of proteins were quantified using Bio-Rad Protein Assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Lysates (~20 µg) were separated by SDS-PAGE (10%) and transferred to PVDF membranes (Bio-Rad Laboratories, Inc.). Subsequently, the membranes were blocked [5% fat-free milk in Tris buffer saline with 0.1% Tween-20 (TBS-T)] for 1 h at room temperature. Following blocking, membranes were incubated with primary antibodies, including ER (dilution, 1:1,000; incubated overnight at 4°C; cat. no. ab108398; Abcam, Cambridge, MA, USA), DNMT1 (dilution, 1:1,000; incubated overnight at 4°C; catalog no. ab13537; Abcam) and GAPDH (dilution, 1:5,000; incubated overnight at 4°C; cat. no. 5174; Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Then, the membrane was washed for 5 min 3 times with TBST. Subsequently, goat anti-rabbit immunoglobulin (for ER, cat. no. ab150077; Alexa Fluor; Abcam) and goat anti-mouse immunoglobulin (for DNMT1, cat. no. ab150113; Alexa Fluor; Abcam) were added at a 1:5,000 dilution and incubated with the membrane at room temperature for 30 min.
Prediction of miRNA-148a targets
TargetScan 5.1 online software (http://www.targetscan.org/; Whitehead Institute for Biomedical Research, Cambridge, MA, USA) was used to predict the target genes of miRNA-148a.
Statistical analysis
Student's t-test was used to determine statistical significance between the groups. SPSS version 17 (IBM Corp., Armonk, NY, USA) was used. P<0.05 was considered to indicate a statistically significant difference.
Results
miRNA-148a is expressed more highly in ER-α positive cells
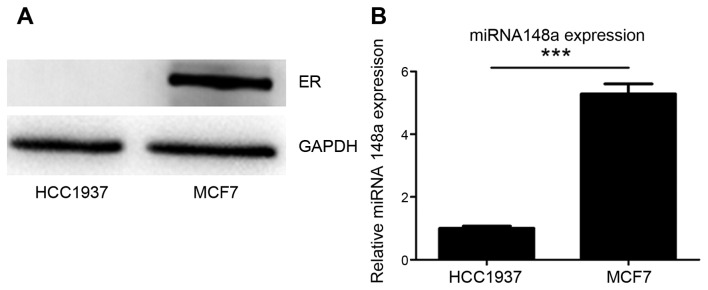
It was confirmed that ER-α expression was negative in HCC1937 cells and positive in MCF7 cells (Fig. 1A). It was also identified that miRNA-148a was more highly expressed in MCF7 cells than in HCC1937 cells (Fig. 1B). These data may indicate an association between ER and miRNA-148a expression in HCC1937 and MCF7 breast cancer cells.
Figure 1.
Expression of ER-α and miRNA-148a in breast cancer cells. (A) ER-α expression was not detected in HCC1937 cells, whereas ER-α was highly expressed in MCF7 cells. (B) The expression of miRNA-148a in MCF7 cells was 5 times higher than HCC1937 cells. ***P<0.001. ER-α, estrogen receptor-α; miRNA, microRNA.
DNMT1 is a target of miRNA-148a
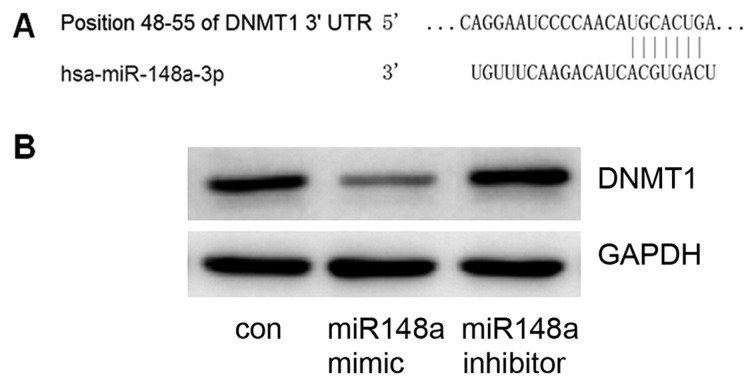
TargetScan was used to predict miRNA-148a target genes; the DNMT1 gene 3′UTR included 7 sequential pairing bases with the 5′ region of miRNA-148a (Fig. 2A), indicating that DNMT1 is a potential target for miRNA-148a. To confirm this prediction, an miRNA-148a mimic or inhibitor was transfected into MCF7 cells. It was confirmed that the miRNA-148a mimic could decrease DNMT1 expression and that the miRNA-148a inhibitor could increase DNMT1 expression (Fig. 2B). These data suggest that DNMT1 is a target gene of miRNA-148a.
Figure 2.
DNMT1 is a target of miRNA-148a. (A) TargetScan was used to predict miRNA-148a target genes. The DNMT1 gene 3′UTR included 7 sequential pairing bases with the 5′ of miRNR-148a, indicating that DNMT1 may be a potential target of miRNA-148a. (B) In order to confirm that DNMT1 is a target, an miRNA-148a mimic or inhibitor was transfected into MCF7 cells. At 48 h after transfection, the detection of DNMT protein levels by western blot analysis indicated that the expression of DNMT1 decreased upon miRNA-148a mimic transfection and increased upon miRNA-148a inhibitor transfection. DNMT1, DNA methyltransferase 1; miRNA, micro RNA; 3′UTR, 3′ untranslated region.
ER-α expression was regulated by DNMT1
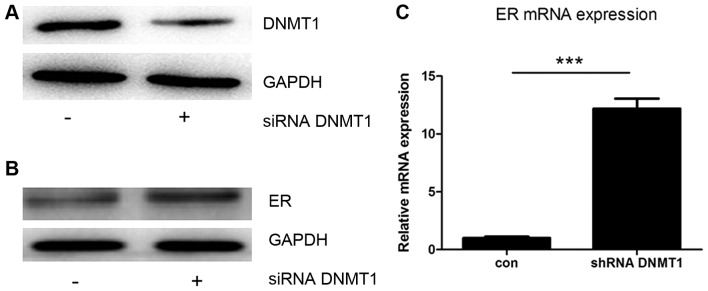
To confirm that DNMT1 can influence ER-α expression in MCF7 cells, siRNA against DNMT1 was used to knock down DNMT1 expression. Transfection with the siRNA caused a decrease in DNMT1 protein expression (Fig. 3A), and an increase in the protein and mRNA levels of ER-α (Fig. 3B and C). These data show that expression of the ER-α gene may be downregulated by the expression of DNMT1.
Figure 3.
DNMT1 regulated ER-α expression in MCF7 cells. (A) siRNA against DNMT1 was transfected into MCF7 cells and the expression of DNMT1 was reduced. At 48 h subsequent to siRNA transfection, the expression of ER-α increased at the (B) protein and (C) mRNA level. DNMT1, DNA methyltransferase 1; ER-α, estrogen receptor-α; siRNA, small interfering RNA. ***P<0.001 vs. control.
miRNA-148a influences ER-α expression
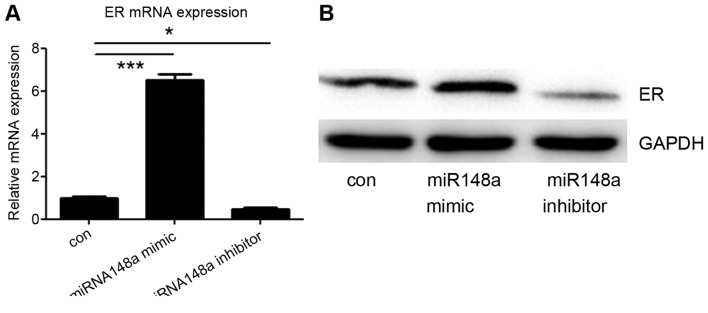
Finally, it was investigated whether miRNA-148a could influence the expression of ER-α in MCF7 cells. An miRNA-148a mimic or inhibitor was transfected into MCF7 cells, and the protein and mRNA levels of ER-α were detected. It was demonstrated that transfection with the miRNA-148a mimic increased ER-α expression, whereas the miRNA-148a inhibitor decreased ER-α expression, including at the mRNA and protein level (Fig. 4A and B). These data suggest that miRNA-148a enhances the expression of ER-α by targeting DNMT1.
Figure 4.
miRNA-148a regulated ER-α expression in MCF7 cells. miRNA-148a mimic and inhibitor were transfected to MCF7 cells. At 48 h after transfection, miRNA-148a mimic increased ER-α expression whereas the miRNA-148a inhibitor decreased ER-α expression. The finding was confirmed at the (A) mRNA and (B) protein level. *P<0.05, ***P<0.001. miRNA, micro RNA; ER-α, estrogen receptor-α.
Discussion
Although breast cancer is a heterogeneous disease, two-thirds of breast cancers share the common feature of being dependent on the presence and interaction of estrogen with the nuclear ER-α protein (2). Previous evidence has demonstrated that ER-α is upregulated in mammary epithelial cells during early stages of breast cancer, and that its overexpression is an important stimulatory factor for proliferation, eventually leading to tumor development (11). ER-α is an important therapeutic target in breast cancer treatment due to its role in orchestrating the expression of genes from growth-associated pathways (12).
There is a range of breast cancer cell lines available for research, including MCF-7, HCC1937, T47D, SK-BR3, MDA-MB-231, MDA-MB-435, HDQ-P1, MCF-10A, MCF-12A. Of these cell lines, MCF-7 and T47D are ER-α positive and the others, including HCC1937, are ER-α negative (13). A previous study investigated the differential expression of miRNAs in ER-α positive or negative cell lines and identified a number of differently expressed miRNAs, including miRNA-148a (14). In the present study, it was confirmed that miRNA-148a was more highly expressed in the ER-α positive MCF-7 cells than in the ER-α negative HCC1937 cells. It was then considered whether the elevated expression of miRNA-148a is associated with the ER-α expression status of MCF-7. A previous study reported that miRNA-152 caused the downregulation of DNMT1, a DNA methyltransferase, resulting in the demethylation of the ER-α gene in human aortic smooth muscle cells (15). As miRNA-152 and miRNA-148a belong to the same miRNA family and their sequences are similar (16), we hypothesized that miRNA-148a may also target DNMT1. This hypothesis was confirmed by the present study.
Methylation is a molecular modification of DNA that is associated with gene function. The concept that different cell types have different patterns of methylation was introduced more than three decades ago (17). A later study reported that DNA methylation patterns are different between tissue types, and between tumors and the surrounding tissue (18). In the context of breast cancer, a study identified that the number of CpG hypermethylated islands increased with the decreasing extent of tumor differentiation, indicating that broad DNA methylation signatures could be used for differentiating and staging breast cancer (19). The DNA methylation reaction is catalyzed by DNMTs including DNMT1, DNMT3a and DNMT3b. DNMT1 is the most abundant DNA methyltransferase in mammalian cells and the key maintenance enzyme for hemimethylated DNA during DNA replication, including for various types of cancer cell (20). Previous studies have demonstrated that miRNAs may affect the promoter methylation in CpG islands by targeting the 3′UTR of DNMTs (21–23). In the present study, it was identified that miRNA-148a targets DNMT1, which may then regulate ER-α expression. These results indicate a potential miRNA-based strategy to modulate ER-α expression in breast cancer cells.
Although the majority of breast cancers are ER-α positive, ER-α negative breast cancer remains a popular area of research in oncology. Triple-negative breast cancer (TNBC) is defined by its lack of ER-α and progesterone-receptor (PR) expression, along with the absence of human epidermal growth factor receptor 2 (HER2) overexpression or gene amplification (24). TNBC thus lacks a recognized target for molecular-oriented therapy. It is a biologically aggressive disease that is commonly associated with distant recurrence, visceral metastases and mortality compared with other breast cancer types (25). Although a number of novel targeted therapies on TNBC are in development, the effort is not producing the expected results (26). miRNAs have been investigated as biomarkers for the diagnosis and prognosis of cancer, and are potential therapeutic tools against breast cancer (27). The present study demonstrated that an miRNA may modulate ER-α expression in breast cancer cells. This provides a novel perspective for investigating the role of miRNAs in treating breast cancer.
In conclusion, it was identified in the present study that miRNA-148a may regulate ER-α expression by inhibiting DNMT1-mediated DNA methylation in breast cancer cells. This represents a potential miRNA-based strategy to modulate ER-α expression, and provides a novel perspective for investigating the role of miRNAs in treating breast cancer.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Giacinti L, Claudio PP, Lopez M, Giordano A. Epigenetic information and estrogen receptor alpha expression in breast cancer. Oncologist. 2006;11:1–8. doi: 10.1634/theoncologist.11-1-1. [DOI] [PubMed] [Google Scholar]
- 3.Cuzick J, Sestak I, Baum M, Buzdar A, Howell A, Dowsett M, Forbes JF. ATAC/LATTE investigators: Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 10-year analysis of the atac trial. Lancet Oncol. 2010;11:1135–1141. doi: 10.1016/S1470-2045(10)70257-6. [DOI] [PubMed] [Google Scholar]
- 4.Martínez-Galán J, Torres-Torres B, Núñez MI, López-Peñalver J, Del Moral R, De Almodóvar Ruiz JM, Menjón S, Concha A, Chamorro C, Ríos S, Delgado JR. Esr1 gene promoter region methylation in free circulating DNA and its correlation with estrogen receptor protein expression in tumor tissue in breast cancer patients. BMC Cancer. 2014;14:59. doi: 10.1186/1471-2407-14-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 7.Yahya SM, Elsayed GH. A summary for molecular regulations of miRNAs in breast cancer. Clin Biochem. 2015;48:388–396. doi: 10.1016/j.clinbiochem.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 8.Yan L, Nass SJ, Smith D, Nelson WG, Herman JG, Davidson NE. Specific inhibition of DNMT1 by antisense oligonucleotides induces re-expression of estrogen receptor-alpha (ER) in ER-negative human breast cancer cell lines. Cancer Biol Ther. 2003;2:552–556. doi: 10.4161/cbt.2.5.469. [DOI] [PubMed] [Google Scholar]
- 9.Xu Q, Jiang Y, Yin Y, Li Q, He J, Jing Y, Qi YT, Li W, Lu B, Peiper SS, et al. A regulatory circuit of mir-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J Mol Cell Biol. 2013;5:3–13. doi: 10.1093/jmcb/mjs049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 11.Huang B, Warner M, Gustafsson JÅ. Estrogen receptors in breast carcinogenesis and endocrine therapy. Mol Cell Endocrinol 418 Pt. 2015;3:240–244. doi: 10.1016/j.mce.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Shoker BS, Jarvis C, Clarke RB, Anderson E, Hewlett J, Davies MP, Sibson DR, Sloane JP. Estrogen receptor-positive proliferating cells in the normal and precancerous breast. Am J Pathol. 1999;155:1811–1815. doi: 10.1016/S0002-9440(10)65498-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Liu R, Fu P, Du F, Hong Y, Yao M, Zhang X, Zheng S. N1-guanyl-1,7-diaminoheptane sensitizes estrogen receptor negative breast cancer cells to doxorubicin by preventing epithelial-mesenchymal transition through inhibition of eukaryotic translation initiation factor 5A2 activation. Cell Physiol Biochem. 2015;36:2494–2503. doi: 10.1159/000430209. [DOI] [PubMed] [Google Scholar]
- 14.de Souza Rocha, Simonini P, Breiling A, Gupta N, Malekpour M, Youns M, Omranipour R, Malekpour F, Volinia S, Croce CM, Najmabadi H, et al. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res. 2010;70:9175–9184. doi: 10.1158/0008-5472.CAN-10-1318. [DOI] [PubMed] [Google Scholar]
- 15.Wang YS, Chou WW, Chen KC, Cheng HY, Lin RT, Juo SH. MicroRNA-152 mediates DNMT1-regulated DNA methylation in the estrogen receptor α gene. PLoS One. 2012;7:e30635. doi: 10.1371/journal.pone.0030635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Song YX, Wang ZN. The microRNA-148/152 family: Multi-faceted players. Mol Cancer. 2013;12:43. doi: 10.1186/1476-4598-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Razin A, Szyf M. DNA methylation patterns. Formation and function. Biochim Biophys Acta. 1984;782:331–342. doi: 10.1016/0167-4781(84)90043-5. [DOI] [PubMed] [Google Scholar]
- 18.Szyf M. DNA methylation signatures for breast cancer classification and prognosis. Genome Med. 2012;4:26. doi: 10.1186/gm325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan PS, Perry MR, Laux DE, Asare AL, Caldwell CW, Huang TH. CpG island arrays: An application toward deciphering epigenetic signatures of breast cancer. Clin Cancer Res. 2000;6:1432–1438. [PubMed] [Google Scholar]
- 20.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- 21.Chen KC, Wang YS, Hu CY, Chang WC, Liao YC, Dai CY, Juo SH. OxLDL up-regulates microRNA-29b, leading to epigenetic modifications of MMP-2/MMP-9 genes: A novel mechanism for cardiovascular diseases. FASEB J. 2011;25:1718–1728. doi: 10.1096/fj.10-174904. [DOI] [PubMed] [Google Scholar]
- 22.Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52:60–70. doi: 10.1002/hep.23660. [DOI] [PubMed] [Google Scholar]
- 23.Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology. 2010;51:881–890. doi: 10.1002/hep.23381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109:25–32. doi: 10.1002/cncr.22381. [DOI] [PubMed] [Google Scholar]
- 25.Anders CK, Carey LA. Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin Breast Cancer. 2009;9(Suppl 2):S73–S81. doi: 10.3816/CBC.2009.s.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomao F, Papa A, Zaccarelli E, Rossi L, Caruso D, Minozzi M, Vici P, Frati L, Tomao S. Triple-negative breast cancer: New perspectives for targeted therapies. Onco Targets Ther. 2015;8:177–193. doi: 10.2147/OTT.S67673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertoli G, Cava C, Castiglioni I. MicroRNAs: New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5:1122–1143. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]