Abstract
Stomach adenocarcinoma (STAD) is one of the leading causes of cancer morbidity and mortality worldwide. The present study aimed to identify the microRNAs associated with STAD survival time. The clinical information and microarray miRNA and mRNA expression profiles of STAD patients were downloaded from The Cancer Genome Atlas. Differential expression (DE) analysis was performed to identify DEmiRNAs and DEmRNAs in STAD. The DEmiRNAs associated with the survival time of patients with STAD were identified through DE analysis, negative correlation pair analysis, miRNA target gene prediction, univariate Cox regression analysis and Kaplan-Meier analysis. The functions of the target genes of the DEmiRNAs were predicted with Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. A total of 355 DEmiRNAs and 1,722 DEmRNAs were identified between STAD and normal tissues. A total of 5 DEmiRNAs were identified to be associated with STAD survival, including 4 risk-associated DEmiRNAs (miR-30a, −143, −145 and −133b) and 1 protective DEmiRNA (miR-135b). The target DEmRNAs were significantly enriched for DNA metabolic process in the biological process GO category, and in KEGG cell cycle signaling pathways. Kaplan-Meier curves indicated that the overall survival time in the miR-30a, −143, −145 and −133b high expression groups was significantly shorter than that in the low expression groups, whereas the survival time was prolonged in the miR-135b high expression group compared with that in the low expression group. Therefore, miR-30a, −143, −145, −133b and −135b may be involved in the tumorigenesis and development of STAD, and may be potential biomarkers for its early diagnosis and prognosis.
Keywords: stomach adenocarcinoma, survival, microRNA, expression profiling, The Cancer Genome Atlas
Introduction
Stomach carcinoma (SC) was the third leading cause of cancer-associated mortality in men and the fifth in women worldwide in 2012 (1). The incidence rate of SC in men is approximately twice as high as that in women. In general, incidence rates of SC are high in Eastern Asia (particularly in Korea, Mongolia, Japan and China) and low in Africa (1). In China, SC is the second and third most commonly diagnosed cancer in men and women, respectively, and is the second leading cause of cancer-associated mortality in men and women, ranking below only lung and bronchial cancer (2).
Stomach adenocarcinoma (STAD) accounts for >95% of all SC cases. Due to the atypical symptoms of the disease in its early stages, >80% of patients are clinically diagnosed at advanced stages, which is associated with a poor outcome (3). The 5-year survival rate for those diagnosed in the early stages of STAD is >90%, whereas for those diagnosed at an advanced stage, it is typically <5% (3,4).
To date, the major etiological factors of STAD have been reported as environmental factors and familial inheritance (5,6). Environmental factors include tobacco and alcohol consumption, and Helicobacter pylori (H. pylori) infection. H. pylori infection has been identified as the most pronounced risk factor for STAD; 90% of new cases of non-cardia STAD worldwide are attributed to chronic H. pylori infection, with a hazard ratio of 5.9 (7).
Familial inheritance of host factors is another reported risk factor for STAD, although it contributes to <10% of STAD cases (7). Mutations of stromelysin-1, Ras homolog family member A, DNA polymerase β and Notch 1 are associated with STAD tumorigenesis (8–11).
The molecular mechanisms of STAD development are unclear. Increasing evidence indicates that microRNAs (miRNAs or miRs) may contribute to the tumorigenesis and development of STAD. For example, the inhibited or unchanged expression of miR-486-5p compared with adjacent non-tumor tissues was previously identified to indicate a poor prognosis for patients with STAD (12); miR-15a and miR-16-1 suppress cell proliferation, monolayer colony formation, invasion and migration via targeting Yes-associated protein 1 in STAD (13); miR-200b-3p is associated with brain metastasis of STAD (14); and miR-107 is upregulated in gastric cancer, and the single nucleotide polymorphism rs2296616 in the promoter of miR-107 is associated with the increased survival of patients with STAD and a decreased STAD susceptibility (15).
In the present study, bioinformatics and univariate Cox regression analyses were applied to identify the miRNAs associated with the overall survival (OS) time of STAD patients, aiming to provide valuable information on potential biomarkers for the diagnosis, prognosis and therapy of STAD.
Materials and methods
Patients and samples
The data from a total of 443 patients with STAD were retrieved from The Cancer Genome Atlas (TCGA) data portal on the 16th of November 2015 (16). The exclusion criteria for patients were as follows: i) Patients had other history of malignancy; and ii) patients had incomplete follow-up records, with data on survival time unavailable. Overall, 423 patients with STAD were included in the study. The clinical records of the 423 patients, the corresponding level 3 raw data of miRNA and mRNA expression from tumors, and the normal control tissue data from the Illumina HiSeq_miRNAseq platform, were downloaded from the TCGA data portal. Clinical information included age, sex, ethnicity, tumor grade, history of other malignancy, survival status and follow-up data.
Identification of differentially expressed (DE) mRNAs and DEmiRNAs in STAD
mRNA expression data was available for 415 of the 423 patients with STAD and 35 of the control datasets. miRNA expression data was available for 400 of the 423 patients with STAD and 46 of the control datasets. mRNAs or miRNAs expressed in <20% of the patients were excluded from the expression dataset. The significance of the difference in the expression of mRNAs and miRNAs between STAD and normal controls was analyzed by the DESeq2 package in R version 3.2.1 (17). The threshold for DEmRNAs and DEmiRNAs was set at a false discovery rate (FDR) <0.001 and |log2 fold change| >1.5.
Pearson correlation coefficient (PCC) analysis between DEmRNAs and DEmiRNAs
In order to analyze the correlation between DEmRNAs and DEmiRNAs, PCCs (18) were calculated for each miRNA-mRNA interaction with a negative correlation through the cor.test function of R. PCC <-0.3 and P<0.05 were set as the thresholds for significant interactions between DEmiRNAs and DEmRNAs.
Target prediction for DEmiRNAs
To obtain the target genes for miRNAs, the DEmiRNAs were queried in the miRanda database (microRNA.org) of experimentally confirmed interactions between target genes and miRNAs. miRNA-target gene pairs obtained from PCC analysis in the present study and from the miRanda search were overlapped, and interactions that were not included in the aforementioned lists were screened.
Analysis of DEmiRNAs associated with survival time of STAD
The association between miRNA expression level and OS was performed using univariate Cox regression, with a threshold of P<0.01. A hazard ratio (HR) >1 indicates miRNAs negatively associated with survival in patients with STAD (i.e., higher expression=shorter OS time), whereas a HR <1 indicates miRNAs positively associated with survival in patients with STAD (i.e., higher expression=longer OS time).
miRNA-mRNA regulatory network
DEmiRNAs associated with STAD survival and the associated target genes were used to construct an miRNA-mRNA regulatory network with Cytoscape version 3.1.0 software (19).
Function and pathways enrichment of target genes of miRNAs
The underlying functions and pathways of the target genes of miRNAs were predicted by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses (20,21). FDR <0.05 was set as the cut-off for GO and KEGG analysis.
Kaplan-meier analysis of the association between 5 DEmiRNAs and survival time
Kaplan-Meier curves for OS analysis were produced to analyze the association between the respective expression level of 5 DEmiRNAs and OS time. The 400 patients with miRNA expression data from the TCGA dataset were segregated into high- and low-expression groups with the cut-off of the median miRNA expression level. Kaplan-Meier curves were drawn, and a log-rank test was performed to determine the survival difference between groups. P<0.05 was considered to indicate a significant difference. Kaplan-Meier and log-rank test analyses were performed by SPSS version 19.0 (IBM Corp., Armonk, NY, USA).
Results
Basic information of the study cohort
Of the 423 patients enrolled in the study, there were follow-up and miRNA data available for 312 patients. Table I shows the basic information of the study cohort.
Table I.
Basic information of the study cohort.
| Patient information | Data |
|---|---|
| Sex | |
| Male | 195 |
| Female | 117 |
| Age, years | |
| Mean | 64.5 |
| Median | 65 |
| Range | 30–90 |
| Ethnicity | |
| Asian | 82 |
| White | 204 |
| African | 6 |
| Tumor grade | |
| G1 | 5 |
| G2 | 107 |
| G3 | 193 |
| GX | 7 |
| Patient survival | |
| Yes | 220 |
| No | 92 |
| Follow-up time, days | |
| Mean | 651 |
| Median | 509 |
| Range | 0–3, 720 |
Identification of DEmRNAs in STAD
mRNA expression data from 415 STAD tissue samples and 35 non-tumor tissue samples were downloaded from TGCA (Table II). From the total 20,531 mRNAs, the mRNAs expressed in <20% of the patients were removed, to produce a list of 16,678 mRNAs, which subsequently underwent differential expression analysis. A total of 1,772 mRNAs, including 1,060 upregulated genes and 712 downregulated genes, were identified as DE based on the criteria of FDR <0.001 and |log2 fold change| >1.5. The top 10 upregulated and downregulated DEmRNAs in STAD are included in Table III.
Table II.
Basic information of mRNA and miRNA expression data.
| Data type | Platform | Genes | Cases | Controls |
|---|---|---|---|---|
| mRNA | Illumina HiSeq_RNASeqV2 | 20,531 | 415 | 35 |
| miRNA | Illumina HiSeq_miRNASeq | 1,046 | 400 | 46 |
miRNA, microRNA.
Table III.
The top 10 upregulated and downregulated differentially expressed mRNAs in stomach adenocarcinoma.
| A, Upregulated mRNAs | |||||
|---|---|---|---|---|---|
| Gene symbol | Gene name | Gene ID | log2 fold change | P-value | False discovery rate |
| FLG | Filaggrin | 2312 | 5.8937 | 1.86×10−47 | 1.44×10−44 |
| RBP2 | Retinol binding protein 2 | 5948 | 5.4607 | 3.51×10−25 | 1.75×10−23 |
| AQP10 | Aquaporin 10 | 89872 | 5.4469 | 2.55×10−32 | 3.28×10−30 |
| MEP1B | Meprin A subunit β | 4225 | 5.4444 | 8.12×10−29 | 6.53×10−27 |
| SYNPO2L | Synaptopodin 2-like | 79933 | 4.7689 | 9.87×10−48 | 8.23×10−45 |
| MAL | Mal T-cell differentiation protein | 4118 | 4.5980 | 8.80×10−22 | 3.06×10−20 |
| ALOX12 | Arachidonate 12-lipoxygenase, 12S type | 239 | 4.5728 | 2.61×10−37 | 6.21×10−35 |
| ENPP3 | Ectonucleotide pyrophosphatase/phosphodiesterase 3 | 5169 | 4.5383 | 1.52×10−34 | 2.43×10−32 |
| TGM3 | Transglutaminase 3 | 7053 | 4.4663 | 3.68×10−16 | 5.66×10−15 |
| ENDOU | Endonuclease, poly(U) specific | 8909 | 4.4587 | 9.78×10−19 | 2.17×10−17 |
| B, Downregulated mRNAs | |||||
| Gene symbol | Gene name | Gene ID | log2 fold change | P-value | False discovery rate |
| CST1 | Cystatin SN | 1469 | −7.4241 | 1.90×10−99 | 1.58×10−95 |
| COL10A1 | Collagen type-X α 1 chain | 1300 | −7.0398 | 5.07×10−104 | 8.45×10−100 |
| ALPP | Alkaline phosphatase, placental | 250 | −6.7971 | 1.35×10−42 | 6.25×10−40 |
| COL11A1 | Collagen type-XI α 1 chain | 1301 | −6.7034 | 2.41×10−70 | 1.00×10−66 |
| IBSP | Integrin-binding sialoprotein | 3381 | −6.3919 | 4.7×10−40 | 1.70×10−37 |
| HOXC10 | Homeobox C10 | 3226 | −6.3265 | 7.66×10−59 | 1.60×10−55 |
| BAAT | Bile acid-CoA:amino acid N-acyltransferase | 570 | −5.8683 | 1.30×10−43 | 6.97×10−41 |
| CST4 | Cystatin S | 1472 | −5.8488 | 5.52×10−42 | 2.36×10−39 |
| PRAME | Preferentially expressed antigen in melanoma | 23532 | −5.8100 | 3.13×10−37 | 7.30×10−35 |
| IGF2BP1 | Insulin like growth factor 2 mRNA binding protein 1 | 10642 | −5.4442 | 5.02×10−29 | 4.08×10−27 |
Identification of DEmiRNAs in STAD
miRNA expression data from 400 STAD tissue samples and 46 non-tumor tissues were downloaded (Table I). From 1,046 miRNAs, miRNAs expressed in <20% of the patients with STAD were removed, to give a total of 355 miRNAs that subsequently underwent differential expression analysis. A total of 43 miRNAs, including 15 upregulated miRNAs and 28 downregulated miRNAs, were identified as DE based on the criteria of FDR <0.001 and |log2 fold change| >1.5 (Table IV).
Table IV.
All identified differentially expressed miRNAs in stomach adenocarcinoma.
| A, Upregulated miRNAs | |||||
|---|---|---|---|---|---|
| miRNA | log2 fold change | P-value | False discovery rate | ||
| hsa-miR-490 | 3.1458 | 6.24×10−15 | 3.63×10−14 | ||
| hsa-miR-1–2 | 3.1339 | 2.77×10−26 | 6.15×10−25 | ||
| hsa-miR-133a-1 | 3.0433 | 7.51×10−26 | 1.57×10−24 | ||
| hsa-miR-133b | 2.7491 | 2.43×10−19 | 2.27×10−18 | ||
| hsa-miR-145 | 2.3263 | 4.71×10−28 | 1.19×10−26 | ||
| hsa-miR-139 | 2.3006 | 2.48×10−42 | 1.76×10−40 | ||
| hsa-miR-143 | 2.0146 | 2.32×10−21 | 3.05×10−20 | ||
| hsa-miR-551b | 2.0124 | 1.05×10−16 | 7.75×10−16 | ||
| hsa-miR-204 | 1.8210 | 4.64×10−10 | 1.62×10−9 | ||
| hsa-miR-203 | 1.6346 | 3.41×10−11 | 1.36×10−10 | ||
| hsa-miR-129–1 | 1.6208 | 3.33×10−9 | 9.92×10−9 | ||
| hsa-miR-195 | 1.5777 | 1.67×10−23 | 2.69×10−22 | ||
| hsa-miR-29c | 1.5755 | 2.27×10−26 | 5.38×10−25 | ||
| hsa-miR-30a | 1.5660 | 8.37×10−22 | 1.19×10−20 | ||
| hsa-miR-486 | 1.5314 | 3.65×10−15 | 2.27×10−14 | ||
| B, Downregulated miRNAs | |||||
| miRNA | log2 fold change | P-value | False discovery rate | ||
| hsa-miR-552 | −5.5020 | 3.04×10−39 | 1.35×10−37 | ||
| hsa-miR-196a-1 | −4.6379 | 5.6×10−70 | 9.93×10−68 | ||
| hsa-miR-196b | −4.3338 | 4.45×10−51 | 5.27×10−49 | ||
| hsa-miR-196a-2 | −4.1434 | 3.00×10−41 | 1.78×10−39 | ||
| hsa-miR-135b | −3.0610 | 1.16×10−43 | 1.03×10−41 | ||
| hsa-miR-935 | −2.6811 | 1.64×10−19 | 1.57×10−18 | ||
| hsa-miR-937 | −2.4858 | 7.67×10−20 | 7.78×10−19 | ||
| hsa-miR-615 | −2.3883 | 4.68×10−17 | 3.53×10−16 | ||
| hsa-miR-483 | −2.3147 | 1.84×10−11 | 7.61×10−11 | ||
| hsa-miR-3648 | −2.3098 | 5.98×10−15 | 3.54×10−14 | ||
| hsa-miR-18a | −2.0652 | 3.4×10−31 | 1.01×10−29 | ||
| hsa-miR-592 | −1.9724 | 6.98×10−16 | 4.76×10−15 | ||
| hsa-miR-21 | −1.8538 | 8.04×10−71 | 2.85×10−68 | ||
| hsa-miR-146b | −1.8327 | 4.90×10−40 | 2.48×10−38 | ||
| hsa-miR-508 | −1.8088 | 7.69×10−11 | 3.00×10−10 | ||
| hsa-miR-301b | −1.8030 | 2.04×10−12 | 9.79×10−12 | ||
| hsa-miR-1304 | −1.7771 | 4.30×10−10 | 1.51×10−9 | ||
| hsa-miR-877 | −1.7532 | 5.21×10−18 | 4.20×10−17 | ||
| hsa-miR-96 | −1.7472 | 2.16×10−20 | 2.39×10−19 | ||
| hsa-miR-183 | −1.7346 | 1.12×10−20 | 1.33×10−19 | ||
| hsa-miR-1254 | −1.7246 | 1.88×10−10 | 6.95×10−10 | ||
| hsa-miR-7–3 | −1.6924 | 7.52×10−12 | 3.22×10−11 | ||
| hsa-miR-1228 | −1.6516 | 1.42×10−12 | 6.91×10−12 | ||
| hsa-miR-3651 | −1.6116 | 1.74×10−9 | 5.57×10−9 | ||
| hsa-miR-181b-2 | −1.5918 | 3.05×10−17 | 2.36×10−16 | ||
| hsa-miR-501 | −1.5581 | 1.54×10−28 | 4.2×10−27 | ||
| hsa-miR-584 | −1.5446 | 1.64×10−14 | 9.12×10−14 | ||
| hsa-miR-335 | −1.5428 | 2.25×10−20 | 2.42×10−19 | ||
miRNA, microRNA; hsa-miR, Homo sapiens microRNA.
Negative correlation analysis between DEmiRNAs and DEmRNAs
The 1,722 DEmRNAs and 43 DEmiRNAs were subjected to a PCC correlation analysis. PCC <-0.3 and P<0.05 were set as the threshold values. A total of 3,115 significantly negatively correlated DEmiRNA-DEmRNA pairs, including 632 DEmRNAs and 30 DEmiRNAs, were identified (data not shown).
Target gene prediction of DEmiRNAs
To identify the targeted genes of DEmiRNAs, the 30 identified DEmiRNAs were queried in the miRanda database. A total of 14 of the miRNAs were available in the miRanda database. DEmiRNA-target genes predicted through miRanda database were overlapped with DEmiRNA-DEmRNA pairs calculated through PCC analysis to obtain 500 DEmiRNA-DEmRNA pairs, including 230 DEmRNAs and 14 DEmiRNAs. hsa-miR-96 and hsa-miR-183 had the highest connectivity with DEmRNAs, regulating 91 and 59 DEmiRNAs, respectively (data not shown).
Identification of miRNAs associated with survival time of STAD
To identify the miRNAs associated with the survival time of patients with STAD, the expression levels of 355 miRNAs and the corresponding survival times of 400 STAD patients were compared with univariate Cox regression analysis. A total of 57 miRNAs associated with survival time, including 55 risk-associated miRNAs and 2 protective miRNAs were identified, as included in Table V. The 57 miRNAs were overlapped with the 14 DEmiRNAs previously mentioned, and this identified 5 DEmiRNAs associated with STAD survival, including 4 risk-associated DEmiRNAs (miR-30a, −143, −145 and −133b) and 1 protective DEmiRNA (miR-135b), as described in Table VI.
Table V.
miRNAs associated with stomach adenocarcinoma survival.
| A, miRNAs negatively associated with patient survival time | |||||
|---|---|---|---|---|---|
| 95% confidence interval | |||||
| miRNA | Coefficient | Hazard ratio | Lower | Upper | P-value |
| hsa-miR-136 | 0.3317 | 1.3933 | 1.1717 | 1.6567 | 1.74×10−4 |
| hsa-miR-328 | 0.3140 | 1.3688 | 1.1778 | 1.5909 | 4.26×10−5 |
| hsa-miR-653 | 0.3128 | 1.3673 | 1.1988 | 1.5595 | 3.12×10−6 |
| hsa-miR-199a-1 | 0.2918 | 1.3388 | 1.1301 | 1.5861 | 7.41×10−4 |
| hsa-miR-199a-2 | 0.2911 | 1.3379 | 1.1263 | 1.5892 | 9.20×10−4 |
| hsa-miR-125a | 0.2880 | 1.3338 | 1.1241 | 1.5826 | 9.65×10−4 |
| hsa-miR-101–1 | 0.2878 | 1.3335 | 1.1009 | 1.6152 | 3.25×10−3 |
| hsa-miR-708 | 0.2869 | 1.3323 | 1.1441 | 1.5514 | 2.22×10−4 |
| hsa-miR-34b | 0.2837 | 1.3280 | 1.1216 | 1.5724 | 9.99×10−4 |
| hsa-miR-199b | 0.2830 | 1.3271 | 1.1154 | 1.5789 | 1.42×10−3 |
| hsa-miR-152 | 0.2807 | 1.3240 | 1.1111 | 1.5777 | 1.70×10−3 |
| hsa-miR-2355 | 0.2770 | 1.3192 | 1.0972 | 1.5861 | 3.21×10−3 |
| hsa-miR-181a-2 | 0.2734 | 1.3144 | 1.1177 | 1.5457 | 9.49×10−4 |
| hsa-miR-28 | 0.2721 | 1.3127 | 1.0948 | 1.5740 | 3.31×10−3 |
| hsa-miR-218–2 | 0.2719 | 1.3124 | 1.1535 | 1.4932 | 3.63×10−5 |
| hsa-miR-100 | 0.2663 | 1.3051 | 1.1539 | 1.4762 | 2.25×10−5 |
| hsa-miR-1271 | 0.2618 | 1.2993 | 1.0884 | 1.5511 | 3.77×10−3 |
| hsa-miR-497 | 0.2608 | 1.2980 | 1.1203 | 1.5039 | 5.16×10−4 |
| hsa-miR-125b-1 | 0.2604 | 1.2974 | 1.1446 | 1.4707 | 4.65×10−5 |
| hsa-miR-99b | 0.2550 | 1.2904 | 1.1082 | 1.5026 | 1.03×10−3 |
| hsa-miR-34c | 0.2535 | 1.2885 | 1.0955 | 1.5156 | 2.20×10−3 |
| hsa-miR-3605 | 0.2500 | 1.2841 | 1.0749 | 1.5340 | 5.85×10−3 |
| hsa-miR-331 | 0.2453 | 1.2780 | 1.0639 | 1.5353 | 8.76×10−3 |
| hsa-miR-887 | 0.2422 | 1.2741 | 1.0819 | 1.5004 | 3.69×10−3 |
| hsa-miR-217 | 0.2409 | 1.2724 | 1.1290 | 1.4340 | 7.88×10−5 |
| hsa-miR-433 | 0.2356 | 1.2657 | 1.0815 | 1.4813 | 3.32×10−3 |
| hsa-miR-758 | 0.2307 | 1.2595 | 1.0937 | 1.4505 | 1.36×10−3 |
| hsa-miR-214 | 0.2243 | 1.2514 | 1.0829 | 1.4462 | 2.38×10−3 |
| hsa-miR-539 | 0.2219 | 1.2484 | 1.0750 | 1.4498 | 3.64×10−3 |
| hsa-miR-130a | 0.2210 | 1.2473 | 1.0699 | 1.4542 | 4.76×10−3 |
| hsa-miR-127 | 0.2206 | 1.2468 | 1.0633 | 1.4619 | 6.61×10−3 |
| hsa-miR-409 | 0.2194 | 1.2453 | 1.0637 | 1.4580 | 6.38×10−3 |
| hsa-miR-216a | 0.2139 | 1.2385 | 1.0854 | 1.4133 | 1.49×10−3 |
| hsa-miR-487a | 0.2122 | 1.2364 | 1.0544 | 1.4498 | 9.00×10−3 |
| hsa-miR-370 | 0.2110 | 1.2349 | 1.0535 | 1.4475 | 9.24×10−3 |
| hsa-miR-99a | 0.2098 | 1.2334 | 1.1090 | 1.3716 | 1.09×10−4 |
| hsa-let-7c | 0.2072 | 1.2302 | 1.0953 | 1.3818 | 4.74×10−4 |
| hsa-miR-337 | 0.2026 | 1.2246 | 1.0689 | 1.4029 | 3.49×10−3 |
| hsa-miR-145 | 0.1971 | 1.2179 | 1.0979 | 1.3509 | 1.94×10−4 |
| hsa-miR-134 | 0.1918 | 1.2114 | 1.0536 | 1.3929 | 7.06×10−3 |
| hsa-miR-30a | 0.1875 | 1.2062 | 1.0511 | 1.3842 | 7.60×10−3 |
| hsa-miR-654 | 0.1867 | 1.2053 | 1.0587 | 1.3721 | 4.77×10−3 |
| hsa-miR-125b-2 | 0.1856 | 1.2040 | 1.0740 | 1.3496 | 1.45×10−3 |
| hsa-miR-143 | 0.1843 | 1.2023 | 1.0633 | 1.3595 | 3.28×10−3 |
| hsa-miR-323b | 0.1701 | 1.1854 | 1.0573 | 1.3291 | 3.57×10−3 |
| hsa-miR-296 | 0.1654 | 1.1798 | 1.0531 | 1.3217 | 4.33×10−3 |
| hsa-miR-133b | 0.1506 | 1.1625 | 1.0688 | 1.2644 | 4.46×10−4 |
| hsa-miR-490 | 0.1483 | 1.1599 | 1.0836 | 1.2415 | 1.93×10−5 |
| hsa-miR-9–2 | 0.1392 | 1.1493 | 1.0433 | 1.2662 | 4.85×10−3 |
| hsa-miR-133a-1 | 0.1388 | 1.1489 | 1.0594 | 1.2460 | 7.91×10−4 |
| hsa-miR-9–1 | 0.1380 | 1.1480 | 1.0419 | 1.2649 | 5.28×10−3 |
| hsa-miR-1–2 | 0.1211 | 1.1287 | 1.0429 | 1.2216 | 2.70×10−3 |
| hsa-miR-1247 | 0.1147 | 1.1216 | 1.0297 | 1.2216 | 8.51×10−3 |
| hsa-miR-675 | 0.1144 | 1.1212 | 1.0390 | 1.2099 | 3.25×10−3 |
| hsa-miR-187 | 0.1136 | 1.1203 | 1.0398 | 1.2070 | 2.81×10−3 |
| B, miRNAs positively associated with patient survival time | |||||
| 95% confidence interval | |||||
| miRNA | Coefficient | Hazard ratio | Lower | Upper | P-value |
| hsa-miR-135b | −0.1446 | 0.8653 | 0.7821 | 0.9574 | 5.06×10−3 |
| hsa-miR-338 | −0.1892 | 0.8276 | 0.7190 | 0.9527 | 8.42×10−3 |
miRNA, microRNA; hsa-miR, Homo sapiens microRNA.
Table VI.
Connectivity of five differentially expressed miRNAs associated with stomach adenocarcinoma survival time.
| 95% confidence interval | ||||||
|---|---|---|---|---|---|---|
| miRNA | Coefficient | Hazard ratio | Lower | Upper | P-value | Connectivity |
| hsa-miR-30a | 0.1875 | 1.2062 | 1.0511 | 1.3842 | 7.60×10−3 | 47 |
| hsa-miR-143 | 0.1843 | 1.2023 | 1.0633 | 1.3595 | 3.28×10−3 | 41 |
| hsa-miR-145 | 0.1971 | 1.2179 | 1.0979 | 1.3509 | 1.94×10−4 | 36 |
| hsa-miR-133b | 0.1506 | 1.1625 | 1.0688 | 1.2644 | 4.46×10−4 | 20 |
| hsa-miR-135b | −0.1446 | 0.8653 | 0.7821 | 0.9574 | 5.06×10−3 | 7 |
miRNA, microRNA; hsa-miR, Homo sapiens microRNA.
Regulatory network construction between DEmiRNAs associated with STAD survival and targeted DEmRNAs
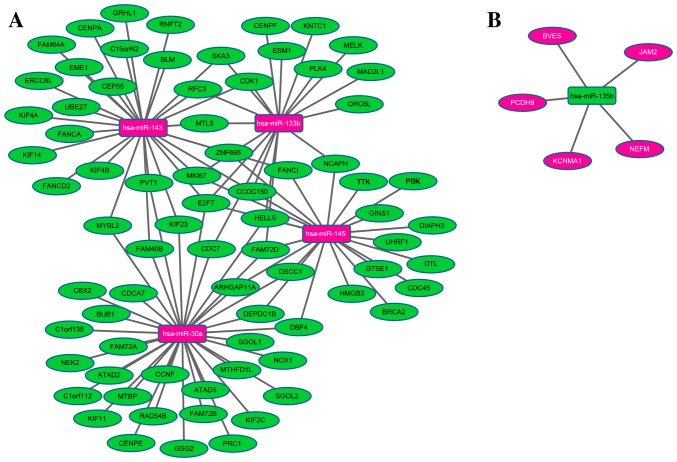
A regulatory network was constructed of the DEmiRNAs associated with STAD survival and their DEmRNA targets. The network comprised a total of 85 nodes and 107 edges. miR-30a, −143, −145, −133b and −135b regulated 34, 28, 22, 18 and 5 DEmRNAs in STAD, respectively, as illustrated in Fig. 1.
Figure 1.
Interaction networks showing DEmiRNAs associated with STAD survival time and their target genes that were differentially expressed in the present study. Interaction networks of (A) the upregulated miRNAs and their downregulated targets, and (B) the downregulated miRNA and its upregulated targets are shown. The central rectangular nodes represent miRNAs, and circular nodes represent target genes; green nodes represent downregulation in STAD, whereas pink nodes represent upregulation. Lines indicate an interaction between the miRNAs and an mRNA. DEmiRNAs, differentially expressed microRNAs; STAD, stomach adenocarcinoma.
Function and pathways enrichment
To gain insight into the biological function and pathways of the target DEmRNAs of the 5 DEmiRNAs associated with STAD survival time, GO and KEGG enrichment analyses were performed. All 80 downregulated DEmRNAs were significantly enriched in the DNA metabolic process of the biological process category; no enrichment in items from the cellular component and molecular function GO categories was identified (Table VII). In addition, 79 of the 80 downregulated DEmRNAs (those with an FDR <0.05) were significantly enriched in 4 KEGG signaling pathways, including Cell cycle, Homologous recombination, Progesterone-mediated oocyte maturation and Oocyte meiosis, as described in Table VII.
Table VII.
Function and pathway enrichment of targeted differentially expressed mRNAs of 5 differentially expressed microRNAs associated with stomach adenocarcinoma survival.
| ID | Terms | P-value | False discovery rate | Associated genes |
|---|---|---|---|---|
| Kyoto encyclopedia of genes and genomes pathway | ||||
| hsa04110 | Cell cycle | 1.53×10−8 | 6.12×10−8 | CDC7, MAD2L1, DBF4, CDC45, TTK, BUB1, CDK1 |
| hsa03440 | Homologous recombination | 3.79×10−7 | 7.58×10−7 | RAD54B, EME1, BRCA2, BLM |
| hsa04914 | Progesterone-mediated oocyte maturation | 1.77×10−5 | 2.36×10−5 | MAD2L1, BUB1, CDK1 |
| hsa04114 | Oocyte meiosis | 1.26×10−4 | 1.26×10−4 | SGOL1, MAD2L1, BUB1, CDK1 |
| Gene Ontology terms biological process | ||||
| GO:0006259 | DNA metabolic process | 1.68×10−4 | 2.14×10−1 | RFC3, CDK1, UBE2T, BRCA2, CENPF, BLM, HMGB3, GINS1, FANCA, UHRF1, DTL, HELLS, FANCD2, ORC6, DBF4, MKI67, CDC45, DSCC1, EME1, TICRR, FANCI, CDC7 |
OS analysis in STAD patients by Kaplan-Meier
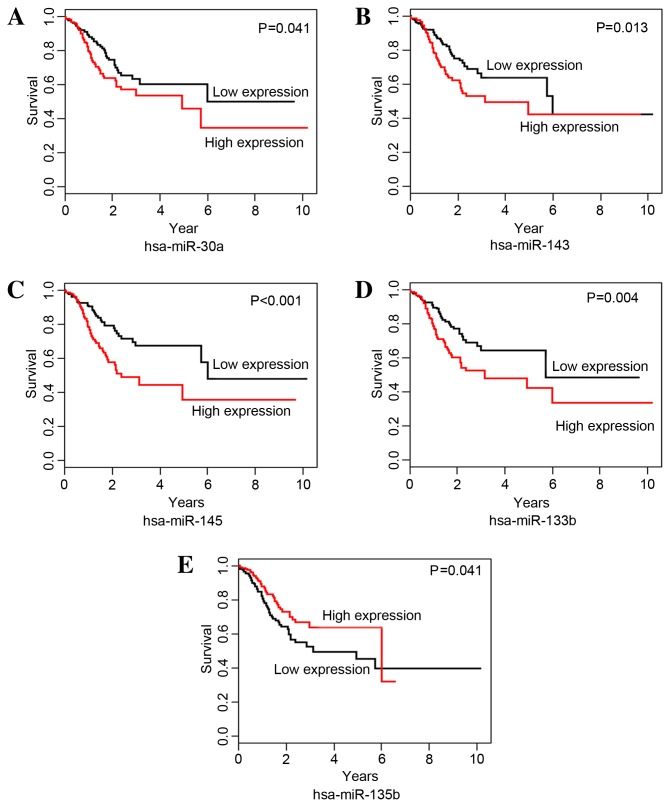
The correlations between DEmiRNAs and STAD survival time were analyzed with Kaplan-Meier curves. miR-30a, −143, −145 and −133b were identified as risk-associated DEmiRNAs; the OS time of the high expression group was significantly shorter than that of the low expression group for miR-30a (P=0.041; Fig. 2A), −143 (P=0.013; Fig. 2B), −145 (P=0.001; Fig. 2C) and −133b (P=0.004; Fig. 2D). hsa-miR-135b was identified as a protective DEmiRNA for STAD survival; the OS time for the high expression group was significantly higher than that of the low expression group for hsa-miR-135b (P=0.041; Fig. 2E).
Figure 2.
Kaplan-Meier survival curves for the association between the expression levels of differentially microRNAs and the overall survival time of patients with stomach adenocarcinoma. Analyses of the associations between expression levels of (A) hsa-miR-30a, (B) hsa-miR-143, (C) hsa-miR-145, (D) hsa-miR-133b and (E) hsa-miR-135b and overall survival time are shown. hsa-miR, Homo sapiens microRNA.
Discussion
In the present study, 5 DEmiRNAs associated with the survival time of patients with STAD were identified, including 4 risk-associated DEmiRNAs (miR-30a, −143, −145 and −133b) and 1 protective DEmiRNA (miR-135b).
miR-30a was upregulated in STAD tumor tissue and was identified as a risk-associated DEmiRNA with regard to survival time. miR-30a targeted 34 DEmRNAs. It was previously reported that miR-30a is overexpressed in a number of types of tumor, including glioma, papillary thyroid carcinoma (PTC), pancreatic cancer, nasopharyngeal carcinoma and variant serous adenocarcinoma (VSAD) (22–27). miR-30a-5p expression is induced by the Wnt/β-catenin pathway and promotes glioma cell invasion by inhibiting the expression of neural cell adhesion molecules (22). miR-30a-5p was significantly increased in serum samples and fine-needle aspiration biopsy samples from a group of patients with malignant PTC compared with patients with benign nodular goiters (23). miR-30a-5p was also demonstrated to be upregulated in the urine of patients with VSAD, and in VSAD tumor tissues and cell lines; following the surgical removal of VSAD, urinary miR-30a was distinctly reduced (24). miR-30a upregulation in stomach signet-ring cell carcinoma compared with stomach tubular adenocarcinoma has also been observed (25). Furthermore, the high expression of the miR-30 family, including miR-30a, −30b or −30c, promotes CD133+ pancreatic cancer cell migratory and invasive abilities, and is associated with resistance to gemcitabine (26). Overexpression of miR-30a also promotes the metastatic and invasive abilities of nasopharyngeal carcinoma tumor cells in vivo and in vitro (27). When taken together, the results of these previous studies and the present study may indicate that miR-30a serves a key role in the tumorigenesis and development of STAD. However, the molecular mechanism of miR-30a in STAD requires further investigation.
In the present study, miR-143 was identified as an upregulated DEmiRNA in STAD. This was consistent with previous reports that miR-143 is upregulated in STAD compared with non-tumor tissue (28). However, other studies have indicated that miR-143 is downregulated in gastric cancer tumor tissues and cell lines, and the dysregulation of miR-143 has been associated with cisplatin resistance, cell growth and lymph node metastasis (29–31). Our future study will aim to validate the expression of miR-143 in patients with STAD with a large sample size of STAD tumor and adjacent non-tumor specimens.
In the present study, miR-135b was downregulated in STAD and identified as a protective DEmiRNA associated with the survival time of patients with STAD. miR-135b in gastric adenocarcinoma samples was previously demonstrated to be downregulated compared with non-atrophic chronic gastritis and intestinal metaplasia samples (32). In addition, miR-135b was observed to be commonly downregulated in glioblastoma stem-like cells (GSCs), and the restoration of miR-135b expression significantly inhibited the proliferation, migration and clonogenic abilities of GSCs (33). It has also been demonstrated that miR-135b overexpression enhances the radio-resistance of U87 cells, a radio-resistant human glioblastoma cell line, by targeting glycogen synthase kinase 3β, whereas miR-135b knockdown reverses the radio-resistance of U87R cells (34). miR-135b is also downregulated in osteosarcoma tissues and cell lines, and the inhibition of miR-135b expression promotes cell proliferation, migration and invasion via the increased expression of c-Myc (35). To the best of our knowledge, the biological function of miR-135b in STAD has not been identified. The mechanism by which miR-135b serves a protective function in the pathogenesis of STAD requires further exploration.
It has been reported that miR-145 and −133b are downregulated in gastric cancer compared with adjacent non-tumor tissue (36,37). However, the opposite result was observed in the present study, as miR-145 and −133b were upregulated in STAD tissue. The abnormal expression of miR-145 and −133b has been associated with cell cycle dysregulation and tumor migration, invasion and metastasis in STAD (38–40). The expression levels of miR-145 and −133b in STAD compared with paired non-tumor controls requires verification with a large sample size in future studies.
In conclusion, 5 potential miRNAs associated with the survival time of patients with STAD were identified through the analysis of microarray expression data from TCGA database. The study had certain limitations. Firstly, the expression levels of miR-30a, −143, −145, −133b and −135b must be further verified in STAD tissue with reverse transcription-quantitative polymerase chain reaction. Secondly, DEmiRNAs associated with STAD survival were identified, but no prognosis model for STAD survival time was established. In our further study, the expression of the 5 identified DEmiRNAs in STAD tissues will be verified with a larger sample size, a prognosis model will be constructed to predict the survival time of patients with STAD based on the survival-associated DEmiRNAs identified in the present study, and the model will be further verified through clinical observation.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network: Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ren G, Cai R, Zhang WJ, Ou JM, Jin YN, Li WH. Prediction of risk factors for lymph node metastasis in early gastric cancer. World J Gastroenterol. 2013;19:3096–3107. doi: 10.3748/wjg.v19.i20.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lagergren J, Andersson G, Talbäck M, Drefahl S, Bihagen E, Härkönen J, Feychting M, Ljung R. Marital status, education, and income in relation to the risk of esophageal and gastric cancer by histological type and site. Cancer. 2016;122:207–212. doi: 10.1002/cncr.29731. [DOI] [PubMed] [Google Scholar]
- 6.Brosens LA, Wood LD, Offerhaus GJ, Arnold CA, Lam-Himlin D, Giardiello FM, Montgomery EA. Pathology and genetics of syndromic gastric polyps. Int J Surg Pathol. 2016;24:185–199. doi: 10.1177/1066896915620013. [DOI] [PubMed] [Google Scholar]
- 7.Plummer M, Franceschi S, Vignat J, Forman D, de Martel C. Global burden of gastric cancer attributable to Helicobacter pylori. Int J Cancer. 2015;136:487–490. doi: 10.1002/ijc.28999. [DOI] [PubMed] [Google Scholar]
- 8.Krishnaveni D, Bhayal AC, Shravan KP, Jyothy A, Pratibha N, Venkateshwari A. Heterozygosity of stromelysin-1 (rs3025058) promoter polymorphism is associated with gastric cancer. Indian J Cancer. 2015;52:251–254. doi: 10.4103/0019-509X.175806. [DOI] [PubMed] [Google Scholar]
- 9.Thutkawkorapin J, Picelli S, Kontham V, Liu T, Nilsson D, Lindblom A. Exome sequencing in one family with gastric- and rectal cancer. BMC Genet. 2016;17:41. doi: 10.1186/s12863-016-0351-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ushiku T, Ishikawa S, Kakiuchi M, Tanaka A, Katoh H, Aburatani H, Lauwers GY, Fukayama M. RHOA mutation in diffuse-type gastric cancer: A comparative clinicopathology analysis of 87 cases. Gastric Cancer. 2016;19:403–411. doi: 10.1007/s10120-015-0493-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan X, Wang H, Luo G, Ren S, Li W, Cui J, Gill HS, Fu SW, Lu Y. Clinical significance of a point mutation in DNA polymerase beta (POLB) gene in gastric cancer. Int J Biol Sci. 2015;11:144–155. doi: 10.7150/ijbs.10692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, Ren C, Han C, Wang D, Chen Y, Fu D. Expression and prognostic value of miR-486-5p in patients with gastric adenocarcinoma. PLoS One. 2015;10:e0119384. doi: 10.1371/journal.pone.0119384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang W, Tong JH, Lung RW, Dong Y, Zhao J, Liang Q, Zhang L, Pan Y, Yang W, Pang JC, et al. Targeting of YAP1 by microRNA-15a and microRNA-16-1 exerts tumor suppressor function in gastric adenocarcinoma. Mol Cancer. 2015;14:52. doi: 10.1186/s12943-015-0323-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minn YK, Lee DH, Hyung WJ, Kim JE, Choi J, Yang SH, Song H, Lim BJ, Kim SH. MicroRNA-200 family members and ZEB2 are associated with brain metastasis in gastric adenocarcinoma. Int J Oncol. 2014;45:2403–2410. doi: 10.3892/ijo.2014.2680. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Lv C, Jin H, Xu M, Kang M, Chu H, Tong N, Wu D, Zhu H, Gong W, et al. A common genetic variation in the promoter of miR-107 is associated with gastric adenocarcinoma susceptibility and survival. Mutat Res. 2014;769:35–41. doi: 10.1016/j.mrfmmm.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Tomczak K, Czerwińska P, Wiznerowicz M. The cancer genome atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benesty J, Chen J, Huang Y, Cohen I. Noise Reduction in Speech Processing. 1st. Vol. 2. Springer; Heidelberg: 2009. Pearson correlation coefficient; pp. 1–4. [DOI] [Google Scholar]
- 19.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashimoto K, Goto S, Kawano S, Aoki-Kinoshita KF, Ueda N, Hamajima M, Kawasaki T, Kanehisa M. KEGG as a glycome informatics resource. Glycobiology. 2006;16:63R–70R. doi: 10.1093/glycob/cwj010. [DOI] [PubMed] [Google Scholar]
- 21.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Dai X, Chen Y, Sun C, Zhu Q, Zhao H, Liu G, Huang Q, Lan Q. MiR-30a-5p is induced by Wnt/β-catenin pathway and promotes glioma cell invasion by repressing NCAM. Biochem Biophys Res Commun. 2015;465:374–380. doi: 10.1016/j.bbrc.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Igci YZ, Ozkaya M, Korkmaz H, Bozgeyik E, Bayraktar R, Ulasli M, Erkilic S, Eraydin A, Oztuzcu S. Expression levels of miR-30a-5p in papillary thyroid carcinoma: A comparison between serum and fine needle aspiration biopsy samples. Genet Test Mol Biomarkers. 2015;19:418–423. doi: 10.1089/gtmb.2015.0062. [DOI] [PubMed] [Google Scholar]
- 24.Zhou J, Gong G, Tan H, Dai F, Zhu X, Chen Y, Wang J, Liu Y, Chen P, Wu X, Wen J. Urinary microRNA-30a-5p is a potential biomarker for ovarian serous adenocarcinoma. Oncol Rep. 2015;33:2915–2923. doi: 10.3892/or.2015.3937. [DOI] [PubMed] [Google Scholar]
- 25.Li FQ, Xu B, Wu YJ, Yang ZL, Qian JJ. Differential microRNA expression in signet-ring cell carcinoma compared with tubular adenocarcinoma of human gastric cancer. Genet Mol Res. 2015;14:739–747. doi: 10.4238/2015.January.30.17. [DOI] [PubMed] [Google Scholar]
- 26.Tsukasa K, Ding Q, Miyazaki Y, Matsubara S, Natsugoe S, Takao S. miR-30 family promotes migratory and invasive abilities in CD133(+) pancreatic cancer stem-like cells. Human Cell. 2016;29:130–137. doi: 10.1007/s13577-016-0137-7. [DOI] [PubMed] [Google Scholar]
- 27.Wang HY, Li YY, Fu S, Wang XP, Huang MY, Zhang X, Shao Q, Deng L, Zeng MS, Zeng YX, Shao JY. MicroRNA-30a promotes invasiveness and metastasis in vitro and in vivo through epithelial-mesenchymal transition and results in poor survival of nasopharyngeal carcinoma patients. Exp Biol Med (Maywood) 2014;239:891–898. doi: 10.1177/1535370214532758. [DOI] [PubMed] [Google Scholar]
- 28.Stánitz E, Juhász K, Tóth C, Gombos K, Natali PG, Ember I. Evaluation of MicroRNA expression pattern of gastric adenocarcinoma associated with socioeconomic, environmental and lifestyle factors in northwestern Hungary. Anticancer Res. 2013;33:3195–3200. [PubMed] [Google Scholar]
- 29.Zhuang M, Shi Q, Zhang X, Ding Y, Shan L, Shan X, Qian J, Zhou X, Huang Z, Zhu W, et al. Involvement of miR-143 in cisplatin resistance of gastric cancer cells via targeting IGF1R and BCL2. Tumour Biol. 2015;36:2737–2745. doi: 10.1007/s13277-014-2898-5. [DOI] [PubMed] [Google Scholar]
- 30.Wu XL, Cheng B, Li PY, Huang HJ, Zhao Q, Dan ZL, Tian DA, Zhang P. MicroRNA-143 suppresses gastric cancer cell growth and induces apoptosis by targeting COX-2. World J Gastroenterol. 2013;19:7758–7765. doi: 10.3748/wjg.v19.i43.7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu WY, Xue XY, Chen ZJ, Han SL, Huang YP, Zhang LF, Zhu GB, Shen X. Potentially predictive microRNAs of gastric cancer with metastasis to lymph node. World J Gastroenterol. 2011;17:3645–3651. doi: 10.3748/wjg.v17.i31.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vidal AF, Cruz AMP, Magalhães L, Pereira AL, Anaissi AK, Alves NC, Albuquerque PJ, Burbano RM, Demachki S, Ribeiro-dos-Santos Â. Hsa-miR-29c and hsa-miR-135b differential expression as potential biomarker of gastric carcinogenesis. World J Gastroenterol. 2016;22:2060–2070. doi: 10.3748/wjg.v22.i6.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lulli V, Buccarelli M, Martini M, Signore M, Biffoni M, Giannetti S, Morgante L, Marziali G, Ilari R, Pagliuca A, et al. miR-135b suppresses tumorigenesis in glioblastoma stem-like cells impairing proliferation, migration and self-renewal. Oncotarget. 2015;6:37241–37256. doi: 10.18632/oncotarget.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao S, Yang Z, Lv R, Zhao J, Wu M, Liao Y, Liu Q. miR-135b contributes to the radioresistance by targeting GSK3β in human glioblastoma multiforme cells. PLoS One. 2014;9:e108810. doi: 10.1371/journal.pone.0108810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z, Zhang G, Li J, Liu J, Lv P. The tumor-suppressive microRNA-135b targets c-myc in osteoscarcoma. PLoS One. 2014;9:e102621. doi: 10.1371/journal.pone.0102621. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Khalili M, Vasei M, Khalili D, Alimoghaddam K, Sadeghizadeh M, Mowla SJ. Downregulation of the genes involved in reprogramming (SOX2, c-MYC, miR-302, miR-145 and P21) in gastric adenocarcinoma. J Gastrointest Cancer. 2015;46:251–258. doi: 10.1007/s12029-015-9695-2. [DOI] [PubMed] [Google Scholar]
- 37.Guo L, Bai H, Zou D, Hong T, Liu J, Huang J, He P, Zhou Q, He J. The role of microRNA-133b and its target gene FSCN1 in gastric cancer. J Exp Clin Cancer Res. 2014;33:99. doi: 10.1186/s13046-014-0099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu T, Zhou X, Wang J, Du Y, Xu J, Huang Z, Zhu W, Shu Y, Liu P. MiR-145, miR-133a and miR-133b inhibit proliferation, migration, invasion and cell cycle progression via targeting transcription factor Sp1 in gastric cancer. FEBS Lett. 2014;588:1168–1177. doi: 10.1016/j.febslet.2014.02.054. [DOI] [PubMed] [Google Scholar]
- 39.Chen JJ, Cai WY, Liu XW, Luo QC, Chen G, Huang WF, Li N, Cai JC. Reverse correlation between MicroRNA-145 and FSCN1 affecting gastric cancer migration and invasion. PLoS One. 2015;10:e0126890. doi: 10.1371/journal.pone.0126890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Zhang X, Zhang Y, Hu Z, Yang D, Wang C, Guo M, Cai Q. Identification of miRNomes in human stomach and gastric carcinoma reveals miR-133b/a-3p as therapeutic target for gastric cancer. Cancer Lett. 2015;369:58–66. doi: 10.1016/j.canlet.2015.06.028. [DOI] [PubMed] [Google Scholar]