Abstract
Carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1), a cell-cell adhesion molecule, has been revealed to perform an important role in tumor progression. Although there are a number of studies on CEACAM1 in patients with breast cancer, there is limited information on the roles of CEACAM1 in breast cancer metastasis. The present study aimed to identify whether CEACAM1 is involved in breast cancer development and to investigate the underlying mechanisms. First, the expression of CEACAM1 was observed in patients with breast cancer, and the association between CEACAM1 expression levels and migration and invasion of breast cancer cells was analyzed. As there are 12 isoforms of CEACAM1, of which CEACAM1-4S dominates in the human breast epithelium, subsequent study focused on CEACAM1-4S as a representative of all the isoforms. Results of the present study demonstrated that CEACAM1-4S suppresses breast cancer cell invasion and migration in a manner that is dependent on the balance between matrix metalloproteinase 2/tissue inhibitor of metalloproteinase 2 and E-/N-cadherin expression. In addition, CEACAM1-4S was likely to cause reversal of epithelial-mesenchymal transition of breast cancer cells through repressing Smad2 and signal transducer and phosphorylation of activator of transcription 3. In conclusion, the present study demonstrated that CEACAM1-4S performs an inhibitory role in breast cancer metastasis, and restoring CEACAM1-4S expression may provide a novel strategy for therapy of patients with metastatic breast cancer.
Keywords: carcinoembryonic antigen-related cell adhesion molecule 1-4S, breast cancer, invasion, migration, balanced regulation, epithelial-mesenchymal transition
Introduction
Breast cancer is one of the leading causes of cancer-associated mortality in females worldwide (1). Breast cancer mortality is largely due to metastasis, which begins with the invasion and migration of cells into the surrounding tissues and vasculature (2). To date, the molecular mechanisms underlying breast cancer metastasis are poorly understood. Carcinoembryonic antigen (CEA)-related cell adhesion molecule 1 (CEACAM1), formerly termed BGP, C-CAM or CD66a, is a member of the CEA family, which belongs to the immunoglobulin superfamily (3). CEACAM1 is a multifunctional molecule with a broad distribution in human epithelia, endothelia and hematopoietic cells (4). A growing body of evidence suggests that CEACAM1 expression is altered in numerous types of cancer. Downregulation of CEACAM1 expression has been reported in various types of malignancies, including colorectal cancer (5), hepatocellular cancer (6), renal carcinoma (7) and prostate cancer (8). Based on these findings, a tumor suppressive role for CEACAM1 was postulated (9,10). Several studies have reported a decreased expression of CEACAM1 in breast cancer (11–13). In addition, CEACAM1 was revealed to have an essential role in human mammary gland morphogenesis (14). Notably, CEACAM1-4S, a major isoform of CEAMCA1, can revert breast cancer cells to a normal morphogenic phenotype (15). These findings indicate that CEACAM1 may perform a pivotal role in breast cancer initiation and progression. However, knowledge on the functions of CEACAM1 in breast cancer remains limited.
The present study aimed to reveal the functions of CEACAM1 in breast cancer. First, the pattern of expression of CEACAM1 was observed, and its association with cancer metastasis was evaluated. As CEACAM1-4S is the main CEACAM1 isoforms and acts as the main modulator in cellular and molecular regulation in human breast epithelia (15), subsequent experiments concentrated on CEACAM1-4S to investigate the effect of CEACAM1-4S on the invasive behavior of breast cancer cells.
Materials and methods
Patients and specimens
A total of 62 breast cancer tissue samples were collected from patients who underwent surgical resection at Shanghai Jiao Tong University Affiliated Sixth People's Hospital (Shanghai, China) between October 2013 and February 2015. All patients were female, with a mean age of 56.6 years (range, 26–78 years). None of the enrolled patients had undergone chemotherapy or radiotherapy prior to surgery. The histological types were evaluated according to the World Health Organization criteria (16). The clinical information of the patients was obtained from medical records. All patients provided informed consent prior to involvement in the present study. The present study was approved by the Ethics Committee of Shanghai Jiao Tong University in accordance with The Declaration of Helsinki.
Cell lines
Human breast cancer cell lines with different metastatic potential (BT549, Hs578T, T-47D, MCF7, MDA-MB-231 and MDA-MB-468) were purchased from the Institute of Biochemistry and Cell Biology at the Chinese Academy of Science (Shanghai, China). MCF10A, a non-invasive human immortal mammary epithelial cell line derived from a patient with fibrocystic breast disease, was obtained from the American Type Culture Collection (Manassas, VA, USA). All cell lines were cultured according to the supplier's protocol. BT-549 and T-47D cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), Hs578T and MCF7 cells were cultured in Dulbecco's modified eagle's medium (Gibco; Thermo Fisher Scientific, Inc.), and MDA-MB-231 and MDA-MB-468 cells were cultured in Leibovitz's L-15 medium (Gibco; Thermo Fisher Scientific, Inc.). MCF10A cells was cultured in mammary epithelial growth media (Lonza Group, Ltd., Basel, Switzerland). All cell lines were cultured at 37°C in a humidified atmosphere containing 95% air and 5% CO2, and media were supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.).
Immunohistochemistry (IHC)
Immunohistochemical staining for CEACAM1 was performed on breast cancer tissue specimens. The tissue samples were sliced 4-µm thick and fixed in 4% (w/v) paraformaldehyde in 0.1 M PBS overnight at 4°C, dehydrated in increasing concentrations of ethanol, and then embedded in paraffin. Briefly, the paraffin-embedded tissue sections were de-waxed, rehydrated in descending concentrations of ethanol, and retrieved in a water bath at 95°C, followed by incubation overnight at 4°C with primary anti-CEACAM1 antibody (1:75; ab49510; Abcam, Cambridge, UK) in a humidified chamber. Next, the sections were incubated with a biotinylated secondary antibody (1:100; K4061; Dako; Agilent Technologies, Inc., Santa Clara, CA, USA) for 1 h at room temperature. Subsequent to rinsing in PBS, the streptavidin-peroxidase complex reagent [StrepABComplex/horseradish peroxidase (HRP) Duet; Dako; Agilent Technologies, Inc.] was added. Finally, sections were visualized with 3,3′-diaminobenzidine, followed by hematoxylin counterstaining for 5 min at room temperature. For negative controls, the primary antibody was omitted. The images were obtained using a microscope (Nikon Eclipse80i; Nikon Corporation, Tokyo, Japan) at ×200 magnification. The intensity of CEACAM1 staining was quantitatively evaluated using Image Pro-Plus 6.0 software (Media Cybernetics, Inc., Rockville, MD, USA).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Quantitative real-time polymerase chain reaction (qPCR) was performed using SYBR® Green Premix Ex Taq™ (Tli RNaseH Plus) kit (RR420A; Takara Bio Inc., Shiga, Japan). The cells were collected, and total RNA was isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The synthesis of complementary DNA was performed with the PrimeScript RT reagent kit (Takara Biotechnology, Shiga, Japan), according to the manufacturer's protocol. qPCR was performed on a thermal cycler ABI7500 (Applied Biosystems; Thermo Fisher Scientific, Inc.) using specific primers for CEACAM1-4S (NCBI GenBank accession no. NM_00104912) (forward, 5′-AAACCAGAGTCTCCCGTCCT-3′; reverse, 5′-TGGAGTGGTCCTGAGCTGCCG-3′). The amplification conditions were as follows: 95°C for 30 sec, 95°C for 5 sec and 64°C for 34 sec, for 40 cycles. In addition, melting curve was generated to validate the specificity of the analysis. For quantitation of CEACAM1-4S mRNA, the 2−ΔΔCq (−ΔCq=CqGAPDH-CqCEACAM1) method (17) was employed. The relative amount of CEACAM1-4S mRNA was calculated by normalization to the housekeeping gene GAPDH. The primer sequences for GAPDH were forward, 5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse, 5′-ATGGTGGTGAAGACGCCAGT-3′.
Cell transfection
Plasmids containing the CEACAM1-4S gene and green fluorescent protein (GFP) reporter gene were constructed by OriGene Technologies, Inc. (Rockville, MD, USA) as previously described by Oliveira-Ferrer et al (18). Empty-vector (PS100071; OriGene Technologies, Inc.) was used as a control. Cultured breast cancer BT549 or Hs578T cells were seeded in 6-well plates at 3×105 cells/well and transfected with 1.25 µg plasmid DNA using Opti-MEM Reduced Serum medium (Invitrogen; Thermo Fisher Scientific, Inc.) and Lipofectamine Plus reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The transfection efficiency was monitored using the GFP reporter gene. The result of the GFP-positive cells counting revealed transfection efficiency ranging between 50 and 75%. At 6 h post-transfection, the cells were washed and incubated in complete growth medium (RPMI-1640 medium for BT549 and Dulbecco's modified Eagle's medium for Hs578T). After 48 h, the cells were lysed for western blot analysis or used in subsequent experiments.
Western blot analysis
The cells were harvested using a curet and centrifuged at 3,500 × g for 10 min at 4°C and then lysed in ice-cold radioimmunoprecipitation assay lysis buffer (P0013B; Beyotime Institute of Biotechnology, Shanghai, China). Equal amounts of protein (30 µg) were separated by SDS-PAGE (10% gel) and subsequently transferred to a polyvinylidene difluoride membrane. Subsequent to blocking with 5% skimmed milk at room temperature for 1 h, the membranes were incubated at 4°C overnight with primary antibodies, including anti-CEACAM1 (1:1,000; ab49510; Abcam), anti-N-cadherin (1:500; sc8424; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), anti-E-cadherin (1:500; sc8426; Santa Cruz Biotechnology, Inc.), anti-vimentin (1:1,000; #3932; Cell Signaling Technology, Inc., Danvers, MA, USA), anti-signal transducer and activator of transcription 3 (STAT3; 1:1,000; #9139; Cell Signaling Technology, Inc.), anti-Smad2/3 (1:1,000; #8685; Cell Signaling Technology, Inc.), anti-phospho-STAT3 (Tyr705; 1:1,000; #4113; Cell Signaling Technology, Inc.), anti-phospho-Smad2 (Ser465/467; 1:1,000; #3108; Cell Signaling Technology, Inc.), anti-matrix metalloproteinase (MMP)2 (1:1,000; #4022; Cell Signaling Technology, Inc.), anti-MMP9 (1:1,000; #3852; Cell Signaling Technology, Inc.), anti-tissue inhibitor of metalloproteinase (TIMP)1 (1:1,000; #8946; Cell Signaling Technology, Inc.), anti-TIMP2 (1:1,000; #5738; Cell Signaling Technology, Inc.) and anti-GAPDH (1:1,000; ab9484; Abcam), followed by incubation at room temperature for 2 h with HRP-conjugated polyclonal secondary antibody (1:5,000; ab6789/ab6721; Abcam). All western blots were visualized using the enhanced plus chemiluminescence assay kit (EMD Millipore, Billerica, MA, USA), according to the manufacturer's protocol.
Transwell invasion assay
Cell invasion was investigated using Matrigel invasion chambers with a pore size of 8 mm (Costar; Corning Life Sciences, Cambridge, MA, USA). Briefly, BT549 or Hs578T cells (4×104 cells per chamber) in serum-free medium were seeded in the upper chamber, and 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific, Inc.) was used as a chemoattractant in the bottom well. After incubation for 24 h at 37°C, the non-invasive cells on the upper surface of the membrane were removed with a cotton swab, and the invasive cells on the bottom side were fixed in 100% methanol at room temperature for 5 min, stained with 1% crystal violet at room temperature for 10 min and counted using a microscope (Nikon Eclipse80i; Nikon Corporation) under ×100 magnification with five fields of view per sample.
Wound healing assay
Wound healing assays were performed to investigate the cell migration. BT549 or Hs578T cells were cultured on 6-well plates (3×105 cells/well) until 100% confluence was reached. The cultures were then scratched to form a wound line using a pipette tip. Subsequent to washing with PBS, the cells were cultured in media containing 1% FBS for an additional 24 h. The wound area was observed by inverted phase contrast microscopy (magnification, ×200; Nikon Corporation). The migration area was calculated by subtracting the unhealed area at 24 h from the initial wound area.
Immunofluorescent staining (IF)
For immunofluorescence, BT549 or Hs578T cells were seeded on coverslips (2×104 cells/well; 24-well plates). The next day, the cells were fixed with 100% cold methanol at room temperature for 10 min and subsequently blocked with 1% bovine serum albumin (ST023; Beyotime Institute of Biotechnology) in PBS at room temperature for 1 h, followed by overnight incubation at 4°C with the primary antibodies: Anti-E-cadherin (1:100; sc8426; Santa Cruz Biotechnology, Inc.) and anti-vimentin (1:200; #3932; Cell Signaling Technology, Inc.). Following washing using PBS three times, the cells were incubated at room temperature for 1 h with Alexa Fluor 594-conjugated secondary antibody (016580084; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) at a dilution of 1:400. The nuclei were stained with DAPI (Invitrogen; Thermo Fisher Scientific, Inc.) at room temperature for 10 min in the dark. Images were captured using an inverted fluorescence microscope (magnification, ×200; Nikon Corporation). For negative controls, the primary antibody incubation step was omitted. The Image Pro-Plus version 6.0 software (Media Cybernetics, Inc.) was used for imaging analysis.
Statistical analysis
All data are expressed as the mean ± standard deviation and represent at least three experiments unless stated otherwise. The statistical significance of the differences between two different groups was determined with Student's t-test or nonparametric Mann-Whitney test using SPSS19.0 statistics software (IBM SPSS, Armonk, NY, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
CEACAM1 expression is inversely associated with metastasis in patients with breast cancer
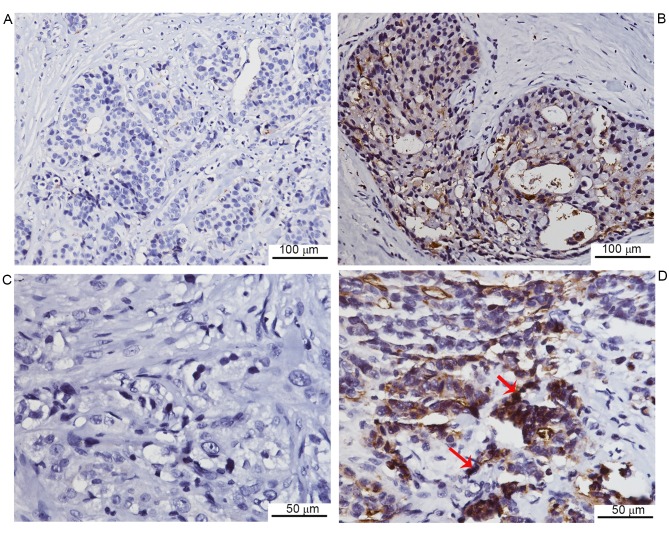
To investigate CEACAM1 expression in breast cancer tissues from patients, immunohistochemical staining was performed. Negative CEACAM1 expression was observed in 13 of 62 cases, while the remaining cases exhibited positive expression (Fig. 1). Furthermore, the CEACAM1 expression was quantitatively assessed, and the association between CEACAM1 expression levels and clinical characteristics was analyzed. It was revealed that CEACAM1 staining was markedly lower in the lymph node metastasis group compared with the non-metastasis group. In addition, the CEACAM1 expression was markedly lower in invasive duct cancer cases compared with cancer cases of other histological types, including lobular, mucinous and tubular types of cancer. No significant association was observed between CEACAM1 expression and other clinical parameters, including age, histological grade and tumor size (Table I). The data indicated that CEACAM1 expression is negatively associated with lymph node metastasis in breast cancer.
Figure 1.
Representative images of CEACAM1 immunohistochemical staining in human breast cancer tissues (n=62). (A) Negative CEACAM1 staining. Original magnification, ×200. (B) Positive CEACAM1 staining. Original magnification, ×200. (C) Negative CEACAM1 staining. Original magnification, ×400. (D) Positive CEACAM1 staining. Original magnification, ×400. Red arrows indicate positive CEACAM1 staining (brown color). CEACAM1, carcinoembryonic antigen-related cell adhesion molecule 1.
Table I.
Association between clinical parameters of the study population and CEACAM1 expression.
| CEACAM1 expression (IOD) | ||||
|---|---|---|---|---|
| Clinical parameters | Number of samples | Median | Range | P-value |
| Histological grade | ||||
| 1 | 12 | 12,489 | 0–69,744 | 0.164 |
| 2 | 25 | 11,805 | 0–58,974 | |
| 3 | 25 | 4,853 | 0–59,528 | |
| Histological type | ||||
| Invasive duct carcinoma | 42 | 6,400 | 0–59,528 | 0.038 |
| Others | 20 | 27,957 | 0–69,744 | |
| Tumor size | ||||
| T1 | 25 | 9,347 | 0–58,974 | 0.363 |
| T2 | 33 | 8,707 | 0–69,744 | |
| T3 | 4 | 2,513 | 0–32,286 | |
| Lymph node metastasis | ||||
| No | 39 | 19,248 | 0–69,744 | 0.017 |
| Yes | 23 | 5,025 | 0–59,528 | |
| Age, years | ||||
| <50 | 15 | 7,884 | 0–59,528 | 0.710 |
| ≥50 | 47 | 8,707 | 0–69,744 | |
| Estrogen receptor status | ||||
| Negative | 26 | 11,321 | 0–55,320 | 0.414 |
| Positive | 36 | 6,400 | 0–69,744 | |
| Progesterone receptor status | ||||
| Negative | 33 | 10,202 | 0–69,744 | 0.960 |
| Positive | 29 | 7,040 | 0–55,320 | |
| Her2 status | ||||
| Negative | 25 | 7,040 | 0–69,744 | 0.834 |
| Positive | 37 | 8,707 | 0–59,528 | |
| Ki-67 expression, % | ||||
| <20 | 24 | 10,078 | 0–69,744 | 0.270 |
| ≥20 | 38 | 6,127 | 0–59,528 | |
Her2, human epidermal growth factor receptor 2; IOD, integrated optical density; CEACAM1, carcinoembryonic antigen-related cell adhesion molecule 1.
CEACAM1-4S mRNA expression is associated with decreased metastatic potential of human breast cancer cell lines
To confirm immunohistochemical findings, a panel of breast cancer cell lines with different metastatic potential was analyzed for CEACAM1 mRNA expression levels by qPCR. Although 12 isoforms of CEACAM1 have been described in humans (19), it is well-accepted that CEACAM1-4S, a short cytoplasmic isoform of CEACAM1, predominates in the human breast epithelium (15,20). Therefore, the following experiments focused on CEACAM1-4S as a representative of CEACAM1. As shown in Fig. 2, there was markedly decreased expression of CEACAM1-4S mRNA in the highly metastatic Hs578T, BT549 and MDA-MB-231 cells compared with the minimally-metastatic MDA-MB-468 and T47D cells as well as the non-invasive breast epithelial MCF10A cells. When normalized to MCF10A, the levels of CEACAM1-4S mRNA in the highly metastatic cell lines exhibited a mean expression 4–5-fold lower compared with the expression in minimally metastatic cell lines, with the exception of MCF7 cells.
Figure 2.

CEACAM1-4S mRNA expression in relation to the metastatic potential of human breast cancer cell lines. CEACAM1-4S mRNA levels were detected in immortal human breast MCF10A cells and breast cancer BT549, Hs578T, MDA-MB-231, MDA-MB-468, T47D, and MCF7 cells by quantitative polymerase chain reaction. Data display the fold change relative to MCF10A. Each experiment was repeated at least three times. CEACAM1-4S, carcinoembryonic antigen-related cell adhesion molecule 1-4S.
It was also noted that MCF7, a less aggressive cell line compared with the other cell lines (21), expressed a low level of CEACAM1-4S, indicating that CEACAM1-4S may not have a critical role in regulation of metastatic potential of MCF7 cells. These data indicated a negative association between CEACAM1-4S mRNA expression and breast cancer metastatic potential. Considering the IHC results from clinical samples, it was hypothesized that CEACAM1-4S may serve an important role in the breast cancer metastasis.
CEACAM1-4S suppresses invasion and migration in human breast cancer cells
To verify the aforementioned hypothesis, a CEACAM1-4S expression vector or an empty vector were transfected into Hs578T and BT549 cells, two highly metastatic cell lines with low levels of endogenous CEACAM1-4S expression (21). The transfection efficiency was evidenced by western blot analysis (Fig. 3A), and Transwell invasion and wound healing assays were then conducted. As shown in Fig. 3B-E, when compared with wild-type or empty-vector controls, CEACAM1-4S-tansfected cells exhibited significantly decreased abilities to invade through the Matrigel-coated membranes and to migrate into the wounded areas. For cells overexpressing CEACAM1-4S, the number of invading cells was a mean 4–5-fold lower compared with controls, and the motilities decreased by >40%. In addition, it was revealed that CEACAM1-4S overexpression had little effect on the proliferation of these two cell lines (data not shown), indicating that all the changes observed in cell invasion and migration were not due to cell proliferation. Therefore, results indicated that CEACAM1-4S inhibits invasion and migration of breast cancer cell.
Figure 3.
Transfection of CEACAM1-4S in breast cancer cells suppressed invasion and migration. (A) Western blot analysis verified the transfection efficiency of CEACAM1-4S in BT549 and Hs578T cells. Transwell invasion assays were performed following CEACAM1-4S overexpression in (B) BT549 and (C) Hs578T cells. Representative images are shown (magnification, ×100), and data are presented as the mean ± standard deviation of three independent experiments. ***P<0.001. Effects of CEACAM1-4S on cell migration were assessed by wound healing assays in (D) BT549 and (E) Hs578T cells following CEACAM1-4S transfection (magnification, ×200). Relative closure of the cell-free areas is presented as the mean ± standard deviation from three separate experiments, **P<0.01; *P<0.05. CEACAM1-4S, carcinoembryonic antigen-related cell adhesion molecule 1-4S.
MMP2 and ITMP2 are involved in the inhibitory effects of CEACAM1-4S on breast cancer invasion and migration
Next, it was investigated whether CEACAM1-4S suppresses breast cancer cell invasion and migration. One possible explanation is that CEACAM1 induces secondary activation of the well-characterized MMPs/TIMPs system that performs a prominent role in cancer invasion (22). Therefore, four main members of the MMP/TIMP family were analyzed by western blot analysis in the present study. The results demonstrated that, compared with corresponding controls, the levels of TIMP2 expression were markedly increased in CEACAM1-4S-transfected BT549 and Hs578T cells, while the levels of MMP2 expression were markedly lower (Fig. 4). In addition, the results revealed that MMP9 and TIMP1 expression were not markedly changed following CEACAM1-4S overexpression in the two cell lines. The data suggested that MMP2/TIMP2, but not MMP1/TIMP1, are involved in CEACAM1-4S-mediated inhibition of cell invasion and migration.
Figure 4.
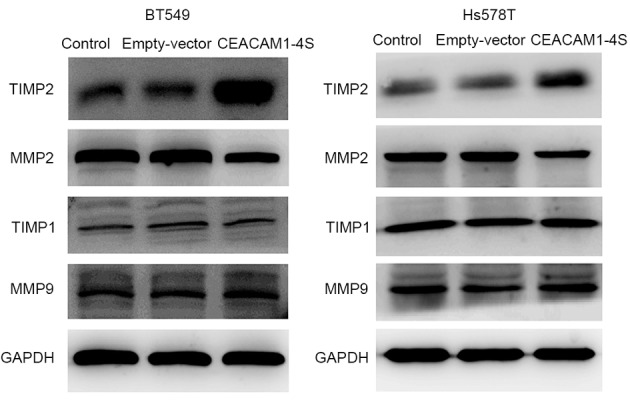
Western blot analysis of the relative expression of MMP2, TIMP2, MMP9 and TIMP1 in CEACAM1-4S-transfected breast cancer cells. Increased TIMP2 expression accompanied by decreased MMP2 was observed in human breast cancer BT549 and Hs578T cells following CEACAM1-4S transfection compared with respective controls. By contrast, no difference in MMP9 and TIMP1 expression was observed among different groups. MMP, matrix metalloproteinase; TIMP, tissue inhibitor of metalloproteinase.
E and N-cadherin are implicated in CEACAM1-4S-induced repression of cell invasion and migration
In an attempt to identify the involvement of other regulatory pathways in the inhibition of cell invasion and migration by CEACAM1-4S, the effect of CEACAM1-4S on the expression of E- and N-cadherin, two crucial molecules for cancer cell metastasis, was analyzed (23,24). As expected, western blot analysis revealed that overexpression of CEACAM1-4S markedly altered the expression of E- and N-cadherin. As shown in Fig. 5A, CEACAM1-4S overexpression in BT549 and Hs578T cells resulted in a substantial increase in E-cadherin expression, whereas N-cadherin expression was markedly decreased. Notably, E- and N-cadherin are widely viewed as markers of epithelial-mesenchymal transition (EMT), an important process during cancer metastasis (25,26). The CEACAM1-4S-induced changes in E- and N-cadherin expression appeared to be in parallel with EMT reversal. Therefore, the expression of vimentin, another critical molecular marker for EMT (25,26), was examined by western blot analysis. The results showed a marked reduction of vimentin expression in CEACAM1-4S-transfected BT549 and Hs578T cells compared with the respective controls (Fig. 5A). The results were in accordance with the aforementioned western blot analysis, showing a downregulation of vimentin accompanied by upregulation of E-cadherin following CEACAM1-4S overexpression in BT549 and Hs578T cells (Fig. 5A). Furthermore, two well-known signal molecules associated with cancer metastasis and EMT, STAT3 and Smad2 (25), were analyzed. It was revealed that CEACAM1-4S overexpression markedly decreased the phosphorylation of STAT3 and Smad2 but had no effect on the levels of total protein levels (Fig. 5B). Results of western blot analysis of vimentin and E-cadherin expression in CECAM1-4S-transfected BT549 and Hs578T cells were confirmed by IF staining (Fig. 5C and D). Taken together, these data demonstrated that the suppressive functions of CEACAM1-4S may be dependent on the regulation of E and N-cadherin as well as EMT in breast cancer cells.
Figure 5.
CEACAM1-4S induces a gene expression pattern consistent with epithelial-mesenchymal transition reversal. (A) Western blot analysis of E-cadherin, N-cadherin and vimentin in CEACAM1-4S-transfected BT549 and Hs578T cells. CEACAM1-4S overexpression led to the induction of E-cadherin and abrogation of N-cadherin and vimentin. (B) Western blot analysis of p-STAT3, p-Smad2, total STAT3 and Smad2/3 in CEACAM1-4S-transfected BT549 and Hs578T cells. Exogenous expression of CEACAM1-4S decreased the phosphorylation of STAT3 and Smad2 but had no marked effect on the levels of total STAT3 and Smad2/3. Immunofluorescence images of controls and breast cancer cells overexpressing CEACAM1-4S. (C) BT549 and (D) Hs578T cells were stained for E-cadherin and vimentin (magnification, ×200). CEACAM1-4S-transfected cells exhibited weaker staining for vimentin and stronger staining for E-cadherin compared with corresponding controls. CEACAM1-4S, carcinoembryonic antigen-related cell adhesion molecule 1-4S; p-, phosphorylated; STAT3, signal transducer and activator of transcription.
Discussion
A previous study by the present authors reported that CEACAM1 is downregulated in breast cancer (12). The present study aimed to address the functional significance of CEACAM1 in breast cancer. First, CEACAM1 expression was examined in 62 tissue samples from patients with breast cancer by IHC staining. The results indicated a negative association between CEACAM1 expression and breast cancer metastasis. It has been reported that the loss of CEACAM1 expression is associated with a poorer prognosis for patients with breast cancer (11), and the findings of the present study indicated that CEACAM1 may have a protective role against breast cancer metastasis. To test this hypothesis, the mRNA expression of CEACAM1-4S, the short cytoplasmic isoform of CEACAM1 that predominates in human breast epithelium (15), was detected in a panel of six breast cancer cell lines with different metastatic potentials. Consistent with IHC results, the data revealed that CEACAM1-4S mRNA expression was associated with the decreased metastatic potential of breast cancer cell lines. The subsequent in vitro experiments demonstrated that the overexpression of CEACAM1-4S significantly inhibited the invasion and migration of BT549 and Hs578T cells. Aberrant expression of CEACAM1 in breast cancer was observed a few years ago (13), however, there is limited information regarding the roles of CEACAM1 in the invasive behavior of breast cancer. However, it has been demonstrated previously that CEACAM1 is able to function as a tumor suppressor that prevents tumorigenicity in a mice model of breast cancer (27). The present study provides evidence that the tumor suppressive activities of CEACAM1 extend to the aggressive breast cancer cell phenotype including increased migratory and invasive abilities.
CEACAM1 has a wide range of biological functions, the majority of which are associated with the hallmarks of cancer, including proliferation, apoptosis, immune evasion, inflammation and angiogenesis (19,28). In 2004, Ebrahimnejad et al (29) reported that CEACAM1 actively contributes to tumor invasion and migration in melanoma. Subsequently, CEACAM1 has been reported to promote tumor invasiveness in thyroid cancer (30). However, these findings were contradictory to previously published studies that identified CEACAM1 as a tumor suppressor (27,31,32), which was also demonstrated in the current study. Therefore, the roles of CEACAM1 in malignancies appear to be conflicting. One of the reasons responsible for this paradox may be that CEACAM1 comprises isoform diversity. Previous studies have demonstrated that CEACAM1-4L promotes colon and hepatocellular cancer cell invasion and migration, while CEACAM1-4S exhibits inhibitory effects. Therefore, there are functional differences between CEACAM1 isoforms (33,34). Considering that CEACAM1-4S is the physiological predominant isoform (15), the present study mimicked the scenario in normal human breast cells by increasing CEACAM1-4S expression in breast cancer cells. In accordance with the aforementioned studies (33,34), results of the present study revealed an inhibitory effect of CEACAM1-4S on cell invasion and migration in breast cancer. Based on the present results and results of previous studies (33,34), it is hypothesized that CEACAM1-4S acts as a tumor suppressor, whereas CEACAM1-4L may be a tumor stimulator. Combined with the findings that different malignancies preferentially express distinct CEACAM1 isoforms (15,33), this partly explains the paradoxical roles attributed to CEACAM1. Additional extensive and intensive studies are required to confirm the biological functions of CEACAM1 in cancer metastasis.
As cancer metastasis is a highly integrated process, which is regulated by a large number of extracellular matrix-associated enzymes, including MMPs and their natural inhibitors, TIMPs (35), experiments detecting changes of these enzymes were performed. High TIMP2 and low MMP2 expression patterns were observed in CEACAM1-4S-transfected breast cancer cells. Therefore, CEACAM1-4S may exhibit the suppressive functions through up and downregulation of the MMP2/TIMP2 balance. In addition, the present results demonstrated that the expression of MMP9 and its natural inhibitor TIMP1 were not markedly affected by CEACAM1-4S overexpression, indicating that the MMP9/TIMP1 balance was not involved in the regulation of cell invasion by CEACAM1-4S. Furthermore, the present data revealed that cadherins may be downstream effectors of CEACAM1-4S. It is well-documented that cadherins control the balance between repression and promotion of cancer cell migration and invasion (23,36–38). Currently, E-cadherin and N-cadherin are the most comprehensively investigated cadherins in cancer, and they balance each other with E-cadherin frequently downregulated in cancer and acts as an invasion suppressor, whereas N-cadherin is an invasion promoter upregulated in the majority of cancer types (23,39). A previous study demonstrated that N-cadherin has an important promoting effect on CEACAM1-mediated migration in LoVo (human colon cancer) cells (24). In the present study, it was revealed that CEACAM1-4S not only regulates N-cadherin, but also E-cadherin, and this regulation was in parallel with EMT reversal. Additional findings that vimentin and intracellular signals p-Smad2 and p-STAT3 were downregulated by CEACAM1-4S further supported a reversal effect of CEACAM1-4S on EMT in breast cancer cells. However, it should be noted that EMT is a complicated process orchestrated by various biological molecules, and whether CEACAM1-4S serves a crucial role in EMT reversal requires further study.
In summary, the present study demonstrated that CEACAM1-4S performs an inhibitory role in breast cancer cell invasion and migration, possibly through regulating the balance between MMP2/TIMP2 and E-/N-cadherins. In addition, evidence that CEACAM1-4S may induce EMT reversal in breast cancer cells was provided. Therefore, restoring CEACAM1-4S expression may provide a novel avenue for therapy of patients with breast cancer.
Acknowledgements
The present study was supported by the National Natural Science Foundation of China (grant nos. 81272479, 814024198, 81572821, 81502490 and 81502491), the Shanghai Committee of Science and Technology (grant no. 14YF1412200), the Program of Shanghai Leading Talents (grant no. 038) and the Program of Shanghai Shen-Kang Hospital Development Center (grant no. SHDC22014004).
Glossary
Abbreviations
- CEACAM1
carcinoembryonic antigen-related cell adhesion molecule 1
- PCR
polymerase chain reaction
- EMT
epithelial-mesenchymal transition
- TIMP
tissue inhibitor of metalloproteinase
- MMP
matrix metalloproteinase
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Weigelt B, Peterse JL, van 't Veer LJ. Breast cancer metastasis: Markers and models. Nat Rev Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 3.Gray-Owen SD, Blumberg RS. CEACAM1: Contact-dependent control of immunity. Nat Rev Immunol. 2006;6:433–446. doi: 10.1038/nri1864. [DOI] [PubMed] [Google Scholar]
- 4.Prall F, Nollau P, Neumaier M, Haubeck HD, Drzeniek Z, Helmchen U, Löning T, Wagener C. CD66a (BGP), an adhesion molecule of the carcinoembryonic antigen family, is expressed in epithelium, endothelium and myeloid cells in a wide range of normal human tissues. J Histochem Cytochem. 1996;44:35–41. doi: 10.1177/44.1.8543780. [DOI] [PubMed] [Google Scholar]
- 5.Neumaier M, Paululat S, Chan A, Matthaes P, Wagener C. Biliary glycoprotein, a potential human cell adhesion molecule, is down-regulated in colorectal carcinomas. Proc Natl Acad Sci USA. 1993;90:10744–10748. doi: 10.1073/pnas.90.22.10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka K, Hinoda Y, Takahashi H, Sakamoto H, Nakajima Y, Imai K. Decreased expression of biliary glycoprotein in hepatocellular carcinomas. Int J Cancer. 1997;74:15–19. doi: 10.1002/(SICI)1097-0215(19970220)74:1<15::AID-IJC3>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 7.Kammerer R, Riesenberg R, Weiler C, Lohrmann J, Schleypen J, Zimmermann W. The tumour suppressor gene CEACAM1 is completely but reversibly downregulated in renal cell carcinoma. J Pathol. 2004;204:258–267. doi: 10.1002/path.1657. [DOI] [PubMed] [Google Scholar]
- 8.Busch C, Hanssen TA, Wagener C, OBrink B. Down-regulation of CEACAM1 in human prostate cancer: Correlation with loss of cell polarity, increased proliferation rate and Gleason grade 3 to 4 transition. Hum Pathol. 2002;33:290–298. doi: 10.1053/hupa.2002.32218. [DOI] [PubMed] [Google Scholar]
- 9.Nittka S, Gunther J, Ebisch C, Erbersdobler A, Neumaier M. The human tumor suppressor CEACAM1 modulates apoptosis and is implicated in early colorectal tumorigenesis. Oncogene. 2004;23:9306–9313. doi: 10.1038/sj.onc.1208259. [DOI] [PubMed] [Google Scholar]
- 10.Estrera VT, Chen DT, Luo W, Hixson DC, Lin SH. Signal transduction by the CEACAM1 tumor suppressor. Phosphorylation of serine 503 is required for growth-inhibitory activity. J Biol Chem. 2001;276:15547–15553. doi: 10.1074/jbc.M008156200. [DOI] [PubMed] [Google Scholar]
- 11.Wang JL, Sun SZ, Qu X, Liu WJ, Wang YY, Lv CX, Sun JZ, Ma R. Clinicopathological significance of CEACAM1 gene expression in breast cancer. Chin J Physiol. 2011;54:332–338. [PubMed] [Google Scholar]
- 12.Yang C, He P, Liu Y, He Y, Yang C, Du Y, Zhou M, Wang W, Zhang G, Wu M, Gao F. Down-regulation of CEACAM1 in breast cancer. Acta Biochim Biophys Sin (Shanghai) 2015;47:788–794. doi: 10.1093/abbs/gmv075. [DOI] [PubMed] [Google Scholar]
- 13.Riethdorf L, Lisboa BW, Henkel U, Naumann M, Wagener C, Loning T. Differential expression of CD66a (BGP), a cell adhesion molecule of the carcinoembryonic antigen family, in benign, premalignant and malignant lesions of the human mammary gland. J Histochem Cytochem. 1997;45:957–963. doi: 10.1177/002215549704500705. [DOI] [PubMed] [Google Scholar]
- 14.Huang J, Hardy JD, Sun Y, Shively JE. Essential role of biliary glycoprotein (CD66a) in morphogenesis of the human mammary epithelial cell line MCF10F. J Cell Sci. 1999;112:4193–4205. doi: 10.1242/jcs.112.23.4193. [DOI] [PubMed] [Google Scholar]
- 15.Kirshner J, Chen CJ, Liu P, Huang J, Shively JE. CEACAM1-4S, a cell-cell adhesion molecule, mediates apoptosis and reverts mammary carcinoma cells to a normal morphogenic phenotype in a 3D culture. Proc Natl Acad Sci USA. 2003;100:521–526. doi: 10.1073/pnas.232711199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tavassoli FA, Devilee P. World Health Organization classification of tumors Pathology and genetics of tumors of the breast and female genital organs. Lyon: IARC Press; 2003. [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Oliveira-Ferrer L, Tilki D, Ziegeler G, Hauschild J, Loges S, Irmak S, Kilic E, Huland H, Friedrich M, Ergün S. Dual role of carcinoembryonic antigen-related cell adhesion molecule 1 in angiogenesis and invasion of human urinary bladder cancer. Cancer Res. 2004;64:8932–8. doi: 10.1158/0008-5472.CAN-04-0505. [DOI] [PubMed] [Google Scholar]
- 19.Beauchemin N, Arabzadeh A. Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev. 2013;32:643–671. doi: 10.1007/s10555-013-9444-6. [DOI] [PubMed] [Google Scholar]
- 20.Kirshner J, Hardy J, Wilczynski S, Shively JE. Cell-cell adhesion molecule CEACAM1 is expressed in normal breast and milk and associates with beta1 integrin a 3D model of morphogenesis. J Mol Histol. 2004;35:287–299. doi: 10.1023/B:HIJO.0000032360.01976.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakrabarti R, Hwang J, Blanco Andres M, Wei Y, Lukačišin M, Romano RA, Smalley K, Liu S, Yang Q, Ibrahim T, et al. Elf5 inhibits the epithelial -mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2. Nat Cell Biol. 2012;14:1212–1222. doi: 10.1038/ncb2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Said AH, Raufman JP, Xie G. The role of matrix metalloproteinases in colorectal cancer. Cancers. 2014;6:366–375. doi: 10.3390/cancers6010366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derycke LD, Bracke ME. N-cadherin in the spotlight of cell-cell adhesion, differentiation, embryogenesis, invasion and signalling. Int J Dev Biol. 2004;48:463–476. doi: 10.1387/ijdb.041793ld. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Di G, Wu CT, Hu X, Duan H. CEACAM1 inhibits cell-matrix adhesion and promotes cell migration through regulating the expression of N-cadherin. Biochem Biophys Res Commun. 2013;430:598–603. doi: 10.1016/j.bbrc.2012.11.107. [DOI] [PubMed] [Google Scholar]
- 25.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA, Williams ED, Thompson EW. Epithelial-mesenchymal and mesenchymal-epithelial transitions in carcinoma progression. J Cell Physiol. 2007;213:374–383. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- 27.Luo WP, Wood CG, Earley K, Hung MC, Lin SH. Suppression of tumorigenicity of breast cancer cells by an epithelial cell adhesion molecule (C-CAM1): The adhesion and growth suppression are mediated by different domains. Oncogene. 1997;14:1697–1704. doi: 10.1038/sj.onc.1200999. [DOI] [PubMed] [Google Scholar]
- 28.Obrink B. Is CEACAM1 a lymphangiogenic switch? Blood. 2007;110:4137–4138. doi: 10.1182/blood-2007-09-111005. [DOI] [Google Scholar]
- 29.Ebrahimnejad A, Streichert T, Nollau P, Horst AK, Wagener C, Bamberger AM, Brümmer J. CEACAM1 enhances invasion and migration of melanocytic and melanoma cells. Am J Pathol. 2004;165:1781–1787. doi: 10.1016/S0002-9440(10)63433-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W, Wei W, Winer D, Bamberger AM, Bamberger C, Wagener C, Ezzat S, Asa SL. CEACAM1 impedes thyroid cancer growth but promotes invasiveness: A putative mechanism for early metastases. Oncogene. 2007;26:2747–2758. doi: 10.1038/sj.onc.1210077. [DOI] [PubMed] [Google Scholar]
- 31.Luo W, Tapolsky M, Earley K, Wood CG, Wilson DR, Logothetis CJ, Lin SH. Tumor-suppressive activity of CD66a in prostate cancer. Cancer Gene Ther. 1999;6:313–321. doi: 10.1038/sj.cgt.7700055. [DOI] [PubMed] [Google Scholar]
- 32.Volpert O, Luo W, Liu TJ, Estrera VT, Logothetis C, Lin SH. Inhibition of prostate tumor angiogenesis by the tumor suppressor CEACAM1. J Biol Chem. 2002;277:35696–35702. doi: 10.1074/jbc.M205319200. [DOI] [PubMed] [Google Scholar]
- 33.Ieda J, Yokoyama S, Tamura K, Takifuji K, Hotta T, Matsuda K, Oku Y, Nasu T, Kiriyama S, Yamamoto N, et al. Re-expression of CEACAM1 long cytoplasmic domain isoform is associated with invasion and migration of colorectal cancer. Int J Cancer. 2011;129:1351–1361. doi: 10.1002/ijc.26072. [DOI] [PubMed] [Google Scholar]
- 34.Kiriyama S, Yokoyama S, Ueno M, Hayami S, Ieda J, Yamamoto N, Yamaguchi S, Mitani Y, Nakamura Y, Tani M, et al. CEACAM1 long cytoplasmic domain isoform is associated with invasion and recurrence of hepatocellular carcinoma. Ann Surg Oncol. 2014;21(Suppl 4):S505–S514. doi: 10.1245/s10434-013-3460-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stamenkovic I. Matrix metalloproteinases in tumor invasion and metastasis. Semin Cancer Biol. 2000;10:415–433. doi: 10.1006/scbi.2000.0379. [DOI] [PubMed] [Google Scholar]
- 36.Cavallaro U, Christofori G. Cell adhesion and signaling by cadherins and Ig-CAMs in cancer. Nat Rev Cancer. 2004;4:118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- 37.Wheelock MJ, Shintani Y, Maeda M, Fukumoto Y, Johnson KR. Cadherin switching. J Cell Sci. 2008;121:727–735. doi: 10.1242/jcs.000455. [DOI] [PubMed] [Google Scholar]
- 38.Theveneau E, Mayor R. Cadherins in collective cell migration of mesenchymal cells. Curr Opin Cell Biol. 2012;24:677–684. doi: 10.1016/j.ceb.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakada M, Nakada S, Demuth T, Tran NL, Hoelzinger DB, Berens ME. Molecular targets of glioma invasion. Cell Mol Life Sci. 2007;64:458–478. doi: 10.1007/s00018-007-6342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]