Abstract
The amount and evolutionary impact of horizontal gene transfer in eukaryotes remain contentious issues. A new phylogenomic study suggests that gene transfer from prokaryotes has contributed significantly to the adaptation and metabolic evolution of Blastocystis, the most widespread human gut eukaryotic parasite.
Eukaryotic cells usually possess complex structures, including sophisticated cytoskeleton and endomembrane systems, which allow many species to feed by phagocytosis upon other cells or particles. Phagocytosis is instrumental for the facility, observed across eukaryotic phyla, to acquire endosymbionts. The best-known examples are mitochondria and chloroplasts, ancient endosymbiotic bacteria evolved into cell organelles, but many other endosymbioses –involving members of the three domains of life- have been described in a vast spectrum of eukaryotic hosts. They provide a variety of new functions, ranging from detoxification to cell defense though, in most cases, endosymbionts are recruited because their metabolic abilities are useful for their hosts [1]. By contrast, prokaryotic cells are structurally simpler and unable to carry out phagocytosis, so that endosymbiosis is much rarer. Nonetheless, prokaryotes can easily gain new, notably metabolic, functions by acquiring genes from more or less distant organisms via horizontal gene transfer (HGT). Although the importance of HGT in prokaryotic evolutionary history is widely recognized [2–4], the frequency and evolutionary role of HGT in eukaryotic evolution remain controversial. Some authors claim that, except for the numerous genes transferred long ago from mitochondria and chloroplast ancestors to the host genome, recent gene acquisitions from prokaryotes are too rare to have broad evolutionary significance in eukaryotes [5]. In a recent issue of Current Biology, Eme et al. [6] present a counterpoint to this view by showing that several strains of the human parasite Blastocystis sp. appear to have acquired up to 2.5% of their genes by HGT from different donors, mostly bacteria.
Blastocystis is a genus of unicellular parasites belonging to the Stramenopiles, a large eukaryotic group exhibiting wide phenotypic diversity. Its very simplified cellular structure has long hindered the recognition of the diversity existing within this genus, but 18S rRNA gene sequence analysis has revealed considerable cryptic diversity. Blastocystis species appear to have low host specificity and infect a variety of animal taxa, both vertebrates and invertebrates. Although its implication in disease remains uncertain, Blastocystis is highly prevalent in humans and up to 9 different lineages -called subtypes ST1 to ST9-, can be distinguished in the human gut upon their 18S rRNA sequences [7]. Complete genome sequences were available for subtypes ST4 and ST7 and Eme et al. have now determined the genome sequence of the highly prevalent ST1 subtype [6]. Living in the intestinal tract, Blastocystis coexists with the extremely diverse gut microbiome, being exposed to multifarious sources of exogenous DNA [8]. Thus, Blastocystis genome sequences represent an excellent material to evaluate the impact of HGT on eukaryotic genomes.
Accurate HGT detection is challenging since many confounding factors can blur phylogenetic signal and lead to false positive and negative results. It is well known that different HGT detection methods very often produce incongruent results, and some of them -notably sequence similarity-based methods- are prone to HGT overestimation [9]. In addition, contamination in the samples used for genome sequencing can lead to spurious assemblages containing genes from mixed origins and producing a high number of false HGTs. This problem is especially important for predatory eukaryotes as the DNA samples usually contain DNA from both the targeted species and their prey. The recent controversy on HGT levels in tardigrades, a group of minute animals, exemplifies this problem. Analysis of the first tardigrade genome sequence (Hypsibius dujardini) suggested that more than 17% of its genes derived from HGT from various sources, mostly bacteria but also plants, fungi, archaea, and viruses [10]. However, a second independently sequenced genome of the same species, this time curated using metagenomic approaches, revealed that those HGT levels were largely inflated due to contamination. Actual HGT appears to account for less than 1-2% of tardigrade genes [11].
With these premises in mind, Eme et al. looked for alien genes using a rigorous phylogeny-based approach upon the 6544 proteins predicted in the Blastocystis ST1 genome. To maximize detection accuracy, they reconstructed phylogenetic trees for all proteins and focused only on well-supported trees where Blastocystis branched far from other stramenopiles. 2.5% of the ST1 protein-coding genes responded to these criteria. They represent 74 gene families for a total of 167 genes (some issued from duplications). Most of them (80%) are of prokaryotic origin and were likely acquired from predominant gut bacterial phyla (Firmicutes, Proteobacteria and Bacteroidetes). More unexpectedly, the remaining HGTs come from eukaryotic donors. But what about contamination issues? Eme et al. provide several lines of evidence to discard them: 1) introns are present in 66 of the 74 gene families, something not expected in contaminant bacterial DNA; 2) HGT candidates (24%) and genes in the complete ST1 genome (26%) share similar frequencies of termination codons created by polyadenylation, a mechanism exclusive to Blastocystis; and 3) HGT candidates in ST1 also occur in other Blastocystis genomes and are flanked by typical Blastocystis genes. Therefore, the genes reported by Eme et al. appear to be bona fide HGT acquisitions. Furthermore, they represent only a low boundary for the actual amount of HGT in Blastocystis, as Eme et al. applied stringent criteria that probably discarded additional HGT cases.
Eme et al. qualified these HGT events as “recent” because transferred genes were not found in other stramenopiles. However, their restricted distribution could also be due to “ancient” transfers followed by massive loss in the rest of stramenopiles. An alternative way to look at this question is to measure the similarity between the transferred genes and those in the potential donors (high similarity indicating recent HGT). In their recent work, Ku and Martin propose to generalize this approach [5]. Through HGT analysis in a number of prokaryotic and eukaryotic genomes, they conclude that recent prokaryote-to-eukaryote HGT is extremely rare and that genes in eukaryotic genomes with >70% amino-acid sequence identity to prokaryotic homologs most likely correspond to assembly or annotation artifacts. Although this approach seems straightforward, it is not devoid of problems, as sequence similarity does not only depend on the timing of the transfer but also on the evolutionary rate of the acceptor and donor lineages. If one of them -or both- is fast-evolving, similarity will decrease rapidly, which might result in falsifying truly “ancient HGT” cases. Blastocystis, as many other parasites, is a very fast-evolving lineage to the point that, on average, the ST1 sequences of the 74 HGT families are only 62% identical to their ST4 and ST7 orthologues. Even so, one of the HGT families, the endoribonuclease L-PSP, breaks the “70% rule” as it exhibits 74% identity to the closest prokaryotic sequences. This suggests that, unfortunately, a general rule is seemingly impossible to be established and case-by-case inspection of HGT events remains necessary to validate or reject them.
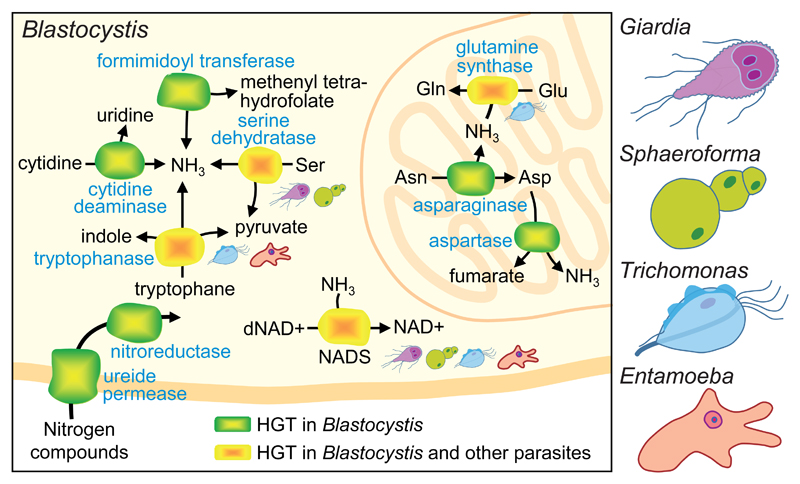
The Blastocystis HGTs appear to be involved in a surprising variety of functions, notably carbohydrate scavenging and metabolism, anaerobic amino acid and nitrogen metabolism, oxygen-stress resistance and pH homeostasis. Many of them probably participate in the interaction with the external environment as they encode membrane or secreted proteins, including some likely related to infection and avoidance of host defenses. A good example concerns the metabolism of fucose, a sugar abundant in the surface of the human intestinal mucosal epithelium, where it acts as an important mediator with gut microbes, including diverse pathogens [12]. This is probably the case for Blastocystis, which possesses five HGTs of bacterial origin involved in the acquisition and metabolism of L-fucose, some of them similar to those of the bacterial pathogen Campylobacter jejuni. Other HGTs of bacterial origin allow Blastocystis to cope with oxidative stress and to detoxify its close environment. Two of them (yqhD and gldA) may degrade methylglyoxal, a toxic molecule produced by other intestinal microbes. Notably, yqhD has been acquired by other mucosal eukaryotic parasites (Entamoeba and Trichomonas) via independent HGT events. Several other convergent acquisitions (Figure 1) indicate a strong selective advantage of the corresponding genes in these parasites, and make them interesting potential clinical targets.
Figure 1. Horizontal gene transfers involved in nitrogen metabolism shared by Blastocystis and other eukaryotic unicellular parasites.
The metabolic diagram, based on Eme et al. [6], shows all enzymes acquired by Blastocystis from different donors. Those that are shared by several other distantly related protist parasites, the excavates Giardia and Trichomonas, the ichthyosporean Sphaeroforma, and the amoebozoan Entamoeba, are highlighted in yellow and the species where the respective enzyme is found are indicated.
Eme et al. provide strong evidence for the crucial role that HGT has played in the adaptation of Blastocystis to the gut environment, with a significant HGT-mediated modification of its metabolism. This discovery adds to the already rich list of cases where HGT -very often from prokaryotic sources- has been shown as an important mechanism for adaptation and metabolic evolution in unicellular eukaryotes. A very common example is the acquisition of enzymes involved in anaerobic metabolism [13–16], but HGT seems also to be at the basis of surprising evolutionary paths followed by some microbial eukaryotes, such as the adaptation to extreme environments [17, 18] or the reversal from parasitism to a free-living lifestyle [19]. HGT is probably not as prevalent in eukaryotes as in prokaryotes, but the increasing panoply of HGT cases detected in all eukaryotic phyla reveals that eukaryotes can take metabolic advantage from prokaryotes, not only by hosting them as endosymbionts, but also by borrowing the genes that they have evolved for their adaptation to an impressive range of habitats and metabolisms.
References
- 1.Gast RJ, Sanders RW, Caron DA. Ecological strategies of protists and their symbiotic relationships with prokaryotic microbes. Trends Microbiol. 2009;17:563–569. doi: 10.1016/j.tim.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Koonin EV, Makarova KS, Aravind L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu Rev Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.López-García P, Zivanovic Y, Deschamps P, Moreira D. Bacterial gene import and mesophilic adaptation in archaea. Nat Rev Microbiol. 2015;13:447–456. doi: 10.1038/nrmicro3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koonin EV. Horizontal gene transfer: essentiality and evolvability in prokaryotes, and roles in evolutionary transitions. 1000Res. 2016;5:1805. doi: 10.12688/f1000research.8737.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ku C, Martin WF. A natural barrier to lateral gene transfer from prokaryotes to eukaryotes revealed from genomes: the 70 % rule. BMC Biol. 2016;14:89. doi: 10.1186/s12915-016-0315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eme L, Gentekaki E, Curtis B, Archibald JM, Roger AJ. Lateral gene transfer in adaptation of the anaerobic parasite Blastocystis to the gut. Curr Biol. 2017 doi: 10.1016/j.cub.2017.02.003. In press. [DOI] [PubMed] [Google Scholar]
- 7.Stensvold CR, Clark CG. Current status of Blastocystis: A personal view. Parasitol Int. 2016;65:763–771. doi: 10.1016/j.parint.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Langille MG, Meehan CJ, Beiko RG. Human microbiome: a genetic bazaar for microbes? Curr Biol. 2012;22:023. doi: 10.1016/j.cub.2011.11.023. [DOI] [PubMed] [Google Scholar]
- 9.Koski LB, Golding GB. The closest BLAST hit is often not the nearest neighbor. J Mol Evol. 2001;52:540–542. doi: 10.1007/s002390010184. [DOI] [PubMed] [Google Scholar]
- 10.Boothby TC, Tenlen JR, Smith FW, Wang JR, Patanella KA, Nishimura EO, Tintori SC, Li Q, Jones CD, Yandell M, et al. Evidence for extensive horizontal gene transfer from the draft genome of a tardigrade. Proc Natl Acad Sci U S A. 2015;112:15976–15981. doi: 10.1073/pnas.1510461112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koutsovoulos G, Kumar S, Laetsch DR, Stevens L, Daub J, Conlon C, Maroon H, Thomas F, Aboobaker AA, Blaxter M. No evidence for extensive horizontal gene transfer in the genome of the tardigrade Hypsibius dujardini. Proc Natl Acad Sci U S A. 2016;113:5053–5058. doi: 10.1073/pnas.1600338113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goto Y, Uematsu S, Kiyono H. Epithelial glycosylation in gut homeostasis and inflammation. Nat Immunol. 2016;17:1244–1251. doi: 10.1038/ni.3587. [DOI] [PubMed] [Google Scholar]
- 13.Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 14.Andersson JO. Gene transfer and diversification of microbial eukaryotes. Annu Rev Microbiol. 2009;63:177–193. doi: 10.1146/annurev.micro.091208.073203. [DOI] [PubMed] [Google Scholar]
- 15.Tsaousis AD, Ollagnier de Choudens S, Gentekaki E, Long S, Gaston D, Stechmann A, Vinella D, Py B, Fontecave M, Barras F, et al. Evolution of Fe/S cluster biogenesis in the anaerobic parasite Blastocystis. Proc Natl Acad Sci U S A. 2012;109:10426–10431. doi: 10.1073/pnas.1116067109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nyvltova E, Stairs CW, Hrdy I, Ridl J, Mach J, Paces J, Roger AJ, Tachezy J. Lateral gene transfer and gene duplication played a key role in the evolution of Mastigamoeba balamuthi hydrogenosomes. Mol Biol Evol. 2015;32:1039–1055. doi: 10.1093/molbev/msu408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schonknecht G, Chen WH, Ternes CM, Barbier GG, Shrestha RP, Stanke M, Brautigam A, Baker BJ, Banfield JF, Garavito RM, et al. Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science. 2013;339:1207–1210. doi: 10.1126/science.1231707. [DOI] [PubMed] [Google Scholar]
- 18.Harding T, Brown MW, Simpson AG, Roger AJ. Osmoadaptative strategy and its molecular signature in obligately halophilic heterotrophic protists. Genome Biol Evol. 2016;8:2241–2258. doi: 10.1093/gbe/evw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu F, Jerlstrom-Hultqvist J, Kolisko M, Simpson AG, Roger AJ, Svard SG, Andersson JO. On the reversibility of parasitism: adaptation to a free-living lifestyle via gene acquisitions in the diplomonad Trepomonas sp. PC1. BMC Biol. 2016;14:016–0284. doi: 10.1186/s12915-016-0284-z. [DOI] [PMC free article] [PubMed] [Google Scholar]