Abstract
The 18 kDa translocator protein (TSPO) is found in mitochondrial membranes and mediates the import of cholesterol and porphyrins into mitochondria. In line with the role of TSPO in mitochondrial function, TSPO ligands are used for a variety of diagnostic and therapeutic applications in animals and humans. Here we present the three-dimensional high-resolution structure of mammalian TSPO reconstituted in detergent micelles in complex with its high-affinity ligand PK11195. The TSPO-PK11195 structure is described by a tight bundle of five transmembrane α-helices that form a hydrophobic pocket accepting PK11195. Ligand-induced stabilization of the structure of TSPO suggests a molecular mechanism for the stimulation of cholesterol transport into mitochondria.
The 18 kDa translocator protein (TSPO) is preferentially expressed in mitochondrial membranes of steroidogenic tissues and its gene family is present in almost all organisms (1–3). TSPO was first described as a peripheral benzodiazepine receptor, a secondary receptor for diazepam (1, 4). TSPO was subsequently found to be responsible for the transport of cholesterol into mitochondria thereby influencing the synthesis of neurosteroids (1, 5). TSPO also plays an important role in apoptosis (6) and stress adaptation (7). Expression of TSPO is strongly upregulated in areas of brain injury and in neuroinflammatory conditions including Alzheimer and Parkinson disease (2). TSPO is located at contact sites between the outer and inner mitochondrial membrane, and was suggested to form together with cyclophilin D, the voltage-dependent anion channel and the adenine nucleotide translocator the mitochondrial permeability transition pore (8).
TSPO ligands have potential diagnostic and therapeutic applications from attenuation of cancer cell proliferation (6) to neuroprotective effects (2). TSPO ligands such as XBD-173 might also be useful in treating anxiety with reduced side effects when compared to traditional benzodiazepine-related drugs (9). The best characterized ligand of TSPO is 1-(2-chlorophenyl)-N-methyl-N-(1-methylpropyl)-3-isoquinolinecarboxamide (PK11195) that binds to TSPO with nanomolar affinity in many species (10–13). PK11195 is used as a biomarker in positron emission tomography to visualize brain inflammation in patients with neuronal damage (2, 10). Moreover, the combination of TSPO ligands PK11195 and Ro5-4864 reduces levels of soluble β-amyloid in mice (14). Biochemical consequences of binding of PK11195 to TSPO include the stimulation of TSPO-mediated transport of cholesterol into mitochondria (12, 15).
Sequence analysis indicated the organization of TSPO into five transmembrane helices (1, 4, 16). Circular dichroism and nuclear magnetic resonance (NMR) studies on isolated peptides supported a predominantly helical structure of mouse TSPO (mTSPO) (17). Electron microscopy further provided a 10 Å resolution density map suggesting that a bacterial TSPO homologue, the tryptophan-rich sensory protein from Rhodobacter sphaeroides, folds into a five-helix bundle, which further associates into a homodimer (18). In addition, previous studies have shown that mammalian TSPO can be overexpressed and purified in a functional form (11). Drug ligands bind to the recombinant protein with a 1:1 stoichiometry and a pharmacological profile similar to that reported for the native protein (11, 12, 19).
We prepared recombinant mTSPO and reconstituted it into dodecylphosphocholine (DPC) micelles. In the absence of PK11195, NMR signals were highly overlapped and clustered into one region (Fig. S1). The low signal dispersion might point to a dynamic nature of the free protein and explain difficulties in structural studies of this protein. However, in the presence of (R)-PK11195, which is used for positron emission tomography (10) and has a higher TSPO affinity than the racemate (20), high quality NMR spectra were obtained (Fig. S1). The combination of Nuclear Overhauser Enhancement (NOE) experiments and relaxation-optimized assignment strategies together with complementary amino acid-specific labeling (please see Supplemental Methods (21)) allowed assignment of most resonances (Figs. S2 and S3). Specifically, 98% of backbone and 95% of the side chain resonances were assigned, including full stereospecific discrimination of all leucine and valine methyl groups. The completeness of the resonance assignment together with the excellent quality of the NOE experiments at high magnetic fields yielded more than 3300 NOE distance restraints and enabled direct detection of 61 TSPO-PK11195 contacts in 3D F1-13C,15N-filtered/edited-NOE experiments (Table S1 and Fig. S3,S4). The large number of medium- and long-range NOE contacts (Fig. S4) defined the structure of residues W5 to S159 of the TSPO-PK11195 complex at high resolution (Figs. 1A,B), while the N- and C-terminal tails remained dynamic (Fig. S5).
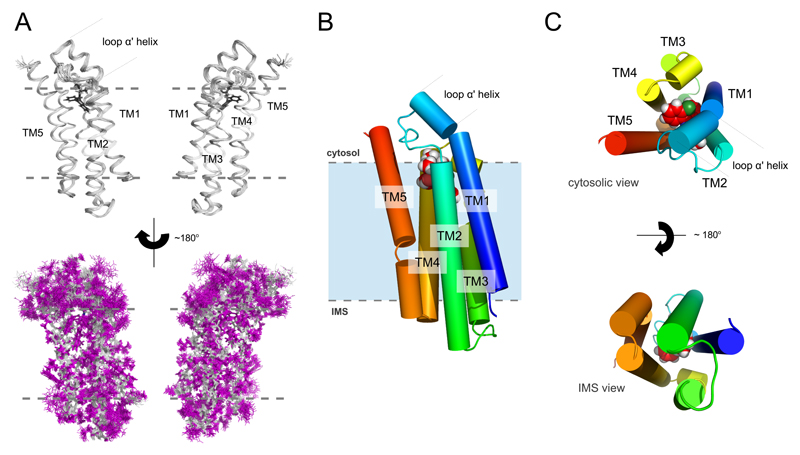
Fig. 1. High-resolution solution structure of the mTSPO-PK11195 complex.
(A) Backbone ribbon trace (upper panel) and all atom view (lower panel) of the 20 lowest-energy structures determined by NMR spectroscopy. The backbone and the side chains are shown in silver and magenta, respectively. The ligand is black. (B) Cylindrical representation of the lowest-energy complex structure with the ligand atoms shown as spheres. (C) Cytosolic and IMS views of the mTSPO-PK11195 complex. Gray dotted lines in (A) and (B) indicate approximate membrane boundaries.
The 3D structure of the mTSPO-PK11195 complex comprises five transmembrane helices (TM1 to TM5) that tightly pack together in the clockwise order TM1-TM2-TM5-TM4-TM3 when viewed from the cytosol (Figs. 1B,C). TM1 is perturbed by P15, while TM2 and TM3 are perturbed by the double-proline motifs 44PP45 and 96PP97, respectively. TM2 and TM3 also contain prolines at positions 51 and 81, respectively, and TM5 has a proline a position 139 that kinks it towards the intermembrane space (IMS). The C-terminus is highly positively charged and exposed to the cytoplasm (Fig. S6), as established by topological analysis (16). In contrast, an equal number of positive and negative charges is present on the IMS side of the receptor (Fig. S6). The charged patches exposed to the solvent might be important for interaction with proteins carrying endogenous TSPO ligands such as cholesterol (22). The loops between the transmembrane helices are short with the exception of residues G28-P45, which connect TM1 and TM2. E29-A35 fold into a short α-helix that is inclined with respect to the long axis of TM1 and positions the TM1-TM2 loop in close proximity to TM5 and the TM3-TM4 loop (Fig. 1). Due to the high sequence conservation of TSPO (Fig. S7), the topology of the mTSPO structure is likely to be retained in other mammalian species as well as bacterial homologues.
TSPO binds drug ligands as a monomer, but can also form dimers and higher-order oligomers (11, 23). The high quality of the NMR spectra suggested that mTSPO is predominantly monomeric when reconstituted into DPC micelles (Figs. S1,S2). To further support the monomeric state of the mTSPO-PK11195 complex, we performed titration experiments using the paramagnetic compound 16-Doxyl-stearic acid. A continuous paramagnetic ring pattern around the protein molecule was observed (Fig. S8), demonstrating that the central parts of all five transmembrane helices contact the hydrophobic tails of the detergent. In addition, the paramagnetic broadening data confirmed the topology of mTSPO and delineated the regions of TSPO that become inserted into membranes (Fig. 1B,C and Fig. S8).
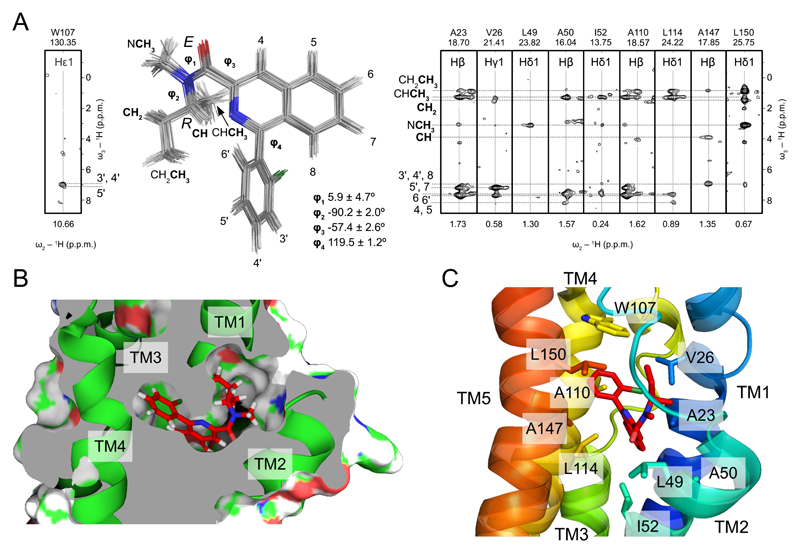
In agreement with the 1:1 stoichiometry of the TSPO-PK11195 interaction (11), only a single set of protein-ligand contacts was observed (Fig. 2A and Table S1). 61 contacts of PK11195 to TSPO described a binding pocket formed by the five transmembrane helices in the upper cytosolic part of the helical bundle (Figs. 2B,C). PK11195 binds to mTSPO with the E-amide rotamer (Figs. 2A,B), although free in solution both the E- and Z-rotameric conformation are possible (24). PK11195 contacts several conserved residues in the binding pocket that is formed by residues A23, V26, L49, A50, I52, W107, L114, A147 and L150 (Figs. 2B,C and Fig. S7). One of these residues, A147, is particularly interesting, as its natural polymorphism to threonine (Ala147Thr polymorphism, rs6971) strongly affects TSPO-binding of second-generation radioligands - but not PK11195 - and thereby the application of these ligands in humans (25). The binding pocket is closed from the cytosolic side by the long loop between TM1 and TM2 (Figs. 1A,B and Fig. S7). Deletion of residues 41-51 in mTSPO as well as site-directed mutagenesis in several species supported the importance of the TM1-TM2 loop for PK11195 binding (19, 26). The mode of PK11195-TSPO binding is likely to be important for other interactions, as PK11195 competes with several small molecules for binding to TSPO (1, 2, 10). In addition, synthetic or endogenous ligands might involve additional binding sites (1, 2, 10), providing a further level of regulation of TSPO function.
Fig. 2. Binding of PK11195 to TSPO.
(A) Superposition of the 20 lowest-energy (R)-PK11195 conformations as found in the TSPO-PK11195 complex structure. Strips from 3D F1-13C,15N-filtered/edited-NOESY-1H-13C-HSQC (right) and 3D F1-13C,15N-filtered/edited-NOESY-1H-15N-HSQC (left) experiments show intermolecular NOEs between PK11195 and selected TSPO residues. Ligand atoms are labeled. Dihedral angles φ1, φ2, φ3, and φ4 are the average values found in the 20 lowest-energy structures ± st.dev. (B) Detailed view of the PK11195 binding cavity. Transmembrane helices are shown as green ribbons, the ligand in a stick representation. (C) TSPO residues that form the ligand binding pocket. The same residues as in (A) are shown in a stick representation. Transmembrane helices are color coded.
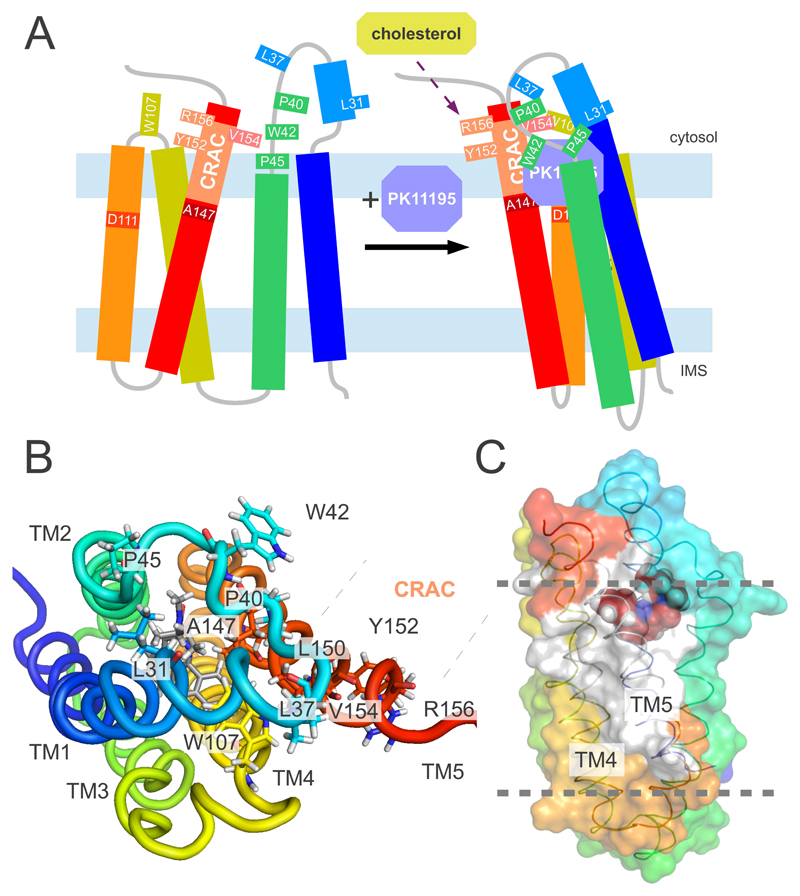
Cholesterol binds with nanomolar affinity to recombinant TSPO (11). The interaction occurs through the cholesterol recognition sequence 147ATVLNYYVWRDNS159 at the carboxylic terminus of TM5 (Fig. 3A) (19, 27). The 3D structure of the TSPO-PK11195 complex reveals that the side chains of Y152, Y153 and R156, which are essential for cholesterol binding (19, 27), are located on the outside of the TSPO structure and point toward the membrane environment (Fig. 3B). They are therefore not involved in binding to PK11195, consistent with the finding that site-directed mutagenesis of these residues inhibited binding to cholesterol but not to PK11195 (19, 27). The location of residues essential for cholesterol binding at the outside of the TSPO structure in combination with the known ability of cholesterol to dimerize suggests that cholesterol binding can modulate oligomerization of TSPO. Indeed, several transporters function as dimers (28). Residues from the cholesterol recognition sequence as well as L112-V115 in TM4 are highly stable as evidenced by hydrogen/deuterium exchange (Fig. 3C and Fig. S9). Thus binding of PK11195 stabilizes the 3D structure of TSPO, in agreement with the pronounced increase in NMR signal dispersion upon addition of PK11195 (Fig. S1). The ligand-induced stabilization of the TSPO structure might provide a mechanism to promote transport of cholesterol (Fig. 3A) (12, 13), consistent with the observation that PK11195 dramatically increased the binding of cholesterol to TSPO polymers (23).
Fig. 3. Model for the ligand-induced modulation of the TSPO structure.
(A) Cartoon representation of the conformation of TSPO in the absence (left) and presence (right) of PK11195. The representation of the ligand-bound state is based on the 3D structure of the mTSPO-PK11195 complex shown in Fig. 1. In agreement with circular dichroism (17), the secondary motifs in the ligand-free state have been assumed to be similar to the one in the mTSPO-PK11195 complex. The 2D correlation spectrum of mTSPO in DPC micelles displayed narrow signal dispersion (Fig. S1), indicating increased mobility and decreased helix packing of the ligand-free structure. The cholesterol recognition amino acid consensus, CRAC, is labeled. Upon ligand binding, the cytosolic entrance to the channel is covered by the lid comprising the TM1-TM2 loop. (B) Atomic resolution view onto the TM1-TM2 lid. The CRAC residues Y152 and R156, which are essential for cholesterol binding (19, 27), are located on the outside of the TSPO structure and are not involved in PK11195 binding. (C) Surface representation of the TSPO-PK11195 complex highlighting the residues that were most protected after 28 h of hydrogen/deuterium exchange (marked in white). The rest of the protein is colored as in (A). Gray dotted lines indicate the approximate membrane boundaries.
The 3D structure of the TSPO-PK11195 complex reveals how the members of this important receptor family are organized at the molecular level and provides a basis for understanding the function of TSPO in physiological and pathological conditions.
Supplementary Material
One Sentence Summary.
A ligand stabilizes a mammalian mitochondrial cholesterol transporter allowing determination of its structure.
Acknowledgments
The 3D structure of the mTSPO-PK11195 complex has been deposited at the ProteinDataBank (accession code: 2mgy). This work was partially supported by the Foundation for Polish Science (Fundacja na rzecz Nauki Polskiej, FNP, http://www.fnp.org.pl/) START and Ventures Programme (to M.J. and L.J.) co-financed by the EU European Regional Development Fund. M.Z was supported the DFG Collaborative Research Center 803, Project A11, and the ERC (ERC grant agreement number 282008).
References and Notes
- 1.Papadopoulos V, et al. Translocator protein (18kDa): new nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol Sci. 2006;27:402. doi: 10.1016/j.tips.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Rupprecht R, et al. Translocator protein (18 kDa) (TSPO) as a therapeutic target for neurological and psychiatric disorders. Nat Rev Drug Discov. 2010;9:971. doi: 10.1038/nrd3295. [DOI] [PubMed] [Google Scholar]
- 3.Fan J, Lindemann P, Feuilloley MG, Papadopoulos V. Structural and functional evolution of the translocator protein (18 kDa) Curr Mol Med. 2012;12:369. doi: 10.2174/1566524011207040369. [DOI] [PubMed] [Google Scholar]
- 4.Braestrup C, Squires RF. Specific benzodiazepine receptors in rat brain characterized by high-affinity (3H)diazepam binding. Proc Natl Acad Sci U S A. 1977;74:3805. doi: 10.1073/pnas.74.9.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veenman L, Papadopoulos V, Gavish M. Channel-like functions of the 18-kDa translocator protein (TSPO): regulation of apoptosis and steroidogenesis as part of the host-defense response. Curr Pharm Des. 2007;13:2385. doi: 10.2174/138161207781368710. [DOI] [PubMed] [Google Scholar]
- 6.Veenman L, Gavish M. The role of 18 kDa mitochondrial translocator protein (TSPO) in programmed cell death, and effects of steroids on TSPO expression. Curr Mol Med. 2012;12:398. doi: 10.2174/1566524011207040398. [DOI] [PubMed] [Google Scholar]
- 7.Frank W, et al. A mitochondrial protein homologous to the mammalian peripheral-type benzodiazepine receptor is essential for stress adaptation in plants. Plant J. 2007;51:1004. doi: 10.1111/j.1365-313X.2007.03198.x. [DOI] [PubMed] [Google Scholar]
- 8.Scarf AM, Ittner LM, Kassiou M. The translocator protein (18 kDa): central nervous system disease and drug design. J Med Chem. 2009;52:581. doi: 10.1021/jm8011678. [DOI] [PubMed] [Google Scholar]
- 9.Rupprecht R, et al. Translocator protein (18 kD) as target for anxiolytics without benzodiazepine-like side effects. Science. 2009;325:490. doi: 10.1126/science.1175055. [DOI] [PubMed] [Google Scholar]
- 10.Owen DR, Matthews PM. Imaging brain microglial activation using positron emission tomography and translocator protein-specific radioligands. Int Rev Neurobiol. 2011;101:19. doi: 10.1016/B978-0-12-387718-5.00002-X. [DOI] [PubMed] [Google Scholar]
- 11.Lacapere JJ, et al. Structural and functional study of reconstituted peripheral benzodiazepine receptor. Biochem Biophys Res Commun. 2001;284:536. doi: 10.1006/bbrc.2001.4975. [DOI] [PubMed] [Google Scholar]
- 12.Krueger KE, Papadopoulos V. Peripheral-type benzodiazepine receptors mediate translocation of cholesterol from outer to inner mitochondrial membranes in adrenocortical cells. J Biol Chem. 1990;265:15015. [PubMed] [Google Scholar]
- 13.Papadopoulos V, Mukhin AG, Costa E, Krueger KE. The peripheral-type benzodiazepine receptor is functionally linked to Leydig cell steroidogenesis. J Biol Chem. 1990;265:3772. [PubMed] [Google Scholar]
- 14.Barron AM et al. Ligand for translocator protein reverses pathology in a mouse model of Alzheimer's disease. J Neurosci. 2013;33:8891. doi: 10.1523/JNEUROSCI.1350-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gatliff J, Campanella M. The 18 kDa translocator protein (TSPO): a new perspective in mitochondrial biology. Curr Mol Med. 2012;12:356. doi: 10.2174/1566524011207040356. [DOI] [PubMed] [Google Scholar]
- 16.Joseph-Liauzun E, Delmas P, Shire D, Ferrara P. Topological analysis of the peripheral benzodiazepine receptor in yeast mitochondrial membranes supports a five-transmembrane structure. J Biol Chem. 1998;273:2146. doi: 10.1074/jbc.273.4.2146. [DOI] [PubMed] [Google Scholar]
- 17.Murail S, et al. Secondary and tertiary structures of the transmembrane domains of the translocator protein TSPO determined by NMR. Stabilization of the TSPO tertiary fold upon ligand binding. Biochim Biophys Acta. 2008;1778:1375. doi: 10.1016/j.bbamem.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 18.Korkhov VM, Sachse C, Short JM, Tate CG. Three-dimensional structure of TspO by electron cryomicroscopy of helical crystals. Structure. 2010;18:677. doi: 10.1016/j.str.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Papadopoulos V. Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus pattern. Endocrinology. 1998;139:4991. doi: 10.1210/endo.139.12.6390. [DOI] [PubMed] [Google Scholar]
- 20.Shah F, Hume SP, Pike VW, Ashworth S, McDermott J. Synthesis of the enantiomers of [N-methyl-11C]PK 11195 and comparison of their behaviours as radioligands for PK binding sites in rats. Nucl Med Biol. 1994;21:573. doi: 10.1016/0969-8051(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 21.Supplementary materials are available on Science Online.
- 22.Liu J, Rone MB, Papadopoulos V. Protein-protein interactions mediate mitochondrial cholesterol transport and steroid biosynthesis. J Biol Chem. 2006;281:38879. doi: 10.1074/jbc.M608820200. [DOI] [PubMed] [Google Scholar]
- 23.Delavoie F, et al. In vivo and in vitro peripheral-type benzodiazepine receptor polymerization: functional significance in drug ligand and cholesterol binding. Biochemistry. 2003;42:4506. doi: 10.1021/bi0267487. [DOI] [PubMed] [Google Scholar]
- 24.Lee YS, Simeon FG, Briard E, Pike VW. Solution structures of the prototypical 18 kDa translocator protein ligand, PK 11195, elucidated with 1H/13C NMR spectroscopy and quantum chemistry. ACS Chem Neurosci. 2012;3:325. doi: 10.1021/cn3000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owen DR, et al. Mixed-affinity binding in humans with 18 kDa translocator protein ligands. J Nucl Med. 2011;52:24. doi: 10.2967/jnumed.110.079459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farges R, et al. Site-directed mutagenesis of the peripheral benzodiazepine receptor: identification of amino acids implicated in the binding site of Ro5-4864. Mol Pharmacol. 1994;46:1160. [PubMed] [Google Scholar]
- 27.Jamin N, et al. Characterization of the cholesterol recognition amino acid consensus sequence of the peripheral-type benzodiazepine receptor. Mol Endocrinol. 2005;19:588. doi: 10.1210/me.2004-0308. [DOI] [PubMed] [Google Scholar]
- 28.Morrison EA, Henzler-Wildman KA. Reconstitution of integral membrane proteins into isotropic bicelles with improved sample stability and expanded lipid composition profile. Biochim Biophys Acta. 2012;1818:814. doi: 10.1016/j.bbamem.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keller R. PhD thesis Diss ETH Nr; 15947. [Google Scholar]
- 30.Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci U S A. 1998;95:13585. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A. 1997;94:12366. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bax A. Multidimensional Nuclear-Magnetic-Resonance Methods for Protein Studies. Curr Opin Struct Biol. 1994;4:738. [Google Scholar]
- 33.Riek R, Wider G, Pervushin K, Wuthrich K. Polarization transfer by cross-correlated relaxation in solution NMR with very large molecules. Proc Natl Acad Sci U S A. 1999;96:4918. doi: 10.1073/pnas.96.9.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wuthrich K. NMR of proteins and nucleic acids. Wiley Interscience; 1986. [Google Scholar]
- 35.Marion D et al. Overcoming the Overlap Problem in the Assignment of H-1Nmr Spectra of Larger Proteins by Use of 3 Dimensional Heteronuclear H-1 N-15 Hartmann-Hahn Multiple Quantum Coherence and Nuclear Overhauser Multiple Quantum Coherence Spectroscopy - Application to Interleukin-1-Beta. Biochemistry. 1989;28:6150. doi: 10.1021/bi00441a004. [DOI] [PubMed] [Google Scholar]
- 36.Shen Y, Bax A. Prediction of Xaa-Pro peptide bond conformation from sequence and chemical shifts. J Biomol NMR. 2010;46:199. doi: 10.1007/s10858-009-9395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bax A, Clore GM, Gronenborn AM. H-1-H-1 Correlation Via Isotropic Mixing of C-13 Magnetization, a New 3-Dimensional Approach for Assigning H-1 and C-13 Spectra of C-13 Enriched Proteins. J Magn Reson. 1990;88:425. [Google Scholar]
- 38.Lundstrom P, et al. Fractional 13C enrichment of isolated carbons using [1-13C]- or [2-13C]-glucose facilitates the accurate measurement of dynamics at backbone Calpha and side-chain methyl positions in proteins. J Biomol NMR. 2007;38:199. doi: 10.1007/s10858-007-9158-6. [DOI] [PubMed] [Google Scholar]
- 39.Zwahlen C, et al. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: Application to a bacteriophage lambda N-peptide/boxB RNA complex. J Am Chem Soc. 1997;119:6711. [Google Scholar]
- 40.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 41.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 43.Farrow NA, et al. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 44.Berjanskii MV, Wishart DS. A simple method to predict protein flexibility using secondary chemical shifts. J Am Chem Soc. 2005;127:14970. doi: 10.1021/ja054842f. [DOI] [PubMed] [Google Scholar]
- 45.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.