Abstract
Plastids, the photosynthetic organelles of eukaryotes, exhibit remarkably stable genome architecture. However, a recent study of microscopic red algae has found new record-sized plastid genomes with unusual architectures. These species form a new branch in the tree of red algae.
More than one billion years ago, a heterotrophic protist engulfed a cyanobacterium which, instead of being digested immediately, established a long-term symbiotic relationship with its predator. This cyanobacterium transformed into a stable cell organelle, the plastid, and allowed the birth of the first photosynthetic eukaryotes. This evolutionary event had planetary consequences since photosynthetic eukaryotes occupied new ecological niches, diversified extraordinarily and became major players in the global carbon cycle. Three eukaryotic phyla are the direct descendants of this cyanobacterial endosymbiosis: the rare and poorly-known glaucophyte algae and the much more widespread and species-rich red algae, green algae and land plats [1]. Both red and green algae have been privileged targets of study for several generations of biologists and their diversity was considered to be fairly well known. Moreover, the increasing availability of genome and transcriptome sequence data for a variety of species has led to the construction of robust multi-gene phylogenies for these algae [2,3]. Plastid genomes have been invaluable as data sources for algal phylogeny studies, such that good knowledge of these organellar genomes has accumulated over the years. When compared with typical contemporary cyanobacterial genomes (encoding between 1,800 and 12,000 proteins), which likely resemble the genome of the cyanobacterial plastid ancestor, plastid genomes appear highly reduced, compact (they encode only between 80 and 230 proteins), and conservative regarding their architecture [4,5]. However, by analyzing six understudied filamentous and unicellular red microalgae (belonging to the classes Stylonematophyceae, Compsopogonophyceae, Porphyridiophyceae, and Rhodellophyceae), Muñoz-Gómez et al. have unveiled plastid genome sequences of unprecedented size and complexity [6].
The largest plastid genomes previously reported are found in green algae like Floydiella and Volvox, which reach sizes of ~500 Kbp [7,8]. Two of the red plastid genomes characterized by Muñoz-Gómez et al. surpass them: Bulboplastis apyrenoidosa (610 Kbp) and, especially, Corynoplastis japonica, which exceeds 1 Mbp. These record sizes do not correlate with increased numbers of protein coding genes (163 and 179, respectively), which remain in the range of other red plastid genomes and are even smaller than those of plastids of much reduced size (e.g., the 205 Kbp plastid genome of Bangiopsis subsimplex contains 192 protein coding genes). Surprisingly, despite being evolutionarily related within the same class Rhodellophyceae, these two species have followed very different evolutionary paths to increase their plastid genome sizes. Whereas the plastid genome of C. japonica has been massively invaded by introns (almost 64% of the genome), that of B. apyrenoidosa has been overrun by insertion sequences of bacterial origin, in addition to a notable number of introns [6]. These observations, accompanied by a high level of gene order rearrangements, break the high conservation of plastid genome architecture paradigm [5].
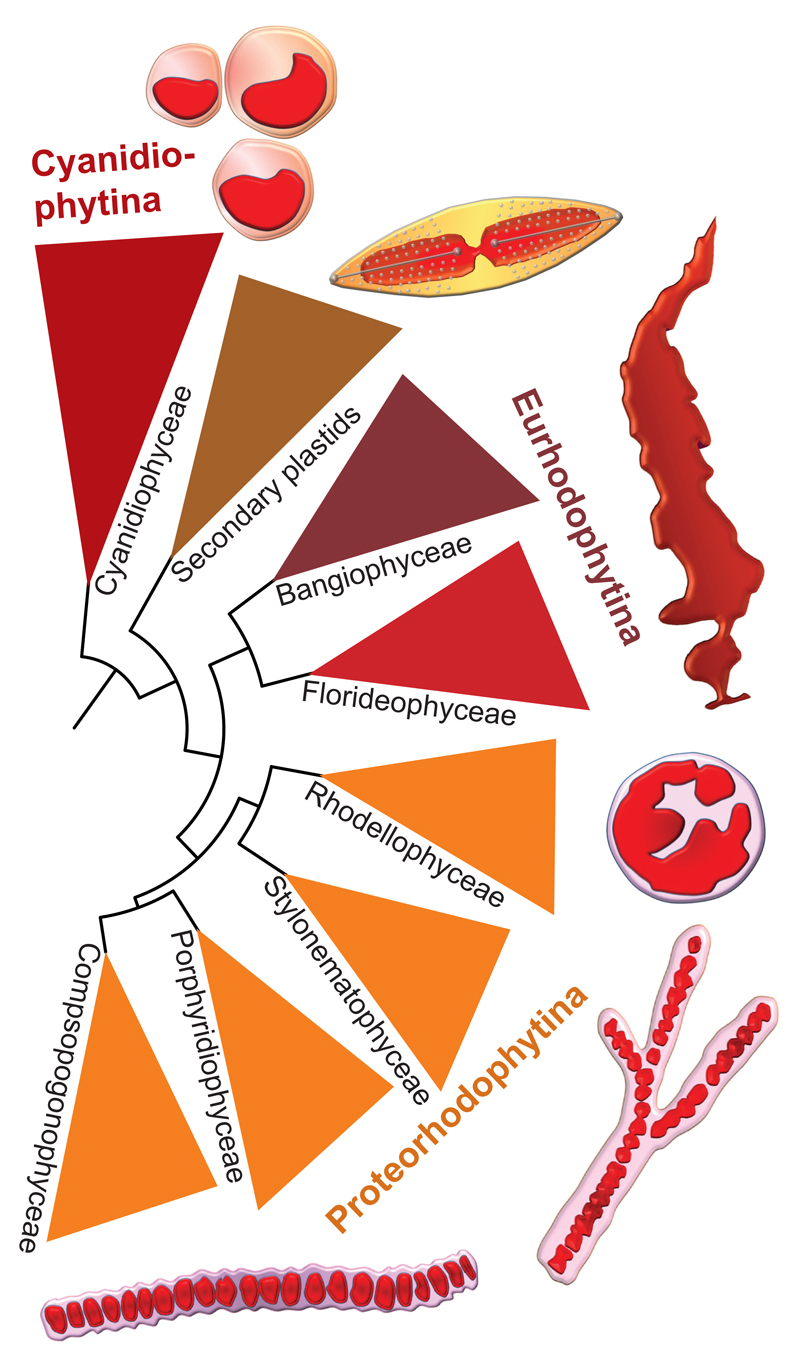
The new red algal plastid genomes have also revealed to be a precious source of phylogenetic information. Trees based on the analysis of 170 proteins encoded in plastid genomes of 37 red algae strongly support that the four classes Stylonematophyceae, Compsopogonophyceae, Porphyridiophyceae, and Rhodellophyceae form a monophyletic group branching deeply in the red algal tree, which Muñoz-Gómez et al. propose to classify as the new subphylum 'Proteorhodophytina' (Figure 1). A very recent phylogenetic study, this time based on a large dataset of 298 nucleus-encoded protein sequences fetched from genome and transcriptome data, also retrieved the monophyly of the four classes, although with weaker resolution of the relationships among them [3]. The congruence between the trees based on plastid- and nucleus-encoded proteins offers a strong argument in favor of the reliability of the new red algal phylogeny. It contradicts previous phylogenies, based on much more reduced sequence datasets, where these four red algal classes emerged as four independent, paraphyletic branches basal to the well-known Florideophyceae and Bangiophyceae, the two classes containing multicellular macroscopic species [9]. According to this previous view of the red algal phylogeny, these red seaweeds would have emerged relatively late during the evolution of red algae, being preceded by several lineages of microscopic species. However, in the new phylogeny, the Florideophyceae and Bangiophyceae branch very deeply as the sister clade of the Proteorhodophytina, opening the possibility for an early origin of red algal seaweeds. Paleontology appears also to support this possibility, notably by the recent discovery of exceptionally preserved ~1.6 billion years old multicellular fossils that exhibit structural similarity with florideophycean seaweeds [10].
Figure 1. The new phylogeny of red algae based on plastid genome data.
This phylogeny, based on Muñoz-Gómez et al. [6], shows the new sub-phylum Proteorhodophytina, composed of unicellular and filamentous microscopic species. Red secondary plastids branch deeply with no close relatives among the known red algal lineages.
The new phylogeny of red algae provides a robust evolutionary framework for the study of one of the most hotly debated questions in evolutionary biology: the origin of secondary red plastids. As explained above, the original cyanobacterial endosymbiosis that endowed eukaryotes with the capacity to carry out photosynthesis, called the 'primary endosymbiosis', gave rise to three eukaryotic phyla: Glaucophyta, Rhodophyta, and Viridiplantae [11,12]. However, photosynthesis spread into many other phyla through secondary and tertiary endosymbioses, namely symbioses involving hosts that acquired different eukaryotic algae as endosymbionts. These endosymbionts can be red (in cryptophytes, haptophytes, alveolates, and stramenopiles) or green (in chlorarachniophytes and euglenids) algae [12]. In contrast with primary plastids, surrounded by two membranes, all these secondary plastids possess at least three membranes. Whereas it is clear that green secondary plastids evolved through two independent acquisitions of green algae in chlorarachniophytes and euglenids, the number of endosymbioses and the identity of the partners involved at the origin of red secondary plastids have been much more controversial issues. The main reason for this comes from the incongruence between the phylogenies based on plastid genes and those based on host nuclear genes. The first support the monophyly of all secondary red plastids as a single group within the red algae, but the second suggests that their different hosts are not monophyletic. When the first phylogenies of red secondary plastids were reconstructed, their monophyly was interpreted as strong evidence for a common origin involving a single endosymbiosis followed by an impressive diversification to give rise to all contemporary groups carrying red secondary plastids. This view, dubbed the 'Chromalveolate hypothesis' [13], began to be eroded by subsequent phylogenetic analyses showing the non-monophyly of the host component of these organisms. To reconcile these contradictory results, it has been proposed that an initial red algal endosymbiosis originated one of the groups endowed with red secondary plastids which, in turn, was acquired by the other groups through an unknown number of subsequent tertiary endosymbioses [14].
Still another alternative would be several independent secondary endosymbioses involving closely related red algae as endosymbionts. Two reasons explain why this hypothesis is much less discussed: red secondary plastid phylogenies appear to support their monophyly and SELMA, the complex molecular machinery that transports proteins from the cytosol into the multiple-membrane red secondary plastids, seems to have a single origin [15]. However, red secondary plastid phylogenies have suffered of the poor taxonomic sampling for many of the major red algal lineages [16], which may lead to a false impression of monophyly of the plastid sequences. The work of Muñoz-Gómez et al. has improved the situation by contributing sequence data for four deep-branching red algal lineages. They also inspected the phylogeny of three groups of red-secondary-plastid-containing algae (cryptophytes, haptophytes, and stramenopiles; the fast-evolving plastids of alveolates were excluded from the analysis) using a dataset of 106 plastid-encoded proteins. Bayesian analysis of this dataset retrieves a tree where all these plastids form a monophyletic group branching very deeply as sister group of a clade containing the Proteorhodophytina and the Eurhodophytina (Bangiophyceae plus Florideophyceae). However, the plastid clade does not receive full statistical support and, moreover, maximum likelihood analysis of the same dataset fails to find its monophyly, indicating that it is very fragile, even in the absence of the problematic alveolate plastids.
Thus, important questions on the origin of the red secondary plastids remain open as these organelles have not yet found their mother lineage among the red algae. Certainly, the most efficient way to address this issue is keeping characterizing the poorly-explored diversity of non-seaweed microscopic red algae. Muñoz-Gómez et al. show that this approach not only provides valuable evolutionary information but can also reserve surprises in terms of genome architecture and function.
References
- 1.Ponce-Toledo RI, Deschamps P, Lopez-Garcia P, Zivanovic Y, Benzerara K, Moreira D. An early-branching freshwater cyanobacterium at the origin of plastids. Curr Biol. 2017;27:386–391. doi: 10.1016/j.cub.2016.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leliaert F, Tronholm A, Lemieux C, Turmel M, DePriest MS, Bhattacharya D, Karol KG, Fredericq S, Zechman FW, Lopez-Bautista JM. Chloroplast phylogenomic analyses reveal the deepest-branching lineage of the Chlorophyta, Palmophyllophyceae class. nov. Sci Rep. 2016;6:25367. doi: 10.1038/srep25367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiu H, Yoon HS, Bhattacharya D. Red algal phylogenomics provides a robust framework for inferring evolution of key metabolic pathways. PLoS Curr. 2016;8 doi: 10.1371/currents.tol.7b037376e6d84a1be34af756a4d90846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janouskovec J, Liu SL, Martone PT, Carre W, Leblanc C, Collen J, Keeling PJ. Evolution of red algal plastid genomes: ancient architectures, introns, horizontal gene transfer, and taxonomic utility of plastid markers. PloS One. 2013;8:e59001. doi: 10.1371/journal.pone.0059001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee J, Cho CH, Park SI, Choi JW, Song HS, West JA, Bhattacharya D, Yoon HS. Parallel evolution of highly conserved plastid genome architecture in red seaweeds and seed plants. BMC Biol. 2016;14:75. doi: 10.1186/s12915-016-0299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muñoz-Gómez SA, Mejía-Franco FG, Durnin K, Colp M, Grisdale CJ, Archibald JM, Slamovits CH.The new red algal subphylum Proteorhodophytina comprises the largest and most divergent plastid genomes known Curr Biol 2017In press [DOI] [PubMed] [Google Scholar]
- 7.Brouard JS, Otis C, Lemieux C, Turmel M. The exceptionally large chloroplast genome of the green alga Floydiella terrestris illuminates the evolutionary history of the Chlorophyceae. Genome Biol Evol. 2010;2:240–256. doi: 10.1093/gbe/evq014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith DR, Lee RW. Low nucleotide diversity for the expanded organelle and nuclear genomes of Volvox carteri supports the mutational-hazard hypothesis. Mol Biol Evol. 2010;27:2244–2256. doi: 10.1093/molbev/msq110. [DOI] [PubMed] [Google Scholar]
- 9.Scott J, Yang EC, West JA, Yokoyama A, Kim HJ, Loiseaux de Goër S, O’Kelly CJ, Orlova E, Kim SY, Park JK, et al. On the genus Rhodella, the emended orders Dixoniellales and Rhodellales with a new order Glaucosphaerales (Rhodellophyceae, Rhodophyta) Algae. 2011;26:277–288. [Google Scholar]
- 10.Bengtson S, Sallstedt T, Belivanova V, Whitehouse M. Three-dimensional preservation of cellular and subcellular structures suggests 1.6 billion-year-old crown-group red algae. PLoS Biol. 2017;15:e2000735. doi: 10.1371/journal.pbio.2000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moreira D, Le Guyader H, Philippe H. The origin of red algae and the evolution of chloroplasts. Nature. 2000;405:69–72. doi: 10.1038/35011054. [DOI] [PubMed] [Google Scholar]
- 12.Archibald JM. The puzzle of plastid evolution. Curr Biol. 2009;19:R81–88. doi: 10.1016/j.cub.2008.11.067. [DOI] [PubMed] [Google Scholar]
- 13.Cavalier-Smith T. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origin and the eukaryote family tree. J Euk Microbiol. 1999;46:347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- 14.Baurain D, Brinkmann H, Petersen J, Rodriguez-Ezpeleta N, Stechmann A, Demoulin V, Roger AJ, Burger G, Lang BF, Philippe H. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol Biol Evol. 2010;27:1698–1709. doi: 10.1093/molbev/msq059. [DOI] [PubMed] [Google Scholar]
- 15.Gould SB, Maier UG, Martin WF. Protein import and the origin of red complex plastids. Curr Biol. 2015;25:R515–521. doi: 10.1016/j.cub.2015.04.033. [DOI] [PubMed] [Google Scholar]
- 16.Yoon HS, Hackett JD, Pinto G, Bhattacharya D. The single, ancient origin of chromist plastids. Proc Natl Acad Sci USA. 2002;99:15507–15512. doi: 10.1073/pnas.242379899. [DOI] [PMC free article] [PubMed] [Google Scholar]