Abstract
Paramagnetic effects provide structure and dynamics information of biomolecules. We developed a robust method that paramagnetically lightens up high-molecular weight proteins through binding of a reporter protein that carries lanthanide tags at distinct locations. Transmission of several independent molecular alignments provides a multitude of paramagnetic restraints for proteins of unknown 3D structure.
Keywords: NMR spectroscopy, Lanthanides, Structural biology, Proteins, Paramagnetic restraints
Lanthanoid tags are used in diverse areas such as NMR spectroscopy[1], X-ray crystallography[2] and luminescence-based measurements.[3] Unpaired electrons in f orbitals of lanthanoids cause a paramagnetic effect. When the lanthanoid is located in a target molecule, the paramagnetic effect results in weak alignment of the molecule in the magnetic field and affects the signals of its NMR spectrum in a distance (paramagnetic relaxation enhancement, PRE), orientation (residual dipolar couplings, RDC) and orientation/distance-dependent way (pseudo-contact shifts, PCS).[4] Since the magnetic moment of electrons is almost three orders of magnitude larger than that of protons, the perturbation is transmitted over long distances.[5] The long-range nature of paramagnetic restraints allows unique insights into the structure and dynamics of biomolecules.[1a, 1b, 6]
In order to obtain solvent-nucleus distance information and thereby structural information about the biomolecule, lanthanoids can be added as soluble complexes to the sample.[7] However, for access to orientation information through RDCs and intra- or intermolecular distance information, lanthanoids have to be attached to a specific site in the molecule.[1c, 8] In case of Ca2+- or Mg2+-containing metalloproteins, this can be achieved by substitution of the natural metal by lanthanoid ions[9], while for most proteins, lanthanoids have to be attached to the target protein by methods such as insertion of lanthanoid binding motifs into protein loops[10], attachment of lanthanoid binding motifs to the N- or C-terminus[4f, 11] or covalent tagging of a small organic molecule to a surface-reactive cysteine.[1c, 12] In addition, diamagnetic proteins lacking native metal-binding sites might be fused with entire paramagnetic protein domains such as zinc finger proteins[13], EF hand motifs[14] or calmodulin-binding peptides loaded with paramagnetic lanthanoids to transmit molecular alignment.[4a]
For the lanthanoid tagging methods it is essential that the modification does not perturb the structure and dynamics of the protein. In addition, covalent tagging to reactive cysteines requires one or a pair (in case of double-legged tags) of unique surface accessible cysteines, a condition that can be difficult to fulfil especially in large proteins, when they contain several cysteine residues. Another problem is that in cases where the 3D structure of the target protein is unknown, it can be difficult to insert a lanthanoid binding loop or select a surface accessible site to attach the tag. The situation is even more challenging, when complementary sets of paramagnetic data are to be acquired from different positions of the tag on the surface of the target protein, which generally requires different single cysteine mutants of the target protein.
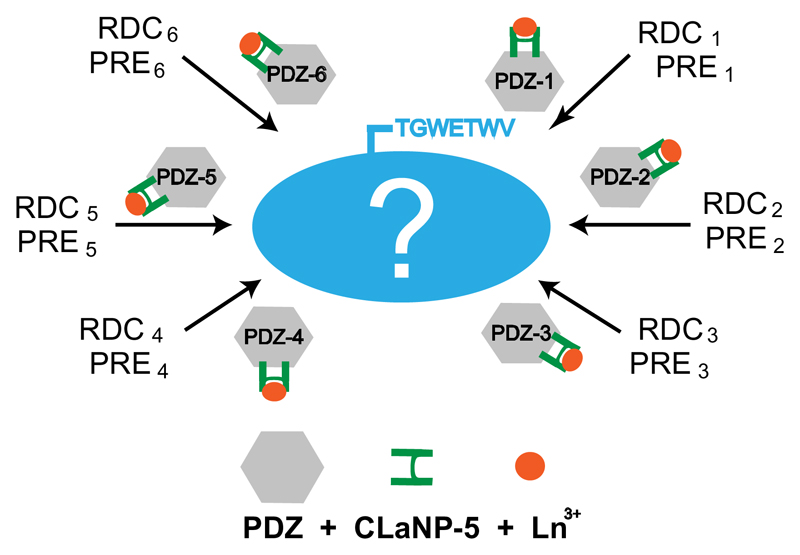
Here we present a strategy that alleviates many of the above mentioned drawbacks and was motivated by the early approaches of fusion with a paramagnetic domain.[4a, 13–14] The central idea is to attach the lanthanoid tag not to the target protein itself, but instead to a reporter protein, which in turns binds and hence transmits the paramagnetic effects to any target that contains a specific recognition sequence (Figure 1). In this way the reporter protein can be tagged in a separate reaction before its mixture/binding with the target protein, such that it can be applied to proteins with multiple surface accessible cysteines. An important advantage of the present strategy is that it is applicable to proteins of unknown 3D structure: as long as there is binding between the target and the reporter protein, the paramagnetic effect is transmitted. In addition, by designing a set of reporter mutants that are tagged on different position on its surface, each mutant can produce independent molecular alignments on the target protein and the same set of reporter mutants can be employed to study different proteins of interest.
Figure 1.
Schematic diagram illustrating the developed method. The lanthanoid tag is not attached to the target protein itself (blue), but instead to a reporter protein (grey), which in turn binds and hence transmits the paramagnetic effect to any target that contains a specific recognition sequence (TGWETWV). By attaching the lanthanoid tag at different positions in PDZ, multiple independent alignments of the target protein are obtained.
The reporter protein that was chosen is the Erbin PDZ domain (Figure S1)[18]. It is a stable, 103-residue, well-characterized protein that does not contain any cysteine and binds with high affinity to the sequence TGWETWV when it is located at the C-terminus of another protein (Figure S2).[19] Six different double-cysteine mutants of the Erbin PDZ domain (PDZ-1 to PDZ-6) were designed (Figure S1). The mutants were then tagged with the lanthanoid tag CLaNP-5[20], which had been preloaded with the paramagnetic lanthanoid Tm3+ or diamagnetic Lu3+. CLaNP-5 attaches to two cysteine residues, resulting in increased rigidity of the tag. Even though the tagging of all PDZ mutants was optimized, tagging efficiency greatly varied (Table S1). In addition, PDZ-4 turned out to be unstable and was excluded from further analysis.
The strategy was tested on the 8 kDa protein ubiquitin and the 42 kDa maltose binding protein[21] (MBP), both with the C-terminal TGWETWV extension. Binding of wild-type PDZ to ubiquitin and MBP was slow on the NMR time scale. In line with the high affinity of the PDZ-TGWETWV interaction, binding to MBP was saturated at a 2:1 excess of PDZ. As the PDZ domain is not labelled with 15N, unbound PDZ does not contribute to the signal in 15N-edited NMR experiments. In addition, chemical shift changes induced by PDZ-binding in ubiquitin and MBP away from the C-terminus were small, indicating that PDZ-binding does not perturb the structure of the protein of interest (Figure S3a and S3b). The data demonstrate formation of well-defined PDZ-protein complexes suitable for NMR.
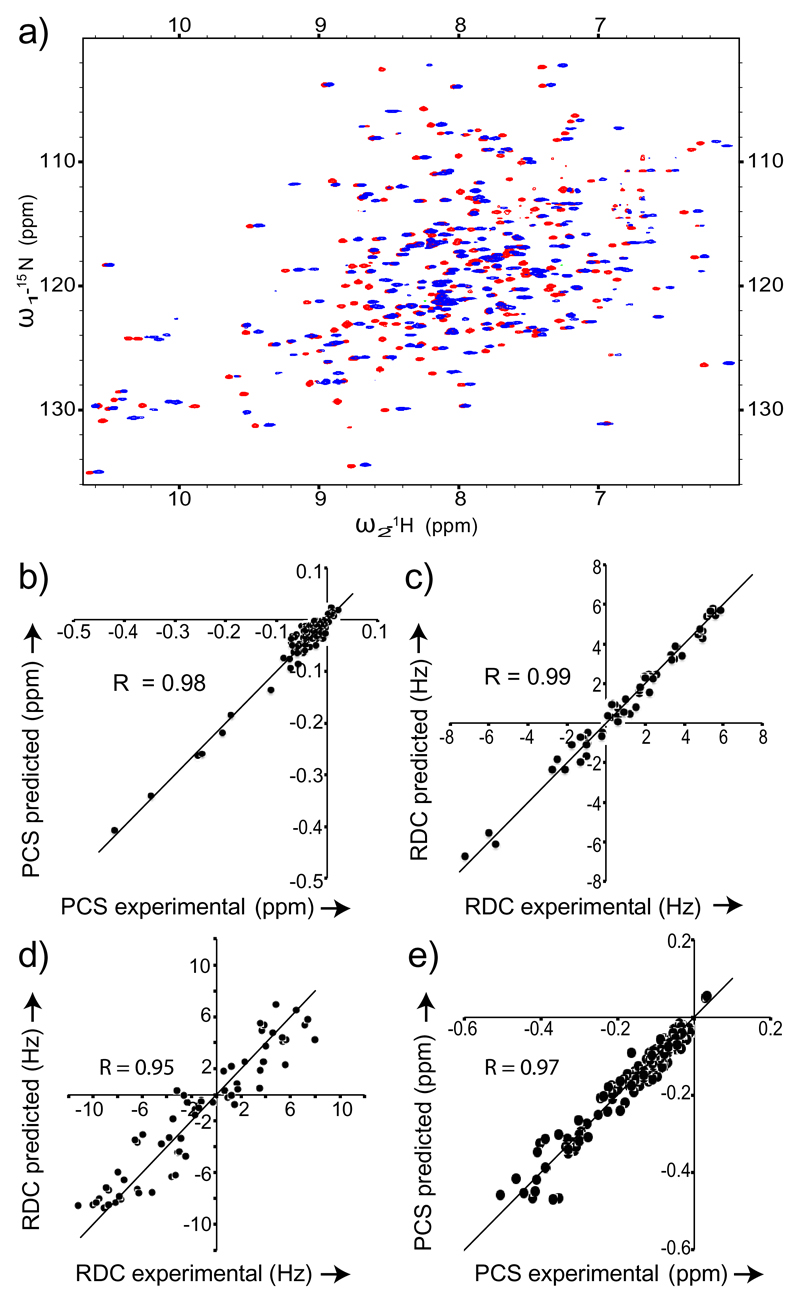
We then added the PDZ mutants, already tagged with CLaNP-5, to 15N-labelled ubiquitinTGWETWV, as well as 15N/2H-labelled or 15N/13C/2H-labeled MBPTGWETW. Binding of the PDZ domains induced RDCs, PCSs and PREs (Figure 2, S3 and S4). 2D 1H-15N HSQC spectra of ubiquitin and MBP were of high quality (Figure 2a and S3b). In case of MBP, we also obtained 3D HNCO experiments (Figure S4g). Thus, the increase in molecular weight due to binding of the PDZ domain (Figure S5) did not interfere with the precise measurement of RDCs and PCSs (Figures 2 and S3e).
Figure 2.
RDCs and PCSs transmitted by PDZ variants to ubiquitinTGWETWV and MBPTGWETWV. a) Superposition of 1H-15N HSQC spectra of MBPTGWETWV bound to PDZ-1 tagged with CLaNP-5,) Correlation between two datasets of PCSs (recorded in identical experimental conditions at 600 MHz) induced by PDZ-1 in ubiquitinTGWETWV. c) RDCs induced in ubiquitinTGWETWV by PDZ-1 binding (1H Larmor frequency of 900 MHz; 37°C) were acquired using the BSD-IPAP HSQC[15] experiment and fit to the 3D structure of ubiquitin (PDB code: 1D3Z) using PALES. d) RDCs observed at 900 MHz were fitted to MBP’s 3D structure (PDB code: 1DMB) using the program PALES.[16] e) Experimental 15N and 1H PCS induced in MBPTGWETWV by PDZ-2 were fit using the program Numbat[17]. In both plots the line indicates y=x. Only cross-peaks not strongly affected by signal overlap were included into the analysis.
At a proton Larmor frequency of 900 MHz, RDCs induced by PDZ-1 in ubiquitin ranged from -7.3 to +5.8 Hz (Figure 2c). The fit of 57 experimental RDCs to the 3D structure of ubiquitin was of very high quality with a Pearson's correlation coefficient of 0.99. At 600 MHz, the RDCs decreased according to the change in field strength by a factor 2.25, but the correlation coefficient to the 3D structure remained at 0.98 (Figure S3c). In case of MBP, experimental RDC induced by PDZ-2 were in the range from -11.2 to 7.9 Hz (Figure 2d and Table 1). The fit of the experimental RDCs and PCSs to the 3D structure of MBP resulted in Pearson correlation coefficients of 0.95 and 0.97, respectively (Figure 2d, e). The lower quality of the RDC fit when compared to ubiquitin is due to the lower precision of the RDC measurement at the increased molecular weight as well as the lower resolution of the 3D structure of MBP. The magnitude of alignment transmitted to MBP varied between different PDZ mutants (Table 1). The small size of RDCs transmitted by PDZ-5 is likely due to a higher flexibility of the CLaNP-5 tag in PDZ-5, where the two cysteine residues/attachment sites are not located in rigid secondary structures (Figure S1). Taken together the data demonstrated that high-quality paramagnetic restraints for both small and large proteins can be obtained by external tagging.
Table 1.
Parameters of the alignment tensors transmitted to the 42 kDa protein MBP by the five different PDZ mutants tagged with CLaNP-5. Alignment tensors and Q-values were obtained by best-fit of the experimental RDC to the 3D structure 1DMB[21a] using the software PALES [16, 22].
| PDZ mutants | Aa (10-4) | Ar (10-4) | αc | β c | γ c | Q value |
|---|---|---|---|---|---|---|
| PDZ-1a | 2.8 | 2.2 | -13.5 | -17.9 | 37.2 | 0.23 |
| PDZ-2b | -3.8 | -1.8 | 42.9 | 239.1 | -8.4 | 0.26 |
| PDZ-3a | -1.2 | -0.5 | 78.9 | -32.8 | 251.1 | 0.29 |
| PDZ-5b | 1.6 | 0.3 | 111.1 | -41.1 | -42.1- | 0.24 |
| PDZ-6b | 2.8 | 1.2 | 90.8 | 22.0 | -1.1 | 0.28 |
[a] Measured at 800 MHz or [b] at 900 MHz; [c] Euler angles.
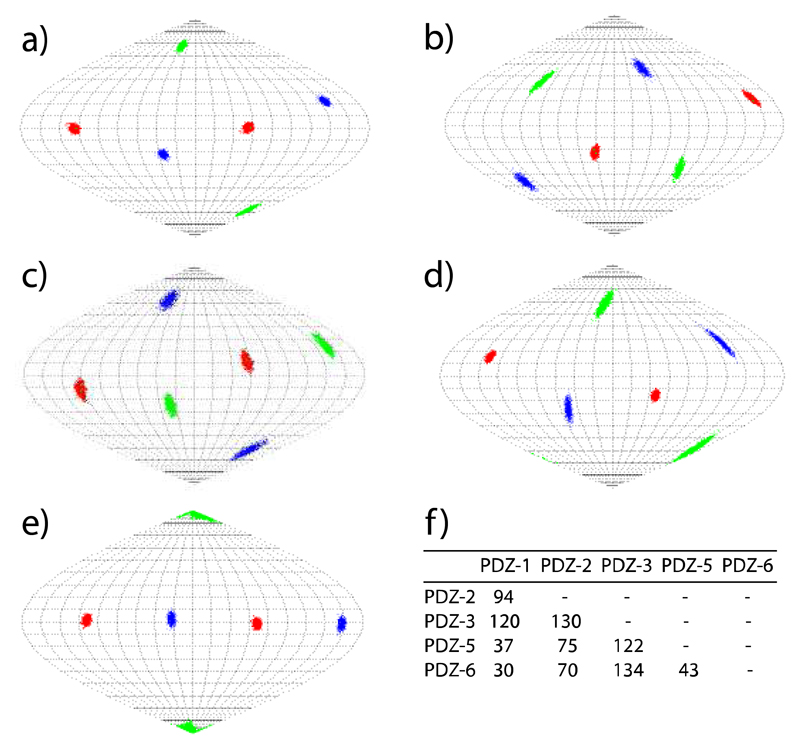
The experimental RDCs were then used to determine the properties of the alignment tensors induced in MBP by the five PDZ variants. Figure 3 illustrates the orientation of the alignment tensors using a Sanson-Flamsteed projection. In agreement with the high quality of the experimental RDCs, the orientations of the three tensor axes were well defined. Moreover, the orientations of the axes differed largely between the PDZ variants. Quantification using a generalized angle in the five-dimensional space spanned by the Saupe matrix revealed that each alignment tensor differed by more than 30° from any of the other four tensors (Figure 3f). For some of the combinations, 5D angles of 90-130° were found. Thus, the use of the different PDZ variants allows access to multiple independent alignments.
Figure 3.
Orientations of the alignment tensor transmitted to the 42 kDa protein MBPTGWETWV by the CLaNP-5 tagged PDZ mutants 1, 2, 3, 5, 6 (a-e). z, y and x axis are shown in red, blue and green in Sanson Flamsteed projections, respectively. The orientation of the alignment tensors was obtained by fitting the experimental RDCs to MBP’s 3D structure (PDB code: 1DMB).[21a] Uncertainties were evaluated by 1000 cycles of the "structural noise Monte-Carlo method".[16, 23] f) 5D angles between the alignment tensors induced in MBP.
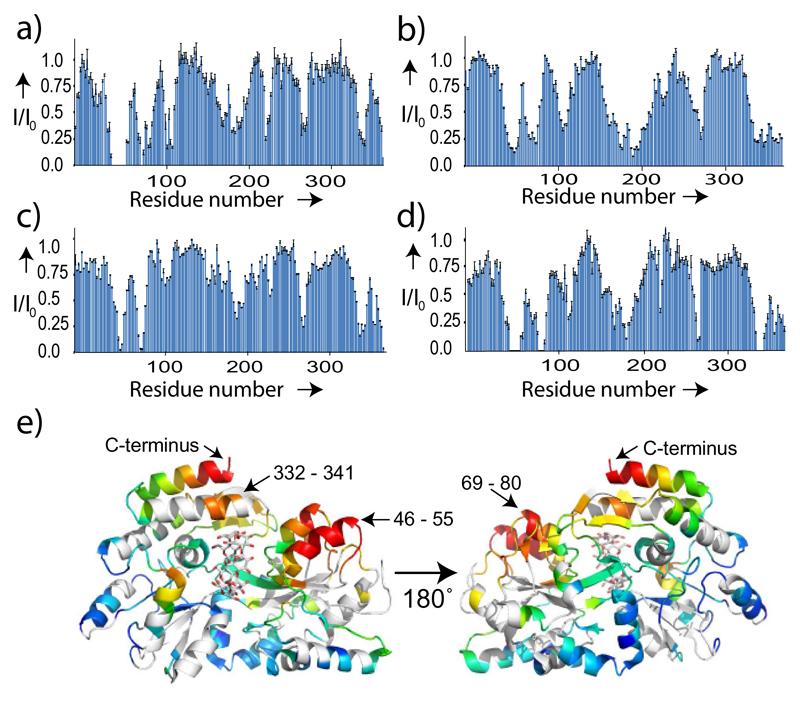
In addition to orientational restraints, lanthanoids caused relaxation of NMR signals of nearby nuclei. For example, when adding PDZ-1 to MBPTGWETWV, residues from 46 to 55 were broadened beyond detection (Figure 4a). The location of these residues in the 3D structure of MBP is close to the C-terminus, where PDZ binds to the peptide sequence TGWETWV (Figure 4e). Comparison of PRE broadening profiles of MBP in complex with different PDZ variants further showed that larger PRE effects were produced by those PDZ variants for which the tagging site is close to the TGWETWV binding pocket. For example, PDZ-1, whose tagging site is just next to several residues that participate in binding to TGWETWV (Figure S1), produced the biggest PRE (Figure 4a), while PDZ-3, in which the tagging site is far away from the binding pocket, caused less PRE (Figure 4b). Importantly, beyond an overall similarity of the PRE effect along the protein sequence, the PRE profiles show distinct sequence-specific features (see for example the PRE broadening at the C-terminus of MBP in case of PDZ-1, PDZ-3 and PDZ-5 in Figure 4). Thus, a wealth of long-range distance information is accessible without internal modification of the target protein.
Figure 4.
External tagging provides access to long-range distance information in the 42 kDa protein MBP. a) - d) PRE profiles in MBPTGWETWV upon addition of CLaNP-5 tagged PDZ-1, PDZ-3, PDZ-5 and PDZ-6, respectively. Error bars were calculated on the basis of the signal-to-noise ratio in the NMR spectra. Only cross-peaks not strongly affected by signal overlap (also with respect to residual diamagnetic signals) were included into the analysis. e) Mapping of the PRE broadening of MBPTGWETWV signals produced by PDZ-1 onto the 3D structure 1DMB. From red (most affected) to blue (unaffected) the distance-dependent decrease in signal intensity is shown. The PDZ recognition sequence, TGWETWV, was located at the C-terminus of MBP.
Due to inter-domain dynamics between the CLaNP-5 carrying PDZ domain and the protein of interest the magnitude of paramagnetic alignment is decreased. For example, the RDCs observed directly within PDZ-1 at 900 MHz were in the range from -31.2 Hz to 18.0 Hz (Figures S6 and S7), while the ones transmitted upon binding to MBPTGWETWV ranged from -9.4 to 7.3 Hz (under identical conditions; Figure S4a). The axial component of the tensor in PDZ-1 was -1.1x10-3 compared to a value of 2.8x10-4 transmitted to MBPTGWETWV (Tables 1 and S2). The presence of inter-domain flexibility was further supported by 15N spin-relaxation measurements on ubiquitinTGWETWV bound to PDZ. The last 3 residues of ubiquitin and the first 2 residues of the sequence TGWETWV were dynamic (Figure S8). Importantly, however, the decrease in paramagnetic effects did not interfere with the determination of high quality RDCs for both ubiquitin and MBP (Figure 2, S3, S4). Moreover, scaling down of PCSs due to inter-domain dynamics has the advantage that cross peaks in the paramagnetic spectrum are only slightly shifted compared to the diamagnetic spectrum (Figure 2a and S3b). This simplifies the assignment of both spectra, in particular for high molecular weight proteins.
In summary, we have presented a robust method that allows access to a large number of RDCs, PCSs and PRE restraints produced from independent molecular alignments. Although in this study a PDZ domain with the lanthanoid tag CLaNP-5 was used, other lanthanoid tags can be attached[4d, 12, 24], as long as the probe mobility is low enough to provide sufficient alignment in the protein of interest, lanthanoid binding motifs might be engineered into the PDZ domain[3, 10] or proteins other than PDZ might be employed, such as calmodulin, which binds with high affinity to short peptide sequences.[4a] Importantly, the method can be applied to proteins of unknown 3D structure and regardless of the number of cysteines. Moreover, the fact that one of the PDZ variants, PDZ-4, was found to be unstable, highlights a further advantage of the method. When the tag is directly attached to the protein of interest, this protein has to be mutated to enable attachment of the lanthanoid tag. As exemplified by PDZ-4, the mutation potentially leads to an unstable protein. In contrast, the present set of PDZ mutants are already known to be stable. Thus, they can just be added to any target protein, as long as it contains the TGWETWV extension, and thereby provide multiple independent molecular alignments. No internal modification of the protein of interest is required that could affect its structure and dynamics.
Supplementary Material
Acknowledgments
This work was supported by the ERC (ERC grant agreement number 282008 to M.Z.) and the Netherlands Organisation for Scientific Research, VICI grant 700.58.441 (W.-M.L. and M.U.).
Contributor Information
Aldo R. Camacho-Zarco, Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37077 Göttingen, Germany
Dr. Francesca Munari, German Center for Neurodegenerative Diseases (DZNE), Göttingen & Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37077 Göttingen, Germany & Center for the Molecular Physiology of the Brain, University Medical Center, Göttingen
Melanie Wegstroth, Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37077 Göttingen, Germany.
Wei-Min Liu, Leiden Institute of Chemistry, Leiden University, Einsteinweg 55, 2333 CC Leiden, The Netherlands.
Prof. Marcellus Ubbink, Leiden Institute of Chemistry, Leiden University, Einsteinweg 55, 2333 CC Leiden, The Netherlands
Dr. Stefan Becker, Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37077 Göttingen, Germany
Prof. Markus Zweckstetter, German Center for Neurodegenerative Diseases (DZNE), Göttingen & Max Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37077 Göttingen, Germany & Center for the Molecular Physiology of the Brain, University Medical Center, Göttingen
References
- [1].a) Otting G. Annu Rev Biophys. 2010;39:387–405. doi: 10.1146/annurev.biophys.093008.131321. [DOI] [PubMed] [Google Scholar]; b) Bertini I, Luchinat C. Curr Opin Chem Biol. 1999;3:145–151. doi: 10.1016/S1367-5931(99)80026-X. [DOI] [PubMed] [Google Scholar]; c) Keizers PM, Ubbink M. Prog Nucl Mag Res Sp. 2011;58:88–96. doi: 10.1016/j.pnmrs.2010.08.001. [DOI] [PubMed] [Google Scholar]; d) Liu WMO, Ubbink M, M Coordination Chemistry Reviews. 2013 [Google Scholar]
- [2].Silvaggi NR, Martin LJ, Schwalbe H, Imperiali B, Allen KN. J Am Chem Soc. 2007;129:7114–7120. doi: 10.1021/ja070481n. [DOI] [PubMed] [Google Scholar]
- [3].Allen KN, Imperiali B. Curr Opin Chem Biol. 2010;14:247–254. doi: 10.1016/j.cbpa.2010.01.004. [DOI] [PubMed] [Google Scholar]
- [4].a) Feeney J, Birdsall B, Bradbury AF, Biekofsky RR, Bayley PM. J Biomol Nmr. 2001;21:41–48. doi: 10.1023/a:1011924017938. [DOI] [PubMed] [Google Scholar]; b) Gaponenko V, Altieri AS, Li J, Byrd RA. J Biomol Nmr. 2002;24:143–148. doi: 10.1023/a:1020948529076. [DOI] [PubMed] [Google Scholar]; c) Wohnert J, Franz KJ, Nitz M, Imperiali B, Schwalbe H. J Am Chem Soc. 2003;125:13338–13339. doi: 10.1021/ja036022d. [DOI] [PubMed] [Google Scholar]; d) Ikegami T, Verdier L, Sakhaii P, Grimme S, Pescatore B, Saxena K, Fiebig KM, Griesinger C. J Biomol Nmr. 2004;29:339–349. doi: 10.1023/B:JNMR.0000032611.72827.de. [DOI] [PubMed] [Google Scholar]; e) Iwahara J, Schwieters CD, Clore GM. J Am Chem Soc. 2004;126:5879–5896. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]; f) Donaldson LW, Skrynnikov NR, Choy WY, Muhandiram DR, Sarkar B, Forman-Kay JD, Kay LE. J Am Chem Soc. 2001;123:9843–9847. doi: 10.1021/ja011241p. [DOI] [PubMed] [Google Scholar]; g) Xu XF, Keizers PHJ, Reinle W, Hannemann F, Bernhardt R, Ubbink M. J Biomol Nmr. 2009;43:247–254. doi: 10.1007/s10858-009-9308-0. [DOI] [PubMed] [Google Scholar]; h) Keizers PHJ, Mersinli B, Reinle W, Donauer J, Hiruma Y, Hannemann F, Overhand M, Bernhardt R, Ubbink M. Biochemistry-Us. 2010;49:6846–6855. doi: 10.1021/bi100598f. [DOI] [PubMed] [Google Scholar]; i) Volkov AN, Worrall JAR, Holtzmann E, Ubbink M. P Natl Acad Sci USA. 2006;103:18945–18950. doi: 10.1073/pnas.0603551103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Blackledge M. Prog Nucl Mag Res Sp. 2005;46:23–61. [Google Scholar]
- [6].a) Tang C, Schwieters CD, Clore GM. Nature. 2007;449:1078–U1012. doi: 10.1038/nature06232. [DOI] [PubMed] [Google Scholar]; b) Hiruma Y, Hass MA, Kikui Y, Liu WM, Olmez B, Skinner SP, Blok A, Kloosterman A, Koteishi H, Lohr F, Schwalbe H, Nojiri M, Ubbink M. J Mol Biol. 2013;425:4353–4365. doi: 10.1016/j.jmb.2013.07.006. [DOI] [PubMed] [Google Scholar]
- [7].Madl T, Bermel W, Zangger K. Angew Chem Int Edit. 2009;48:8259–8262. doi: 10.1002/anie.200902561. [DOI] [PubMed] [Google Scholar]
- [8].a) Pintacuda G, Park AY, Keniry MA, Dixon NE, Otting G. J Am Chem Soc. 2006;128:3696–3702. doi: 10.1021/ja057008z. [DOI] [PubMed] [Google Scholar]; b) John M, Pintacuda G, Park AY, Dixon NE, Otting G. J Am Chem Soc. 2006;128:12910–12916. doi: 10.1021/ja063584z. [DOI] [PubMed] [Google Scholar]
- [9].a) Lee L, Sykes BD. Biochemistry-Us. 1981;20:1156–1162. doi: 10.1021/bi00508a017. [DOI] [PubMed] [Google Scholar]; b) Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. P Natl Acad Sci USA. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Bertini I, Gelis I, Katsaros N, Luchinat C, Provenzani A. Biochemistry-Us. 2003;42:8011–8021. doi: 10.1021/bi034494z. [DOI] [PubMed] [Google Scholar]
- [10].Barthelmes K, Reynolds AM, Peisach E, Jonker HRA, DeNunzio NJ, Allen KN, Imperiali B, Schwalbe H. J Am Chem Soc. 2011;133:808–819. doi: 10.1021/ja104983t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Martin LJ, Hahnke MJ, Nitz M, Wohnert J, Silvaggi NR, Allen KN, Schwalbe H, Imperiali B. J Am Chem Soc. 2007;129:7106–7113. doi: 10.1021/ja070480v. [DOI] [PubMed] [Google Scholar]
- [12].Su XC, Otting G. J Biomol Nmr. 2010;46:101–112. doi: 10.1007/s10858-009-9331-1. [DOI] [PubMed] [Google Scholar]
- [13].Gaponenko V, Dvoretsky A, Walsby C, Hoffman BM, Rosevear PR. Biochemistry-Us. 2000;39:15217–15224. doi: 10.1021/bi001381w. [DOI] [PubMed] [Google Scholar]
- [14].Ma C, Opella SJ. J Magn Reson. 2000;146:381–384. doi: 10.1006/jmre.2000.2172. [DOI] [PubMed] [Google Scholar]
- [15].Yao LS, Ying JF, Bax A. J Biomol Nmr. 2009;43:161–170. doi: 10.1007/s10858-009-9299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zweckstetter M. Nat Protoc. 2008;3:679–690. doi: 10.1038/nprot.2008.36. [DOI] [PubMed] [Google Scholar]
- [17].Schmitz C, Stanton-Cook MJ, Su XC, Otting G, Huber T. J Biomol Nmr. 2008;41:179–189. doi: 10.1007/s10858-008-9249-z. [DOI] [PubMed] [Google Scholar]
- [18].Skelton NJ, Koehler MFT, Zobel K, Wong WL, Yeh S, Pisabarro MT, Yin JP, Lasky LA, Sidhu SS. J Biol Chem. 2003;278:7645–7654. doi: 10.1074/jbc.M209751200. [DOI] [PubMed] [Google Scholar]
- [19].Laura RP, Witt AS, Held HA, Gerstner R, Deshayes K, Koehler MFT, Kosik KS, Sidhu SS, Lasky LA. J Biol Chem. 2002;277:12906–12914. doi: 10.1074/jbc.M200818200. [DOI] [PubMed] [Google Scholar]
- [20].Keizers PHJ, Saragliadis A, Hiruma Y, Overhand M, Ubbink M. J Am Chem Soc. 2008;130:14802–14812. doi: 10.1021/ja8054832. [DOI] [PubMed] [Google Scholar]
- [21].a) Sharff AJ, Rodseth LE, Quiocho FA. Biochemistry-Us. 1993;32:10553–10559. doi: 10.1021/bi00091a004. [DOI] [PubMed] [Google Scholar]; b) Gardner KH, Zhang XC, Gehring K, Kay LE. J Am Chem Soc. 1998;120:11738–11748. [Google Scholar]
- [22].Cornilescu G, Marquardt JL, Ottiger M, Bax A. J Am Chem Soc. 1998;120:6836–6837. [Google Scholar]
- [23].Zweckstetter M, Bax A. J Biomol Nmr. 2002;23:127–137. doi: 10.1023/a:1016316415261. [DOI] [PubMed] [Google Scholar]
- [24].a) Liu WM, Keizers PHJ, Hass MAS, Blok A, Tirnmer M, Sarris AJC, Overhand M, Ubbink M. J Am Chem Soc. 2012;134:17306–17313. doi: 10.1021/ja307824e. [DOI] [PubMed] [Google Scholar]; b) Haussinger D, Huang JR, Grzesiek S. J Am Chem Soc. 2009;131:14761–14767. doi: 10.1021/ja903233w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.