Abstract
Background
In vivo optical imaging of neural activity provides important insights into brain functions at the single-cell level. Cranial windows and virally delivered calcium indicators are commonly used for imaging cortical activity through two-photon microscopes in head-fixed animals. Recently, head-mounted one-photon microscopes have been developed for freely behaving animals. However, minimizing tissue damage from the virus injection procedure and maintaining window clarity for imaging can be technically challenging.
New method
We used a wide-diameter glass pipette at the cortical surface for infusing the viral calcium reporter AAV-GCaMP6 into the cortex. After infusion, the scalp skin over the implanted optical window was sutured to facilitate postoperative recovery. The sutured scalp was removed approximately two weeks later and a miniature microscope was attached above the window to image neuronal activity in freely moving mice.
Results
We found that cortical surface virus infusion efficiently labeled neurons in superficial layers, and scalp skin suturing helped to maintain the long-term clarity of optical windows. As a result, several hundred neurons could be recorded in freely moving animals.
Comparison with existing methods
Compared to intracortical virus injection and open-scalp postoperative recovery, our methods minimized tissue damage and dura overgrowth underneath the optical window, and significantly increased the experimental success rate and the yield of identified neurons.
Conclusion
Our improved cranial surgery technique allows for high-yield calcium imaging of cortical neurons with head-mounted microscopes in freely behaving animals. This technique may be beneficial for other optical applications such as two-photon microscopy, multi-site imaging, and optogenetic modulation.
Keywords: Cranial window, scalp skin suture, virus infusion, cortical neuron ensemble, miniaturized microscopy, calcium imaging
1. Introduction
Understanding cortical processing related to vital behavioral functions requires the ability to assess the activity of large populations of cells within the temporal context of behavior. Current methods to study in vivo large-scale neuronal activity during active behavior possess distinct strengths and weaknesses. For example, chronic multi-array electrode recording has high temporal resolution and is applicable to freely moving animals, but sorting out the identity of recorded cells is challenging (Murakami et al., 2014; Rossant et al., 2016; Sul et al., 2011). In vivo two-photon imaging directly reveals the activity of specifically labeled neurons, but it requires animal head-fixation and restricts certain behavioral expression (Chen et al., 2013; Kuhlman et al., 2013; Minderer et al., 2016; Peters et al., 2014).
Recently developed miniature one-photon microscopes can achieve both cell-type specificity and recording in freely moving animals with genetically encoded calcium indicators (Ghosh et al., 2011; Ziv et al., 2013). This technique usually requires the insertion of a small graded-index (GRIN) optical lens inside the brain, and has been successfully used to study the function of deep brain structures such as the hippocampus (Okuyama et al., 2016; Ziv et al., 2013), amygdala (Grewe et al., 2017), striatum (Barbera et al., 2016) and hypothalamus (Jennings et al., 2015). However, the cerebral cortex located at the top of the brain has not yet been extensively studied through this technique (Pinto and Dan, 2015).
An important factor to consider for the application of this head-attached microscope is the trade-off between optical access and tissue damage. Direct insertion of a GRIN lens into the cortex from the pial surface will destroy superficial layer cortical neurons and their projections, which play important roles in integrating columnar and inter-areal neural signals (Harris and Shepherd, 2015; Petersen and Crochet, 2013). While it is possible to obtain a side view of cortical neurons through an inserted prism probe, which rotates the imaging plane 90 degrees from the direction of probe insertion (Andermann et al., 2013; Murayama et al., 2007), this method still leads to significant tissue damage adjacent to the imaged neurons. To avoid any direct damage to cortical neurons, one option is to image through a cranial window implanted above the cortex, as has been done in many two-photon imaging applications (Cao et al., 2015; Fu et al., 2012; Holtmaat et al., 2009; Yang et al., 2010). However, cranial window surgeries can be challenging to conduct successfully, due to the brain dura and pia responding to surgery damage with overgrowth and inflammation. The yield of clear windows is considered dependent on individual operators and sometimes unpredictable (Holtmaat et al., 2009).
The optical clarity of cranial windows is especially important for imaging cortical neurons with head-attached one-photon microscopes, because light-scattering can severely limit optical resolution of cell bodies in this imaging mode. Classic cranial window surgery protocols include the removal of a piece of the scalp, leaving the newly implanted window to recover unprotected by skin (Cao et al., 2013; Goldey et al., 2014; Holtmaat et al., 2009). In addition, viruses encoding calcium indicators are typically injected intracortically in the same surgical procedure as the implantation of the cranial window (Chen et al., 2013; Lowery and Majewska, 2010; Resendez et al., 2016). Variations in intracortical injection-induced damage and viral spread increase unpredictability in the experimental yield. Here, we describe an improved cranial surgery technique that uses scalp skin suturing and cortical surface viral infusion to reduce procedural inconsistencies and enhance the success rate of superficial-layer cortical neuron imaging by head-mounted microscopes in freely moving animals.
2. Materials and Method
2.1 Mouse strain
Procedures involving animal subjects have been approved by the Institutional Animal Care and Use Committee (IACUC) at the National Institute of Mental Health, Maryland; and LifeSource Biomedical Services, NASA Ames Research Center, California. C57BL/6 wild type mice were used for all experiments.
2.2 Pre-Surgery preparation
Mice were first anesthetized with Avertin (1.5% solution given at 0.01 ml/g, i.p.) and then mounted in a stereotaxic frame. Ear bars and a nose cone/tooth mount were used to hold the animal’s head securely. The eyes were protected with ophthalmic ointment and covered by a piece of black paper to prevent irritation and damage from surgical lights. Mice were pre-treated with dexamethasone (0.2 mg/kg, s.c.) and carprofen (5 mg/kg, s.c.) to prevent surgery-related pain, swelling and inflammation.
2.3 Craniotomy
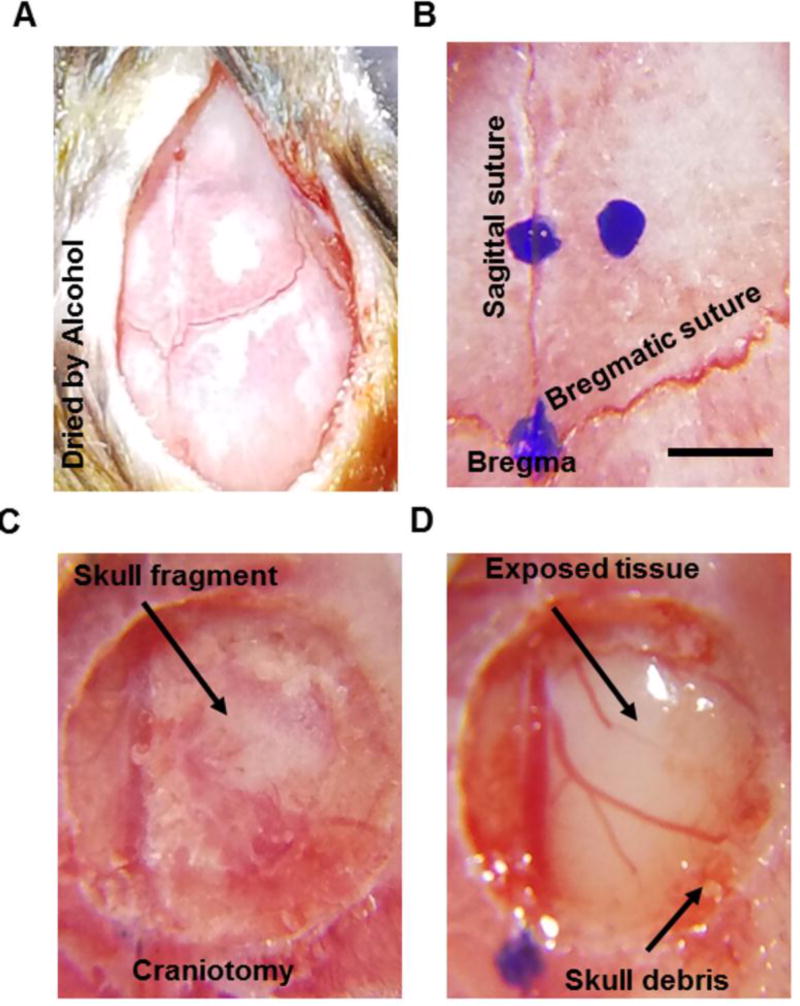
The fur between the eyes and ears was trimmed and the skin was disinfected by 3 alternate swabs of 70% ethanol and betadine. A ~ 1.5 cm incision in the scalp was made using a sterile surgical blade or scissors. Critical step: It is important not to remove the scalp. Bulldog clamps could be used if needed to keep the skin flaps away from the surgery site. To prevent tissue growth and to stabilize the coverslip, periosteum was wiped away and scraped off from the skull by a scalpel blade and the skull was cleaned and dried by 100% alcohol, especially around the target craniotomy region (Fig. 1A). After the skull is dry, the center of the desired glass window implant relative to bregma was marked using a stereotaxic instrument (Fig. 1B). A high-speed microdrill (typically between 7000–10,000 rpm) with a 0.5 mm burr was used to create a circular craniotomy (slightly larger than 3mm) in the skull around the desired viral injection location (Fig. 1C, Coordinates: AP1.7 ML 0.8). Then the craniotomy region was moistened and the bone island was lifted gently from the brain. The bone island should be separated cleanly with no bleeding (Fig 1D). Critical step: skull debris (Fig. 1D.) around the edge of the window should be carefully removed with fine forceps to prevent bleeding during coverslip installation (Fig. 2A).
Figure 1. Cranial window surgery procedures.
A, Example image illustrates the skull where periosteum tissue was removed by scraping with a blade and the surface was dried by 100% alcohol. B. Example image showing marks of the center of the glass window and spatial reference points on skull sutures. C. Example image showing a 3.5mm circular craniotomy region of interest with bone island still attached to the skull. The arrow points at the bone island. D. Example image showing the cranial window with the bone island removed. The arrows point to the exposed brain tissue in the center and the skull debris around the edge of the window. Scale bar: 1mm.
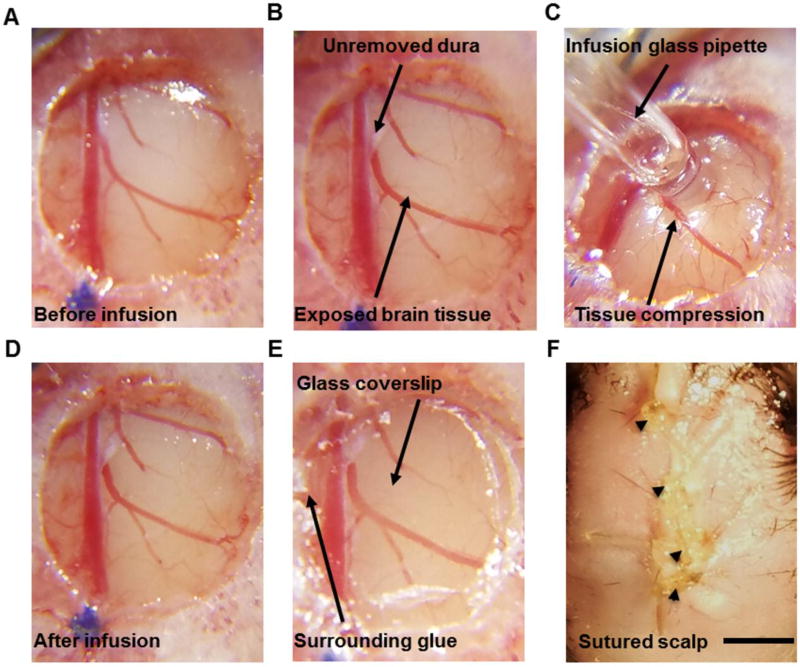
Figure 2. Cortical surface-based virus infusion method.
A, Example image shows a cleared brain surface where skull debris around the window edge were carefully removed. B, Example image shows the infusion target area where a piece of dura (~ 1 mm2) was removed using a pair of fine forceps. The upper arrow points to the residual dura, and the lower arrow points to exposed brain tissue. C, Example image showing cortical surface-based viral infusion using a 0.3–0.5 mm (tip size) glass pipette. D, Example image showing no bleeding or blood vessel disturbance after surface infusion. E, Application of cyanoacrylate adhesive around the edges of the glass coverslip to fix the coverslip to the surrounding bone. F, The scalp skin was sutured over the implanted window. Arrowheads point at the four suture knots. Scale bar: 1mm.
2.4 Glass pipette preparation
To make a wide-tip pipette (tip diameter between 0.3–0.5 mm) for viral infusion, a glass pipette (Sutter Instrument, Item#: BF120-94-10) was pulled by a micropipette puller (Sutter Instrument, model: P87) and then cut by a diamond blade. If the tip is not even and smooth, it should be polished by a grinder or Dremel. Critical step: a smooth, even pipette tip will provide a tighter seal with the brain tissue surface for the viral infusion.
2.5 Cortical pial surface infusion vs. intracortical injection
After skull debris were removed, a pair of fine forceps or a 30g needle was used to remove 1 mm2 of dura around the injection area (Fig. 2B). Reminder: dura removal will become difficult approximately 10 minutes after the cranial window is opened. If the dura is not removed, a higher titer and higher volume of virus would be necessary for sufficient virus expression. Serial dilution experiments should be performed to establish optimal virus volume and concentration for specific brain regions and cell types of interest.
For viral infusion, the prepared glass pipette was attached (tip size:0.3–0.5 mm) to a 20 µl microsyringe. 0.6 µl virus (Penn vector core, AAV2/9.syn.GCaMP6s.WPRE.SV40, titer diluted to 4×1012 copies/ml) was drawn into the glass pipette through negative pressure. Using the stereotaxic arm, the glass pipette tip was lowered toward the brain until it touched the intact pia mater (Coordinates: AP 1.7 ML 0.8). To seal the pipette against the brain surface, the glass pipette was slowly lowered again by about 400–500 µm to press upon but not break through the pia. 2–3 minutes later, virus infusion was started at 60 nl/min (Fig. 2C), until 0.6 µl virus was infused (around 10 minutes). Usually, no bleeding was observed during or after surface infusion (Fig. 2D). Critical step: a good seal between the pial surface and the glass pipette will help the virus infuse into the cortical tissue.
For intracortical injections, the pulled glass pipette (tip size 10 µm) was directly connected with a 20 µl microsyringe pump and 0.6 µl virus was drawn up through negative pressure. The glass pipette was lowered by a manipulator until it touched the intact pia mater. The tip of the pipette was inserted into the brain (Coordinates: AP 1.7 ML 0.8) at 2 µm per second to 100–150 µm beneath the pial surface. 2–3 minutes later, 0.6 µl of virus was slowly injected at 60 nl/min. Backflow of virus to the brain surface may occur at this shallow injection depth.
2.6 Cranial window implantation
After viral infusion or injection is complete, 0.6 µl saline was added atop the brain surface to keep it moist, then a dry, pre-sanitized 3 mm glass coverslip (Thomas Scientific, Catalog#, 64-0720, Thickness: 0.15 mm) was laid over the brain within the craniotomy. Gentle pressure on the coverslip was applied to keep the coverslip at the same level as the surrounding skull. A cranial cap was then created with Krazy glue (KG484), starting from the edges of the cranial window and extending over the rest of the skull. Self-curing liquid (Ce Tray Plastic, Fast Set) was used to hasten the hardening of the glue. (Fig. 2E).
2.7 Sutured scalp vs open scalp condition
After cranial window implantation, sterile saline or Ringer's solution was added to the scalp tissue and skull surface for rehydration, allowing the skin to be more easily stretched back over the skull and sutured. Critical step: the scalp was sutured over the installed cranial window (Fig. 2F) for improved postoperative recovery. In the open-scalp condition, the scalp skin was removed in the initial surgical preparation.
Animals were treated with ketoprofen (5mg/kg) intraperitoneally to reduce pain and moved to a warm recovery cage until ambulatory. Single housing was adopted post-surgery to protect the surgery site.
2.8 Baseplate attachment for miniature microscope installation
1–2 weeks after cranial window implantation, animals were retrieved to check the recovery of the cranial window, as well as viral expression levels. Following the pre-operative steps, the animal was anesthetized and set in the stereotaxic frame. The scalp over the cranial window was completely removed using a scalpel or surgery scissors. There may be a regrowth of periosteum beneath the scalp, over the skull and cranial window. This periosteum was removed using fine forceps (e.g. FST, Dumont #5) and the entire skull was cleaned and dried by 100% ethanol to prepare for the baseplate installation.
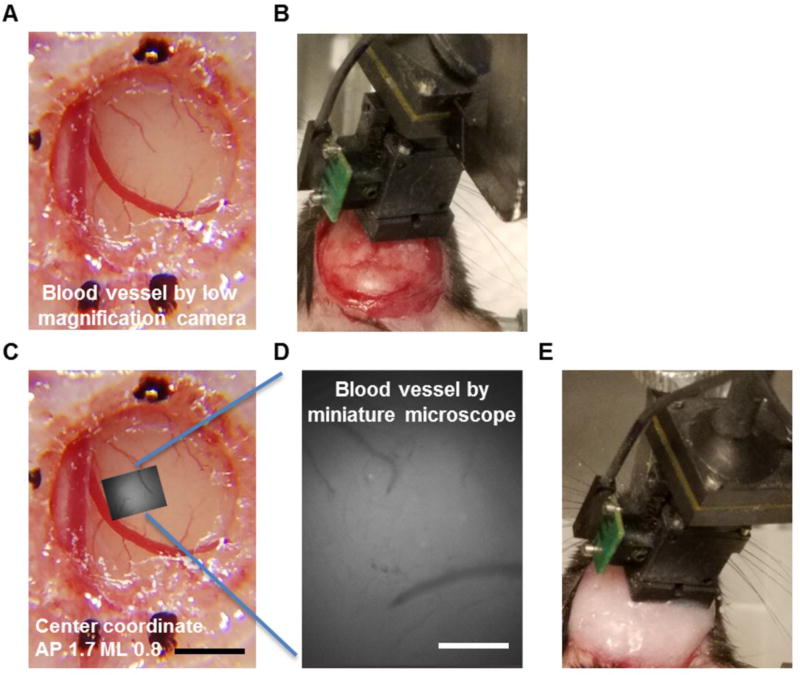
The cranial window was examined and gently cleaned with Ringer’s solution and lens paper. The glass coverslip should be firmly fixed to the surrounding skull, and there should be no blood beneath the window. The brain surface blood vessels should appear clean and sharp in a good cranial window surgery (Fig. 3A). To locate the cranial coordinates of the imaged area, a picture of the blood vessels (Fig. 3A) was taken by a Samsung cellphone (Samsung, galaxy S7 edge) with a magnification lens (Arbitron, 15X).
Figure 3. Installation of the baseplate for the head-attached miniature microscope.
A, Example image shows clear and sharp blood vessel patterns inside the cranial window 10 days after window surgery and virus infusion. The sutured scalp has already been removed. B, Example image shows the positioning of a miniature microscope above the cranial window. The baseplate is attached to the microscope and has not yet been glued onto the skull. C and D, The brain surface area imaged through the miniature microscope (D) was matched to the global blood vessel pattern taken by a low-magnification camera (C) to locate the spatial coordinates of the imaged region. E, The baseplate was glued to the skull and the microscope was attached on animal’s head after baseplate installation. Scale bar in A and C: 1 mm and scale bar in D: 200 µm.
Next, a miniaturized one-photon microscope (Inscopix, nVista) was connected to the data acquisition computer. The microscope was attached to a baseplate and secured on the stereotaxic micromanipulator by a gripper. It was gradually lowered towards the coverslip until there was approximately a 2 mm gap between the coverslip and the microscope lens (Fig. 3B). The microscope LED was then turned on. The position of the microscope was adjusted through the manipulator arm until the LED light was right on top of the coverslip. The microscope was lowered again until a clear blood vessel pattern was observed through the acquisition software (Inscopix, nVista Acquisition Software). Critical step: The microscope objective and the cranial window must be parallel to one another to facilitate finding the optimal plane of focus. Snapshots were taken by the microscope, so that the spatial coordinates of the imaged area could be determined by matching the blood vessel pattern inside the microscope field of view to that across the entire cranial window (Fig. 3C and 3D).
The miniature microscope focus was adjusted by changing the microscope’s distance to the cranial window surface with the stereotaxic manipulator arm, and a live ΔF/F visualization of the tissue (Inscopix, nVista Acquisition Software) was used to quickly determine if there were labeled cells within the tissue. After the focus was set, a reference image was saved to serve as a foundational location to return to in future imaging sessions. Critical point: the position of the microscope should not be adjusted after this point.
Dental acrylic or cyanoacrylate glue was used to permanently fix the baseplate to the acrylic cap covering the animal’s skull around the cranial window. Any gap between the baseplate and the cap was bridged with the acrylic or adhesive to build a solid foundation for the microscope, allowing for secure re-attachment in subsequent imaging sessions (Fig. 3E). The adhesive could experience some volume shrinkage in the following day or two over the course of curing, which could change the focal plane in the tissue. Critical step: the dental acrylic or cyanoacrylate glue should be gradually and carefully added to the gap between the baseplate and the skull layer-by-layer, and self-curing liquid should be used in each layer to hasten hardening and decrease future adhesive shrinkage. Once the adhesive had fully hardened, a final reference fluorescence image was acquired through the acquisition software to document the final location of the field of view in the tissue. Finally, the microscope was removed and the implanted cranial window was protected by a baseplate cover (Inscopix). The animal was placed in a warm recovery chamber until it awoke and was returned to its home cage. Single housing was used for all animals with implanted baseplates to protect the implant.
2.9 Large-scale Ca2+ imaging of cortical neurons in freely moving mice
Calcium imaging was performed in freely moving mice in a Y-maze using the head-attached microscope (Inscopix; 2 mm diameter objective lens; LED power: 0.6 – 1.0 mW; camera resolution: 1440 x1080 pixels). Images were acquired at 30 Hz using nVista Acquisition Software (Inscopix). At the beginning of each imaging session, the protective baseplate cover of the previously implanted baseplate was removed and the microscope was attached in its place. The imaging field of view was approximately 900 × 600 µm2 at 0.65 µm/pixel resolution and the imaging depth was selected by adjusting the focus of the microscope until clear cell signals were observed in the online ΔF/F images.
2.10 Cell detection
Calcium imaging videos were analyzed by using the Mosaic data analysis software (Inscopix) and custom-written scripts in Matlab following published algorithms (Hyvarinen and Oja, 2000; Mukamel et al., 2009). Raw videos were first down-sampled by four-fold along spatial dimensions to reduce file size and noise. The mean fluorescence intensity of each pixel during a recording session was calculated as F0 and changes in pixel intensity at time t were expressed as (Ft - F0)/F0 or ΔF/F0. To extract active neuron signals, principal component and independent component analysis (PCA-ICA) was applied to the spatial-temporal data matrices of ΔF/F0 using CellSort and fastICA toolboxes (Matlab) (Hyvarinen and Oja, 2000; Mukamel et al., 2009). This analysis decomposes a spatiotemporal data matrix into independent components based on the skewness of data distribution. Each component has a characteristic spatial filter over the imaged area and a corresponding temporal signal during the imaging period. The spatial filter and the temporal signal of each component were graphically displayed and inspected by human observers who were blind to the experimental conditions of each video. If the spatial filter for a component overlapped with the dark shadows cast by blood vessels in the F0 image, this component was likely contributed by blood flow and was therefore rejected. With this method, we detected neurons and extracted their spatial locations and corresponding calcium traces from the images, in both scalp-closed and opened preparations.
2.11 Histology and GFAP staining
Labeling of astrocytes was performed with antibodies against glial fibrillary acidic protein (GFAP, Thermo Fisher Scientific, 13–0300) using a standard immunostaining protocol (Gan and Lichtman, 1998). The immunolabeled sections were imaged with a standard confocal fluorescent microscope (Zeiss LSM 780) at constant settings between experimental groups. To compare the GFAP expression levels in different groups, the average fluorescent intensity of GFAP-labeled images was calculated for each subject and divided by the highest subject value, and then compared between the experimental groups.
2.12 Quantification and statistical analysis
To compare differences in window clarity between open and closed-scalp preparations, five human observers who were blind to the surgical conditions were instructed to rank the quality of windows based on the presence of opaque white membranes underneath the coverslip, the proportion of membrane-covered area, and the sharpness of blood vessels. The average rankings were compared between open-scalp and closed-scalp preparations. Two independent tests were carried out using batches of animals prepared by two different surgeons at NIMH and Inscopix, respectively.
All data are presented by mean ±SEM; the student t-test or ranksum test was used to measure the difference between different two groups. * indicates p<0.05 and *** indicates p<0.001.
3. Results
3.1 Closed-scalp post-operative recovery improves the clarity of chronic cranial windows
To image cortical neuronal activity by head-attached one-photon microscopes over time, high-quality cranial windows lasting days to weeks are necessary. With traditional open-skull cranial window surgery methods, surgery-related inflammation and dura thickening could reduce the clarity of implanted windows after a few days (Holtmaat et al., 2009). Anti-inflammatory drugs, such as dexamethasone and carprofen, have been used to decrease such responses (Cao et al., 2013; Golshani and Portera-Cailliau, 2008), but the efficacy of these treatments is still variable. Alternatively, the thinned-skull cranial window technique, which was considered less invasive due to the preservation of some skull and sutured skin over the window, has been used for imaging cortical neurons with two-photon microscopy (Fu et al., 2012; Yang et al., 2010). But this technique requires thinning of the skull to an even layer of ~20 µm repeatedly over time, which is surgically challenging and behaviorally interruptive. The skull remnant also reduces the optical clarity of cranial windows for imaging deeper brain structures.
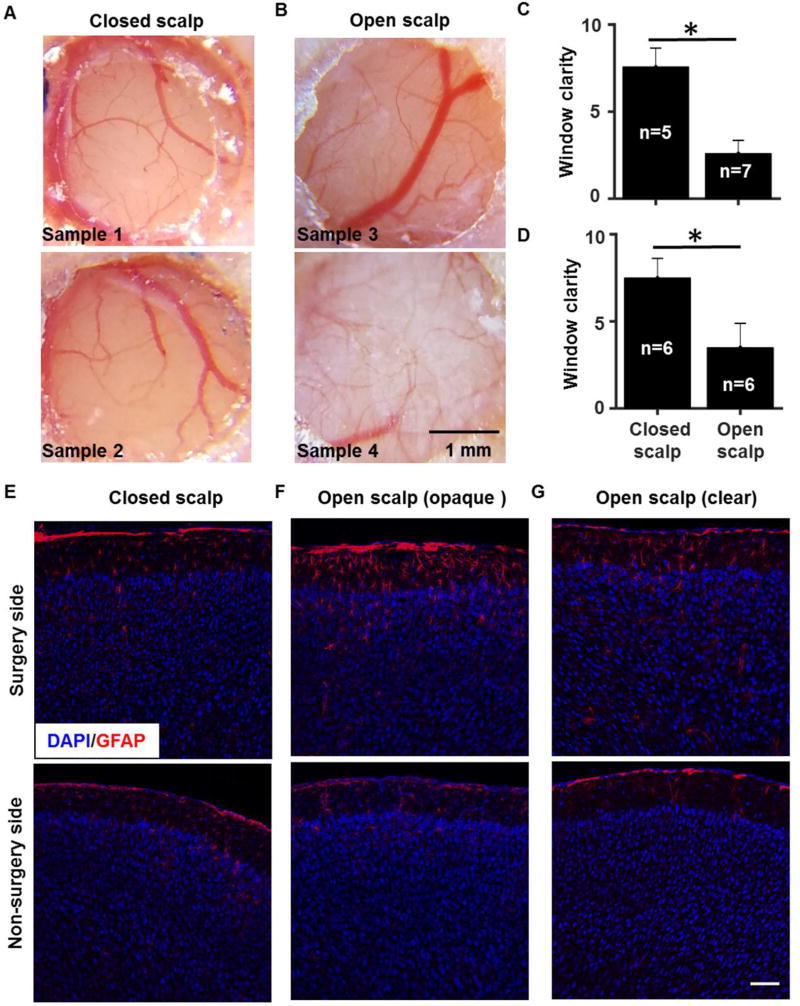
To search for a simple and robust surgery procedure to produce high-clarity cranial windows, we examined whether keeping the scalp closed after the open-skull cranial window surgery may help protect the brain and improve long-term window clarity. We compared results between open-scalp and closed-scalp postoperative conditions, and found that scalp suturing after cranial window surgery appeared to decrease connective tissue growth underneath the window, leading to sharper images of brain blood vessel patterns (Fig. 4A and 4B). To quantify window clarity, images of cranial windows were ranked based on the sharpness of blood vessels and the extent of opaque connective tissue inside the window by human observers who were blind to the surgery conditions. As shown in Figure 4C and 4D, in two independent cohorts of animals tested in two separate facilities, scalp suturing after cranial window surgery consistently increased the window clarity compared to the open-scalp condition (cohort 1: p < 0.05, n = 5 mice for open scalp, n = 7 mice for closed scalp; cohort 2: p < 0.05, n = 6 mice for open scalp and n = 6 mice for closed scalp).
Figure 4. Closed-scalp post-operative recovery improves the clarity of cranial windows.
A, Example images showing cranial windows two weeks after surgery in the closed-scalp recovery condition. B, Example images showing cranial windows two weeks after surgery in the open-scalp recovery condition. C and D, Bar graphs illustrating that the window clarity in the closed-scalp condition was increased compared to that in the open-scalp condition. Two independent surgery tests were carried out by separate research groups in Maryland (C) and California (D) using the same protocol. The clarity score for each brain was averaged from the rankings given by the same five human observers who were blind to the surgery conditions. Nonparametric ranksum test, p < 0.05, n = 5 mice for open scalp, n = 7 mice for closed scalp in (C); p < 0.05, n = 6 mice for open scalp, n = 6 mice for closed scalp in (D). E, F and G, Example confocal fluorescent images of coronal cortical sections labeled with anti-GFAP antibody (red) and nuclei counterstain DAPI (blue) in a closed-scalp animal (E), an open-scalp animal with an opaque cranial window (F), and an open-scalp animal with a clear cranial window (G). Scale bar in A and B: 1 mm and scale bar in E–G: 100 µm.
Under traditional open-scalp cranial window conditions, it has been reported that increased astrocytic activation as indicated by glial fibrillary acidic protein (GFAP) expression may occur between 2–14 days after surgery in association with variations in surgical preparations (Holtmaat et al., 2009; Xu et al., 2007). We performed GFAP immunostaining 14 days after surgery and found that animals recovering with the scalp sutured appeared to have clear cranial windows and similar GFAP expression level underneath the window compared to the un-operated side of the brain (Fig. 4E, n = 3 mice). By contrast, in the open-scalp condition, excessive GFAP expression was observed underneath an opaque cranial window (Fig. 4F, n = 4 mice), whereas little GFAP increase was observed underneath a clear window (Fig. 4G, n = 3 mice). These results suggest that a closed scalp after surgery may help to reduce inflammatory reactions, thereby improving the clarity of chronic windows for long-term functional imaging studies.
3.2 Virus infusion at the pial surface labels neurons in the superficial cortical layers
Besides the clarity of the cranial window, the neuronal labeling method is another important factor for successful imaging of cortical neurons with a head-mounted one-photon microscope. The focal plane of this type of microscope is typically 150~200 µm beneath the objective lens (Ghosh et al., 2011). In mouse cortex, layer I is approximately 100 µm thick and is mainly composed of axons and dendrites derived from neuronal soma located in layers II to VI. Although a one-photon microscope can focus on the soma of layer II/III neurons, excessive labeling of layer I neuropil could generate a large amount of out-of-focus fluorescence and obscure the signals emitted by the cell soma. An efficient and consistent cell labeling method that would preferentially target superficial rather than deep layer neurons would reduce the extent of neuropil labeling and increase the efficiency of extracting single-cell activity of layer II/III neurons from a calcium movie data taken by a head-mounted one-photon microscope.
We designed a new neuronal labeling method based on viral infusion at the cortical surface, and compared it to the traditional intracortical injection method. For surface-based infusion, a large glass pipette (0.3–0.5 mm in tip diameter) was loaded with calcium indicator virus (AAV2/9-hSyn-GCaMP6s), and pressed against the cortical pial surface without breaking into the tissue. The virus was slowly infused into the brain with a syringe pump. Intracortical injection was conducted with a sharp glass pipette (10 µm in tip diameter) directly inserted into the brain to target layer II/III neurons (100–150 µm below the pia). As shown in Figure 5A, surface infusion preferentially labeled neurons in cortical layer II/III. In contrast, intracortical injections, despite being targeted to layer II/III, labeled both superficial and deep layers of cortex, probably due to downward spread of virus from the tip (Fig. 5B).
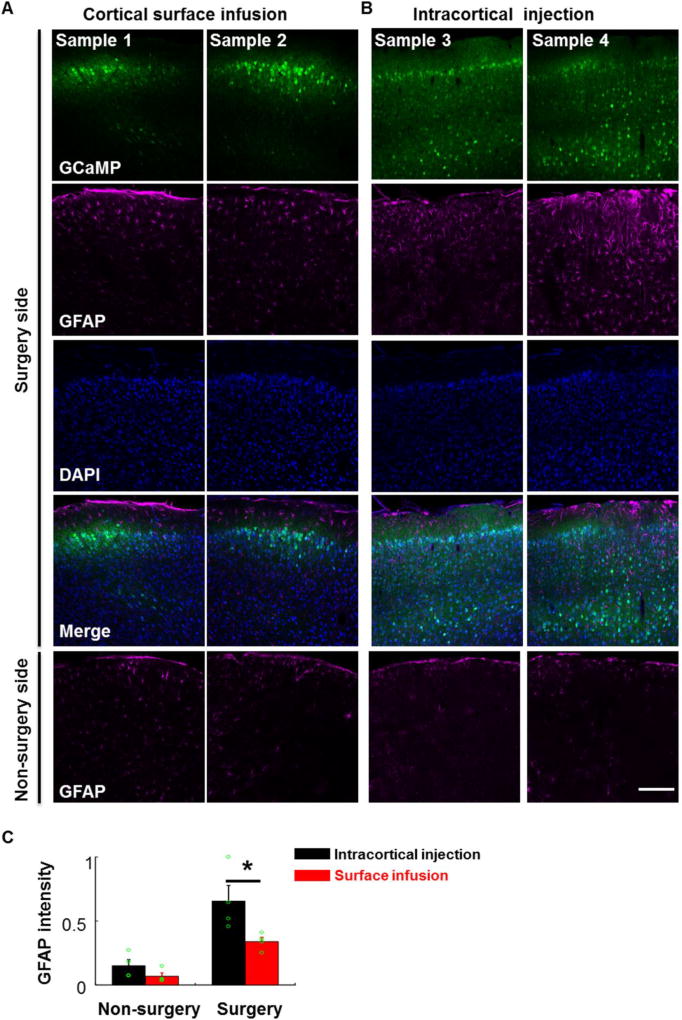
Figure 5. Cortical surface-based viral infusion mainly labeled neurons in the superficial layers.
A, Confocal fluorescent images of coronal cortical sections showing that cortical surface-based viral infusion (AAV2/9-hSyn-GCaMP6s) mainly labeled neurons in the superficial layers of cortex, and the level of GFAP expression at 14 days post infusion was low. First row: GCaMP6, second: GFAP, third: DAPI, fourth: merged 3-channel images, all in the surgery side. Fifth: GFAP staining in the non-surgery side. B, Intracortical viral injection labeled neurons in both superficial and deep layers of the cortex and led to higher GFAP staining in the surgery side. Scale bar in A and B: 200 µm. C, Bar graph showing GFAP intensity under the cortical surface infusion and intracortical injection conditions (n = 4 mice for each group, t-test, p < 0.05, intracortical injection vs. surface infusion in the surgery side).
We also performed GFAP immunostaining 14 days after surgery and found that GFAP expression levels in the operated hemisphere side appeared somewhat higher than the un-operated hemisphere side (Fig 5A, B and C). Importantly, cortical surface viral infusion significantly minimized the increase in GFAP expression compared to intracortical injection (p < 0.05, n = 4 mice for each group). These results suggest that cortical surface infusion may help to reduce brain damage by avoiding direct glass pipette insertion into the brain tissue.
3.3 High-yield imaging of cortical neuron activity with head-mounted one-photon microscopes in freely moving mice
By suturing the scalp back over the implanted cranial window after cortical surface virus infusion, we consistently obtained clear optical windows for longitudinal imaging with a head-attached miniature microscope starting ten days after surgery (8 out of 8 mice tested in Fig 6; Fig. 6A).
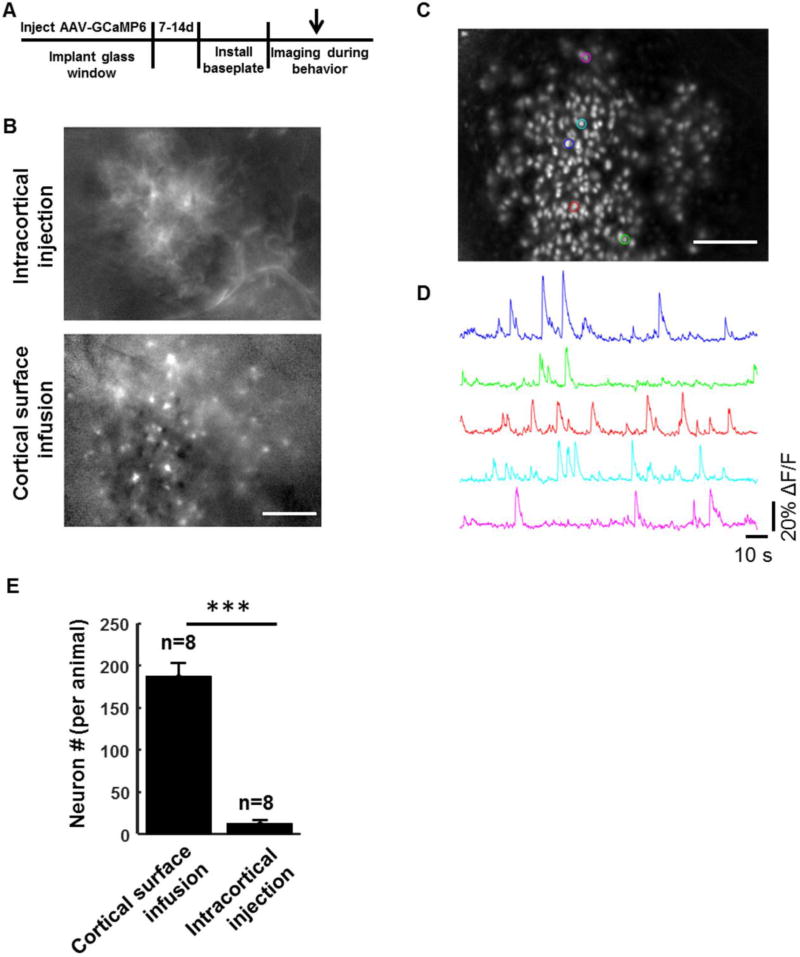
Figure 6. Detection of individual cortical neuron activity in freely moving animals with miniaturized head-attached microscopes.
A, Schematic drawing of the timeline for labeling and imaging cortical neuron activity in behaving mice. B, Single-frame ΔF/F images showed patchy fluorescence and few single cells in animals labeled by intracortical viral injection (Upper), and numerous individual cells in animals labeled by cortical surface infusion (Lower). C, A representative standard deviation projection of 15,000 ΔF/F image frames collected over a period of 5 minutes in the secondary motor cortex of a freely moving mouse in a Y-maze. D, Example traces representing calcium transients from ROIs with matched colors in C. E, Bar graph showing number of detected neurons with calcium transients under the cortical surface infusion and intracortical injection conditions (t-test, p < 0.001, n = 8 mice for each group). Scale bar: 200µm.
With the intracortical viral injection method, only a few single cells could be visualized in calcium activity (ΔF/F) videos – most of the cortical activity appeared as patchy neuropil fluorescence extending over tens to hundreds of microns (Fig. 6B, supplementary video I). In contrast, with the cortical surface labeling method, many individual cells could be clearly visualized in in vivo calcium videos (Fig. 6B, supplementary video II). We performed Principal Component – Independent Component (PCA-ICA) analysis on the calcium movie data (5 minutes in duration) to detect and extract the locations of individual cells (Fig. 6C) and their corresponding calcium dynamics (Fig. 6D). On average, 187±15 neurons (n = 8 animals) were detected in an imaging field of view of 950x600 µm2 under the surface virus infusion condition, but only 12±4 neurons were detected under the intracortical injection condition (n = 8 animals) (Fig. 6E, p<0.001). Taken together, our results suggest that using a cortical surface infusion technique to label neurons and a postsurgical scalp suture method for cranial window recovery can greatly increase the yield of calcium imaging by head-mounted one-photon microscopes in freely moving mice.
4. Discussion
The cerebral cortex receives multisensory inputs and transforms these signals for decision making, behavioral control and other vital cognitive functions. Superficial cortical layers (Layer II/III) are thought to play an important role in integrating various inputs from cortical and subcortical regions and have been subjected to extensive studies via in vivo two-photon imaging approaches in head-fixed animals (Harris and Shepherd, 2015; Peters et al., 2014; Petersen and Crochet, 2013). Recently developed head-mounted miniature one-photon microscopes have the ability to image neuronal activity when animals are conducting naturalistic, freely moving behaviors (Ghosh et al., 2011; Ziv et al., 2013). However, due to limitations of traditional cranial window surgeries and neuron labeling methods, it has been challenging to obtain high animal yield to perform longitudinal studies of layer II/III neuron functions with head-mounted microscopes.
Chronic cranial windows are broadly used to study the morphology of neurons (Fu et al., 2012; Holtmaat et al., 2009; Mastwal et al., 2014; Yang et al., 2010), to examine gene expression patterns in individual neurons (Cao et al., 2015; Wang et al., 2006), for photo-stimulation through optogenetic methods (Carrillo-Reid et al., 2016; Silasi et al., 2013), and to image neural activity through genetically encoded calcium indicators such as GCaMP (Andrew et al., 2014; Chen et al., 2013; Manago et al., 2016). However, due to inflammation and tissue regrowth after surgery, success rates of clear windows vary and tend to be low (Holtmaat et al., 2009; Yang et al., 2010). Here, we systematically compared window clarity between open-scalp and closed-scalp preparations and found that closing the scalp above the window during the first week of post-surgical recovery period significantly increased the window clarity. This improvement may be attributed to the protective effects of the scalp on the underlying skull and muscle tissues, such as prevention of tissue desiccation and infection, maintenance of a biological milieu conducive for wound healing, and buffering of mechanical impact to the window implant. Without scalp protection, excessive inflammatory reaction and growth of connective tissue are more likely to occur, which can significantly reduce the clarity of implanted optical windows.
The pattern of neuronal labeling is another important factor for resolving the activities of individual neurons using a one-photon microscope above the cranial window. Traditional neuronal labeling methods typically involve the insertion of a glass pipette deep into the cortical tissue for intracortical viral injection. If a small amount of virus is injected, only a sparse population of cortical neurons will be labeled and population-level, behavior-related neuronal activity might be missed. On the other hand, if a large amount of virus is injected, the virus will spread broadly across cortical laminae. When both superficial layers and deep layers are labeled, excessive neuropil signals, particularly those produced by apical dendrites and axons in layer I, can mask the cell body signals of layer II/III cortical neurons under one-photon microscopy. We show that with a cortical surface-based infusion method, viruses spread downwards from the pial surface, efficiently labeling superficial layer neurons while avoiding deep layer neurons. In addition, the large diameter pipette tip increased the lateral coverage of neuronal labeling. Therefore, our surface infusion method balanced labeling depth and volume to preferentially and efficiently label neurons in the superficial layers of cortex for functional imaging. These improvements increased the number of individual neurons detected by miniature microscope imaging in the superficial layers of cortex in an efficient and reproducible manner.
While the current study focused on methods to increase both animal and cell yield for imaging superficial cortical neurons in freely moving animals, our methods may be further modified to extend the depth and the breadth of imaged neurons. For example, with layer-specific transgenic driver lines, neuronal labeling can be restricted to subpopulations of deep layer cortical neurons (Bareyre et al., 2005; Gerfen et al., 2013). Although the soma of these neurons will be beyond the optical reach of a lens positioned above the cortex, the apical dendritic trunks of these deep-layer neurons may be feasibly imaged through a cranial window and provide a proxy of their activity (Hill et al., 2013). In addition, future studies may combine large optical windows with movable miniature microscope placements to allow for multi-site imaging in the same animal (Allen et al., 2017; Goldey et al., 2014). Lastly, our cranial window technique may be also used to improve the experimental yield for in vivo two-photon imaging in head-fixed animals, or for optogenetic modulation of cortical neurons.
5. Conclusions
Our studies demonstrate a cortical surface-based viral infusion method to preferentially and efficiently label superficial layer cortical neurons, alongside a closed-scalp post-operative recovery method to improve the clarity of chronic cranial windows. Taken together, these methods significantly improve the efficiency and consistency of animal preparations to study cortical neuronal functions by one-photon miniaturized microscopy in freely moving animals.
Supplementary Material
HIGHLIGHTS.
Cortical surface virus infusion efficiently labels superficial layer neurons
Scalp skin suturing after cranial window implants improves long-term optical clarity
High yield neuronal ensemble imaging is achieved with head-mounted miniscopes
Acknowledgments
This research was supported by the National Institute of Mental Health (NIMH) Division of Intramural Research Programs ZIA MH002897 to KHW, NIMH postdoctoral fellowships (XL, WZ, SM and QL), and the Inscopix Inc (VC and SO). We thank other members of Wang lab for scientific discussion, and Dale Voelker and Zhouyang Ma for helping to rank the clarity of cranial windows.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. SO and VC are paid employees at Inscopix; SO is Director of Science at Inscopix; and VC is Content and Training Manager at Inscopix. This does not alter the authors’ adherence to the journal’s policies and all the authors have abided by the statement of ethical standards for manuscripts submitted to the Journal of Neuroscience Methods.
References
- Allen WE, Kauvar IV, Chen MZ, Richman EB, Yang SJ, Chan K, Gradinaru V, Deverman BE, Luo L, Deisseroth K. Global Representations of Goal-Directed Behavior in Distinct Cell Types of Mouse Neocortex. Neuron. 2017;94:891–907000000. doi: 10.1016/j.neuron.2017.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andermann ML, Gilfoy NB, Goldey GJ, Sachdev RN, Wolfel M, McCormick DA, Reid RC, Levene MJ. Chronic cellular imaging of entire cortical columns in awake mice using microprisms. Neuron. 2013;80:900–13. doi: 10.1016/j.neuron.2013.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew JP, Simon XC, Takaki K. Emergence of reproducible spatiotemporal activity during motor learning. Nature. 2014 doi: 10.1038/nature13235. [DOI] [PubMed] [Google Scholar]
- Barbera G, Liang B, Zhang L, Gerfen CR, Culurciello E, Chen R, Li Y, Lin DT. Spatially Compact Neural Clusters in the Dorsal Striatum Encode Locomotion Relevant Information. Neuron. 2016;92:202–13. doi: 10.1016/j.neuron.2016.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bareyre FM, Kerschensteiner M, Misgeld T, Sanes JR. Transgenic labeling of the corticospinal tract for monitoring axonal responses to spinal cord injury. Nat Med. 2005;11:1355–60. doi: 10.1038/nm1331. [DOI] [PubMed] [Google Scholar]
- Cao VY, Ye Y, Mastwal S, Ren M, Coon M, Liu Q, Costa RM, Wang KH. Motor Learning Consolidates Arc-Expressing Neuronal Ensembles in Secondary Motor Cortex. Neuron. 2015;86:1385–92. doi: 10.1016/j.neuron.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao VY, Ye Y, Mastwal SS, Lovinger DM, Costa RM, Wang KH. In vivo two-photon imaging of experience-dependent molecular changes in cortical neurons. Journal of visualized experiments : JoVE. 2013 doi: 10.3791/50148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo-Reid L, Yang W, Bando Y, Peterka DS, Yuste R. Imprinting and recalling cortical ensembles. Science (New York, N.Y.) 2016;353:691–4. doi: 10.1126/science.aaf7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K, Kim DS. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Yu X, Lu J, Zuo Y. Repetitive motor learning induces coordinated formation of clustered dendritic spines in vivo. Nature. 2012;483:92–5. doi: 10.1038/nature10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan WB, Lichtman JW. Synaptic segregation at the developing neuromuscular junction. Science. 1998;282:1508–11. doi: 10.1126/science.282.5393.1508. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Paletzki R, Heintz N. GENSAT BAC cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron. 2013;80:1368–83. doi: 10.1016/j.neuron.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh KK, Burns LD, Cocker ED, Nimmerjahn A, Ziv Y, Gamal AE, Schnitzer MJ. Miniaturized integration of a fluorescence microscope. Nature methods. 2011;8:871–8. doi: 10.1038/nmeth.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldey GJ, Roumis DK, Glickfeld LL, Kerlin AM, Reid RC, Bonin V, Schafer DP, Andermann ML. Removable cranial windows for long-term imaging in awake mice. Nat Protoc. 2014;9:2515–38. doi: 10.1038/nprot.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golshani P, Portera-Cailliau C. In vivo 2-photon calcium imaging in layer 2/3 of mice. Journal of visualized experiments : JoVE. 2008 doi: 10.3791/681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe BF, Grundemann J, Kitch LJ, Lecoq JA, Parker JG, Marshall JD, Larkin MC, Jercog PE, Grenier F, Li JZ, Luthi A, Schnitzer MJ. Neural ensemble dynamics underlying a long-term associative memory. Nature. 2017;543:670–5. doi: 10.1038/nature21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KD, Shepherd GM. The neocortical circuit: themes and variations. Nat Neurosci. 2015;18:170–81. doi: 10.1038/nn.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DN, Varga Z, Jia H, Sakmann B, Konnerth A. Multibranch activity in basal and tuft dendrites during firing of layer 5 cortical neurons in vivo. Proceedings of the National Academy of Sciences. 2013;110 doi: 10.1073/pnas.1312599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat A, Bonhoeffer T, Chow DK, Chuckowree J, De Paola V, Hofer SB, Hubener M, Keck T, Knott G, Lee WC, Mostany R, Mrsic-Flogel TD, Nedivi E, Portera-Cailliau C, Svoboda K, Trachtenberg JT, Wilbrecht L. Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat Protoc. 2009;4:1128–44. doi: 10.1038/nprot.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvarinen A, Oja E. Independent component analysis: algorithms and applications. Neural Networks. 2000;13:411–30. doi: 10.1016/s0893-6080(00)00026-5. [DOI] [PubMed] [Google Scholar]
- Jennings JH, Ung RL, Resendez SL, Stamatakis AM, Taylor JG, Huang J, Veleta K, Kantak PA, Aita M, Shilling-Scrivo K, Ramakrishnan C, Deisseroth K, Otte S, Stuber GD. Visualizing hypothalamic network dynamics for appetitive and consummatory behaviors. Cell. 2015;160:516–27. doi: 10.1016/j.cell.2014.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman SJ, Olivas ND, Tring E, Ikrar T, Xu X, Trachtenberg JT. A disinhibitory microcircuit initiates critical-period plasticity in the visual cortex. Nature. 2013;501:543–6. doi: 10.1038/nature12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery RL, Majewska AK. Intracranial injection of adeno-associated viral vectors. JoVE (Journal of Visualized Experiments) 2010:e2140-e. doi: 10.3791/2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manago F, Mereu M, Mastwal S, Mastrogiacomo R, Scheggia D, Emanuele M, De Luca MA, Weinberger DR, Wang KH, Papaleo F. Genetic Disruption of Arc/Arg3.1 in Mice Causes Alterations in Dopamine and Neurobehavioral Phenotypes Related to Schizophrenia. Cell reports. 2016;16:2116–28. doi: 10.1016/j.celrep.2016.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastwal S, Ye Y, Ren M, Jimenez DV, Martinowich K, Gerfen CR, Wang KH. Phasic dopamine neuron activity elicits unique mesofrontal plasticity in adolescence. J Neurosci. 2014;34:9484–96. doi: 10.1523/JNEUROSCI.1114-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minderer M, Harvey CD, Donato F, Moser EI. Neuroscience: Virtual reality explored. Nature. 2016 doi: 10.1038/nature17899. [DOI] [PubMed] [Google Scholar]
- Mukamel EA, Nimmerjahn A, Schnitzer MJ. Automated analysis of cellular signals from large-scale calcium imaging data. Neuron. 2009;63:747–60. doi: 10.1016/j.neuron.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Vicente MI, Costa GM, Mainen ZF. Neural antecedents of self-initiated actions in secondary motor cortex. Nat Neurosci. 2014;17:1574–82. doi: 10.1038/nn.3826. [DOI] [PubMed] [Google Scholar]
- Murayama M, Perez-Garci E, Luscher HR, Larkum ME. Fiberoptic system for recording dendritic calcium signals in layer 5 neocortical pyramidal cells in freely moving rats. J Neurophysiol. 2007;98:1791–805. doi: 10.1152/jn.00082.2007. [DOI] [PubMed] [Google Scholar]
- Okuyama T, Kitamura T, Roy DS, Itohara S, Tonegawa S. Ventral CA1 neurons store social memory. Science. 2016;353:1536–41. doi: 10.1126/science.aaf7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AJ, Chen SX, Komiyama T. Emergence of reproducible spatiotemporal activity during motor learning. Nature. 2014;510:263–7. doi: 10.1038/nature13235. [DOI] [PubMed] [Google Scholar]
- Petersen CC, Crochet S. Synaptic computation and sensory processing in neocortical layer 2/3. Neuron. 2013;78:28–48. doi: 10.1016/j.neuron.2013.03.020. [DOI] [PubMed] [Google Scholar]
- Pinto L, Dan Y. Cell-type-specific activity in prefrontal cortex during goal-directed behavior. Neuron. 2015;87:437–50. doi: 10.1016/j.neuron.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resendez SL, Jennings JH, Ung RL, Namboodiri VMK, Zhou ZC, Otis JM, Nomura H, McHenry JA, Kosyk O, Stuber GD. Visualization of cortical, subcortical and deep brain neural circuit dynamics during naturalistic mammalian behavior with head-mounted microscopes and chronically implanted lenses. Nat Protoc. 2016;11:566–97. doi: 10.1038/nprot.2016.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant C, Kadir SN, Goodman DF, Schulman J, Hunter ML, Saleem AB, Grosmark A, Belluscio M, Denfield GH, Ecker AS, Tolias AS, Solomon S, Buzsaki G, Carandini M, Harris KD. Spike sorting for large, dense electrode arrays. Nat Neurosci. 2016;19:634–41. doi: 10.1038/nn.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silasi G, Boyd JD, LeDue J, Murphy T. Improved methods for chronic light-based motor mapping in mice: automated movement tracking with accelerometers, and chronic EEG recording in a bilateral thin-skull preparation. Frontiers in neural circuits. 2013;7:123. doi: 10.3389/fncir.2013.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sul JH, Jo S, Lee D, Jung MW. Role of rodent secondary motor cortex in value-based action selection. Nature neuroscience. 2011;14:1202–8. doi: 10.1038/nn.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Majewska A, Schummers J, Farley B, Hu C, Sur M, Tonegawa S. In vivo two-photon imaging reveals a role of arc in enhancing orientation specificity in visual cortex. Cell. 2006;126:389–402. doi: 10.1016/j.cell.2006.06.038. [DOI] [PubMed] [Google Scholar]
- Xu HT, Pan F, Yang G, Gan WB. Choice of cranial window type for in vivo imaging affects dendritic spine turnover in the cortex. Nat Neurosci. 2007;10:549–51. doi: 10.1038/nn1883. [DOI] [PubMed] [Google Scholar]
- Yang G, Pan F, Parkhurst CN, Grutzendler J, Gan W-B. Thinned-skull cranial window technique for long-term imaging of the cortex in live mice. Nat Protoc. 2010;5:201–8. doi: 10.1038/nprot.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Burns LD, Cocker ED, Hamel EO, Ghosh KK, Kitch LJ, El Gamal A, Schnitzer MJ. Long-term dynamics of CA1 hippocampal place codes. Nat Neurosci. 2013;16:264–6. doi: 10.1038/nn.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.