Abstract
Diagnostics play a significant role in health care. In the developing world and low-resource regions the utility for point-of-care (POC) diagnostics becomes even greater. This need has long been recognized, and diagnostic technology has seen tremendous progress with the development of portable instrumentation such as miniature imagers featuring low complexity and cost. However, such inexpensive devices have not been able to achieve a resolution sufficient for POC detection of pathogens at very small scales, such as single-cell parasites, bacteria, fungi, and viruses. To this end, expansion microscopy (ExM) is a recently developed technique that, by physically expanding preserved biological specimens through a chemical process, enables super-resolution imaging on conventional microscopes and improves imaging resolution of a given microscope without the need to modify the existing microscope hardware. Here we review recent advances in ExM and portable imagers, respectively, and discuss the rational combination of the two technologies, that we term expansion mini-microscopy (ExMM). In ExMM, the physical expansion of a biological sample followed by imaging on a mini-microscope achieves a resolution as high as that attainable by conventional high-end microscopes imaging non-expanded samples, at significant reduction in cost. We believe that this newly developed ExMM technique is likely to find widespread applications in POC diagnostics in resource-limited and remote regions by expanded-scale imaging of biological specimens that are otherwise not resolvable using low-cost imagers.
Keywords: Expansion microscopy, miniature imager, portable, low-cost, point-of-care
Introduction
Diagnostics play a significant role worldwide by providing proper and timely healthcare to patients [1]. The role of diagnostics and in particular, the need for point-of-care (POC) diagnostics, is even more critical in the developing world and low-resource regions [1–3]. This has been recognized for long, and diagnostic technology has seen tremendous progress with the development of, in particular, imaging-based strategies.
Although conventional optical microscopy provides high-end capacities in terms of resolution, often times their bulky sizes, expense, and need for alignment and adjustment cannot satisfy the need of portable observations. Recent advances in POC diagnostics have further challenged existing microscopy techniques where in situ and cost-effective detection is required in addition to sufficient sensitivity and resolution. For example, compact image sensors are widely applicable to POC analysis at mammalian cell level [4–9]. The mini-microscopy has several key advantages over conventional microscopy in undeveloped regions [10], including: i) simplicity, light weight, and compactness: a mini-microscope can be easily constructed with simple design, making the system compact and portable, well suited for POC diagnostics; ii) cost-effectiveness: the main components of a mini-microscope include off-the-shelf items such as a light-emitting diode (LED) and an image sensor, making it affordable in POC diagnostics. Nevertheless, while the mini-microscopes are widely applicable in portable POC diagnosis at the scale of mammalian cells, it remains challenging for their utility in detecting disease-causing pathogens (e.g. parasites, bacteria, viruses, and fungi) due to the limited resolution of their equipped low-end optics.
Expansion microscopy (ExM) is a recently developed technique that enables super-resolution imaging (e.g. below the diffraction limit) on conventional microscopes and improves imaging resolution of a given microscope without the need to modify the existing microscope hardware. ExM relies on a very simple principle, i.e. embedding a biological sample in a matrix of swellable polymer, which chemically anchors fluorescent labels already pre-applied to the specimen, and then physically expands the specimen by swelling the specimen-matrix composite to ~4–5 times in linear dimension [11]. Variations of ExM have been further developed recently to permit the simple, powerful anchoring and separation of proteins and nucleic acids of biological samples [12,13].
As such, the rational combination of ExM and mini-microscopy may potentially push forward a new initiative in POC diagnostics, which we term expansion mini-microscopy (ExMM) [14]. We have thus demonstrated that even a low-cost mini-microscope could image biological samples (both mammalian and bacterial cells) at unprecedentedly high resolution comparable to conventional microscopy without expansion, indicating that the integration of the original version of ExM with a mini-microscope through our ExMM technology can result in resolution that rivals that of a conventional microscope with a hundred-to-thousand-fold reduction in the cost. Here in this Opinion Article, we review recent advances in ExM and portable imagers, respectively, and discuss the preliminary findings of their combinatory form, the ExMM. We finally conclude with future perspectives on the potential application of ExMM in POC diagnostics.
Expansion Microscopy (ExM)
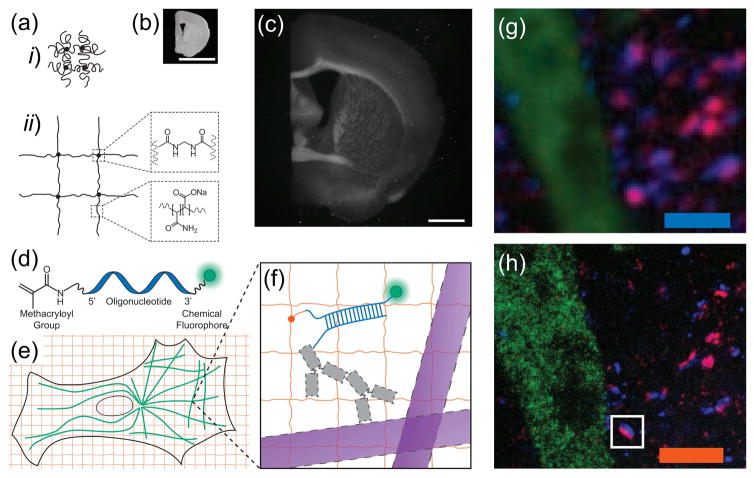
Conventional imaging of specimens with high spatial precision requires expensive equipment, because precise magnification with a minimum of aberration and a maximum of sensitivity requires high-end lenses and cameras, as well as precision alignment. We recently developed a new strategy depending primarily on chemistry to do the magnification – physically instead of optically [11]. We discovered that, by synthesizing a swellable polyelectrolyte hydrogel network directly within a specimen of interest, and subsequently dialyzing the sample in a medium of lower osmolarity (i.e. pure water), it could be physically expanded (Fig. 1a–c). Specifically, we used sodium polyacrylate as the hydrogel material, which when synthesized in high salt is compact, but when the salt is diluted undergoes swelling due to electrostatic repulsion (Fig. 1ai–ii). By staining a sample with a trifunctional label comprised of an antibody, a polymer-linking group, and a fluorophore, we were able to anchor the fluorophore to the hydrogel network (Fig. 1d–f); by enzymatically digesting the endogenous structure, we were able to render the sample mechanically homogeneous. Water, then, swells the sample (Fig. 1b versus 1c). Embedding preserved biological specimens in hydrogels for microscopy imaging purposes has a long history, dating back to 1995 [15], but expansion microscopy uses a specific property of hydrogels – in particular polyelectrolyte gels – namely the massive swelling of such hydrogel upon exposure to pure water, to physically move apart components of a biological specimen, so that nanoscale structures can be resolved.
Figure 1. The ExMM Concept.
(a) Schematic of (i) collapsed polyelectrolyte network, showing crosslinker (dot) and polymerchain (line), and (ii) expanded network after H2O dialysis. (b) Photograph of fixed mouse brain slice. (c) Photograph, post-ExM, of the sample (b) under side illumination. (d) Schematic of label that can be anchored to the gel at site of a biomolecule. (e) Schematic of microtubules (green) and polymer network (orange). (f) The label of (d), hybridized to the oligo-bearing secondary antibody top (top gray shape) bound via the primary (bottom gray shape) to microtubules (purple), is incorporated into the gel (orange lines) via the methacryloyl group (orange dot) and remains after proteolysis (dotted lines). Scale bars, (b) and (c) 5 mm. Schematics are not to scale. (g) Confocal fluorescence images of a Thy1-YFP mouse brain slice, stained with presynaptic (anti-Bassoon, blue) and postsynaptic (anti-Homer1, red) markers, in addition to antibody to GFP (green), (g) pre- versus (h) post-expansion. Scale bars (g) 2.5 μm; (h) 2.5 μm. Adapted with permission from Ref. [11].
Since the sample preparation procedures involve enzymatic homogenization of the mechanical characteristics of the tissue-polymer composite, this technology design enables isotropic and uniform expansion to occur [11]. Comparing images pre- versus post-expansion, using conventional (structured illumination-based) super-resolution microscopes, in fixed cultured HEK cells as well as in mouse brain slice, we confirmed that this expansion process was isotropic [11], and thus a ~4.5-fold expansion enabled an effective resolution of 300 nm (the diffraction limit of the lens used) divided by 4.5, or approximately 60 nm, approaching that attainable with classical super-resolution microcopy methods – but without requiring the hardware [16,17]. ExM has been utilized in super-resolution imaging of a variety of biological specimens using conventional optical microscopy, including for example, cultured mammalian cells [11], brain tissues [11], and cancerous tissues [18]. As shown in Fig. 1, g and h, a Thy1-YFP-H mouse cortex slice was stained with antibodies against yellow fluorescent protein (YFP, green), as well as the pre- and post-synaptic scaffolding proteins Bassoon (blue) and Homer1 (red). The post-ExM image (Fig. 1h) showed clear demarcations between the Bassoon/Homer1 signals while in the pre-ExM image (Fig. 1g) the staining formed overlapping spots at each synapse due to blur by diffraction.
Further Development of ExM
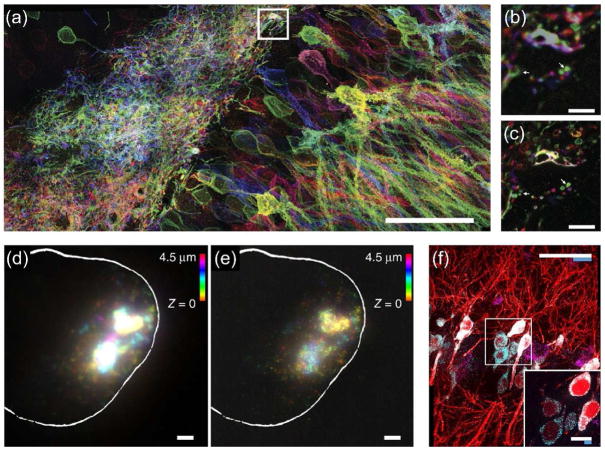
ExM is a chemistry that is easily customized for new applications. The original ExM method was unable to retain native proteins in the hydrogel and used specially designed reagents not widely available [11]. We subsequently developed a variant of ExM, named protein-retention ExM (proExM) [13], where proteins, instead of labels, are anchored to the swellable hydrogel network, through the use of a commercially available crosslinker. Antibodies can be delivered to samples post-expansion if a gentle mechanical homogenization technique is used. Alternatively, we demonstrated simple protocols in which fluorescent signals from both genetically engineered fluorescent proteins and exogenously labeled secondary antibodies, directly conjugated to the hydrogel networks, could be well preserved even when subjected to otherwise non-specific proteolytic digestion. For example, a post-proExM confocal image of a slice of mouse hippocampus expressing Brainbow 3.0 (a transgenic labeling strategy that enables stochastic expression of different combinations of fluorescent proteins in the brain, to be revealed by later staining with fluorescent antibodies) clearly indicated the retention of the fluorescent proteins (Fig. 2a), which showed much higher resolution than the image obtained pre-expansion (Fig. 2c versus 2b). As such, proExM is a simple extension of the original ExM method and is already beginning to enable applications in imaging of protein biomarkers in complex biological samples at high resolution, such as the analysis of neural connections and circuits in the mouse brain [19]. In parallel to our efforts developing proExM, two other groups independently published very similar strategies that expand proteins away from each other, enabling either post-expansion antibody staining [20], or selective retention of fluorescent proteins and antibodies that were administered pre-expansion [21] – which both highlights the rapid spread of expansion microscopy, and also suggests that protein-retention forms of expansion microscopy are indeed simple to implement and robust.
Figure 2. The concepts of proExM and ExFISH/smFISH.
(a) Maximum intensity projection of high-resolution confocal microscopy stack following expansion of neurons expressing Brainbow3.0. (b) Pre-expansion confocal image showing one optical section of the boxed region in f. (c) Post-expansion image of b. Scale bars: (a) 5 μm, (b) 5 μm, (c) 50 μm. Adapted with permission from Ref. [13]. (d) smFISH image of X-inactive specific transcript (XIST) long non-coding RNA (lncRNA) in the nucleus of an HEK293 cell before expansion (white line denotes nuclear envelope in d–e). (e) As in f, using ExFISH. (f) Confocal image of mouse hippocampal tissue, showing single RNA puncta. Inset, one plane of the boxed region (red, YFP protein; cyan, YFP mRNA; magenta, Gad1 mRNA). Scale bars: (d) 2 μm, (e) 2 μm, (c) 50 μm, inset 10 μm. Adapted with permission from Ref. [12].
Another direction for expansion microscopy is to extend the biomolecules interrogated beyond proteins. In particular, the ability to image nucleic acids at high precision in biological tissues is of tremendous interest for identifying specific microorganism strains and cellular and tissue states in both normal and pathological settings, which could not be achieved using the original form of ExM. We recently developed another strategy that allows for ExM of RNAs [12]. We innovated a small molecule linker that allows RNA molecules to be covalently anchored to the swellable polymer network: one part of the linker is an alkylating reagent that binds guanine, and the other part can link to the polymer network. Thus, this linker can bind RNA (and DNA) to the swellable polymer. Post-expansion, fluorescent in situ hybridization (FISH) imaging of RNAs can be subsequently performed with single-molecule precision at high yield, and great specificity, in both cultured mammalian cells (Fig. 2e versus 2d) and intact brain tissue (Fig. 2f). Such post-expansion FISH (ExFISH) methodologies can potentially enable further amplification of signals via techniques such as hybridization chain reaction (HCR), because expanding RNAs away from each other decrowds them and makes room for space-requiring reactions such as HCR; the decrowding of RNAs enabled by expansion microscopy may also help track RNAs over many rounds of hybridization as utilized in multiplexed FISH imaging of dozens to hundreds of transcripts at once [22–26]. ExFISH thus allows for super-resolution imaging of nucleic acids with conventional microscopy in specimens of importance to biology, medicine, and diagnostics. As a testament to the tunability of ExM, a recent variant of ExFISH was published in which polymer-anchorable probes were applied to samples, which were then expanded [27]; the modular, simple architecture of ExM protocols suggests that in addition to a set of universally applicable protocols, a diversity of more specialized protocols can be rapidly developed for use in specific fields of diagnostics.
Portable Microscopy
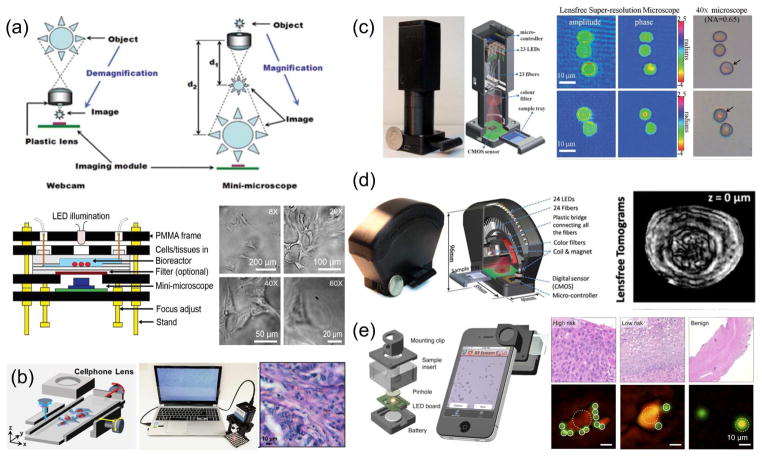
Miniature imagers have become useful tools in modern laboratories. They have found many application niches, ranging from the display of physical phenomena to the monitoring of cell behaviors. In particular, many strategies have been devised for the fabrication of mini-microscopes for observing biological samples. For example, Zhu et al. developed a fluorescence microscope encased in a printed box (5.5×3.5×2.4 cm3; 28 g), which could easily attach a smartphone as a display, recording, and wireless transmission unit [28]. Schaefer et al. developed an automated and low-cost optical mini-microscope with an autofocusing algorithm [29]. This microscope platform was capable of capturing and integrating multiple images in a large composed image through image analysis software. We have previously shown that simple inversion of the lens of a webcam created a low-cost, compact microscope capable of imaging at 20–60X magnifications (6×6×4 cm3; 40 g), which enabled observation of biological samples in real time (Fig. 3a) [30,31]. Dong et al. on the other hand, integrated optical microscopy, multi-angle illumination imaging, and a powerful post-processing algorithm for image synthesis in the frequency domain to develop a compact (8×8×16 mm3; 250g) Fourier ptychographic microscope (FPscope; Fig 3b) [32].
Figure 3. Examples of portable microscopes.
(a) Mini-microscope fabricated by reversing the lens of a webcam and its use to observe hepatic cancer spheroids. Adapted with permission from Ref. [30] and Ref. [31]. (b) Portable microscope using flipped cellphone lenses and a Fourier ptychographic algorithm to stitch images captured from a sample illuminated by different angles with an LED array and its use in observation of histopathological samples. Reproduced with permission from Ref. [32]. (c) Lens-free portable microscope to obtain holographic images and its use to image cells infected with malaria parasites Reproduced with permission from Ref. [39]. (d) Lens-free portable tomographic microscope and its use to observe an egg of parasitic worm. Reproduced with permission from Ref. [40]. (e) Portable microscope adapted to a smart phone, based on the reconstruction of diffraction patterns and its use to analyze cervical cancer. Reproduced with permission from Ref. [41].
More recently, various POC microscopy strategies with more sophisticated hardware designs and/or algorithms have also been proposed to achieve higher resolutions required to inspect bacteria [33,34] or even sometimes viruses [35] and DNA strands [36,37]. For example, Miller et al. developed a relatively small (7.5×13×18 cm3; 1000 g) and low-cost bright-field/fluorescence “global microscope” for POC applications and validated its use for visual confirmation of the presence of Mycobacterium tuberculosis in field-collected clinical sputum samples [38]. Powered by two AA batteries and equipped with LED illumination, this microscope was capable of submicron resolution (0.8 μm) at 1000X magnification. Bishara et al. designed a compact (5×5×20 cm3; less than 100 g), lens-free, on-chip imager capable of digitally collecting and reconstructing holographic images to produce images with a submicron resolution (Fig. 3c) [39]. They validated the performance of this device in POC applications by observing malaria parasites (Plasmodium falciparum) in blood smear samples. Isikman et al. (Fig. 3d) developed a lens-free field-portable tomographic microscope (96×89×40 mm3; 110 g) that obtained holograms and validated the device by observing differently sized microparticles and eggs from the parasitic flatworm Hymenolepsis nana [40]. Im et al. developed a digital diffraction-based POC imager that enabled the screening of cancerous and/or precancerous cells and the detection of human papillomavirus (HPV) in cervical samples (Fig. 3e) [41]. In addition, Wei et al. designed a mobile phone microscope that was capable of imaging DNA strands in a high-throughput manner using a combination of dark-field imaging, thin-film interference filters, and a computation framework [36]. Single DNA molecules of various lengths were successfully imaged at a sizing accuracy of <1 kilobases. More recently, this setup was further adopted to perform DNA sequencing and in situ mutation analysis, which enabled accurate identification of pathogens (or specific mutations) at low cost [37].
With the success in the use of portable microscopes for POC applications, more frequent integration of the technology with other compatible ones have become prevalent. On the one hand, it is common to integrate the miniature imagers to smartphones [28,42,43] and the recently available wearable devices such as the Google Glass [44]; on the other hand, combination of the mini-microscopy with distinct strategies such as microfluidics for sample preparation and expansion [42], is rapidly emerging.
Expansion Mini-Microscopy (ExMM)
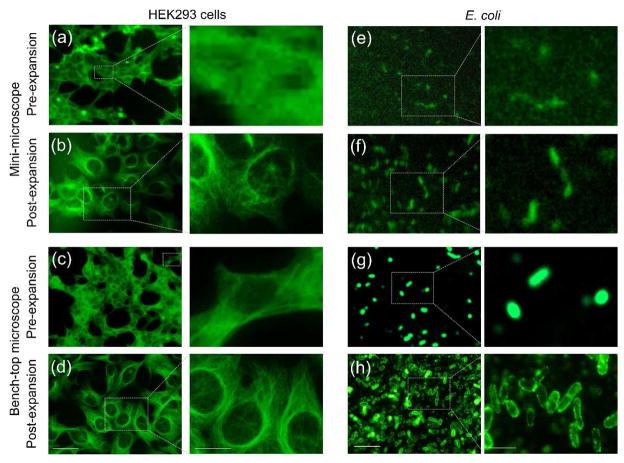
As aforementioned, while inexpensive mini-microscopy features ultralow cost for extensive use in low-resource settings and remote areas, without additional instrumentation or sophisticated algorithms (including that required in certain cases of portable microscopy), it has hardly been able to achieve a resolution sufficient for POC detection of pathogens at very small scales, such as subcellular structures, single-cell parasites, bacteria, and viruses. An effective and convenient strategy in addressing the insufficient resolution of mini-microscopes without the need for redesigning the hardware would be to adapt the ExM technology – and we recently proved that this strategy we term as ExMM – is feasible [14]: post-ExMM images of HEK293 cells labeled for microtubules showed dramatical improvement in observation of these subcellular structures than those pre-expansion, typical with the low-end optics of the mini-microscopes (Fig. 4b versus 4a). More importantly, the post-ExMM images were comparable to those captured with a benchtop microscope equipped with high-end optics and camera (Fig. 4, c and d), if not better than pre-ExM images (Fig. 4c).
Figure 4. The concept of ExMM.
(a–d) ExMM imaging of tubulin for HEK293 cells. (a, b) Pre- and post-expansion images obtained from mini-microscope. (c, d) Pre- and post-expansion images obtained from bench-top microscope. Scale bars: (left) 100 μm; (right) 50 μm. (e–h) ExMM imaging of E. coli. (e, f) Pre- and post-expansion images obtained from mini-microscope. (g, h) Pre- and post-expansion images obtained from bench-top microscope. Scale bars: (left) 5 μm; (right) 2 μm. Adapted with permission from Ref. [14].
In additional preliminary experiments related to POC diagnostics, we labeled a model pathogen Escherichia coli (E. coli) with a monoclonal antibody that targets lipopolysaccharides on the membrane of the bacteria. The bacteria were then embedded in the hydrogel and expanded approximately 4.5 times in dimension. It was noted that, the presence of E. coli could only be imaged using the mini-microscope in post-ExMM samples (Fig. 4e), whereas no deterministic signals were detected from the bacteria in their original unexpanded state since these bacteria are tiny in size with individual cells measuring about only 0.5 μm (width) by 2 μm (length). The use of polyclonal antibodies further improved the signal-to-noise ratio (Fig. 4f). Similarly, the bacterial cells showed much clearer morphology with the bench-top microscope after ExM, as indicated by the well-stained lipopolysaccharides on their membranes (Fig. 4h versus 4g). It is also estimated that, the chemicals involved in the specimen expansion process can be used at extremely low cost due to the ultralow volume involved in microscopy analysis, down to below ¢20 at a volume of <10 μL according to our prior calculations [45]. More importantly, while the chemicals are disposable, the slightly more expensive mini-microscope ($5–100) is not consumable and can be used indefinitely for a large number of assays.
Perspectives
The potential applications of ExMM in POC diagnostics have only been demonstrated in E. coli detection at present. Nevertheless, we see broad potential for the role for ExMM in this arena. For instance, in POC diagnostics for malaria, despite the increasing availability of antigen-based rapid tests, microscopy on stained blood smears remains the gold-standard [46]. We envision that ExMM has the potential for widespread utility in POC diagnostics following the further integration of the newly developed proExM and ExFISH and possibly advanced forms of other portable imagers.
ExMM still faces a few immediate challenges, including insufficient physical magnification, signal intensity, and ergonomics. First, the current form of ExM is only capable of a physical magnification of approximately 4.5X [11]. However, small-angle x-ray diffraction determinations suggest that several polymers, similar to the sodium polyacrylate used in the original protocol, exhibit a mesh size of 1–2 nm [47]. Therefore, greater physical expansion factors seem attainable, which would result in higher magnifications and better resolution with minimal instrumentation requirements. Second, due to physical expansion, the density of the labeled fluorophores is inevitably diluted during the expansion process, leading to a reduced signal-to-noise ratio, especially as related to mini-microscopy equipped with both low-end optics and imaging sensors. This issue may be addressed by the HCR expansion of the post-labeling signals [12] or through the use of labels with improved binding capability pre-expansion (e.g., polyclonal versus monoclonal antibodies [45]). Finally, current ExM protocols are aimed at scientists, with protocol complexities comparable to immunostaining or in situ hybridization, and require many steps, whereas for diagnostics one would want to have potentially a very simple protocol with a small number of steps. We propose the further development of a streamlined ExMM kit that conveniently integrates (a) a disposable device to process all the steps involved in ExM (i.e., labeling, gelation, and expansion); (b) the mini-microscope systems; and (c) the algorithms that enable automated image processing. This integration should simplify the use of ExMM-POC diagnostics for biological specimens of interest by reducing the amount of labor and technical expertise required [45]. With this step forward, we are optimistic regarding the future success of ExMM in providing ultralow-cost and high-resolution imaging for POC diagnostics in resource-limited regions.
Highlights.
Expansion microscopy enables super-resolution optical imaging through physical expansion of specimens.
Expansion microscopy has been further developed to enable protein and nucleic acid imaging
Miniature and portable microscopy shows advantage in point-of-care diagnostics
Combination of expansion microscopy and mini-microscopy allows for high-resolution point-of-care diagnostics at low cost
Acknowledgments
The authors acknowledge funding from the Office of Naval Research Young National Investigator Award, the National Institutes of Health (AR057837, DE021468, D005865, AR068258, AR066193, EB022403, EB021148, Transformative Award 1R01MH103910, 1U01MH106011, Director’s Pioneer Awards 1DP1NS087724, and 5DP1HD086071), the New York Stem Cell Foundation-Robertson Award, and the Presidential Early Career Award for Scientists and Engineers (PECASE). Dr. Zhang acknowledges the National Cancer Institute of the National Institutes of Health Pathway to Independence Award (K99CA201603). Dr. Trujillo-de Santiago and Dr. Alvarez gratefully acknowledge the funding provided by Consejo Nacional de Ciencia y Tecnología (CONACyT), and Fundación México in Harvard. This research has been partially funded by the Tec de Monterrey and MIT Nanotechnology Program, and the MIT International Science and Technology Initiatives (MISTI). E.S.B. is a co-inventor on a patent on ExM. E.S.B. is a co-founder of Expansion Technologies, Inc., which aims to provide ExM in kit and service form to the community. All expansion microscopy procedures are available freely in protocol form at http://expansionmicroscopy.org.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gubala V, Harris LF, Ricco AJ, Tan MX, Williams DE. Point of care diagnostics: status and future. Analytical Chemistry. 2011;84:487–515. doi: 10.1021/ac2030199. [DOI] [PubMed] [Google Scholar]
- 2.Pai NP, Vadnais C, Denkinger C, Engel N, Pai M. Point-of-care testing for infectious diseases: diversity, complexity, and barriers in low-and middle-income countries. PLoS Medicine. 2012;9:e1001306. doi: 10.1371/journal.pmed.1001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee WG, Kim Y-G, Chung BG, Demirci U, Khademhosseini A. Nano/Microfluidics for diagnosis of infectious diseases in developing countries. Advanced Drug Delivery Reviews. 2010;62:449–457. doi: 10.1016/j.addr.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozcan A, Demirci U. Ultra wide-field lens-free monitoring of cells on-chip. Lab on a Chip. 2008;8:98–106. doi: 10.1039/b713695a. [DOI] [PubMed] [Google Scholar]
- 5.Greenbaum A, Zhang Y, Feizi A, Chung P-L, Luo W, Kandukuri SR, Ozcan A. Wide-field computational imaging of pathology slides using lens-free on-chip microscopy. Science Translational Medicine. 2014;6:267ra175–267ra175. doi: 10.1126/scitranslmed.3009850. [DOI] [PubMed] [Google Scholar]
- 6.Coskun AF, Sencan I, Su T-W, Ozcan A. Wide-field lensless fluorescent microscopy using a tapered fiber-optic faceplate on a chip. Analyst. 2011;136:3512–3518. doi: 10.1039/c0an00926a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cybulski JS, Clements J, Prakash M. Foldscope: origami-based paper microscope. Plos One. 2014;9:e98781. doi: 10.1371/journal.pone.0098781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh KK, Burns LD, Cocker ED, Nimmerjahn A, Ziv Y, El Gamal A, Schnitzer MJ. Miniaturized integration of a fluorescence microscope. Nature Methods. 2011;8:871–878. doi: 10.1038/nmeth.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mudanyali O, Tseng D, Oh C, Isikman SO, Sencan I, Bishara W, Oztoprak C, Seo S, Khademhosseini B, Ozcan A. Compact, light-weight and cost-effective microscope based on lensless incoherent holography for telemedicine applications. Lab on a Chip. 2010;10:1417–1428. doi: 10.1039/c000453g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim SB, Bae H, Koo K-i, Dokmeci MR, Ozcan A, Khademhosseini A. Lens-free imaging for biological applications. Journal of Laboratory Automation. 2012;17:43–49. doi: 10.1177/2211068211426695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Chen F, Tillberg PW, Boyden ES. Expansion microscopy. Science. 2015;347:1260088. doi: 10.1126/science.1260088. This paper denotes the invention of the groundbreaking technology of expansion microscopy, where the authors showed nanoscale imaging using conventional diffraction-limited optical microscopy through physical expansion of biological specimens. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Chen F, Wassie AT, Cote AJ, Sinha A, Alon S, Asano S, Daugharthy ER, Chang J-B, Marblestone A, Church GM. Nanoscale imaging of RNA with expansion microscopy. Nature Methods. 2016;13:679–684. doi: 10.1038/nmeth.3899. The authors extend the appliation of the original expansion microscopy to high-resolution imaging of nucleic acids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Tillberg PW, Chen F, Piatkevich KD, Zhao Y, Yu C-CJ, English BP, Gao L, Martorell A, Suk H-J, Yoshida F. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nature Biotechnology. 2016;34:987–992. doi: 10.1038/nbt.3625. The authors extend the appliation of the original expansion microscopy to high-resolution imaging of proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14••.Zhang YS, Chang J-B, Alvarez MM, Trujillo-de Santiago G, Aleman J, Batzaya B, Krishnadoss V, Ramanujam AA, Kazemzadeh-Narbat M, Chen F. Hybrid Microscopy: Enabling Inexpensive High-Performance Imaging through Combined Physical and Optical Magnifications. Scientific Reports. 2016;6:22691. doi: 10.1038/srep22691. This paper provides the first example of the combination of expansion microscopy with mini-microscopy to image biological samples at high resolution with low cost. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Germroth PG, Gourdie RG, Thompson RP. Confocal microscopy of thick sections from acrylamide gel embedded embryos. Microscopy Research and Technique. 1995;30:513–520. doi: 10.1002/jemt.1070300608. [DOI] [PubMed] [Google Scholar]
- 16.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nature Methods. 2006;3:793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hein B, Willig KI, Hell SW. Stimulated emission depletion (STED) nanoscopy of a fluorescent protein-labeled organelle inside a living cell. Proceedings of the National Academy of Sciences. 2008;105:14271–14276. doi: 10.1073/pnas.0807705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bucur O, Zhao Y, Boyden E, Beck AH. Physical expansion of tissue microarrays for high-resolution imaging of normal and cancer samples with conventional microscopy. Cancer Research. 2016;76:4229–4229. [Google Scholar]
- 19.Crittenden JR, Tillberg PW, Riad MH, Shima Y, Gerfen CR, Curry J, Housman DE, Nelson SB, Boyden ES, Graybiel AM. Striosome–dendron bouquets highlight a unique striatonigral circuit targeting dopamine-containing neurons. Proceedings of the National Academy of Sciences. 2016;113:11318–11323. doi: 10.1073/pnas.1613337113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20•.Ku T, Swaney J, Park J-Y, Albanese A, Murray E, Cho JH, Park Y-G, Mangena V, Chen J, Chung K. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nature Biotechnology. 2016;34:973–981. doi: 10.1038/nbt.3641. This paper reports a variation of expansion microscopy based on a similar physical expansion principle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Chozinski TJ, Halpern AR, Okawa H, Kim H-J, Tremel GJ, Wong RO, Vaughan JC. Expansion microscopy with conventional antibodies and fluorescent proteins. Nature Methods. 2016;13:485–488. doi: 10.1038/nmeth.3833. This paper has generalized the use of conventional antibodies and fluorescence proteins in expansion microscopy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lubeck E, Cai L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nature Methods. 2012;9:743–748. doi: 10.1038/nmeth.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nature Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moffitt JR, Hao J, Wang G, Chen KH, Babcock HP, Zhuang X. High-throughput single-cell geneexpression profiling with multiplexed error-robust fluorescence in situ hybridization. Proceedings of the National Academy of Sciences. 2016;113:11046–11051. doi: 10.1073/pnas.1612826113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shah S, Lubeck E, Zhou W, Cai L. In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron. 2016;92:342–357. doi: 10.1016/j.neuron.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsanov N, Samacoits A, Chouaib R, Traboulsi A-M, Gostan T, Weber C, Zimmer C, Zibara K, Walter T, Peter M. smiFISH and FISH-quant–a flexible single RNA detection approach with superresolution capability. Nucleic Acids Research. 2016:gkw784. doi: 10.1093/nar/gkw784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu H, Yaglidere O, Su T, Tseng D, Ozcan A. Cost-effective and compact wide-field fluorescent imaging on a cell-phone. Lab on a Chip. 2011;11:315–322. doi: 10.1039/c0lc00358a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaefer S, Boehm S, Chau K. Automated, portable, low-cost bright-field and fluorescence microscope with autofocus and autoscanning capabilities. Applied Optics. 2012;51:2581–2588. doi: 10.1364/AO.51.002581. [DOI] [PubMed] [Google Scholar]
- 30.Kim S, Koo K, Bae H, Dokmeci M, Hamilton G, Bahinski A, Kim S, Ingber D, Khademhosseini A. A minimicroscope for in situ monitoring of cells. Lab on a Chip. 2012;12:3976–3982. doi: 10.1039/c2lc40345e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Zhang Y, Ribas J, Nadhman A, Aleman J, Selimović Š, Lesher-Perez S, Wang T, Manoharan V, Shin S, Damilano A, et al. A cost-effective fluorescence mini-microscope for biomedical applications. Lab on a Chip. 2015;15:3661–3669. doi: 10.1039/c5lc00666j. Using the webcam-based mini-microscope, the authors describe portable imaging of biological samples at low cost. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong S, Guo K, Nanda P, Shiradkar R, Zheng G. FPscope: a field-portable high-resolution microscope using a cellphone lens. Biomedical Optics Express. 2014;5:3305–33010. doi: 10.1364/BOE.5.003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greenbaum A, Sikora U, Ozcan A. Field-portable wide-field microscopy of dense samples using multi-height pixel super-resolution based lensfree imaging. Lab on a Chip. 2012;12:1242–1245. doi: 10.1039/c2lc21072j. [DOI] [PubMed] [Google Scholar]
- 34.Greenbaum A, Akbari N, Feizi A, Luo W, Ozcan A. Field-portable pixel super-resolution colour microscope. PLOS ONE. 2013;8:p.e76475. doi: 10.1371/journal.pone.0076475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLeod E, Dincer T, Veli M, Ertas Y, Nguyen C, Luo W, Greenbaum A, Feizi A, Ozcan A. Highthroughput and label-free single nanoparticle sizing based on time-resolved on-chip microscopy. ACS Nano. 2015;9:3265–3273. doi: 10.1021/acsnano.5b00388. [DOI] [PubMed] [Google Scholar]
- 36.Wei Q, Luo W, Chiang S, Kappel T, Mejia C, Tseng D, Chan RYL, Yan E, Qi H, Shabbir F. Imaging and sizing of single DNA molecules on a mobile phone. ACS Nano. 2014;8:12725–12733. doi: 10.1021/nn505821y. [DOI] [PubMed] [Google Scholar]
- 37.Kühnemund M, Wei Q, Darai E, Wang Y, Hernández-Neuta I, Yang Z, Tseng D, Ahlford A, Mathot L, Sjöblom T. Targeted DNA sequencing and in situ mutation analysis using mobile phone microscopy. Nature Communications. 2017;8:13913. doi: 10.1038/ncomms13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller A, Davis G, Oden Z, Razavi M, Fateh A, Ghazanfari M, Abdolrahimi F, Poorazar S, Sakhaie F, Olsen R, et al. Portable, battery-operated, low-cost, bright field and fluorescence microscope. PLOS ONE. 2010;5:p.e11890. doi: 10.1371/journal.pone.0011890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bishara W, Sikora U, Mudanyali O, Su T, Yaglidere O, Luckhart S, Ozcan A. Holographic pixel superresolution in portable lensless on-chip microscopy using a fiber-optic array. Lab on a Chip. 2011;11:1276–1279. doi: 10.1039/c0lc00684j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isikman S, Bishara W, Sikora U, Yaglidere O, Yeah J, Ozcan A. Field-portable lensfree tomographic microscope. Lab on a Chip. 2011;11:2222–2230. doi: 10.1039/c1lc20127a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Im H, Castro C, Shao H, Liong M, Song J, Pathania D, Fexon L, Min C, Avila-Wallace M, Zurkiya O, et al. Digital diffraction analysis enables low-cost molecular diagnostics on a smartphone. Proceedings of the National Academy of Sciences. 2015;112:5613–5618. doi: 10.1073/pnas.1501815112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hutchison J, Erikson R, Sheen A, Ozanich R, Kelly R. Reagent-free and portable detection of Bacillus anthracis spores using a microfluidic incubator and smartphone microscope. Analyst. 2015;140:6269–6276. doi: 10.1039/c5an01304f. [DOI] [PubMed] [Google Scholar]
- 43.Huang H, Wei K, Zhao Y. 2016 IEEE 29th International Conference on Micro Electro Mechanical Systems (MEMS) IEEE; 2016. Variable focus smartphone based microscope using an elastomer liquidlens. [Google Scholar]
- 44.Zhang Y, Busignani F, Ribas J, Aleman J, Rodrigues T, Shaegh S, Massa S, Rossi C, Taurino I, Shin S, et al. Google Glass-Directed Monitoring and Control of Microfluidic Biosensors and Actuators. Scientific Reports. 2016;6:22237. doi: 10.1038/srep22237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bochyńska A, Hannink G, Grijpma D, Buma P. Tissue adhesives for meniscus tear repair: an overview of current advances and prospects for future clinical solutions. Journal of Materials Science: Materials in Medicine. 2016;27:1–18. doi: 10.1007/s10856-016-5694-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pirnstill CW, Coté GL. Malaria diagnosis using a mobile phone polarized microscope. Scientific reports. 2015;5:13368. doi: 10.1038/srep13368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen Y, Ramon O, Kopelman IJ, Mizrahi S. Characterization of inhomogeneous polyacrylamide hydrogels. Journal of Polymer Science Part B: Polymer Physics. 1992;30:1055–1067. [Google Scholar]