Abstract
HECT E3 ubiquitin ligases are responsible for many human disease phenotypes, and are promising drug targets. However, screening assays for HECT E3 inhibitors are inherently complex, requiring upstream E1 and E2 enzymes as well as ubiquitin, ATP, and detection reagents. Intermediate ubiquitin thioesters and a complex mixture of polyubiquitin products provide further opportunities for off-target inhibition and to increase the complexity of the assay. We have developed UbFluor, a novel ubiquitin thioester which bypasses the E1 and E2 enzymes and undergoes the direct transthiolation with HECT E3 ligases. The release of fluorophore upon transthiolation allows fluorescence polarization (FP) detection of HECT E3 activity. In the presence of inhibitors, HECT E3 activity is ablated, and thus no reaction and no change in FP are observed. We have adapted this assay for high-throughput screening of small molecules against HECT E3 ligases and have proven its utility in the discovery of HECT E3 ligase inhibitors.
Keywords: ubiquitin, HECT E3 ligase, high-throughput screen, fluorescence polarization, thioester
INTRODUCTION
The ubiquitin (Ub) system is among the largest and most complex sets of enzymes in the eukaryotic genome (Ciechanover and Schwartz, 1998). E1, E2, E3, and deubiquitinase (DUB) enzymes collectively regulated the degradation, and in some cases the localization or activity, of thousands of protein substrates in essentially all eukaryotic life processes (Varshavsky, 2012). Among these enzymes, Homologous to E6AP Carboxyl Terminus (HECT) E3 ubiquitin ligases (28 known) are a chemically tractable and biologically relevant class of evolutionarily conserved enzymes (Scheffner at al., 1995; Huibregtse et al., 1995). HECT E3s act by means of a catalytic cysteine residue, which participates in the formation of an obligate HECT E3~Ub thioester (~ denotes thioester bond) prior to the isopeptide ligation of Ub onto a substrate lysine residue (Scheffner et al., 1995; Verdecia et al., 2003). Mutation, deletion, or overexpression of HECT E3s is known to cause a variety of diseases or abnormal phenotypes in both humans and model organisms (Scheffner and Kumar, 2014). Genetic inactivation of some HECTs is lethal (Boase et al., 2011; Fouladkou et al., 2010), while others are tolerated but result in phenotypes ranging from bone mass increases to intellectual and developmental disabilities (Jiang et al., 1998; Yamashita et al., 2005). HECT E3s are upregulated in several cancers (Bernassola et al., 2008; Nguyen Huu et al., 2008), and are also hijacked by viruses during infections (Yasuda et al., 2003). Thus, discovery of small-molecule modulators of HECT ligases will allow treatment of diseases by reversing misregulated E3 ligase activity, restoring normal ubiquitination of substrate proteins. Accordingly, these enzymes are promising drug targets.
However, due to the complexity of ubiquitin system assays, the study of HECT E3s is difficult, impeding progress towards FDA-approved inhibitors or activators of these enzymes. Ubiquitin must be activated by an E1 enzyme in an ATP-dependent manner, forming an E1~Ub thioester (Schulman and Harper, 2009). Ubiquitin is subsequently transthiolated onto an E2 enzyme, and the resultant E2~Ub thioester can then finally react with an E3 enzyme and ubiquitin can be transferred to a substrate lysine residue (Varshavsky, 2012). In the case of HECT E3 ligases, Ub is covalently transferred to the HECT catalytic cysteine as a HECT E3~Ub thioester before isopeptide ligation onto the substrate (Huibregtse et al., 1995). Due to the presence of lysine residues on substrates, E3 enzymes, and ubiquitin itself, a variety of ubiquitinated products are produced by these reactions. Discovering compounds which specifically inhibit E3 ligases is difficult because hits in a high-throughput screen may actually be targeting the upstream enzymes or their thioester intermediates. Furthermore, measuring the activity of E3 ligases by the observation of these products can be challenging, requiring additional detection agents or low-throughput gel-based imaging techniques.
To address the issues of both complexity and detection, we developed the chemical probe UbFluor, which is comprised of ubiquitin conjugated via its C-terminus to a fluorescein thiol (FluorSH) through a thioester bond (Krist et al., 2016). This thioester exactly mimics the chemistry of a native E2~Ub thioester. UbFluor can be recognized by HECT E3 ligases and undergo a transthiolation reaction in which Ub is transferred onto the HECT catalytic cysteine, forming a HECT E3~Ub thioester with concomitant release of FluorSH (Figure 1). In previous work, we have demonstrated that this reaction accurately recapitulates the native activity of HECT E3 ligases. The release of FluorSH alters the apparent molecular weight of the fluorescent reporter, allowing real-time monitoring of this transthiolation reaction by fluorescence polarization (FP). FP assays are readily adapted to 384-well plates, allowing high-throughput monitoring of HECT E3 ligase activity and thus high-throughput screening (HTS) of prospective HECT E3 ligase inhibitors.
Figure 1.
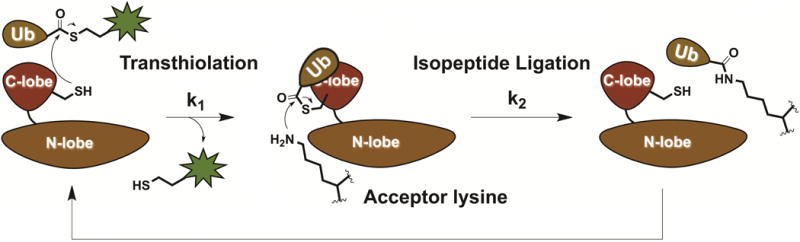
HECT E3 ligase reacts with UbFluor. This initial transthiolation (k1) results in a HECT E3~Ub thioester and the release of FluorSH. HECT E3~Ub is subsequently cleared by isopeptide bond ligation, regenerating free HECT E3. Thus, defects in transthiolation or isopeptide ligation will both be detected by the UbFluor assay MT reaction conditions, in which there is excess of UbFluor over enzyme.
In previously published work, we have shown that HECT E3 ligase reactions with UbFluor are dependent on the HECT catalytic cysteine, as well as on other residues critical for HECT E3-Ub binding interactions and HECT catalysis (Krist et al., 2016). Known mutations which inhibit the activity of HECT E3s can partially or completely abolish HECT reactivity with UbFluor, giving us confidence that small molecule inhibitors of HECT ligases would do the same. We have also shown that UbFluor can quantify the activity of the isolated catalytic HECT domain, rather than requiring the full-length enzyme, which can be difficult to express in large quantities. To establish HTS conditions, we have shown that iodoacetamide (0.5 mM, 0.5% DMSO) can inhibit HECT domain reactivity towards UbFluor by reacting with the HECT catalytic cysteine. Thus, iodoacetamide acts as a positive control for HECT E3 inhibition. Using 0.5% DMSO as a negative control, the percent inhibition of a screening compound can easily be calculated by normalizing the FP measurement of the positive and negative controls to 100% and 0% inhibition, respectively.
Modulation of the UbFluor to HECT E3 ligase ratio allows the monitoring of different aspects of HECT E3 ligase activity (Krist et al., 2016). With an excess of E3 over UbFluor (single turnover conditions, ST), each molecule of HECT E3 will encounter at most 1 molecule of UbFluor on average. Thus, the observed reaction rate will be solely dependent on the initial transthiolation of UbFluor by the HECT E3. In the reverse case of UbFluor in excess over E3 (multiple turnover conditions, MT), each HECT E3 must on average consume more than 1 molecule of UbFluor. Under MT conditions, the overall rate of UbFluor consumption is dependent on the combined rate of UbFluor transthiolation and subsequent isopeptide ligation of Ub to regenerate the free HECT E3 ligase. Therefore, screening of prospective inhibitors of HECT E3s under MT conditions will allow the detection of inhibitors of both transthiolation and/or isopeptide ligation. Accordingly, we have selected MT conditions as our preferred regime for HTS of HECT E3 inhibitors. In principle, it is possible to design a screen under ST conditions if only inhibitors of transthiolation are desired.
Prior to screening a library of compounds against a HECT E3, it is essential to optimize the reaction conditions such that a statistically meaningful separation is observed between the positive and negative controls. HTS assays are often evaluated by their Z-factor (Z′), a calculated value between 0 and 1 that measures the separation between two sets of control values in terms
| Equation 1. Z-factor or Z′ |
of standard deviations (Equation 1) (Zhang et al., 1999). An acceptable Z′ of 0.5 is equal to 12 standard deviations of difference between positive and negative controls. For some HECTs, low innate reactivity of the HECT domain under MT conditions results in low Z′ unless the concentration of HECT is increased to counteract its low reactivity. It is not necessarily possible to predict the reactivity of a particular HECT ligase with UbFluor a priori; optimal conditions must often be determined empirically. In our experience, full length ezyme is more active than isolated HECT domain. In forthcoming work, we will report data from HTS screens; in these screens, we routinely achieved Z′ > 0.7.
Once screening conditions are established, UbFluor allows rapid identification of hit compounds via simple conversion of FP values to percent inhibition. Subsequently, UbFluor can also be used for dose response studies to further evaluate the potency and quality of prospective inhibitors. As in any screen, hits must then be evaluated in orthogonal assays. In our case, we have used western blot and in-gel fluorescence assays to confirm the activity and specificity of our HECT E3 ligase inhibitors in the native cascade.
This protocol describes the determination of screening conditions, the use of UbFluor for HTS, dose response studies using UbFluor, and data analysis. A protocol for the synthesis of UbFluor was previously published.
STRATEGIC PLANNING
Prior to undertaking HTS screens, it is essential to establish screening conditions, select an appropriate library of screening compounds, obtain sufficient quantities of HECT ligase and UbFluor, and ensure that suitable automation of reagent handling can be accomplished. The enzyme construct must be considered carefully, as the activity and ease of expression of different HECTs varies considerably. In some cases, a HECT domain with low innate reactivity may become more active in the presence of its proximal WW domain(s), while in other cases the full-length ligase may be autoinhibited. Care must be taken to ensure consistent quality of HECT domain or ligase, as inconsistency of ligase expression or purification may result in variations in the screening data. The UbFluor assay has been developed specifically for use in 384-well plates, with typical reaction volumes of 25 μL, typical HECT E3 concentrations ranging from 0.1–0.5 μM, and typical UbFluor concentration ranging from 0.5–4 μM. With suitable automation, at least 3,000 compounds can be screened in one day. Because UbFluor is a thioester, free thiols such as dithiothreitol (DTT) or β-mercaptoethanol (BME) must not be present in the screening buffers or enzyme stock solutions.
BASIC PROTOCOL 1: DETERMINATION OF HTS CONDITIONS
Once a HECT E3 ligase of interest has been identified, the ligase and UbFluor must be produced or obtained. Screening conditions must then be empirically determined by preliminary FP experiments. While it is not necessary to use automated liquid handling systems for these preliminary experiments, they can be useful proving grounds for the use of automation. Likewise, an automated multi-plate reader is not required for conditioning experiments, but such a reader is strongly recommended for screening experiments. To ensure high Z′, workable timeframe, and MT reaction conditions, we have established the following necessary criteria for HTS conditions: at least 4-fold excess of UbFluor over HECT, and at least 50% consumption of UbFluor by the goal endpoint time of the assay. We have used 8 hours as our assay endpoint, but in principle this can be altered depending upon limitations of reagent availability, time and instrumentation. For the determination of conditions, we test three initial concentrations of HECT ligase in triplicate using positive and negative controls iodoacetamide (0.5 mM) and DMSO (0.5%), respectively. The reactions also contain final concentrations of 50 mM HEPES (pH 7.5), 150 mM NaCl, and 6 μM Tween-20. If necessary, iterative rounds of testing are performed. Increasing HECT ligase concentration will speed up the reaction in the case of slow or indistinguishable reaction progress. Reduction of HECT ligase concentration will identify the minimal necessary concentrations of HECT and UbFluor in order to conserve reagents.
Materials
REAGENTS
DMSO
Iodoacetamide (100 mM stock solution in DMSO, freshly prepared from solid the same day)
HECT ligase stock solution (either catalytic HECT domain, HECT domain plus WW domains, or full length enzyme; commercially available or obtainable by previously published procedures from E.coli expression)
10X UbFluor Assay Buffer: 1.5 M NaCl, 500 mM HEPES pH 7.5 (see recipe)
Tween-20
UbFluor stock solution (synthesis available from previously published work)
EQUIPMENT AND MATERIALS
384-well plate (low volume, low binding, Corning #3820)
TTP Labtech Mosquito liquid handler or Labcyte Echo 550 liquid handler
Integra VIAFILL rapid reagent dispenser or comparable liquid handler (Optional)
Centrifuge for 384-well plates (Eppendorf Centrifuge 5810 or comparable)
Corning plate shaker platform (CHECK FOR NAME DETAILS)
Tecan Infinite M1000 Pro plate reader or comparable
Procedure for determination of HTS conditions
-
Using the Mosquito or Echo, add 3 wells each of 125 nL DMSO or 100 mM Iodoacetamide in DMSO to a 384-well plate. Repeat for each condition to be tested, as multiple conditions are easily tested in parallel.
For an initial scan of conditions, we typically test three equally-spaced concentrations of HECT ligase with 5-fold excess of UbFluor: 0.1, 0.3, and 0.5 μM HECT with 0.5, 1.5, and 2.5 μM UbFluor. Our reactions also typically contain the following components at the given final concentrations: 150 mM NaCl, 6 μM Tween-20, 50 mM HEPES pH 7.5.
Prepare 100 μL of a 1.67X solution of HECT ligase in UbFluor assay buffer with 6 μM Tween-20. Repeat for each condition to be tested. E.g. to test 0.5 μM HECT ligase, prepare a solution with 0.833 μM HECT ligases, 150 mM NaCl, 6 μM Tween-20, 50 mM HEPES pH 7.5.
-
Add 15 μL of HECT ligase solution to wells in 384-well plate containing iodoacetamide and DMSO.
Use of the VIAFILL reagent dispenser at this step is optional.
Centrifuge plate at 1500 × g for 1 minute.
Shake plate at 850 rpm for 5 minutes.
Centrifuge plate at 1500 × g for 1 minute.
Incubate plate at RT for 30 minutes.
-
Prepare 70 μL of a 2.5X solution of UbFluor from stock solution in UbFluor assay buffer with 6 μM Tween-20. Repeat for each condition to be tested.
E.g. if desired [UbFluor] for the above 0.5 μM HECT is 2.5 μM UbFluor, prepare a solution with 6.25 μM UbFluor, 150 mM NaCl, 6 μM Tween-20, 50 mM HEPES pH 7.5.
-
Add 10 μL of UbFluor solution to wells containing the corresponding concentration of HECT domain.
Use of the VIAFILL reagent dispenser at this step is optional.
Repeat spin/shake/spin cycle above (steps 4–6).
Read FP of all wells immediately using the M1000 plate reader (470 nm excitation wavelength, 530 nm emission wavelength).
-
Read FP of all wells every hour for 8 hours or until the desired endpoint time of the HTS assay. Keep wells covered and protected from light between readings.
This step can be automated using the plate reader software. However, if it is not reasonable to occupy the plate reader for this length of time, FP readings can be taken manually at intervals until the desired endpoint time is reached.
-
Using a simple arithmetic conversion of mP to percent UbFluor consumed, determine whether at least 50% of the UbFluor in the DMSO-containing reactions has been consumed by the desired assay endpoint time. Additionally, confirm that less than 5% of UbFluor has been consumed in the iodoacetamide-containing reactions.
If you have no previous reference points for 0% and 100% UbFluor consumption, you may simply incubate UbFluor at the desired assay concentration in the presence and absence of 1 mM β-mercaptoethanol for 30 minutes, then measure the FP value using the M1000 plate reader. We have found values of ~100 mP for unreacted UbFluor (0% consumed) and ~0 mP for cleaved UbFluor (100% consumed), but these may vary depending upon instrument and calibration.
Repeat steps 1–13 if necessary until conditions are identified in which 50% of UbFluor is consumed by the desired HTS assay endpoint time.
Repeat steps 1–12 using the final identified conditions with 16 wells each of iodoacetamide and DMSO.
-
Calculate Z′ from 16-well experiment. Z′ > 0.5 is desirable. Longer assay time or increased HECT concentration will generally increase Z′.
Inconsistency within the positive and negative controls will lower an assay’s Z′. If there is large variation in the polarization values of either the positive or negative controls, check for possible errors in liquid handling. Low HECT or UbFluor purity may also contribute to variations that result in lower Z′.
BASIC PROTOCOL 2: HTS AGAINST HECT E3 LIGASE WITH UBFLUOR
After appropriate assay conditions have been determined, further planning is needed to ensure an efficient and successful HTS campaign. Production of HECT E3 ligase and UbFluor should be completed prior to screening, in order to allow for sufficient quality control of these materials prior to use in HTS. In addition to the production of consistently active HECT E3 ligase and UbFluor, selection of an appropriate screening library is an essential component of HTS project design. Because HECT E3 ligases contain a catalytic cysteine, it is important to consider whether the goal is to discover covalent or noncovalent inhibitors of the HECT ligase to be screened. The UbFluor assay is amenable to the use of drug-like compounds and fragment compounds alike, again depending on the preference of the user. In our experience, the hit rates range from 1–2.5% for drug-like compounds (50 μM compound), and from 5–9% for fragment compounds (200 μM compound final concentration). Libraries of both types are available commercially from many vendors. The UbFluor assay allows for rapid screening of compound libraries, accommodating up to 5,000 compounds per day with the potential for even more. As a result, meticulous and consistent record-keeping is essential for subsequent analysis of screening data. This protocol describes the procedure for the screening of 16×384-well plates, to be repeated on successive days until the entire selected compound library has been screened. In order to establish comfort with instrumentation and data management, it may be beneficial to start with a smaller number of plates per day and increase throughput thereafter.
Materials
REAGENTS
DMSO
Iodoacetamide (100 mM stock solution in DMSO, freshly prepared from solid the same day)
Screening compounds in DMSO stock solution
HECT ligase stock solution (either catalytic HECT domain, HECT domain plus WW domains, or full length enzyme; commercially available or obtainable by previously published procedures from E.coli expression)
10X UbFluor Assay Buffer: 1.5 M NaCl, 500 mM HEPES pH 7.5 (see recipe)
Tween-20
UbFluor stock solution (synthesis available from previously published work)
EQUIPMENT AND MATERIALS
384-well plate (low volume, low binding, Corning #3820)
TTP Labtech Mosquito liquid handler (optional)
Labcyte Echo 550 liquid handler
Integra VIAFILL rapid reagent dispenser or comparable liquid handler (Optional)
Centrifuge for 384-well plates (Eppendorf Centrifuge 5810 or comparable)
Corning plate shaker platform (CHECK FOR NAME DETAILS)
Tecan Infinite M1000 Pro plate reader or comparable
Procedure for HTS against HECT E3 ligase with UbFluor
-
Using the Mosquito or Echo, add 125 nL DMSO or 100 mM Iodoacetamide in DMSO to alternating wells of columns 1 and 24 of several 384-well plates.
The number of plates required depends on the number of hit compounds identified by HTS.
-
Using the Mosquito or Echo, add 8 amounts (100, 75, 50, 25, 12.5, 5, 2.5, 0 nL, yielding 40, 30, 20, 10, 5, 2, 1, 0 μM) of each hit compound in DMSO to columns 2–23 of the plates. Two compounds can be added to each column, one in rows A-H and the second in rows I-P.
Compounds stored in 10 mM DMSO will yield the above final compound concentrations; adjust accordingly for different stock or desired final concentrations. In principle, a larger or smaller number of concentrations may be interrogated.
Prepare a 1.67X solution of HECT ligase in UbFluor assay buffer with 6 μM Tween-20.
-
Using the VIAFILL reagent dispenser, add 15 μL of HECT ligase solution to columns 1–24 of each 384-well plate.
If centrifuge or plate shaker are not large enough to accommodate all plates at once, it can be helpful to perform steps 4–7 for groups of 4 plates.
Centrifuge plates at 1500 × g for 1 minute.
Shake plates on the plate shaker at 850 rpm for 5 minutes.
Centrifuge plates at 1500 × g for 1 minute.
Incubate plates at RT for 30 minutes.
Prepare a 2.5x solution of UbFluor from stock solution in UbFluor assay buffer with 6 μM Tween-20.
Using the VIAFILL reagent dispenser, add 10 μL of UbFluor solution to columns 1–24 of all plates.
Repeat spin/shake/spin cycle above (steps 5–7).
Read FP of columns 1–24 of all plates immediately using the M1000 plate reader with automated plate stacker (470 nm excitation wavelength, 530 nm emission wavelength).
-
Read FP of all plates every 2 hours for 8 hours or until the desired endpoint time of the HTS assay. Keep plates covered and protected from light between readings.
This step can be automated using the plate reader software. Intermediate readings every 2 hours can be eliminated if necessary, but provide the opportunity for additional quality control.
Use the FP value averages of the DMSO and iodoacetamide controls to calculate 0% and 100% inhibition thresholds, respectively, for each individual plate.
Calculate the percent inhibition of all 8 concentrations of each compound using the 0% and 100% thresholds from step 14 for the corresponding plate.
Plot the percent inhibition of the 8 concentrations of each compound as a function of the log of compound concentration.
Using a 4-parameter logistic fit, determine the maximum inhibition (Ymax), IC50, and Hill slope (n) for the dose response curve of each compound.
-
Eliminate from consideration any compounds which have a Hill slope greater than 2.5 or a ymax less than 50%.
Hill slope can be used to characterize stoichiometric binding of a noncovalent inhibitor to an enzyme. An ideal Hill slope is 1. High Hill slopes indicate aggregation, denaturation, nonspecific binding, or other bulk phenomena not considered useful for specific inhibition of an enzyme.
Rank all remaining compounds by IC50. If desired, manually remove compounds with chemical structures indicative of nonspecific inhibitory modes of action. Select the most potent compounds of interest for validation in orthogonal assays.
BASIC PROTOCOL 3: DOSE RESPONSE OF HIT COMPOUNDS AGIAINST HECT E3 LIGASE WITH UBFLUOR
Validation of hits from HTS begins with confirmation of dose-dependent inhibition. The UbFluor assay can be readily utilized for dose-response studies with minimal alterations to the HTS protocol (Basic Protocol 2). The results of these studies can first and foremost confirm the reproducibility of hits from UbFluor-based HTS and determine their potency. Furthermore, the Hill slope of a dose-response curve can also provide evidence for (or against) stoichiometric binding of a noncovalent inhibitor to the HECT ligase. An ideal Hill slope of 1 indicates stoichiometric binding, but variation between 0.5 and 1.5 is generally considered acceptable (Shoichet, 2006). Accordingly, the rapid generation of a large data set of dose responses using the simple UbFluor assay facilitates the identification of the most promising hit compounds, which can then be validated in orthogonal assays. Furthermore if the same library of chemical compounds is screened against multiple ligases simultaneous dose response curves can provide means to identify selective inhibitors.
Materials
REAGENTS
DMSO
Iodoacetamide (100 mM stock solution in DMSO, freshly prepared from solid the same day)
Screening compounds in DMSO stock solution
HECT ligase stock solution (either catalytic HECT domain, HECT domain plus WW domains, or full length enzyme; commercially available or obtainable by previously published procedures from E.coli expression)
10X UbFluor Assay Buffer: 1.5 M NaCl, 500 mM HEPES pH 7.5 (see recipe)
Tween-20
UbFluor stock solution (synthesis available from previously published work)
EQUIPMENT AND MATERIALS
384-well plate (low volume, low binding, Corning #3820)
TTP Labtech Mosquito liquid handler (optional)
Labcyte Echo 550 liquid handler
Integra VIAFILL rapid reagent dispenser or comparable liquid handler (Optional)
Centrifuge for 384-well plates (Eppendorf Centrifuge 5810 or comparable)
Corning plate shaker platform (CHECK FOR NAME DETAILS)
Tecan Infinite M1000 Pro plate reader or comparable
Procedure for dose response of hit compounds against HECT E3 ligase with UbFluor
-
Using the Mosquito or Echo, add 125 nL DMSO or 100 mM Iodoacetamide in DMSO to alternating wells of columns 1 and 24 of several 384-well plates.
The number of plates required depends on the number of hit compounds identified by HTS.
-
Using the Mosquito or Echo, add 8 amounts (100, 75, 50, 25, 12.5, 5, 2.5, 0 nL, yielding 40, 30, 20, 10, 5, 2, 1, 0 μM) of each hit compound in DMSO to columns 2–23 of the plates. Two compounds can be added to each column, one in rows A-H and the second in rows I-P.
Compounds stored in 10 mM DMSO will yield the above final compound concentrations; adjust accordingly for different stock or desired final concentrations. In principle, a larger or smaller number of concentrations may be interrogated.
Prepare a 1.67X solution of HECT ligase in UbFluor assay buffer with 6 μM Tween-20.
-
Using the VIAFILL reagent dispenser, add 15 μL of HECT ligase solution to columns 1–24 of each 384-well plate.
If centrifuge or plate shaker are not large enough to accommodate all plates at once, it can be helpful to perform steps 4–7 for groups of 4 plates.
Centrifuge plates at 1500 × g for 1 minute.
Shake plates on the plate shaker at 850 rpm for 5 minutes.
Centrifuge plates at 1500 × g for 1 minute.
Incubate plates at RT for 30 minutes.
Prepare a 2.5x solution of UbFluor from stock solution in UbFluor assay buffer with 6 μM Tween-20.
Using the VIAFILL reagent dispenser, add 10 μL of UbFluor solution to columns 1–24 of all plates.
Repeat spin/shake/spin cycle above (steps 5–7).
Read FP of columns 1–24 of all plates immediately using the M1000 plate reader with automated plate stacker (470 nm excitation wavelength, 530 nm emission wavelength).
-
Read FP of all plates every 2 hours for 8 hours or until the desired endpoint time of the HTS assay. Keep plates covered and protected from light between readings.
This step can be automated using the plate reader software. Intermediate readings every 2 hours can be eliminated if necessary, but provide the opportunity for additional quality control.
Use the FP value averages of the DMSO and iodoacetamide controls to calculate 0% and 100% inhibition thresholds, respectively, for each individual plate.
Calculate the percent inhibition of all 8 concentrations of each compound using the 0% and 100% thresholds from step 14 for the corresponding plate.
Plot the percent inhibition of the 8 concentrations of each compound as a function of the log of compound concentration.
Using a 4-parameter logistic fit, determine the maximum inhibition (Ymax), IC50, and Hill slope (n) for the dose response curve of each compound.
-
Eliminate from consideration any compounds which have a Hill slope greater than 2.5 or a ymax less than 50%.
Hill slope can be used to characterize stoichiometric binding of a noncovalent inhibitor to an enzyme. An ideal Hill slope is 1. High Hill slopes indicate aggregation, denaturation, nonspecific binding, or other bulk phenomena not considered useful for specific inhibition of an enzyme.
Rank all remaining compounds by IC50. If desired, manually remove compounds with chemical structures indicative of nonspecific inhibitory modes of action. Select the most potent compounds of interest for validation in orthogonal assays.
BASIC PROTOCOL 4: SINGLE POINT VALIDATION OF HIT COMPOUNDS AGAINST HECT E3 LIGASE IN THE NATIVE UBIQUITINATION CASCADE
Secondary validation of HTS hits continues with western blot-based assays to test hit compound activity in the native cascade. Because UbFluor is a non-native system, it is essential to confirm that prospective inhibitors can inhibit HECT E3s when Ub is received in the form of an E2~Ub thioester. This essential validation experiment is a preliminary indication of whether a hit compound has the potential to be a useful HECT E3 inhibitor. However, since western blots are low-throughput and time-consuming relative to UbFluor assays, this secondary validation is only suitable once hit compounds have been identified, confirmed, and filtered as directed in Basic Protocols 2 and 3.
Materials
REAGENTS
HECT ligase (either catalytic HECT domain, HECT domain plus WW domains, or full length enzyme; commercially available or obtainable by previously published procedures from E. coli expression)
DMSO
Iodoacetamide (100 mM stock solution in DMSO, freshly prepared from solid the same day)
Hit compounds in DMSO stock solution (identified using Basic Protocols 1–3)
10X UbFluor Assay Buffer: 1.5 M NaCl, 500 mM HEPES pH 7.5 (see recipe)
40 mM MgCl2 in H2O
Ubiquitin (Sigma-Aldrich)
UBA1 E1 enzyme (commercially available or obtainable from E. coli expression)
E2 ubiquitin conjugating enzyme (commercially available or obtainable from E. coli expression)
Adenosine triphosphate (ATP; Sigma-Aldrich)
6X Laemmli SDS-PAGE loading buffer (see recipe)
SDS-PAGE running buffer (see recipe)
7.5% SDS-polyacrylamide gel (commercially available or obtainable from standard lab procedures)
Western blot transfer buffer (see recipe)
TBS-T (see recipe)
Blotting-grade blocker
Anti-ubiquitin antibody (rabbit)
Goat anti-rabbit secondary antibody with HRP conjugate
Bio-Rad Clarity Western ECL Substrate
EQUIPMENT AND MATERIALS
Eppendorf 1.5 mL centrifuge tubes or equivalent
Eppendorf tabletop microcentrifuge or equivalent
VWR Advanced Mini Block Heater or equivalent
Bio-Rad Mini-PROTEAN Tetra Cell SDS-PAGE system with Protein Blotting Modules or equivalent
Bio-Rad nitrocellulose western blotting membrane or equivalent
Bio-Rad blot absorbent filter paper or equivalent
Rocking platform
Bio-Rad ChemiDoc XRS+ Molecular Imager
Procedure for dose response of hit compounds against HECT E3 ligase in the native ubiquitination cascade
-
Calculate volume of each enzyme or compound stock solution required for a reaction containing the following final concentrations: 300 nm HECT E3, 50 μM compound or 0.5 mM iodoacetamide, 300 nM E2 enzyme, 100 nM UBA1 (E1 enzyme), 5 μM Ub, 4 mM ATP, 4 mM MgCl2. For each gel available, calculate needed amounts of each reagent for up to 12 compounds and two control tubes (0.5% DMSO, 0.5 mM iodoacetamide).
Typical reaction volume is 10–30 μL. Required volumes will depend on the concentration of enzyme and compound stock solutions. Final reagent concentrations may require optimization for each HECT ligase depending on ligase activity.
-
Combine 10X UbFluor Assay Buffer, HECT E3, and compound (or iodoacetamide solution or DMSO for control reactions) in Eppendorf tubes. If any additional volume of water is needed to achieve the final reaction conditions calculated in step 1, it should be added here as well.
Volumes added will vary depending on stock solution concentrations.
Incubate HECT E3 and compound mixtures from step 2 for 30 minutes at RT.
-
Add MgCl2, Ub, UBA1, and E2 enzyme to the tubes as calculated in step 1.
These reagents may be added individually or as a master mix.
Add ATP to the tubes as calculated in step 1. Mix by gently pipetting the mixture and centrifuge the tubes.
-
Incubate the tubes 60 minutes at RT.
Reaction time may require optimization for each HECT ligase. If necessary, aliquots of each reaction can be quenched at multiple time points ranging from 15 minutes to 2 hours.
Quench reactions using 6x Laemmli buffer. Mix thoroughly and heat at 95°C for 3 minutes.
-
Run quenched reactions on a 7.5% SDS-polyacrylamide gel for 35 minutes at 200 V using the Mini-Protean SDS-PAGE apparatus with SDS-PAGE running buffer.
Gel running time and settings may depend on SDS-PAGE apparatus and user preferences.
-
Using the Protein Blotting Modules, filter paper, and Western blot transfer buffer, transfer the reaction mixtures from the gel to a nitrocellulose membrane at 100 V for 1 hour.
It may be helpful to cut off the bottom portion of the gel containing free ubiquitin, as free ubiquitin may cause high background signal during imaging of the blotted membrane.
Incubate the blotted nitrocellulose membrane for 1 hour at RT with 5% blotting-grade blocker in TBS-T.
Incubate the membrane for 16 hours at 4°C with 5 mL of a 1000:1 dilution of anti-ubiquitin antibody in 5% blotting-grade blocker in TBS-T.
Wash the membrane for 3 × 5 minutes with 5 mL TBS-T.
Incubate the membrane for 1 hour at RT with 3000:1 dilution of goat anti-rabbit antibody in 5% blotting-grade blocker in TBS-T.
Wash the membrane for 3 × 5 minutes with 5 mL TBS-T.
Develop the membrane for 2–4 minutes with 5 mL Clarity Western ECL Substrate (2.5 mL of each included substrate and luminol enhancer solution).
-
Image the membrane using the “blots > chemi” function of the Bio-Rad ChemiDoc XRS+ Molecular Imager.
A high-quality inhibitor should exhibit at least 50% inhibition of poly-ubiquitinated product formation, as evidenced by reduced intensity of free polyubiquitin chains and/or autoubiquitinated HECT ligase. Total polyubiquitinated products can be quantified by the ChemiDoc XRS+ software or by gel imaging programs such as ImageJ.
BASIC PROTOCOL 5: DOSE RESPONSE OF HIT COMPOUNDS AGAINST HECT E3 LIGASE IN THE NATIVE UBIQUITINATION CASCADE
Upon identification of compounds which inhibit the HECT E3 of interest in the native cascade using basic protocol 4, it is necessary to demonstrate that this inhibition is reproducible and dose dependent. This protocol can accommodate fewer compounds per day, and thus should only be performed after initial results confirming inhibition.
Materials
REAGENTS
HECT ligase (either catalytic HECT domain, HECT domain plus WW domains, or full length enzyme; commercially available or obtainable by previously published procedures from E. coli expression)
DMSO
Iodoacetamide (100 mM stock solution in DMSO, freshly prepared from solid the same day)
Hit compounds in DMSO stock solution (identified using Basic Protocols 1–3)
10X UbFluor Assay Buffer: 1.5 M NaCl, 500 mM HEPES pH 7.5 (see recipe)
40 mM MgCl2 in H2O
Ubiquitin (Sigma-Aldrich)
UBA1 E1 enzyme (commercially available or obtainable from E. coli expression)
E2 ubiquitin conjugating enzyme (commercially available or obtainable from E. coli expression)
Adenosine triphosphate (ATP; Sigma-Aldrich)
6X Laemmli SDS-PAGE loading buffer (see recipe)
SDS-PAGE running buffer (see recipe)
7.5% SDS-polyacrylamide gel (commercially available or obtainable from standard lab procedures)
Western blot transfer buffer (see recipe)
TBS-T (see recipe)
Blotting-grade blocker
Anti-ubiquitin antibody (rabbit)
Goat anti-rabbit secondary antibody with HRP conjugate
Bio-Rad Clarity Western ECL Substrate
EQUIPMENT AND MATERIALS
Eppendorf 1.5 mL centrifuge tubes or equivalent
Eppendorf tabletop microcentrifuge or equivalent
VWR Advanced Mini Block Heater or equivalent
Bio-Rad Mini-PROTEAN Tetra Cell SDS-PAGE system with Protein Blotting Modules or equivalent
Bio-Rad nitrocellulose western blotting membrane or equivalent
Bio-Rad blot absorbent filter paper or equivalent
Rocking platform
Bio-Rad ChemiDoc XRS+ Molecular Imager
Procedure for dose response of hit compounds against HECT E3 ligase in the native ubiquitination cascade
-
Calculate volume of each enzyme or compound stock solution required for a reaction containing the following final concentrations: 300 nM HECT E3, 5–50 μM compound or 0.5 mM iodoacetamide, 300 nM E2 enzyme, 100 nM UBA1 (E1 enzyme), 5 μM Ub, 4 mM ATP, 4 mM MgCl2. For each compound, calculate the amounts needed for 6 tubes (5, 10, 20, 30, 40, 50 μM compound) and two control tubes (0.5% DMSO, 0.5 mM iodoacetamide).
Typical reaction volume is 10–30 μL. Required volumes will depend on the concentration of enzyme and compound stock solutions. Final reagent concentrations may require optimization for each HECT ligase depending on ligase activity.
-
Combine 10x UbFluor Assay Buffer, HECT E3, and compound (or iodoacetamide solution or DMSO for control reactions) in Eppendorf tubes. If any additional volume of water is needed to achieve the final reaction conditions calculated in step 1, it should be added here as well.
Volumes added will vary depending on stock solution concentrations.
Incubate HECT E3 and compound mixtures from step 2 for 30 minutes at RT.
-
Add MgCl2, Ub, UBA1, and E2 enzyme to the tubes as calculated in step 1.
These reagents may be added individually or as a master mix.
Add ATP to the tubes as calculated in step 1. Mix by gently pipetting the mixture and centrifuge the tubes.
-
Incubate the tubes 60 minutes at RT.
Reaction time may require optimization for each HECT ligase. If necessary, aliquots of each reaction can be quenched at multiple time points ranging from 15 minutes to 2 hours.
Quench reactions using 6x Laemmli buffer. Mix thoroughly and heat at 95°C for 3 minutes.
-
Run quenched reactions on a 7.5% SDS-polyacrylamide gel for 35 minutes at 200 V using the Mini-Protean SDS-PAGE apparatus with SDS-PAGE running buffer.
Gel running time and settings may depend on SDS-PAGE apparatus and user preferences.
-
Using the Protein Blotting Modules, filter paper, and Western blot transfer buffer, transfer the reaction mixtures from the gel to a nitrocellulose membrane at 100 V for 1 hour.
It may be helpful to cut off the bottom portion of the gel containing free ubiquitin, as free ubiquitin may cause high background signal during imaging of the blotted membrane.
Incubate the blotted nitrocellulose membrane for 1 hour at RT with 5% blotting-grade blocker in TBS-T.
Incubate the membrane for 16 hours at 4°C with 5 mL of a 1000:1 dilution of anti-ubiquitin antibody in 5% blotting-grade blocker in TBS-T.
Wash the membrane for 3 × 5 minutes with 5 mL TBS-T.
Incubate the membrane for 1 hour at RT with 3000:1 dilution of goat anti-rabbit antibody in 5% blotting-grade blocker in TBS-T.
Wash the membrane for 3 × 5 minutes with 5 mL TBS-T.
Develop the membrane for 2–4 minutes with 5 mL Clarity Western ECL Substrate (2.5 mL of each included substrate and luminol enhancer solution).
-
Image the membrane using the “blots > chemi” function of the Bio-Rad ChemiDoc XRS+ Molecular Imager.
A high-quality inhibitor should exhibit dose-dependent inhibition of poly-ubiquitinated product formation, as evidenced by reduced intensity of free polyubiquitin chains and/or autoubiquitinated HECT ligase. Total polyubiquitinated products can be quantified by the ChemiDoc XRS+ software or by gel imaging programs such as ImageJ.
BASIC PROTOCOL 6: EXCLUSION OF E1/E2 TRANSTHIOLATION BY HIT COMPOUNDS
Once identified hit compounds have been shown to inhibit HECT E3 ligase activity in the native ubiquitination cascade, it is important to confirm that hit compounds are specific to the HECT ligase. Compounds which nonspecifically target thioesters will appear to inhibit HECT E3 ligases, but in fact are not useful drug candidates or chemical probes because they may also target E1 and E2 enzymes or their respective thioesters. Other compounds may nonspecifically inhibit enzyme activity through denaturing or the formation of aggregates. Accordingly, it is essential to confirm that prospective HECT E3 inhibitors do not inhibit E1 or E2 enzyme activity. This is judged by the formation of a E2~Ub thioester. Using stable fluorescein-conjugated ubiquitin (FUb, not to be confused with UbFluor), the presence or absence of this thioester can be determined by in-gel fluorescence imaging. Importantly, if the compound inhibits the formation of HECT E3~Ub thioester or isopeptide ligation, but does not inhibit the formation of E1~Ub, or E2~Ub thioesters, most likely this compound acts via specific inhibitory mechanisms.
Materials
REAGENTS
DMSO
Iodoacetamide (100 mM stock solution in DMSO, freshly prepared from solid the same day)
Hit compounds in DMSO stock solution (identified using Basic Protocols 1–4)
10X UbFluor Assay Buffer: 1.5 M NaCl, 500 mM HEPES pH 7.5 (see recipe)
40 mM MgCl2 in H2O
“FUb”: Ubiquitin, Fluorescein-labeled (LifeSensors)
UBA1 E1 enzyme (commercially available or obtainable from E. coli expression)
E2 ubiquitin conjugating enzyme (commercially available or obtainable from E. coli expression)
Adenosine triphosphate (ATP; Sigma-Aldrich)
Non-reducing 6X Laemmli SDS-PAGE loading buffer (see recipe)
SDS-PAGE running buffer (see recipe)
15% SDS-polyacrylamide gel (commercially available or obtainable from standard lab procedures)
EQUIPMENT AND MATERIALS
Eppendorf 1.5 mL centrifuge tubes or equivalent
Eppendorf tabletop microcentrifuge or equivalent
Bio-Rad Mini-PROTEAN Tetra Cell SDS-PAGE system or equivalent
GE Healthcare Typhoon 9400
Procedure for exclusion of E1/E2 transthiolation by hit compounds
-
Calculate volume of each enzyme or compound stock solution required for a reaction containing the following final concentrations: 50 μM compound (or 0.5 mM iodoacetamide/0.5% DMSO), 600 nM E2 enzyme, 100 nM UBA1 (E1 enzyme), 5 μM FUb, 4 mM ATP, 4 mM MgCl2. In addition to one tube for each compound, include positive and negative control tubes (0.5 mM iodoacetamide, 0.5% DMSO).
Typical reaction volume is 15 μL. Required volumes will depend on the concentration of enzyme and compound stock solutions.
-
Combine 10x UbFluor Assay Buffer, UBA1, E2 enzyme, FUb, MgCl2, and compound (or iodoacetamide/DMSO for control reactions) in Eppendorf tubes. If any additional volume of water is needed to achieve the final reaction conditions calculated in step 1, it should be added here as well.
Volumes added will vary depending on stock solution concentrations.
Incubate mixtures from step 2 for 30 minutes at RT.
Add ATP to the tubes as calculated in step 1. Mix by gently pipetting the mixture and centrifuge the tubes.
Incubate the tubes 30 minutes at RT.
-
Quench reactions using non-reducing 6x Laemmli buffer and mix thoroughly.
Do not heat reactions or use Laemmli buffer containing dithiothreitol (DTT) or β-mercaptoethanol (BME). These may cleave/hydrolyze all E2~FUb thioester and make confirmation or exclusion of inhibition impossible.
-
Run quenched reactions on a 15% SDS-polyacrylamide gel for 45 minutes at 200 V using the Mini-Protean SDS-PAGE apparatus with SDS-PAGE running buffer.
Gel running time and settings may depend on SDS-PAGE apparatus and user preferences.
-
Image the gel using the Typhoon 9400 (Excitation wavelength 488 nm, emission wavelength 526 nm).
Compounds that are inert to E1 and E2 enzymes will produce a visible E2~FUb thioester band comparable to the DMSO lane. Such compounds may be suitable specific inhibitors of HECT E3 ligases. Compounds which inhibit E1 and/or E2 enzyme activity will exhibit reduction or complete ablation of the E2~FUb band, indicating that they are nonspecific and cannot be considered specific inhibitors of HECT E3 ligases.
BASIC PROTOCOL 7: DETERMINATION OF TRANSTHIOLATION OR ISOPEPTIDE LIGATION INHIBITION BY HIT COMPOUNDS
Specific inhibitors of HECT E3 ligases may target a number of different aspects of the HECT ligase mechanism. By observing the ability of a HECT ligase to consume E2~Ub thioester, we can divide prospective inhibitors into two major categories. If an inhibitor prevents the transfer of ubiquitin between the catalytic cysteines of the E2 and the HECT E3, it is judged to be an inhibitor of transthiolation. In the presence of such an inhibitor, gel-based analysis of reactions using FUb will show limited or no consumption of E2~Ub thioester. Conversely, if an inhibitor allows transthiolation but is shown in basic protocols 4–5 to exhibit robust inhibition of overall HECT ligase activity, it is considered to be an inhibitor of isopeptide ligation, which broadly includes all activities downstream of the initial transthiolation event. In the presence of such an inhibitor, E2~Ub thioester will be consumed by the HECT E3 at a rate similar to HECT E3 incubated without inhibitor. Thus, this protocol describes an experiment that allows useful initial characterization of promising hit compounds, providing direction for further mechanistic study.
Materials
REAGENTS
HECT ligase (either catalytic HECT domain, HECT domain plus WW domains, or full length enzyme; commercially available or obtainable by previously published procedures from E. coli expression)
DMSO
Iodoacetamide (100 mM stock solution in DMSO, freshly prepared from solid the same day)
Hit compounds in DMSO stock solution (identified using Basic Protocols 1–5)
10X UbFluor Assay Buffer: 1.5 M NaCl, 500 mM HEPES pH 7.5 (see recipe)
40 mM MgCl2 in H2O
“FUb”: Ubiquitin, Fluorescein-labeled (LifeSensors)
UBA1 E1 enzyme (commercially available or obtainable from E. coli expression)
E2 ubiquitin conjugating enzyme (commercially available or obtainable from E. coli expression)
Adenosine triphosphate (ATP; Sigma-Aldrich)
Discharge assay buffer
Non-reducing 6X Laemmli SDS-PAGE loading buffer (see recipe)
SDS-PAGE running buffer (see recipe)
15% SDS-polyacrylamide gel (commercially available or obtainable from standard lab procedures)
EQUIPMENT AND MATERIALS
Eppendorf 1.5 mL centrifuge tubes or equivalent
Eppendorf tabletop microcentrifuge or equivalent
Thermo Fisher 0.5 mL Zeba spin desalting columns
Bio-Rad Mini-PROTEAN Tetra Cell SDS-PAGE system or equivalent
GE Healthcare Typhoon 9400
Procedure for determination of transthiolation or isopeptide ligation inhibition by hit compounds
-
Calculate volume of each enzyme or compound stock solution required for a 25 μL reaction containing the following final concentrations: 50 μM compound (or 0.5 mM iodoacetamide/0.5% DMSO), 600 nM HECT E3 ligase. In addition to one tube for each compound, include positive and negative control tubes (0.5 mM iodoacetamide, 0.5% DMSO). Two hit compounds can be tested on a single gel along with positive and negative controls. Hereafter, these mixtures are referred to as HECT preincubation reactions.
Required volumes will depend on the concentration of enzyme and compound stock solutions.
-
Calculate volume of each enzyme or compound stock solution required for a 25 μL reaction containing the following final concentrations: 100 nM UBA1 (E1 enzyme), 4 μM E2 enzyme, 5 μM FUb, 4 mM ATP, 4 mM MgCl2. 1 tube of this mixture, hereafter referred to as E1/E2 reaction, is required for the 4 tubes in step 1.
Required volumes will depend on the concentration of enzyme and compound stock solutions.
-
Combine 10x UbFluor Assay Buffer, HECT E3, and compound (or iodoacetamide/DMSO for control reactions) in Eppendorf tubes as calculated for the HECT preincubation reactions in step 1. If any additional volume of water is needed to achieve the final reaction conditions calculated in step 1, it should be added as well.
Volumes added will vary depending on stock solution concentrations.
-
Combine 10x UbFluor Assay Buffer, UBA1, E2 enzyme, FUb, MgCl2, and ATP in Eppendorf tube(s) as calculated for the E1/E2 reaction in step 2. If any additional volume of water is needed to achieve the final reaction conditions calculated in step 2, it should be added as well.
Volumes added will vary depending on stock solution concentrations.
-
Incubate mixtures from steps 3–4 for 30 minutes at RT.
During incubation, equilibrate Zeba spin columns as directed in product instruction manual.
-
Cool HECT preincubation mixtures to 4°C.
All subsequent steps until SDS-PAGE (step 14) should be performed on ice or in a cold room.
Dilute E1/E2 reaction with 125 μL of cold (4°C) discharge assay buffer.
Desalt E1/E2 reaction with a Zeba column equilibrated with discharge assay buffer at 4°C.
Using a second Zeba column, desalt E1/E2 reaction a second time at 4°C.
-
For each reaction (including controls), prepare 3 labeled Eppendorf tubes containing 2 μL non-reducing 6x Laemmli buffer.
These must be ready for rapid quenching of each reaction at 3 time points. Typically, the time points are 0.1 minute, 1 minute, and 10 minutes.
-
At 4°C, add 25 μL E1/E2 reaction to each HECT preincubation mixture.
Ensure that tubes from step 10 and all needed pipettors/tips for liquid transfer are ready prior to adding the E1/E2 mixture, especially if the first quenching time point is less than 1 minute.
-
At 3 pre-determined intervals, quench a 10 μL aliquot of each combined reaction from step 11 in the pre-prepared non-reducing 6x Laemmli buffer tubes from step 10.
Typically, the 3 time points are 0.1 minute, 1 minute, and 10 minutes. Optimization may be required for some systems.
-
Run quenched reactions on a 15% SDS-polyacrylamide gel for 45 minutes at 200 V using the Mini-Protean SDS-PAGE apparatus with SDS-PAGE running buffer.
Gel running time and settings may depend on SDS-PAGE apparatus and user preferences.
-
Image the gel using the Typhoon 9400 (Excitation wavelength 488 nm, emission wavelength 526 nm).
The 3 DMSO control time points should show a decrease in E2~FUb thioester over time, likely showing complete disappearance by 10 minutes. The 3 iodoacetamide control time points should show minimal change in the amount of E2~FUb thioester. Compounds which inhibit transthiolation will prevent consumption of E2~FUb thioester, for a gel profile resembling that of the iodoacetamide control. Compounds which inhibit isopeptide ligation will not affect E2~FUb consumption and should show a decrease in E2~FUb comparable to the DMSO control. Gel imaging can also reveal the formation of the HECT E3~FUb thioester. This becomes possible if HECT E3~FUb thioester is sufficiently stable to hydrolysis under given reaction conditions. However, in the case of HECT domains or HECT ligases which autoubiquitinate, one needs to run SDS PAGE under reducing and non-reducing conditions to distinguish between HECT E3~FUb thioester and HECT E3 with a single FUb conjugated to a HECT lysine via an isopeptide bond.
REAGENTS AND SOLUTIONS
Use ddH2O from Thermo Scientific Nanopure or equivalent purification system
10X UbFluor Assay Buffer: 1.5 M NaCl, 500 mM HEPES pH 7.5
1.5 M NaCl
500 mM HEPES
Adjust pH to 7.5 by addition of solid NaOH pellets and/or dropwise addition of 1 M NaOH
6X Laemmli SDS-PAGE loading buffer
0.42 M sodium dodecyl sulfate
0.86 mM bromophenol blue
50% glycerol
50 mM Tris (added as 0.5 M Tris pH 6.8 solution)
5% β-mercaptoethanol
SDS-PAGE running buffer
25 mM Tris
192 mM glycine
3.5 mM sodium dodecyl sulfate
Western blot transfer buffer
125 mM Tris
192 mM glycine
20% methanol
TBS-T
50 mM Tris
150 mM NaCl
Adjust pH to 7.6 by addition of 1 M HCl
Non-reducing 6X Laemmli SDS-PAGE loading buffer
0.42 M sodium dodecyl sulfate
0.86 mM bromophenol blue
50% glycerol
50 mM Tris (added as 0.5 M Tris pH 6.8 solution)
Discharge assay buffer
25 mM HEPES
100 mM NaCl
25 mM ethylene diamine tetraacetic acid (EDTA)
Adjust pH to 7.5 by addition of solid NaOH pellets and/or dropwise addition of 1 M NaOH
COMMENTARY
Background Information
Ubquitination of protein substrates is an essential posttranslational process best known for targeting proteins to the proteasome for degradation (Hershko et al., 1980). Ubiquitination is also known to regulate protein localization and activity by nondegradative mechanisms (Hicke, 2001). Ubiquitin is conjugated to protein substrates via a three-step hierarchical enzyme cascade comprised of E1, E2, and E3 enzymes (Varshavsky, 2012). Of these classes, E3 ubiquitin ligases are by far the largest, containing over 600 known members. The size of the E3 class rivals that of kinases (~500 known), proteases (~600 known), ion channels (~450 known) and GPCRs (~800 known), two of the most commonly targeted enzyme classes in the pharmaceutical industry. However, despite the size and importance of the E3 enzyme class, no FDA-approved drugs have yet been designed to specifically target these enzymes. The only known FDA approved drug that target E3 ligases are teratogenic drugs thalidomide, lenalidomide, and pomalidomide that act by hijacking cullin-RING E3 ligase Cereblon and redirect its substrate specificity to transcripton factors Ikaros/Aiolos which are important for the survival of multiple myeloma. In this regard thalidomide and its analogues are the first FDA approved PROTAC drugs.
Nonetheless, considerable work has demonstrated the biological significance of E3 ligases. Of note, the HECT class of E3s represent a chemically tractable subset of E3 ligases (Scheffner et al., 1993). Considerable biological research has revealed that many HECT E3s, including Nedd4-1 and HUWE1, are essential to mammalian development, with their deletions resulting in embryonic lethality (Fouladkou et al., 2010; Kon et al., 2012). Deletions of other HECT E3s result in disease phenotypes ranging from developmental disabilities to immune syndromes, as well as increased bone mass or blood pressure (Jiang et al., 1998; Lohr et al., 2010; Shu et al., 2013). In other cases, HECT E3 activity is a driving force in oncogenic signaling pathways. For example, both WWP1 and ITCH are known to degrade the tumor-suppressor kinase LATS1 in the Hippo pathway, leading to increased growth and proliferation of some breast cancers (Salah et al., 2014; Yeung et al., 2013). Similarly, E6AP is known to degrade the tumor suppressor protein p53 in HPV-16/18 induced cervical cancer (Beer-Romero et al., 1997). HECT E3s are also thought to be involved in the development of neurodegenerative disorders such as Alzheimer’s disease and Parkinson’s disease (Tardiff et al., 2013; Tofaris et al., 2011). Finally, the HECT E3 Nedd4-1 is known to be hijacked during viral infections, including HIV and Ebola (Dussart et al., 2005; Yasuda et al., 2003). Accordingly, the HECT class of E3 ligases present a well-characterized set of potential drug targets for which small-molecule modulators are needed.
Small molecule inhibitors and activators of HECT E3s would be useful probes for substrate identification using quantitative proteomic methods and for the validation of these enzymes as drug targets (Ordureau et al., 2015). However, few small-molecule modulators of HECT E3s have been discovered (Kathman et al., 2015; Mund et al., 2014). Assays for these and other E3 ligases are necessarily complex, due to the dependence of E3s on upstream E1 and E2 enzymes for the activation and transfer of ubiquitin. HECT E3s, in contrast to most E3s, bear a catalytic cysteine residue, which reacts with an E2~Ub thioester and forms an obligate HECT E3~Ub thioester bond prior to the final isopeptide ligation of Ub onto a substrate lysine residue (Huibregtse et al., 1995). Since HECTs and other E3s require the activity of the upstream E1 and E2 enzymes, assays for discovering E3 inhibitors generally must contain these enzymes, as well as detection reagents, creating complexity and the potential for a multitude of off-target effects. Thus, there is a need for simplified HECT E3 ligase assays for biochemical study and drug discovery.
Accordingly, we designed UbFluor, a chemical probe which chemically mimics the E2~Ub thioester and contains a fluorescent reporter (Krist et al., 2016). The Ub portion of UbFluor interacts with the ubiquitin-binding site on the C-lobe of the HECT catalytic domain, utilizing the same residues as the native E2~Ub thioester. This interaction allows the HECT catalytic cysteine to react with the UbFluor thioester bond, leading to the formation of a catalytically active HECT E3~Ub thioester and release of the fluorescein thiol FluorSH (Figure 1). This reaction can be easily monitored by FP, as the release of the fluorophore reduces the apparent fluorescent molecular weight from ~9 kDa to ~450 Da. The smaller free FluorSH tumbles more rapidly in solution than the intact UbFluor, resulting in a lower FP reading. Thus, UbFluor represents a further improvement on the UbMES-based bypassing system developed previously (Park et al., 2015). In previous work, we have validated the relevance of the UbFluor assay in biochemical point mutants of HECT E3 ligases. The simplicity and throughput of the UbFluor assay make it useful for drug discovery. The discovery, optimization, and use of HECT E3 inhibitors will be reported in the near future. Additionally, other classes of E3 ligases, such as RING-between-RING (RBR) and bacterial E3 ligases, are known to act in an analogous manner to HECT E3s, relying on a catalytic cysteine to bear ubiquitin as a thioester (Hicks and Galán, 2010; Wenzel and Klevit, 2012; Zhang et al., 2006). In principle, the UbFluor assay can be used to screen for inhibitors and activators of these clinically relevant E3 ligases.
Critical Parameters
Since the UbFluor thioester is susceptible to cleavage by thiols, enzyme and assay buffers must be free of common thiols such as dithiothreitol (DTT) and β-mercaptoethanol (BME). The reactivity of UbFluor towards nucleophiles means they should also be absent from any screening compounds. UbFluor is stable in the presence of 100 mM lysine, but it is possible that other free amines may react with UbFluor.
Some small molecules and biological reagents can also cause an undesired increase in fluorescence polarization over time. We observed that bovine serum albumin increases polarization of UbFluor in a time-dependent manner.
Fluorescent small molecules or fluorescence-quenching small molecules may also interfere with FP measurements and should be eliminated from screening libraries wherever possible.
Basic protocols 5 and 6 use stable fluorescein-conjugated ubiquitin (FUb, commercially available from LifeSensors) for in-gel visualization. This is not to be confused with UbFluor. To our knowledge, FUb behaves very similarly to wild-type, unlabeled ubiquitin in the described reactions.
Basic protocols 5 and 6 use gel-based techniques to assess the presence or absence of an E2~FUb thioester. It is essential to use non-reducing 6x Laemmli SDS-PAGE loading buffer to quench and load these reactions for SDS-PAGE. DTT or BME will cleave any present thioester and thus prevent visualization of the amount of E2~FUb thioester in the reaction.
Troubleshooting
HECT does not appear to react with UbFluor.
-Increase concentration of both HECT and UbFluor or increase reaction time.
-Confirm activity of HECT in orthogonal ubiquitination assay. Some enzymes or enzyme constructs have low basal activity. It may be necessary to use a different construct; for example, the deletion or mutation of an autoregulatory C2 domain or the presence of a proximal WW domain may increase the reactivity of a HECT towards UbFluor.
Iodoacetamide positive control wells show an increase in polarization
-We have found a slight increase over several hours to occur normally.
-Larger, more pronounced increases may be due to exposure to light or air. Use a blank plate or plate cover to protect wells of plate between readings.
-Lowering the iodoacetamide concentration will mitigate rising polarization; it is important to confirm that full inhibition of HECT activity is still achieved at lower iodoacetamide concentration.
-In extreme cases, using UbFluor with catalytically inactive HECT or no enzyme may provide a more suitable control.
Screening compound appears to inhibit HECT ligase only at later time points
-Check whether polarization decreases over time, then subsequently increases again, yielding a large apparent increase in percent inhibition over time; such compounds are not useful inhibitors and should be excluded from further validation.
DMSO and iodoacetamide controls in basic protocol 6 do not show discernable difference in the amount of E2~FUb consumed.
-Adjust reaction times: longer reaction times if DMSO-treated reaction shows minimal activity, shorter reaction times if iodoacetamide-treated reaction shows >50% E2~FUb consumption.
-Adjust reaction temperatures: heat reactions to RT if DMSO-treated reaction shows minimal activity, pre-cool reactions in ice/salt bath if iodoacetamide-treated reaction shows >50% E2~FUb consumption.
Anticipated Results
Basic protocol 1 will identify suitable HTS reaction conditions for HECT E3 ligase consumption of UbFluor. Initial conditions screening will be followed by confirmation of Z′, which is expected to exceed 0.7 based on results obtained to date. Basic protocol 2 will screen a library of compounds against a HECT ligase using the UbFluor assay. Basic protocol 2 will likely furnish initial hits at a rate of 0.5–5% depending on the HECT ligase selected, the nature of the screened library, and the percent inhibition cutoff for hit selection. Basic protocol 3 will produce dose response curves for hit compounds, indicating the potency of these compounds and other dose curve parameters. The data produced by basic protocol 3 will thus characterize the quality of hit compound activity and provide direction for subsequent secondary validation studies of the identified prospective inhibitors. It is expected that since UbFluor acts as an unnatural substrate of HECT E3 ligases, the UbFluor assay will enrich for substrate-competitive inhibitors and allosteric inhibitors, but not inhibitors of the enzyme-substrate complex. It is also expected that inhibitors of both HECT E3 transthiolation and isopeptide ligation can be discovered using the UbFluor assay. Future reports of discovered inhibitors will confirm these hypotheses.
Basic protocol 4 will confirm whether discovered inhibitors from basic protocols 2 and 3 inhibit HECT E3 ligase in the native ubiquitination cascade. In our experience, approximately 33% of inhibitors discovered in the UbFluor assay can be translated to the native cascade. Basic protocol 5 will further confirm the robustness and dose-dependency of hits validated in basic protocol 4. Basic protocol 6 will determine whether inhibitors validated in basic protocol 4 are specific to the HECT E3 or have off-target effects on E1 and/or E2 enzymes. At least 50% of prospective inhibitors that reach basic protocol 6 are specific to HECT E3 activity. Basic protocol 7 allows the user to distinguish between inhibitors of E2/E3 transthiolation and inhibitors of isopeptide ligation by the HECT E3. At least 90% of inhibitors studied to date have targeted transthiolation, but this distribution could in principle be altered by adding a known HECT E3 substrate, choosing a particular HECT E3 construct more capable of substrate ubiquitination, or screening a class of molecules known to disrupt E3-substrate interactions.
Time Considerations
UbFluor provides a simple two-component system for the screening of prospective inhibitor libraries against HECT E3 ligases. By reducing the complexity of HECT E3 screening assays, the cost and time of this process can be reduced.
Once HECT ligases and UbFluor are prepared, determination of appropriate conditions should require 2–6 experiments, 1–3 of which can be run per day depending on desired assay time. This includes the determination of Z′.
HTS of at least 5,000 compounds per day is feasible. Assuming comfort with instrumentation and data handling, a screening campaign of 50,000 compounds can be undertaken in 10 working days. Subsequent data analysis should require 3–5 days. Automation using Microsoft Excel macros or other programs will accelerate data analysis, but may require time and expert consultation to establish. Initial time spent establishing analysis platforms will reduce the time required for the analysis of subsequent screens, a key consideration if multiple HTS campaigns are planned.
Dose response studies will take 2–5 days depending on the hit rate and number of concentrations. Data analysis of dose response studies and hit selection will take 3–7 days. Again, automation of data analysis will accelerate data processing. However, it may be desirable to manually select compounds based on structural properties once dose curves are generated, which can add 1–3 days depending on available software and number of hits.
Validation of up to 25 hit compounds by basic protocol 4 can be reasonably accomplished in 2 days. Dose responses for 4 compounds can be accomplished in 2 days. Determination of E1/E2 off-target activity by basic protocol 5 requires 1 day, and up to 25 compounds can be easily tested together. Determination of transthiolation activity by basic protocol 6 can accommodate 2 compounds and can be accomplished in 1 day. An experienced user can run basic protocols 5 and 6 in parallel in 1 day. In the case of HECT ligase constructs that the user is unfamiliar with, 1–5 additional days may be required to identify optimal E2 and HECT E3 concentrations and reaction times for basic protocols 4–7. For larger numbers of hit compounds, time required should be increased accordingly.
Acknowledgments
Funding from Northwestern University, the H-Foundation Pilot Project Award, the Chicago Biomedical Consortium, and the CLP Cornew Innovation Award is greatly acknowledged. A. V. S. is a Pew Scholar in the Biomedical Sciences, supported by the Pew Charitable Trusts. D. T. K. has been supported by National Institute of General Medical Sciences Training Grant 5T32GM008382. D. T. K. is supported by an NU Nicholson fellowship. A. V. S. is supported by R01GM115632-01. This work was supported by the Northwestern University High Throughput Analysis Laboratory.
LITERATURE CITED
- Beer-Romero P, Glass S, Rolfe M. Antisense targeting of E6AP elevates p53 in HPV-infected cells but not in normal cells. Oncogene. 1997;14:595–602. doi: 10.1038/sj.onc.1200872. [DOI] [PubMed] [Google Scholar]
- Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Boase NA, Rychkov GY, Townley SL, Dinudom A, Candi E, Voss AK, Tsoutsman T, Semsarian C, Melino G, Koentgen F, Cook DI, Kumar S. Respiratory distress and perinatal lethality in Nedd4-2-deficient mice. Nat Comm. 2011;2:287. doi: 10.1038/ncomms1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Schwartz AL. The ubiquitin-proteasome pathway: The complexity and myriad functions of proteins death. Proc Natl Acad Sci USA. 1998;95:2727–2730. doi: 10.1073/pnas.95.6.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussart S, Douaisi M, Courcoul M, Bessou G, Vigne R, Decroly E. APOBEC3G ubiquitination by Nedd4-1 favors its packaging into HIV-1 particles. J Mol Biol. 2005;345:547–558. doi: 10.1016/j.jmb.2004.10.067. [DOI] [PubMed] [Google Scholar]
- Fouladkou F, Lu C, Jiang C, Zhou L, She Y, Walls JR, Kawabe H, Brose N, Henkelman RM, Huang A, Bruneau BG, Rotin D. The ubiquitin ligase Nedd4-1 is required for heart development and is a suppressor of thrombospondin-1. J Biol Chem. 2010;285:6770–6780. doi: 10.1074/jbc.M109.082347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A, Heller H, Haas AL, Rose IA. Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc Natl Acad Sci USA. 1980;77:1783–1786. doi: 10.1073/pnas.77.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L. Protein regulation by monoubiquitination. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- Hicks SW, Galán JE. Hijacking the Host Ubiquitin Pathway: Structural Strategies of Bacterial E3 Ubiquitin Ligases. Curr Op Microbiol. 2010;13:41. doi: 10.1016/j.mib.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse J, Scheffner M, Beaudenon S, Howley P. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y-h, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, Beaudet AL. Mutation of the Angelman Ubiquitin Ligase in Mice Causes Increased Cytoplasmic p53 and Deficits of Contextual Learning and Long-Term Potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- Kathman SG, Span I, Smith AT, Xu Z, Zhan J, Rosenzweig AC, Statsyuk AV. A Small Molecule That Switches a Ubiquitin Ligase From a Processive to a Distributive Enzymatic Mechanism. J Am Chem Soc. 2015;137:12442–12445. doi: 10.1021/jacs.5b06839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon N, Zhong J, Qiang L, Accili D, Gu W. Inactivation of arf-bp1 induces 53 activation and diabetic phenotypes in mice. J Biol Chem. 2012;287:5102–5111. doi: 10.1074/jbc.M111.322867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krist DT, Park S, Boneh GH, Rice SE, Statsyuk AV. UbFluor: a mechanism-based probe for HECT E3 ligases. Chem Sci. 2016;7:5587–5595. doi: 10.1039/c6sc01167e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr NJ, Molleston JP, Strauss KA, Torres-Martinez W, Sherman EA, Squires RH, Rider L, Chikwava KR, Cummings OW, Morton DH, Puffenberger EG. Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. Am J Hum Genet. 2010;86:447–453. doi: 10.1016/j.ajhg.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mund T, Lewis MJ, Maslen S, Pelham HR. Peptide and small molecule inhibitors of HECT-type ubiquitin ligases. Proc Natl Acad Sci USA. 2014;111:16736–16741. doi: 10.1073/pnas.1412152111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Huu NS, Ryder WD, Zeps N, Flasza M, Chiu M, Hanby AM, Poulsom R, Clarke RB, Baron M. Tumour-promoting activity of altered WWP1 expression in breast cancer and its utility as a prognostic indicator. J Pathol. 2008;216:93–102. doi: 10.1002/path.2385. [DOI] [PubMed] [Google Scholar]
- Ordureau A, Münch C, Harper JW. Quantifying Ubiquitin Signaling. Mol Cell. 2015;58:660–676. doi: 10.1016/j.molcel.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Krist DT, Statsyuk AV. Protein ubiquitination and formation of polyubiquitin chains without ATP, E1 and E2 enzymes. Chem Sci. 2015;6:1770–1779. doi: 10.1039/c4sc02340d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salah Z, Itzhaki E, Aqeilan RI. The ubiquitin E3 ligase ITCH enhances breast tumor progression by inhibiting the Hippo tumor suppressor pathway. Oncotarget. 2014;5:10886–10900. doi: 10.18632/oncotarget.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Kumar S. Mammalian HECT ubiquitin-protein ligases: biological and pathophysiological aspects. Biochim Biophys Acta. 2014;1843:61–74. doi: 10.1016/j.bbamcr.2013.03.024. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Nuber U, Huibregtse J. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- Schulman BA, Harper JW. Ubiquitin-like protein acivation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoichet BK. Interpreting Steep Dose-Response Curves in Early Inhibitor Discovery. J Med Chem. 2006;49:7274–7277. doi: 10.1021/jm061103g. [DOI] [PubMed] [Google Scholar]
- Shu L, Zhang H, Boyce BF, Xing L. Ubiquitin E3 ligase Wwp1 negatively regulates osteoblast function by inhibiting osteoblast differentiation and migration. J Bone Miner Res. 2013;28:1925–1935. doi: 10.1002/jbmr.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff DF, Jui NT, Khurana V, Tambe MA, Thompson ML, Chung CY, Kamadurai HB, Kim HT, Lancaster AK, Caldwell KA, Caldwell GA, Rochet JC, Buchwald SL, Lindquist S. Yeast reveal a “druggable” Rsp5/Nedd4 Network that Ameliorates α–Synuclein Toxicity in Neurons. Science. 2013;342:979–983. doi: 10.1126/science.1245321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tofaris GK, Kim HT, Hourez R, Jung JW, Kim KP, Goldberg AL. Ubiquitin ligase Nedd4 promotes alpha-synuclein degradation by the endosomal-lysosomal pathway. Proc Natl Acad Sci USA. 2011;108:17004–17009. doi: 10.1073/pnas.1109356108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The ubiquitin system, an immense realm. Annu Rev Biochem. 2012;81:167–176. doi: 10.1146/annurev-biochem-051910-094049. [DOI] [PubMed] [Google Scholar]
- Verdecia MA, Joazeiro CA, Wells NJ, Ferrer JL, Bowman ME, Hunter T, Noel JP. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol Cell. 2003;11:249–259. doi: 10.1016/s1097-2765(02)00774-8. [DOI] [PubMed] [Google Scholar]
- Wenzel DM, Klevit RE. Following Ariadnes thread: a new perspective on RBR ubiquitin ligases. BMC Biol. 2012;10 doi: 10.1186/1741-7007-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Ying SX, Zhang GM, Li C, Cheng SY, Deng CX, Zhang YE. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell. 2005;121:101–113. doi: 10.1016/j.cell.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda J, Nakao M, Kawaoka Y, Shida H. Nedd4 regulates egress of Ebola virus-like particles from host cells. J Virol. 2003;77:9987–9992. doi: 10.1128/JVI.77.18.9987-9992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung B, Ho KC, Yang X. WWP1 E3 Ligase Targets LATS1 for Ubiquitin-Mediated Degradation in Breast Cancer Cells. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0061027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Scr. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Higashide WM, McCormick BA, Chen J, Zhou D. The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol Microbiol. 2006;62:786–793. doi: 10.1111/j.1365-2958.2006.05407.x. [DOI] [PubMed] [Google Scholar]


