Abstract
Importance
Four assays have been registered with the FDA to detect PD-L1 to enrich for patient response to anti-PD-1/PD-L1 therapies. The tests use four separate PD-L1 antibodies on two separate staining platforms and have their own scoring systems which raises questions about their similarity and potential cross-utilization.
Objective
We compared the performance of four PD-L1 platforms, including two FDA-cleared assays and two laboratory developed tests (LDTs).
Design
Four serial histology sections from 90 archival NSCLCs were distributed to three sites that performed the following IHCs: 1) 28-8 antibody on Dako Link 48; 2) 22c3 antibody on Dako Link 48; 3) SP142 antibody on Ventana Benchmark; and 4) E1L3N antibody on Leica Bond. Slides were scanned and scored by thirteen pathologists by estimating the percentage of malignant and immune cells expressing PD-L1. Intraclass correlation coefficients (ICC) and paired and mixed effects statistical analyses were performed to compare antibodies and pathologists scoring of tumor and immune cells.
Results
The SP142 Ventana assay was an outlier with a significantly lower mean score of PD-L1 expression in both tumor and immune cells. Pairwise comparisons showed the 28-8 and E1L3N were not significantly different, but that 22c3 showed a slight but statistically significant reduction in tumor cell labeling. Evaluation of ICC between antibodies to quantify inter-assay variability using the average of thirteen pathologists scores for tumor shows very high concordance between antibodies for tumor cell scoring (0.813) and lower levels of concordance for immune cell scoring (0.277). When examining inter-pathologists variability for any single antibody, the concordance between pathologists’ reads for tumor ranged from ICC of 0.83 to 0.88 for each antibody while the ICC from immune cells for each antibody ranged from 0.17 to 0.23.
Conclusions
The assay using the SP142 antibody is a clear outlier detecting significantly less tumor cell and immune cell PD-L1 expression. Antibody 22c3 shows slight yet statistically significantly lower staining than either 28-8 or E1L3N, but this significance is only detected when using the average of thirteen pathologist scores. Pathologists show excellent concordance when scoring tumor cells stained with any antibody, but poor concordance for scoring immune cell staining.
Keywords: Non-small cell lung cancer, PD-L1, immunohistochemistry
Introduction
Response to check-point inhibitor immunotherapy has been exceptional 1–3, The checkpoint inhibitor ligand PD-L1 is the target for one FDA approved therapy (Atezolizumab) and its receptor, PD-1 is the target for two others (Nivolumab and Pembrolizumab). In registrational trials, each of these drugs has been tested with a companion diagnostic assay that has been independently designed and is based on a combination of a unique antibody with a custom designed assays using proprietary reagents, protocols and thresholds defining “elevated” PD-L1 expression. This has led to a challenge for pathologists who seek to provide companion diagnostic testing, but do not necessarily know which therapeutic will be selected by the oncologist for any given patient
Historically, immunohistochemistry (IHC) has been used to determine the presence or absence of a given protein. In combination with morphology, this assists pathologist in classifying a tumor. IHC assays are optimized by vendors to provide a binary outcome from what is inherently a continuous variable. Companion diagnostic tests are the exception to this approach for IHC since a continuous value, or at least a threshold value is required an expression beyond a threshold number of cells is tightly linked to prescription of a drug. The best examples of this are in breast cancer where estrogen receptor must be expressed in greater than 1% of cells to be considered positive4.
For PD-L1 there are three drug-specific tests that are FDA approved as either companion (Pembrolizumab) or complementary (Atezolizumab/Nivolumab) diagnostics, which use three different antibodies and three sets of assay conditions. They are Nivolumab using the Dako/Agilent 28-8 assay, Pembrolizumab using the Dako/Agilent 22c3 assay and Atezolizumab using the Ventana/Roche SP142 assay. This is a very different approach than that taken historically, where, using the example of estrogen receptor, a handful of common antibodies are used in either FDA approved assays or laboratory developed tests (LDTs) to give a result that can predict response to therapy for around a dozen drugs that inhibit or otherwise modulate estrogen receptor mediated signaling in breast cancer. This raises a new problem for pathologists. Specifically, should they be more concerned about accurate measurement of the target protein or should they focus on the “assay” result as appears to now be required by the FDA in companion diagnostic testing for PD-L1 where 3 separate assays are approved for the same protein.
This problem presents two issues, a theoretical issue and a practical issue. The first is; do each of the assays equally assess the amount of PD-L1 present in the tissue? While this is an important issue, the FDA does not require proof of the number of molecules expressed as compared to some analytic standard. A more practical issue is; are these agency approved assays equivalent as approved and can the assays be cross utilized? That is, can any assay be used for any drug or are the assays and prescribed scoring methods specific to the therapeutic with which they were developed. To address this practical question, two main efforts have begun to compare the assay in the US. The first, labeled the “Blueprint” is a comparison of 39 cases read by 3 industry pathologists comparing the 3 approved tests and a fourth assay which is an investigational use only assay from Astra Zeneca and Ventana based on the SP-263 antibody. This study showed concordance between 3 of the 4 assays with the SP142 assay as an outlier (Hirsch et al, 2016 in press). This study was considered a pilot study and, as such, was not statistically powered, nor was it multi-institutional.
The second US-based study is reported here. This study, sponsored by the NCCN and funded by BMS, sought to provide level 1 evidence for biomarker testing5,6 by using a statistically powered, prospective design in a multi-institutional setting. The primary objective was to compare the performance of available antibodies/assays/test platforms for ability to accurately and reliably measure PD-L1. In the absence of patient outcome data across therapeutic products, we focused on direct comparison between the 4 assays to: 1. understand antibody/test properties and performance relative to one another; 2. evaluate differences in assessment of PD-L1 on tumor cell surface vs. immune infiltrates; and 3. compare result interpretation between pathologists across assays. While limited by the use of untreated patients, the level 1 evidence produced herein is not evidence for prediction or clinical accuracy of the assays, but rather for assay concordance and pathologist concordance in assessment of each assay.
Methods
Case selection
A series of 90 surgically resected NSCLC cases (stages I–III), adenocarcinoma and squamous cell carcinomas were obtained from the Yale School of Medicine Department of Pathology Archives from 2008–2010 summarized in supplemental table 1.
Immunohistochemistry
Four 5 micron sections were cut from each case at Yale and sent to the following institutions for staining as follows: Assay 1) University of Colorado for 22c3 on the Dako Link 48 platform, Assay 2) University of Colorado for 28-8 on the Dako Link 48 platform; Assay 3) The Mayo Clinic (Rochester) for SP-142 on the Ventana Benchmark Platform; and Assay 4) Yale University for Cell Signaling Technology E1L3N on the Leica Bond Platform (as an LDT). Although the Ventana assay is now approved, it was not at the time of staining, so the protocol that was used is now technically an LDT, but that protocol is essentially identical to the current approved protocol with the exception of three steps representing different incubation times as shown in supplemental table 2. All other conditions were identical to the approved test and the appearance of the slides is comparable to those using the approved protocol. For E1L3N, the staining procedure for the LDT can be found in supplemental table 3.
Pathologist Scoring
The stained slides were all sent to University of Colorado for scanning by a Leica Aperio scanner and placement into a database viewable by internet connection. A template for scoring was constructed on the REDCAP database. Pathologist scored the images conveyed by the internet which allowed visualization of the entire slide with full zoom capacity from the equivalent of a 1X objective to a 40X objective. Instructions were provided to 16 pathologists at 8 institutions and a deadline was set for completion of scoring of 90 cases with 4 slides per case representing each stain/platform pair. Since each system has its own scoring protocol, we designed a unified scoring method for both tumor proportion scores (TPS) and Immune cell proportion scores (ICPS) that could be used to calculate a score that fits into the categorical scoring system for each IUO or FDA assay, including the Astra Zeneca/Ventana SP263 test, even though that assay was not tested in this exercise. As in those assays, the score of TPS or ICPS is based on membrane and cytoplasmic staining of any intensity. The scoring system is summarized in supplemental table 4.
Statistical Analysis
The pathologist reading was recorded on a six point scale, each value corresponding to a range of the tumor percentage: the original score of category A is negative or <1% of tumor, the score of B is 1% to 4%, the score of C is 5% to 9%, the score of D is 10% to 24%, the score of E is 25% to 49%, and the score of F is 50% or more. The same statistical analyses were performed for TPS and ICPS. For assay comparison, average readings from the 13 pathologists were plotted for each antibody by cases. Paired Wilcoxon signed-rank test was used to compare the antibody pairs for readings from individual pathologists. Mixed-effects linear model was used to evaluate the statistical significance in difference between antibodies treating effects from pathologists as random effects. To assess concordance of the antibodies, intra-class correlation (ICC) coefficients were calculated (among the 4 antibodies, and among 3 of the 4 except SP142) using the average readings of pathologists for the 90 cases as well as each individual pathologists’ readings. Sample size justification based on ICC was conducted prior to data collection. Assuming four antibodies, we calculated the statistical power that 90 slides can achieve to differentiate an almost perfect agreement (ICC=0.85 or greater) from a moderate (ICC=0.5) or strong (ICC=0.7) agreement. Taking into account the fact that about 35% of the readings will be positive, 90 slides in total can achieve 87.9% power at a significance level 0.05 to differentiate ICC=0.85 vs ICC=0.7. An ICC is interpreted as follows: below 0.3 indicates poor agreement; between 0.3 and 0.7 indicates fair to moderate agreement; between 0.7 and 0.8 indicates strong agreement; and greater than 0.8 indicates almost perfect agreement. Analyses were also performed to quantify the concordance of readings between the pathologists: In both the original 6 levels and three levels (<1%, 1–49%, and >=50%), ICC values between pathologists were calculated for each antibody 7. Variance of the pathologist readings were decomposed to contributions from antibodies and pathologists using Analysis of Variance (ANOVA). Furthermore, the original readings were dichotomized using the cutoff of >50% and the cutoff of >1% to assess the concordance between pathologists for binary tumor evaluation. The Fleiss Kappa coefficient and Kendall concordance coefficient were calculated to evaluate the agreement and concordance of the 13 pathologists’ binary assessment for each antibody. The kappa coefficient is interpreted as poor to fair if <=0.4, as moderate if >0.4 and <=0.6, as substantial if >0.6 and <=0.8, and almost perfect if >0.8. Strength of the Kendall concordance coefficient was interpreted similarly to that of ICC 8,9. Statistical analysis was completed using SAS software (version 9.4, SAS Inc., Cary, NC) and MATLAB (version 2014b, The Mathworks Inc., Natick, MA) based on the prescribed experimental design for objective 1 of the NCCN/BMS study.
Results
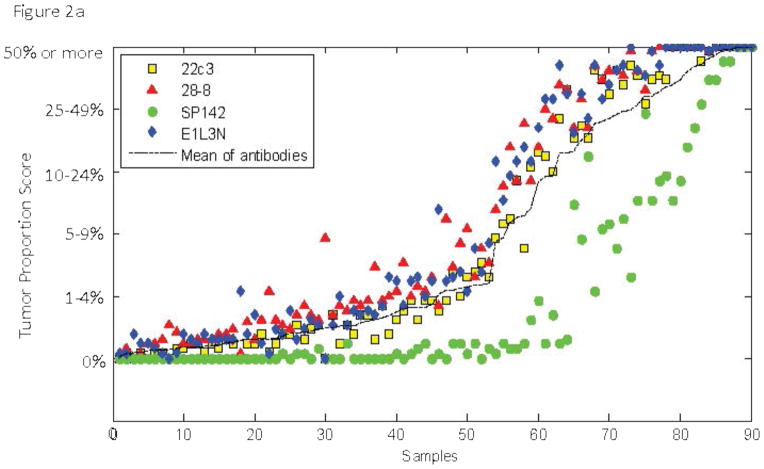
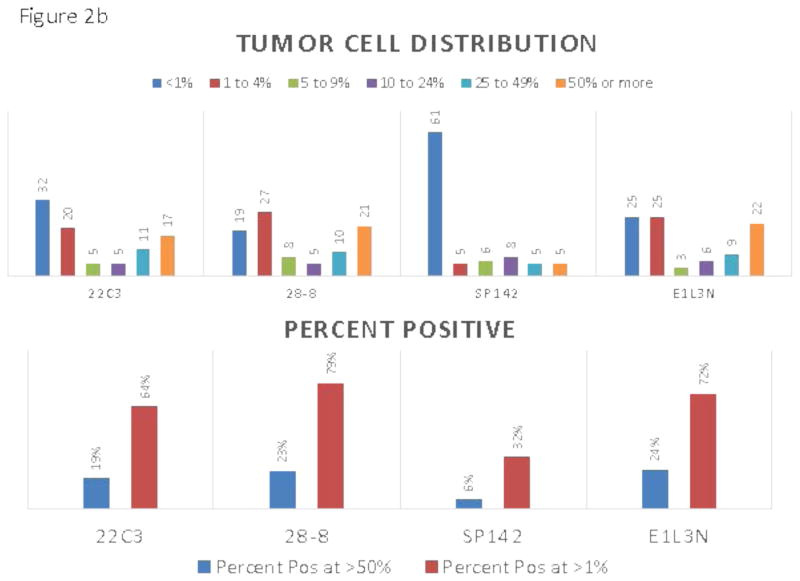
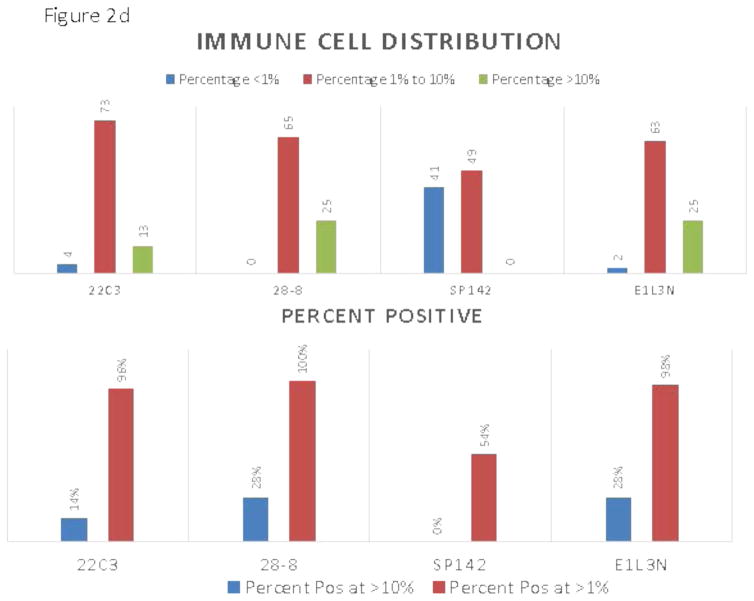
The appearance of the images was similar to that seen previously in PD-L1 IHC work 10–12 showing predominantly membranous staining. The 4 assays appeared largely similar, although one of the 4 assays was substantially lighter in staining intensity (figure 1). Despite extending the deadline, only 13 of the 16 pathologists from 7 of the 8 institutions participating in the study correctly completed the scoring exercise. Figure 2a and c shows a comparison of the TPS and ICPS for each case using the average of 13 pathologists in a continuous percentage score, even though each pathologist entered a categorical score as shown in supplemental table 2. Figure 2b and d shows the scoring results by percentages of patients in each categorical scoring class for each assay for both TPS and ICPS and the percentage positive using only 50% and 1% cut-points to generate a binary score for TPS, and the 10% and 1% cut-points for ICPS.
Figure 1.
IHC images from a representative region with both tumor cell and immune cell staining. Each assay is labeled in the inset and scale bars = 200 μm.
Figure 2.
A) Average sores for tumor cells obtained by averaging the reads of all 13 pathologists for each of the 90 slides. Inset shows the color code for each assay. B) Frequency distributions for tumor cells each assay color coded for scoring category and summarized for percent positive at the >50% and >1% level. C) Average sores for immune cells obtained by averaging the reads of all 13 pathologists for each of the 90 slides. D) Frequency distributions immune cells for each assay color coded for scoring category and summarized for percent positive at the >10% and >1%
To assess inter-assay variability, we first determined the average score for 13 pathologists for each antibody assay and then compared each antibody to show the mean difference for each antibody in pairwise comparisons and then tested for significance using the Wilcoxon signed rank test and a mixed effects model. Table 1 shows the average difference and statistical significance of each for both TPS and ICPS. Only the 28-8 assay and the E1L3N assay were not statistically significantly different by this method and SP142 has the greatest magnitude of difference compared to the other three antibody assays. The intraclass correlation coefficient (ICC) is perhaps a better method to compare these assays. Again using the average of 13 pathologists’ scores we found that the ICC for TPS and ICPS were 0.81 and 0.27, respectively, which increased to 0.97 and 0.80 when excluding SP142 (supplemental table 5).
Table 1.
Pairwise assay comparison
| Tumor Cell Scores | Immune Cell Scores | |||||
|---|---|---|---|---|---|---|
| Average of 13 pathologists | Mixed effects | Average of 13 pathologists | Mixed effects | |||
| Mean (Std) | p-value* | p-value** | Mean (Std) | p-value | p-value | |
| 22c3 – 28-8 | −0.300 (0.393) | <0.001 | 0.0004 | −0.127 (0.164) | <.0001 | 0.0014 |
| 22c3 - Sp142 | 0.970 (1.000) | <.0001 | <.0001 | 0.535 (0.288) | <.0001 | <.0001 |
| 22c3 - E1L3N | −0.246 (0.372) | <.0001 | 0.0024 | −0.128 (0.189) | <.0001 | 0.0022 |
| 28-8 - Sp142 | 1.270 (1.081) | <.0001 | <.0001 | 0.662 (0.294) | <.0001 | <.0001 |
| 28-8 - E1L3N | 0.055 (0.415) | 0.205 | 0.280 | −0.001 (0.194) | 0.961 | 0.9733 |
| Sp142 - E1L3N | −1.216 (1.121) | <.0001 | <.0001 | −0.664 (0.333) | <.0001 | <.0001 |
Wilcoxon signed rank test
paired t-test incorporating random effects of pathologists
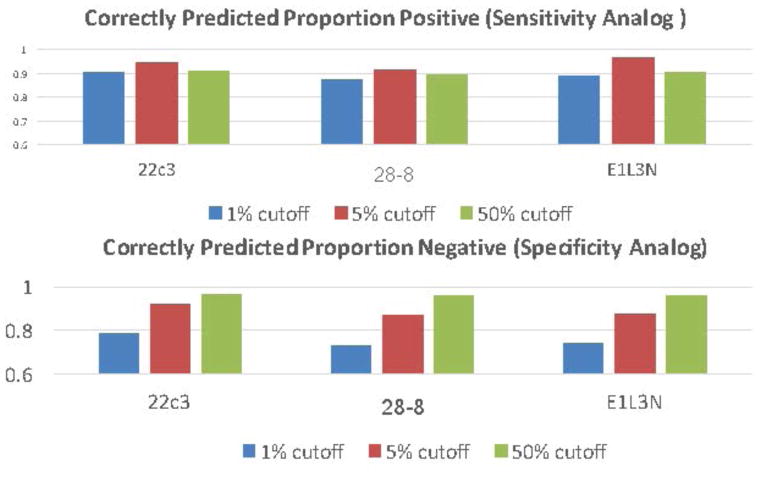
While it is interesting to use the average of 13 pathologists’ scores to compare the assays, the scoring of individual pathologists is more important since, in practice, a case is usually only examined by a single pathologist. The ICC for each pathologist and each antibody assay was measured to assess inter-pathologist variability in scoring both tumor and immune cells. Table 2 shows the ICCs for each antibody assay for both tumor cell scoring (table 2a) and immune cell scoring (table 2b). The concordance between pathologists’ reads for tumor cells was associated with an ICC between 0.83 and 0.88 for each antibody. In contrast, the ICC from immune cells was markedly decreased and in the range between 0.17 and 0.23. A second important variable to determine for comparison of pathologist scores is concordance around the drug prescription decision cut-point. At the time of this submission, there are FDA approved cut-points at >50% and another at >1%. Table 2c shows the concordance, as measured by the Fleiss Kappa statistic for the average of all 4 antibody assays at the >50% cut-point is 0.75 and at the >1% cut-point is 0.54. The present study does not have outcome information for anti-PD-1/L1 therapies. As such, the sensitivity and specificity of the assay could not be determined. However, in efforts to evaluate the ability of any given pathologist to correctly assess each assay, we defined the median pathologist’s score as “truth” and calculated the correctly predicted proportion of positive cases as an analogue for sensitivity and a correctly predicted proportion negative as an analogue for specificity. Figure 3 shows these statistics as each of 3 possible cut-points, >1%, >5% and >50%.
Table 2.
ICC for the Pathologist Scores and Concordance Statistics
| ICC for pathologists by each antibody in Tumor Cells | |||||
| 22c3 | 28-8 | SP142 | E1L3N | Summary | |
| All, N=90 | 0.882 | 0.832 | 0.869 | 0.859 | 0.86(0.02) |
| ICC for pathologists by each antibody in Immune Cells | |||||
| 22c3 | 28-8 | SP142 | E1L3N | Summary | |
| All, N=90 | 0.207 | 0.172 | 0.185 | 0.229 | 0.19(0.03) |
| Cutoff at >50% | Cutoff at >1% | |||
|---|---|---|---|---|
| Fleiss Kappa | Kendall Concordance | Fleiss Kappa | Kendall Concordance | |
| Average of all 4 assays | 0.749 | 0.775 | 0.537 | 0.612 |
Figure 3.
The top chart shows the correctly predicted proportion of positive cases and cut-points color coded including >1%, >5% and >50%. The bottom chart shows the correctly predicted proportion of negative cases at each cutoff.
Discussion
We found that the SP142 assay is associated with statistically significantly lower levels of PD-L1 staining than the other 3 assays for both TPS and ICPS. The 22c3 assay also shows statistically significant lower levels of PD-L1 expression compared to both 28-8 and E1L3N, but this slightly lower level of PD-L1 staining is only detected when an average of 13 pathologists’ scores are used. Also, we found that pathologists are highly concordant for each assay with ICC’s in the 0.8 range for TPS across any single assay, but poorly concordant for ICPS with ICC’s in the 0.2 range. This suggests that IHC may be a good method for assessment of PD-L1 in tumor cells, but is probably inadequate for assessment of immune cell expression, independent of which assay is selected. In tumor cells, we found higher concordance at the 50% cut-point than at the 1% cut-point. The 1% cut-point may require the use of automated systems or pathologist training regimens to improve assay precision.
Since we used a unified scoring system, it allowed us to do an assessment of the pathologists’ ability to score at various TPS levels. The absence of “truth” or response to therapy data limits our observations, but definition of a surrogate for truth, the median pathologists score, allowed us to further dissect where pathologists agree, and where they do not, in a more “real world” manner. In definition of companion diagnostic tests, high assay sensitivity is required for the identification of every patient that may benefit. This approach favors a lower cut-point to increase the percentage of patients that are treated. However if too many patients, that are predicted by the test to respond, do not respond, either the test, the drug or both are more likely to fail. As such we have generated a surrogate for sensitivity by calculating the percentage of times a single pathologist would call the test positive if they exceeded the median score of all pathologists at each cut-point. This data shows that, as designed, these assays, as read by our pathologist group, have a 90–95% sensitivity for any of the tested cut-points to predict a positive test. However, we also used the same approach to see the proportion of pathologists that are lower than the median score. This surrogate for specificity shows that, for each assay, the 1% cutpoint has between 70–80% specificity, compared to >90% for the 5% cut-point and >95% for the 50% cut- point. Although this is only a theoretical estimate of the potential sensitivity and specificity, the model shows that high specificity requires a high cut-point while high sensitivity can be obtained across all thresholds.
A key limitation of this effort is the lack of outcome data since these patients were not treated with PD-1/L1 axis therapies. As such, we can only evaluate this work in the context of assay comparisons and not clinical concordance. However, we note that the distribution of PD-L1 expression at the 50% and 1% cut-points closely reflected the percentages of the population considered positive in Keynote and Checkmate studies 13,14. It is also important to stress that this study is a comparison of the assays as performed in the laboratories stated using the best possible practices. Recently it was shown that the antibodies, from the perspective of interaction with the PD-L1 epitope, are most likely only subtly different if different at al15. Thus the variation seen in this study is most likely a function of the recipe or protocol for each assay. Therefore, another limitation of this study is that the assay used for SP142 on the Ventana platform is not identical to that now approved by the FDA. We believe the difference is minimal and note the similarity in appearance of the images seen in our study with those shown in the “Blueprint” study(Hirsch et al, 2016, in press). Further, the assays differences, as shown in supplemental table 2, appear to be minimal, although since the solutions are proprietary, we cannot exclude the possibility that these small differences result in large effects and are the cause of the lower levels of expression seen in this study.
In summary, this study represents level 1 biomarker evidence for the comparison of these biomarkers. We have shown that the SP142 assay is a clear outlier and that pathologists are much better at reading TPS than ICPS. We have also shown that there appears to be minimal difference between the other 3 assays tested here, which could have implications for assay choices in individual pathologist labs where there is financial pressure to validate only a single PD-L1 assay. We hope that these observations will lead to future clinical concordance studies in patients treated with PD-1 axis therapies.
Supplementary Material
Acknowledgments
This study was funded by Bristol-Myers Squibb in collaboration with the National Comprehensive Cancer Network (NCCN) Oncology Research Program, with special acknowledgements to Kimary Kulig for the concept and design of the study, as well as Patricia Esposito and Nabil Chehab for its execution The sponsors played a role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication, and as such are included in the author list and acknowledgments as per BMS and NCCN policy. Drs Rimm and Wistuba had full access to all the data in the study and Dr. Rimm takes responsibility for the integrity of the data and the accuracy of the data analysis.
The following authors disclose the following potential conflicts of interest:
Rimm: consultant; Astra Zeneca, Agendia, Bethyl Labs, Biocept, BMS, Cell Signaling Technology, ClearSight, Genoptix/Novartis, Merck, OptraScan, Perkin Elmer, and Ultivue.. Research grant support: Cepheid, Genoptix, Gilead Sciences, Pierre Fabre, and Perkin Elmer.
Janis M. Taube: Consultant: BMS, AZ, Merck. Research funding: BMS.
Julia A. Bridge: Cepheid, Inc., NCCN-Peregrine
Robert Anders: Research support & advisory boards: Bristol Myers Squibb, Adaptive Biotechnologies, Five Prime Diagnostics, National Institutes of Health, Stand Up 2 Cancer, Fibrolamellar Cancer Foundation
Fred Hirsch: Advisory boards: Genentech/Roche, BMS, Merck, AstraZeneca, Novartis, Pfizer, Ventana, HTG.
Ignacio I. Wistuba: Honoraria: Genentech/Roche, Bristol-Myers Squibb, Boehringer Ingelheim, AstraZeneca, Pfizer, Ariad, HTG Molecular, Asuragen. Research Grants: Genentech/Roche, Bristol- Myers Squibb, HTG Molecular, Asuragen.
Cory Batenchuk and Virginia Burns are employed by BMS.
References
- 1.Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012 Jun 28;366(26):2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012 Jun 28;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herbst RS, Soria JC, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014 Nov 27;515(7528):563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. Arch Pathol Lab Med. 2010 Jun;134(6):907–922. doi: 10.5858/134.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayes DF, Bast RC, Desch CE, et al. Tumor marker utility grading system: a framework to evaluate clinical utility of tumor markers. J Natl Cancer Inst. 1996 Oct 16;88(20):1456–1466. doi: 10.1093/jnci/88.20.1456. [DOI] [PubMed] [Google Scholar]
- 6.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009 Nov 4;101(21):1446–1452. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cicchetti D. Guidelines, criteria, and rules of thumb for evaluating normed and standardized assessment instruments in psychology. Psychological Assessment. 1994;6(4):284–290. [Google Scholar]
- 8.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 9.Legendre P. Species Associations: The Kendall Coefficient of Concordance Revisited. Journal of Agricultural, Biological and Environmental Statistics. Journal of Agricultural Biological and Environmental Statistics. 2005;10(2):226–245. [Google Scholar]
- 10.Phillips T, Simmons P, Inzunza HD, et al. Development of an automated PD-L1 immunohistochemistry (IHC) assay for non-small cell lung cancer. Appl Immunohistochem Mol Morphol. 2015 Sep;23(8):541–549. doi: 10.1097/PAI.0000000000000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taube JM, Anders RA, Young GD, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Science translational medicine. 2012 Mar 28;4(127):127ra137. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Velcheti V, Schalper KA, Carvajal DE, et al. Programmed death ligand-1 expression in non-small cell lung cancer. Lab Invest. 2014 Jan;94(1):107–116. doi: 10.1038/labinvest.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015 May 21;372(21):2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 14.Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med. 2015 Oct 22;373(17):1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaule P, Smithy JW, Toki M, et al. A Quantitative Comparison of Antibodies to Programmed Cell Death 1 Ligand 1. JAMA oncology. 2016 Aug 18; doi: 10.1001/jamaoncol.2016.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.