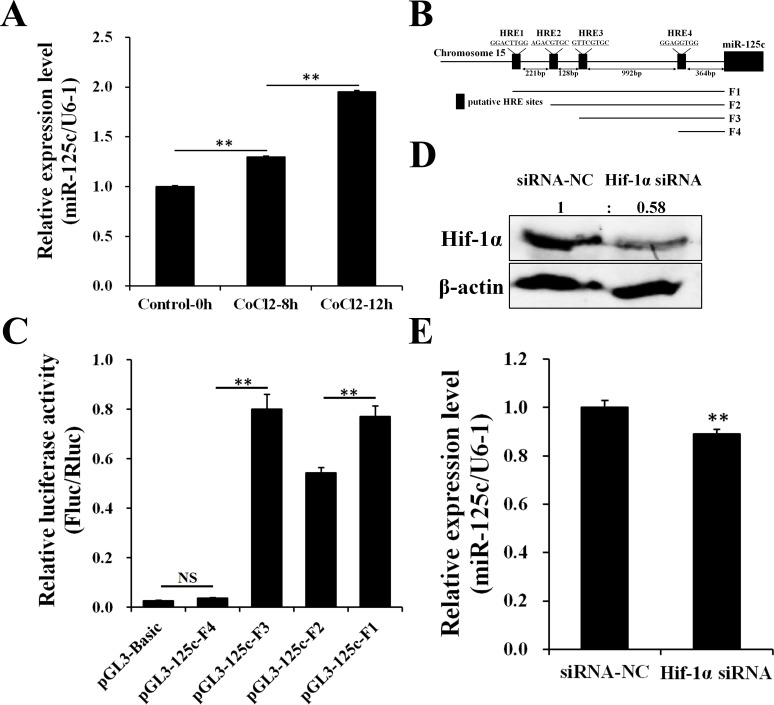
Figure 1. Hif-1α contributes to the transcription of miR-125c.
(A) Expression of miR-125c in ZF4 cells treated with 100 µM CoCl2 for 0 h, 8 h and 12 h before being collected. U6-1 is used as the endogenous control. Values represent means ± S.D. (n = 3, **P < 0.01). (B) A schematic depiction of the miR-125c promoter region. Location and sequence information of four putative HREs (HRE1, HRE2, HRE3 and HRE4) are indicated. A series of promoter fragments (F1, F2, F3 and F4) carrying different HRE motifs were amplified. (C) The luciferase reporters harboring different HREs was cotransfected with the pRL-TK Renilla luciferase reporter (internal control) and pCMV-Myc-Hif-1α into HeLa cells. Luciferase activities were detected after stimulating with 100 μM CoCl2 for 4 h, and Firefly luciferase expression was normalized to Renilla luciferase. Results are presented as mean ± S.E. (n = 3, **P < 0.01, NS, not significant). (D) Protein detection of Hif-1α in ZF4 cells transfected with Hif-1α siRNA (siRNA-NC, negative control siRNA) and treated with 100 μM CoCl2 for 12 h before harvest (hypoxia-simulating). β-actin is used to normalize protein levels. Numbers indicate quantification of the Hif-1α band densities relative to β-actin. (E) Expression of miR-125c in ZF4 cells transfected with Hif-1α siRNA and treated with 100 μM CoCl2 for 12 h before harvest. U6-1 is used as the endogenous control. Values represent means ± S.D. (n = 3, **P < 0.01).