Abstract
There is a genetic susceptibility for nonalcoholic fatty liver disease (NAFLD). To examine the role of genetic factors in the disease, a Bayesian analysis was performed to model gene relationships in NAFLD pathogenesis. The Bayesian analysis indicated a potential gene interaction between the TM6SF2 and PNPLA3 genes. Next, to explore the underlying mechanism at the cellular level, we evaluated the additive effects between the TM6SF2 E167K and PNPLA3 I148M polymorphisms on lipid metabolism. Hepa 1-6 cells were transfected with a control vector or with overexpression vectors for TM6SF2/PNPLA3-wild type, TM6SF2-mutant type, PNPLA3-mutant type, or TM6SF2/PNPLA3-mutant type. Commercial kits were used to measure triglyceride and total cholesterol levels in each of the five groups. The mRNA and protein expression levels of sterol regulatory element-binding transcription factor 1c and fatty acid synthase were analyzed using real-time PCR and western blotting. The triglyceride and total cholesterol contents were significantly different among the groups. The triglyceride and total cholesterol contents and the sterol regulatory element-binding transcription factor 1c and fatty acid synthase mRNA and protein expression levels were significantly higher in the TM6SF2/PNPLA3-mutant type group than in the TM6SF2-mutant type group or the PNPLA3-mutant type group. The TM6SF2 E167K and PNPLA3 I148M polymorphisms may have additive effects on lipid metabolism by increasing the expression of sterol regulatory element-binding transcription factor 1c and fatty acid synthase.
Keywords: nonalcoholic fatty liver disease, additive effect, TM6SF2, PNPLA3, bayesian network
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) is the most common chronic liver disease in the world, affecting 13.48–31.79% of the general population [1]. In recent years, the incidence of NAFLD has been higher than that of viral hepatitis and alcoholic liver disease. NAFLD can be histologically categorized as simple fatty liver, which is usually considered a benign lesion, or as nonalcoholic steatohepatitis [2]. The latter pathological process can dramatically confer increased risks of liver cirrhosis and hepatocellular carcinoma [3] and is characterized by liver inflammation and hepatocyte injury in addition to hepatic steatosis with or without fibrosis [2, 4].
There is a genetic susceptibility for NAFLD [5]. Accordingly, the role of genetic factors in NAFLD pathogenesis has been gaining greater attention. Recent studies, including a pilot study by our group, have shown that the TM6SF2 E167K [6–8] and PNPLA3 I148M [9–11] polymorphisms are both associated with the development of NAFLD. Interestingly, Kozlitina and colleagues observed that inhibition of TM6SF2 significantly reduced the expression of PNPLA3, a protein that plays an important role in triglyceride synthesis [6]. Furthermore, Goffredo et al. determined that the TM6SF2 E167K, PNPLA3 I148M, and GCKR rs1260326 single nucleotide polymorphisms had a joint effect in determining intrahepatic fat accumulation in a cohort of obese children, including Caucasians, African Americans, and Hispanics [12]. These results indicated that the TM6SF2 and PNPLA3 polymorphisms might have additive effects in regulating lipid metabolism. However, the underlying molecular mechanism remains unclear.
The present study evaluated the additive effects of the TM6SF2 E167K and PNPLA3 I148M polymorphisms on lipid metabolism using bioinformatics approaches and then further explored the underlying mechanism at the cellular level.
RESULTS
The bayesian network and potential interaction between the TM6SF2 and PNPLA3 genes
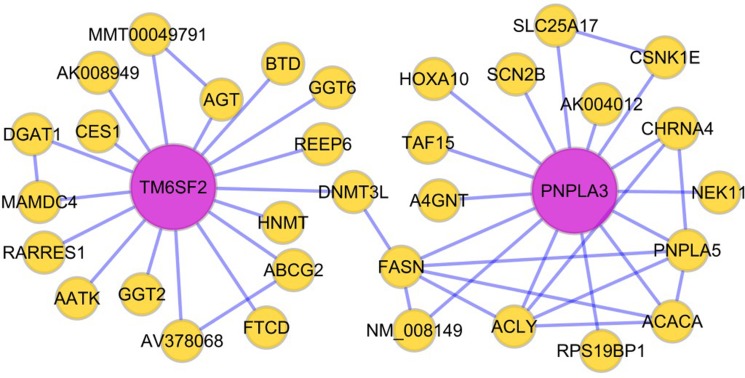
We performed bioinformatics analysis and data mining, and ultimately constructed a Bayesian network to model NAFLD pathogenesis. Table 1 describes the relevant source papers used to construct the Bayesian network, including research articles with liver expression data from both mice and humans. Figure 1 shows a visualization of a potential gene interaction in the Bayesian network focusing on the TM6SF2 and PNPLA3 genes, which are two important regulators of liver fat metabolism. Our data demonstrate that multiple genes related to lipid metabolism are connected with TM6SF2 and PNPLA3. Based on the network shown in Figure 1, we hypothesized that the TM6SF2 and PNPLA3 genes may interact with each other through DNA (cytosine-5)-methyltransferase 3-like (DNMT3L) and FASN.
Table 1. The liver expression data of mouse and human and construction of the bayesian network.
| Tissue | Species | eSNP data | Co-expr. modules | Bayesian networks | Dataset |
|---|---|---|---|---|---|
| Liver | Human | yes | yes | yes | 427 individuals [13] |
| Human | yes | yes | no | 1,008 obese patients [14] | |
| Mouse | - | yes | yes | C57BL/6J × A/J mouse cross [15] | |
| Mouse | - | yes | yes | C57BL/6J × C3H ApoE –/– mouse cross [16, 17] | |
| Mouse | - | yes | yes | C57BL/6J × C3H wild-type mouse cross [13] | |
| Mouse | - | yes | yes | C57BL/6J × BTBR Lepob mouse cross [18] |
Figure 1. Hypothesized Bayesian network indicating linkage between the TM6SF2 and PNPLA3 genes.
The lines in the network represent transcriptional regulation between genes. The TM6SF2 and PNPLA3 genes may interact with each other through DNMT3L and FASN. Abbreviations: DNMT3L: DNA (cytosine-5)- methyltransferase 3-like; FASN: fatty acid synthase.
Effects of the TM6SF2 E167K and PNPLA3 I148M variants on triglyceride and total cholesterol contents in hepa 1-6 cells
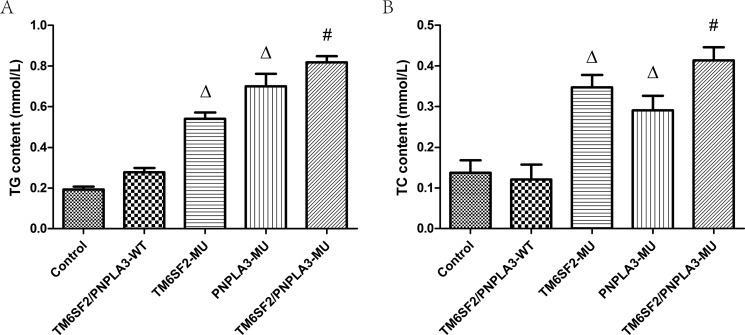
Figure 2 shows the triglyceride and total cholesterol contents for the five groups. According to the one-way ANOVA, there were differences in both the triglyceride (F = 169.48, P < 0.01) and total cholesterol (F = 44.79, P < 0.01) contents among the groups. Further LSD t-tests revealed that the triglyceride and total cholesterol contents of both the TM6SF2-MU group and the PNPLA3-MU group were higher than both the control group and the TM6SF2/PNPLA3-WT group (all P < 0.01). Moreover, significant differences in the triglyceride (P < 0.01) and total cholesterol (P = 0.035) contents were observed between the TM6SF2/PNPLA3-MU group and the TM6SF2-MU group. Similarly, there were differences in both the triglyceride (P = 0.002) and total cholesterol (P = 0.001) contents between the TM6SF2/PNPLA3-MU group and PNPLA3-MU group.
Figure 2. Effects of the TM6SF2 E167K and PNPLA3 I148M variants on triglyceride and total cholesterol contents.
The TM6SF2 E167K and PNPLA3 I148M variants increased triglyceride (TG) and total cholesterol (TC) contents. ΔP < 0.01 vs. TM6SF2/PNPLA3-WT group, #P < 0.01 vs. either the TM6SF2-MU group or the PNPLA3-MU group.
Effects of the TM6SF2 E167K and PNPLA3 I148M variants on the expression of genes involved in triglyceride metabolism
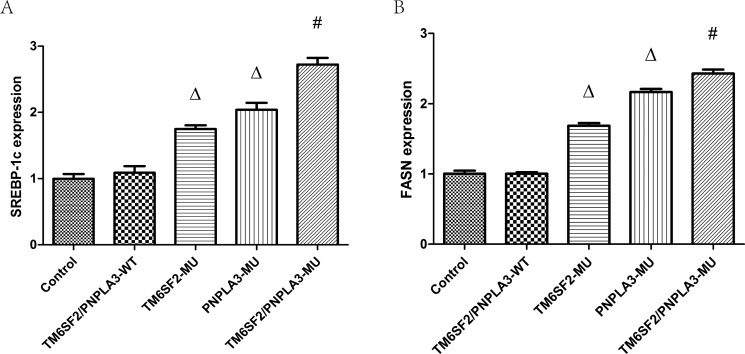
To investigate how the TM6SF2 E167K and PNPLA3 I148M variants mechanistically affect lipid content, we determined and analyzed levels of SREBP-1c and FASN mRNAs and proteins, which are involved in hepatic lipid synthesis and metabolism. Our data revealed that the expression of SREBP-1c and FASN mRNAs were significantly different among the five groups (FSREBP-1c = 194.64, P < 0.01; FFASN = 696.21, P < 0.01). As shown in Figure 3, the expression levels of both SREBP-1c and FASN mRNA were higher in the TM6SF2-MU group and the PNPLA3-MU group than in the TM6SF2/PNPLA3-WT group (all P < 0.01). Furthermore, the SREBP-1c and FASN mRNA expression levels in the TM6SF2/PNPLA3-MU group were significantly higher than both the TM6SF2-MU group and the PNPLA3-MU group (all P < 0.01).
Figure 3. Effects of the TM6SF2 E167K and PNPLA3 I148M variants on SREBP-1c and FASN mRNA expression levels.
The TM6SF2 E167K and PNPLA3 I148M variants significantly upregulated the SREBP-1c and FASN mRNA expression levels. ΔP < 0.01 vs. TM6SF2/PNPLA3-WT group, #P < 0.01 vs. either the TM6SF2-MU group or the PNPLA3-MU group. Abbreviations: SREBP-1c: sterol regulatory element-binding transcription factor 1c; FASN: fatty acid synthase.
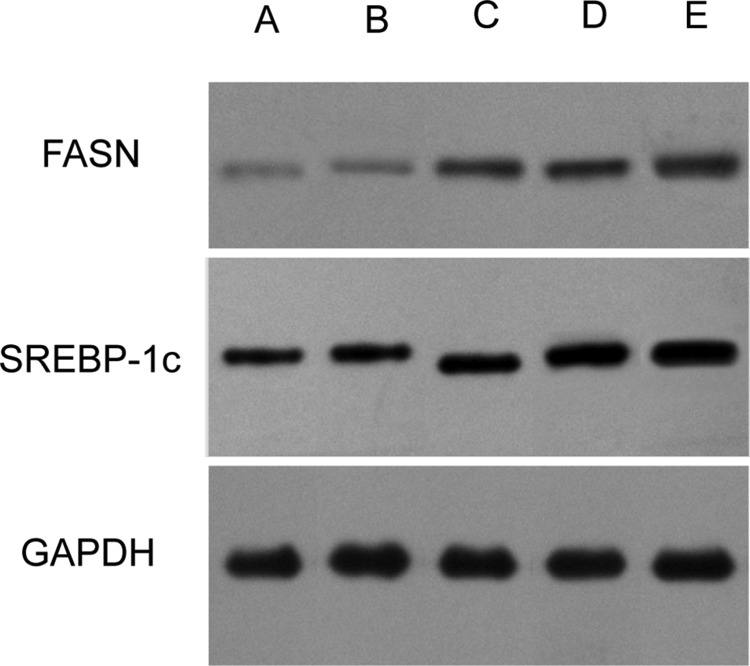
Consistent with the RT-PCR results, the western blotting analysis (Figure 4) revealed that the SREBP-1c and FASN protein levels were also significantly different among the five groups (FSREBP-1c = 474.64, P < 0.01; FFASN = 509.54, P < 0.01). As expected, the mutant forms of TM6SF2 E167K and PNPLA3 I148M caused a substantial increase of the levels of SREBP-1c and FASN proteins compared to the wild type (all P < 0.01). In addition, our data demonstrated that the SREBP-1c and FASN protein levels in the TM6SF2/PNPLA3-MU group were significantly higher than in both the TM6SF2-MU group (P SREBP-1c < 0.01, P FASN < 0.01) and the PNPLA3-MU group (P SREBP-1c = 0.005, P FASN < 0.01). Together, the results suggested that the TM6SF2 E167K and PNPLA3 I148M variants might increase hepatic lipid content by increasing SREBP-1c and FASN levels.
Figure 4. Effects of the TM6SF2 E167K and PNPLA3 I148M variants on SREBP-1c and FASN protein expression levels.
(A) the TM6SF2/PNPLA3-control group; (B) the TM6SF2/PNPLA3-wild type group; (C) the TM6SF2-mutant type group; (D) the PNPLA3-mutant type group; (F) the TM6SF2/PNPLA3-mutant type group. Abbreviations: SREBP-1c: sterol regulatory element-binding transcription factor 1c; FASN: fatty acid synthase; GAPDH: glyceraldehyde 3-phosphate dehydrogenase.
DISCUSSION
The aim of this study was to investigate whether there are additive effects of the TM6SF2 E167K and PNPLA3 I148M polymorphisms on lipid metabolism. The major finding of our research is that the TM6SF2 E167K and PNPLA3 I148M variants may increase hepatic lipid content by increasing the expression of SREBP-1c and FASN.
TM6SF2 and PNPLA3 are two important genes that were identified as conferring susceptibility to NAFLD in a genome-wide association study and an exome-wide association study of multiethnic, population-based cohorts [6, 9]. Both are involved in very low-density lipoprotein (VLDL) secretion and lipid droplet remodeling. The human TM6SF2 gene is located on chromosome 19 and encodes a protein of 351 amino acids [19]. The TM6SF2 protein is an EXPERA domain P-containing enzyme homolog, which was predicted to possess similar catalytic activity as sterol isomerases according to a computational protein sequence analysis [20]. Mahdessian et al. demonstrated that the TM6SF2 protein is predominantly located in both the endoplasmic reticulum (ER) and the ER-Golgi intermediate compartment of hepatocytes in a protein subcellular localization study [19]. Several studies provided evidence that TM6SF2, which is highly expressed in the liver and small intestine [6, 21], can be regarded as a “switch” mediating VLDL secretion [6, 19, 22]. The PNPLA3 gene encodes a polypeptide chain of 481 amino acids and is located on the long arm of chromosome 22 [23]. PNPLA3 is also highly expressed on the ER and in lipid membranes of metabolically relevant tissues (i.e., liver and adipose tissue) [24]. Pingitore and colleagues reported that the PNPLA3 protein, which has a conserved patatin domain, displays predominant triglyceride hydrolase activity and mild lysophosphatidic acid acyltransferase activity [23].
Various studies have established strong links between the TM6SF2 E167K polymorphism and the developmental and progressive stages of NAFLD [6–8]. Similarly, studies have demonstrated an association between the PNPLA3 I148M polymorphism and the development of NAFLD [9–11]. However, NAFLD is a complex disease influenced by many factors, including genetics, diet, and gut microbiota [5, 25]. Neither the TM6SF2 E167K polymorphism nor the PNPLA3 I148M polymorphism alone can fully account for the molecular genetic mechanisms of NAFLD or disease risk. Analysis of gene-gene interactions is essential to understand the etiology of complex diseases and to identify genes responsible for disease susceptibility. In the present study, we provided evidence that the TM6SF2 and PNPLA3 genes may interact with each other through DNMT3L and FASN in a Bayesian analysis. Moreover, the results of our cellular experiments showed that the TM6SF2/PNPLA3-MU group had significantly higher triglyceride and total cholesterol contents than both the TM6SF2-MU group and the PNPLA3-MU group. Our finding is consistent with those of recent studies [12, 26]. Wang et al. found that the TM6SF2 E167K and PNPLA3 I148M variants were the most important risk alleles for NAFLD and indicated that they had an additive effect on NAFLD in a Chinese cohort [26]. Together, these findings indicate that the TM6SF2 E167K and PNPLA3 I148M polymorphisms have a potential additive effect in NAFLD.
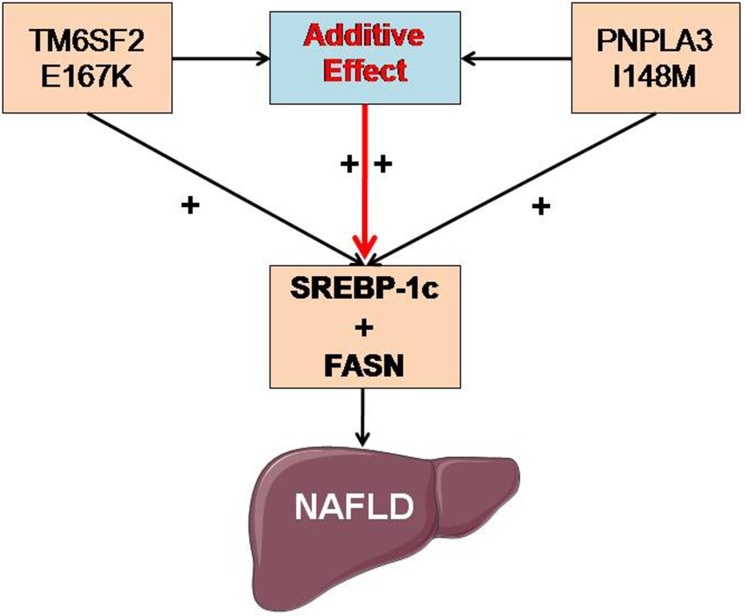
However, the specific underlying mechanism of the above additive effect has not yet been reported. To the best of our knowledge, this study is the first to provide data to understand the proposed additive effect mechanistically. SREBP-1c is a master modifier of fatty acid and triglyceride metabolism, and it is synthesized on the ER membrane in its precursor form [27–28]. As a metabolic regulator, SREBP-1c activates the transcription of target genes, including FASN, acetyl-CoA carboxylase, and stearoyl-CoA desaturase, and consequently, it enhances the synthesis of fatty acids and the accumulation of triglycerides [29]. Our data showed that SREBP-1c and FASN mRNA and protein expression levels were significantly increased in the TM6SF2/PNPLA3-MU group compared to both the TM6SF2-MU group and the PNPLA3-MU group. Considering these findings, we hypothesize that the additive effects of the TM6SF2 E167K and PNPLA3 I148M polymorphisms on NAFLD may be associated with upregulation of SREBP-1c and FASN expression (Figure 5). In the future, the role of DNMT3L in the additive effects of the TM6SF2 E167K and PNPLA3 I148M polymorphisms on lipid metabolism should be further elucidated. In addition, studies at the cellular and individual levels are required to understand the underlying mechanisms involved in this process further.
Figure 5. Potential molecular mechanism of the additive effects of the TM6SF2 E167K and PNPLA3 I148M polymorphisms on NAFLD.
The additive effects of the TM6SF2 E167K and PNPLA3 I148M polymorphisms on NAFLD may be associated with upregulating the expression of SREBP-1c and FASN. Abbreviations: SREBP-1c: sterol regulatory element-binding transcription factor 1c; FASN: fatty acid synthase; NAFLD: nonalcoholic fatty liver disease.
MATERIALS AND METHODS
Establishment of the bayesian network
The Bayesian algorithm is an attractive method that can provide formalism for statistical reasoning about partial beliefs under circumstances of uncertainty [29]. Here, a Bayesian network was constructed to model gene relationships in NAFLD pathogenesis according to previously established methods [30–31]. The network incorporated liver expression data from mice and humans. A Bayesian analysis was performed to explore potential interactions between the TM6SF2 gene and the PNPLA3 gene. These genes were reported to be important modifiers involved in the regulation of triglyceride metabolism.
Hepa 1-6 cell culture
Hepa 1–6 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal bovine serum (FBS; Hyclone, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco®, USA). The cells were incubated at a temperature of 37°C in a humidified atmosphere with 5% CO2. The cells were treated when they reached approximately 80% confluence.
Construction of lentiviral vectors and transfection studies
The present study examined the following five groups: (1) the control group, (2) the TM6SF2/PNPLA3-wild type (WT) group (TM6SF2 EE + PNPLA3 II/IM), (3) the TM6SF2-mutant type (MU) group (TM6SF2 EK/KK + PNPLA3 II/IM), (4) the PNPLA3-mutant type (MU) group (TM6SF2 EE + PNPLA3 MM), and (5) the TM6SF2/PNPLA3-mutant type (MU) group (TM6SF2 EK/KK + PNPLA3 MM). The lentiviral plasmids for the experimental groups and control group were constructed by Shanghai Genechem Co., Ltd. For each group, the plasmids were transfected into 293T cells after the cells were cultured for 24 h in DMEM with 10% FBS. The 293T cells were incubated with the transfection complexes (expression plasmid, packaging plasmid, and transfection reagent) for 48–72 h. The lentivirus was then concentrated and transfected into Hepa 1–6 cells. The success of the transfection process in each group was validated by real-time polymerase chain reaction (RT-PCR) and western blotting, as well as by measuring the percentage of Hepa 1–6 cells with green fluorescent protein.
Biochemical indicator assay
After transfection, the Hepa 1–6 cells were centrifuged, and the supernatants were collected. The concentrations of biochemical indicators (triglyceride and total cholesterol) were measured in each group using commercial kits following the manufacturer’s recommended instructions (Sigma-Aldrich, St. Louis, USA).
Extraction of total RNA and quantitative RT-PCR
The expression levels of sterol regulatory element-binding transcription factor 1c (SREBP-1c) and fatty acid synthase (FASN), two factors associated with lipid metabolism, were analyzed using RT-PCR. Total RNA was isolated from the Hepa 1–6 cells using Trizol reagent (Invitrogen, USA) following the manufacturer’s protocol. Complementary DNA (cDNA) synthesis was performed using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher: K1622, USA).
SYBR® Green Realtime PCR Master Mix (Toyobo: QPK-201, Japan) was used for RT-PCR following the manufacturer’s instructions. For standardization, the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene was used as an internal control. The specific primer sequences were designed and synthesized by the Springen Biotechnology and are shown in Table 2. The RT-PCR parameters consisted of denaturation at 95°C for 5 minutes, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 20 s, and extension at 72°C for 40 s. The relative amounts of the above genes were obtained using the comparative threshold cycle (CT) method [32].
Table 2. Primer sequences for RT-PCR.
| Gene | Primer | Sequences (5′–3′) |
|---|---|---|
| SREBP-1c (163bp) |
Forward Reverse |
GCTCCCTAGGAAGGGCCGTA ACTTCACCTTCGATGTCGGTC |
| FASN (183bp) |
Forward Reverse |
CAGAGCAGCCATGGAGGAG GACAGGTCCTTCAGCTTGCC |
| GAPDH (202bp) |
Forward Reverse |
CATCTTCTTTTGCGTCGCCA TTAAAAGCAGCCCTGGTGACC |
Abbreviations: RT-PCR: Real Time PCR; SREBP-1c: sterol regulatory element-binding transcription factor 1c; FASN: fatty acid synthase; GAPDH: glyceraldehyde 3-phosphate dehydrogenase.
Western blotting
Total protein was extracted from the Hepa 1–6 cells using radioimmunoprecipitation assay buffer (Sigma-Aldrich, USA). The Bradford method was used to determine protein concentration following the manufacturer’s protocol, and the proteins were then frozen at –70°C until analysis. Antibodies against SREBP-1c (Santa Cruz Biotechnology, Inc., SC-13551, USA), FASN (Cell Signaling Technology, Inc., 3180S, USA), and GAPDH (Keygen Biotech, Co. Ltd., KGAA002, China) were used. The expression levels of the SREBP-1c and FASN proteins were normalized relative to the corresponding GAPDH (endogenous reference) level in each lane. The western blots were analyzed using Gel-Pro Analyzer Version 4.5 Software (Media Cybernetics, USA).
Statistical analysis
Statistical significance was determined using one-way analysis of variance (ANOVA) followed by the least significant difference (LSD) t-test. The statistical comparisons among the groups were made using the Statistical Package for the Social Sciences (SPSS version 17.0, SPSS Inc., Chicago, USA) software. All continuous variables were expressed as the mean ± standard deviation (SD) of at least three independent experiments. P < 0.05 was considered statistically significant.
CONCLUSIONS
The TM6SF2 E167K and PNPLA3 I148M polymorphisms may have additive effects on lipid metabolism and the development of NAFLD by upregulating the expression of SREBP-1c and FASN. Understanding how these additive effects contribute to lipid metabolism and NAFLD may help provide a new scientific basis for NAFLD pathogenesis and shed light on genetic predictors for early diagnosis and primary prevention of the disease. Further investigation is needed to elucidate these underlying mechanisms.
Footnotes
Author contributions
Lizhen Chen and Yongning Xin conceived and designed the experiments. Lizhen Chen, Shuixian Du, and Linlin Lu carried out the cell experiments. Lizhen Chen and Zhonghua Lin performed the Bayesian analysis. Lizhen Chen drafted and wrote the manuscript. Zhonghua Lin, Wenwen Jin, Doudou Hu, Xiangjun Jiang, and Yongning Xin revised the manuscript. Shiying Xuan approved the final version.
CONFLICTS OF INTEREST
None.
GRANT SUPPORT
This study was supported by the Key Research Project of Shandong Province (2016GSF201217), the Medical and Health Technology Development Project of Shandong Province (2015WS0321), and the Qingdao, Shinan District Science and Technology Development Project Fund (2015-6-014-YY, 2016-3-016-YY).
REFERENCES
- 1.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of non-alcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 2.LaBrecque DR, Abbas Z, Anania F, Ferenci P, Khan AG, Goh KL, Hamid SS, Isakov V, Lizarzabal M, Peñaranda MM, Ramos JF, Sarin S, Stimac D, et al. and Review Team. and World Gastroenterology Organisation World Gastroenterology Organisation global guidelines: Nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. J Clin Gastroenterol. 2014;48:467–473. doi: 10.1097/MCG.0000000000000116. [DOI] [PubMed] [Google Scholar]
- 3.Watanabe S, Hashimoto E, Ikejima K, Uto H, Ono M, Sumida Y, Seike M, Takei Y, Takehara T, Tokushige K, Nakajima A, Yoneda M, Saibara T, et al. and Japanese Society of Gastroenterology. and Japan Society of Hepatology Evidence-based clinical practice guidelines for nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. J Gastroenterol. 2015;50:364–377. doi: 10.1007/s00535-015-1050-7. [DOI] [PubMed] [Google Scholar]
- 4.Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, Charlton M, Sanyal AJ, and American Gastroenterological Association. and American Association for the Study of Liver Diseases. and American College of Gastroenterologyh The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American Gastroenterological Association, American Association for the Study of Liver Diseases, and American College of Gastroenterology. Gastroenterology. 2012;142:1592–1609. doi: 10.1053/j.gastro.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 5.European Association for the Study of the Liver (EASL) European Association for the Study of Diabetes (EASD) European Association for the Study of Obesity (EASO) EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. J Hepatol. 2016;64:1388–1402. doi: 10.1016/j.jhep.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Kozlitina J, Smagris E, Stender S, Nordestgaard BG, Zhou HH, Tybjærg-Hansen A, Vogt TF, Hobbs HH, Cohen JC. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu YL, Reeves HL, Burt AD, Tiniakos D, McPherson S, Leathart JB, Allison ME, Alexander GJ, Piguet AC, Anty R, Donaldson P, Aithal GP, Francque S, et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat Commun. 2014;5:4309. doi: 10.1038/ncomms5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mancina RM, Sentinelli F, Incani M, Bertoccini L, Russo C, Romeo S, Baroni MG. Transmembrane-6 superfamily member 2 (TM6SF2) E167K variant increases susceptibility to hepatic steatosis in obese children. Dig Liver Dis. 2016;48:100–101. doi: 10.1016/j.dld.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buch S, Stickel F, Trépo E, Way M, Herrmann A, Nischalke HD, Brosch M, Rosendahl J, Berg T, Ridinger M, Rietschel M, McQuillin A, Frank J, et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet. 2015;47:1443–1448. doi: 10.1038/ng.3417. [DOI] [PubMed] [Google Scholar]
- 11.Xu J, Xin YN, Lü WH, Lin ZH, Zhang DD, Zhang M, Dong QJ, Jiang XJ, Xuan SY. Polymorphism rs738409 in PNPLA3 is associated with inherited susceptibility to non-alcoholic fatty liver disease. [Article in Chinese] Zhonghua Gan Zang Bing Za Zhi. 2013;21:619–23. doi: 10.3760/cma.j.issn.1007-3418.2013.08.013. https://www.ncbi.nlm.nih.gov/pubmed/24119744. [DOI] [PubMed] [Google Scholar]
- 12.Goffredo M, Caprio S, Feldstein AE, D’Adamo E, Shaw MM, Pierpont B, Savoye M, Zhao H, Bale AE, Santoro N. Role of TM6SF2 rs58542926 in the pathogenesis of nonalcoholic pediatric fatty liver disease: A multiethnic study. Hepatology. 2016;63:117–125. doi: 10.1002/hep.28283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, Zhu J, Millstein J, Sieberts S, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenawalt DM, Dobrin R, Chudin E, Hatoum IJ, Suver C, Beaulaurier J, Zhang B, Castro V, Zhu J, Sieberts SK, Wang S, Molony C, Heymsfield SB, et al. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 2011;21:1008–1016. doi: 10.1101/gr.112821.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derry JM, Zhong H, Molony C, MacNeil D, Guhathakurta D, Zhang B, Mudgett J, Small K, El Fertak L, Guimond A, Selloum M, Zhao W, Champy MF, et al. Identification of genes and networks driving cardiovascular and metabolic phenotypes in a mouse F2 intercross. PLoS One. 2010;5:e14319. doi: 10.1371/journal.pone.0014319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang SS, Schadt EE, Wang H, Wang X, Ingram-Drake L, Shi W, Drake TA, Lusis AJ. Identification of pathways for atherosclerosis in mice: integration of quantitative trait locus analysis and global gene expression data. Circ Res. 2007;101:e11–30. doi: 10.1161/CIRCRESAHA.107.152975. [DOI] [PubMed] [Google Scholar]
- 17.Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, Drake TA, Lusis AJ. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tu Z, Keller MP, Zhang C, Rabaglia ME, Greenawalt DM, Yang X, Wang IM, Dai H, Bruss MD, Lum PY, Zhou YP, Kemp DM, Kendziorski C, et al. Integrative analysis of a cross-loci regulation network identifies App as a gene regulating insulin secretion from pancreatic islets. PLoS Genet. 2012;8:e1003107. doi: 10.1371/journal.pgen.1003107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahdessian H, Taxiarchis A, Popov S, Silveira A, Franco-Cereceda A, Hamsten A, Eriksson P, van’t Hooft F. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc Natl Acad Sci USA. 2014;111:8913–8918. doi: 10.1073/pnas.1323785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez-Pulido L, Ponting CP. TM6SF2 and MAC30, new enzyme homologs in sterol metabolism and common metabolic disease. Front Genet. 2014;5:439. doi: 10.3389/fgene.2014.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smagris E, Gilyard S, BasuRay S, Cohen JC, Hobbs HH, BasuRay S, Cohen JC, Hobbs HH. Inactivation of Tm6sf2, a gene defective in fatty liver disease, impairs lipidation but not secretion of very low density lipoproteins. J Biol Chem. 2016;291:10659–10676. doi: 10.1074/jbc.M116.719955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen LZ, Xia HH, Xin YN, Lin ZH, Xuan SY. TM6SF2 E167K variant, a novel genetic susceptibility variant, contributing to nonalcoholic fatty liver disease. J Clin Transl Hepatol. 2015;3:265–270. doi: 10.14218/JCTH.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pingitore P, Pirazzi C, Mancina RM, Motta BM, Indiveri C, Pujia A, Montalcini T, Hedfalk K, Romeo S. Recombinant PNPLA3 protein shows triglyceride hydrolase activity and its I148M mutation results in loss of function. Biochim Biophys Acta. 2014;1841:574–580. doi: 10.1016/j.bbalip.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 24.He S, McPhaul C, Li JZ, Garuti R, Kinch L, Grishin NV, Cohen JC, Hobbs HH. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem. 2010;285:6706–6715. doi: 10.1074/jbc.M109.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leung C, Rivera L, Furness JB, Angus PW. The role of the gut microbiota in NAFLD. Nat Rev Gastroenterol Hepatol. 2016;13:412–425. doi: 10.1038/nrgastro.2016.85. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Liu Z, Wang K, Wang Z, Sun X, Zhong L, Deng G, Song G, Sun B, Peng Z, Liu W. Additive effects of the risk alleles of PNPLA3 and TM6SF2 on non-alcoholic fatty liver disease (NAFLD) in a Chinese population. Front Genet. 2016;7:140. doi: 10.3389/fgene.2016.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmed MH, Byrne CD. Modulation of sterol regulatory element binding proteins (SREBPs) as potential treatments for non-alcoholic fatty liver disease (NAFLD) Drug Discov Today. 2007;12:740–747. doi: 10.1016/j.drudis.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Lei T, Huang JF, Wang SB, Zhou LL, Yang ZQ, Chen XD. The link between fibroblast growth factor 21 and sterol regulatory element binding protein 1c during lipogenesis in hepatocytes. Mol Cell Endocrinol. 2011;342:41–47. doi: 10.1016/j.mce.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Pearl J. Probabilistic reasoning in intelligent systems: networks of plausible inference. San Mateo (CA): Morgan Kaufmann Pub; 1988. [Google Scholar]
- 30.Imoto S, Goto T, Miyano S. Estimation of genetic networks and functional structures between genes by using Bayesian networks and nonparametric regression. Pac Symp Biocomput. 2002:175–86. [PubMed] [Google Scholar]
- 31.Toyoshiba H, Yamanaka T, Sone H, Parham FM, Walker NJ, Martinez J, Portier CJ. Gene interaction network suggests dioxin induces a significant linkage between aryl hydrocarbon receptor and retinoic acid receptor beta. Environ Health Perspect. 2004;112:1217–1224. doi: 10.1289/txg.7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]