Abstract
Upon mild liver injury, new hepatocytes originate from pre-existing hepatocytes. However, if hepatocyte proliferation is impaired, a manifestation of severe liver injury, biliary epithelial cells (BECs) contribute to new hepatocytes through BEC dedifferentiation into liver progenitor cells (LPCs), also termed oval cells or hepatoblast-like cells (HB-LCs), and subsequent differentiation into hepatocytes. Despite the identification of several factors regulating BEC dedifferentiation and activation, little is known about factors involved in the regulation of LPC differentiation into hepatocytes during liver regeneration. Using a zebrafish model of near-complete hepatocyte ablation, we here show that Bmp signaling is required for BEC conversion to hepatocytes, particularly for LPC differentiation into hepatocytes. We found that severe liver injury led to the upregulation of genes involved in Bmp signaling, including smad5, tbx2b, and id2a, in the liver. Bmp suppression did not block BEC dedifferentiation into HB-LCs; however, the differentiation of HB-LCs into hepatocytes was impaired due to the maintenance of HB-LCs in an undifferentiated state. Later Bmp suppression did not affect HB-LC differentiation, but increased BEC number through proliferation. Notably, smad5, tbx2b, and id2a mutants exhibited similar liver regeneration defects as those observed in Bmp-suppressed livers. Moreover, BMP2 addition promoted the differentiation of a murine LPC cell line into hepatocytes in vitro. Conclusions: Bmp signaling regulates BEC-driven liver regeneration via smad5, tbx2b and id2a: it regulates HB-LC differentiation into hepatocytes via tbx2b and BEC proliferation via id2a. Our findings provide insights into promoting innate liver regeneration as a novel therapy.
Keywords: liver progenitor cells, oval cells, biliary epithelial cells, dedifferentiation, smad5
Introduction
As a highly regenerative organ, the liver can undergo either hepatocyte- or biliary-driven regeneration. In the former case, upon partial hepatectomy or mild liver injury, hepatocytes proliferate to restore the lost liver mass (1). However, in the latter case, upon severe liver injury in which hepatocyte proliferation is compromised, biliary epithelial cells (BECs) dedifferentiate into liver progenitor cells (LPCs), also called oval cells or hepatoblast-like cells (HB-LCs), and subsequently give rise to hepatocytes (2). Previously, controversies existed regarding the relative contribution of BEC-driven liver regeneration (3); however, recent studies in zebrafish (4, 5) and mice (6) have resolved this controversy by showing the extensive contribution of BECs to hepatocytes upon severe liver injury. In mice, hepatocyte-specific Mdm2 deletion completely blocks hepatocyte proliferation, additionally induces hepatocyte senescence and apoptosis, and subsequently elicits oval cell activation. These oval cells later give rise to hepatocytes, leading to a full liver recovery (6). In the zebrafish studies, the near-complete ablation of hepatocytes elicits the extensive contribution of BECs to hepatocytes through the dedifferentiation of BECs into HB-LCs and subsequent differentiation of the HB-LCs into hepatocytes (4, 5)
Oval cells are frequently observed in diseased livers and their number positively correlates with disease severity (7). Since patients suffering from severe liver diseases have limited treatment options and present with an abundance of hepatic oval cells, promoting the differentiation of oval cells into hepatocytes is deemed an effective therapeutic strategy. Developing such therapies requires a deeper understanding of the mechanisms by which oval cells differentiate into hepatocytes in vivo. Although several factors, such as FGF7 (8) and TWEAK (9), which can induce oval cell activation in vivo, have been identified, factors that regulate oval cell differentiation into hepatocytes in vivo are unknown, mainly due to the lack of an animal model for BEC-driven liver regeneration. However, the recent reports of zebrafish and mouse liver injury models, in which BECs extensively contribute to regenerated hepatocytes, present an opportunity to investigate the mechanisms underlying oval cell differentiation into hepatocytes.
Using the zebrafish BEC-driven liver regeneration model combined with targeted chemical screening, we recently reported on the role of bromodomain extraterminal (BET) proteins in BEC dedifferentiation into HB-LCs and the proliferation of newly-generated hepatocytes (10). Using the same zebrafish model, we now show the essential role of Bmp signaling in HB-LC differentiation into hepatocytes. Bmp signaling plays important roles in early liver development, such as hepatoblast specification and proliferation (11, 12). Despite its clear role in early liver development, the role of Bmp signaling in liver regeneration has not been clearly defined. Current literature provides confounding results that BMP2 (13) and BMP4 (14) negatively while BMP7 (15) positively regulate hepatocyte-driven liver regeneration after partial hepatectomy in rodents. Moreover, there is no report on the role of Bmp signaling in BEC-driven liver regeneration. Given the important role of Bmp signaling in early liver development and its positive effect on regeneration of other organs, including the heart (16), we hypothesized that Bmp signaling might regulate BEC-driven liver regeneration. Our finding that the hepatic expression of several genes implicated in Bmp signaling, including smad5, was upregulated during BEC-driven liver regeneration further supported our hypothesis. In this study, we report that Bmp signaling regulates two distinct steps of BEC-driven liver regeneration: (1) HB-LC differentiation into hepatocytes, and (2) the proliferation of newly-generated BECs.
Experimental procedures
Zebrafish lines
Experiments were performed with approval of the Institutional Animal Care and Use Committee at the University of Pittsburgh. We used smad5m169, tbx2bc144, and id2apt661 mutant and the following transgenic lines: Tg(fabp10a:rasGFP)s942, Tg(Tp1:VenusPEST)s940, Tg(Tp1:H2B-mCherry)s939, Tg(ubb:loxP-EGFP-loxP-mCherry)cz1701, Tg(Tp1:CreERT2)s959, Tg(fabp10a:CFP-NTR)s931, Tg(hs:dnBmpr1)w30, Tg(fabp10a:CreERT2)pt602, Tg(WRE:d2GFP)kyu1, and Tg(Tp1:mAGFP-gmnn)s707. Their full names and references are listed in Table S1.
Hepatocyte ablation and DMH1 treatment
Hepatocyte ablation was performed by treating Tg(fabp10a:CFP-NTR) larvae with 10 mM Mtz in egg water supplemented with 0.2% DMSO and 0.2 mM 1-phenyl-2-thiourea, as previously described (17). To suppress Bmp signaling, 10 μM DMH1 (Tocris, Bristol, UK) was used.
RNAseq analysis
Over 100 livers were manually dissected for each condition (three non-ablating controls at 4.25, 5.25, and 6 days post-fertilization (dpf) and four regenerating livers at A18h, R6h, R12h, and R24h); total RNA was extracted from the dissected livers using the RNeasy Mini Kit (Qiagen, Valencia, CA). This RNA preparation was repeated three times and three-replicate RNA samples were mixed. These mixed samples were processed for single-end deep-transcriptome sequencing using the Illumina HiSeq 2000 platform, of which service was provided from Tufts University Core Facility. Galaxy was used to analyze the sequencing reads.
Additional methods are available in Supporting Information.
Results
Bmp signaling is required for BEC-driven liver regeneration
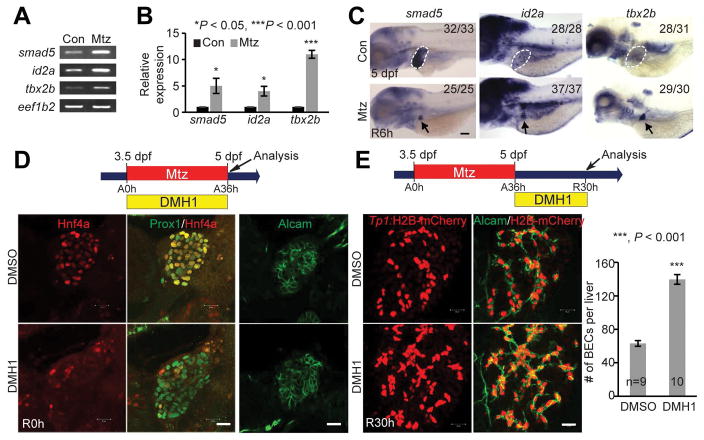
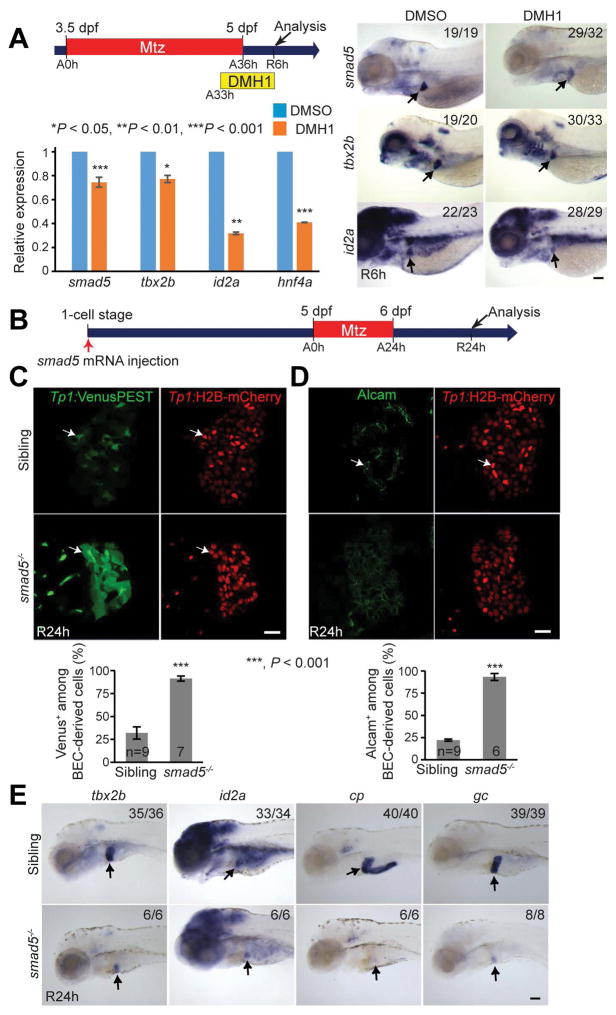
We have established a zebrafish liver injury model in which BECs extensively contribute to hepatocytes (4). Specifically, Tg(fabp10a:CFP-NTR) fish express nitroreductase (NTR) under the hepatocyte-specific fabp10a promoter; the treatment of metronidazole (Mtz), which is converted into a cytotoxic drug by NTR, results in hepatocyte-specific ablation in the transgenic fish. In this model, severe hepatocyte ablation induces the dedifferentiation of BECs into HB-LCs, which then differentiate into hepatocytes, thereby leading to a full liver recovery. To understand the molecular mechanisms underlying BEC-driven liver regeneration, we performed RNAseq analyses and compared gene expression profiles between control and regenerating livers at multiple time points during the regeneration. Through this analysis and subsequent validation with RT-PCR and whole-mount in situ hybridization (WISH) (Fig. S1), we found that genes implicated in the Bmp signaling pathway, such as smad5, id2a, and tbx2b, were upregulated in regenerating livers at regeneration (R) 6h compared with controls (Fig. 1A–1C). Bmp signaling plays an essential role in hepatoblast specification and liver growth during liver development (11, 12), but its role in liver regeneration has not been clearly defined. Thus, to determine whether Bmp signaling was required for BEC-driven liver regeneration, we applied the selective Bmp inhibitor, DMH1, which has been widely used in zebrafish. During BEC-driven liver regeneration, a hepatoblast/hepatocyte marker, Hnf4a, is induced in BECs and the expression of Prox1, a marker for hepatoblasts, hepatocytes, and BECs, is also upregulated in BECs (4). In addition, the expression of Alcam, a good marker of zebrafish BECs (18), is sustained in HB-LCs, but disappears from HB-LCs when these cells differentiate into hepatocytes (4). DMH1 treatment from ablation (A) 0h greatly repressed Hnf4a, but not Prox1 or Alcam, expression, at R0h (Fig. 1D). Intriguingly, DMH1 treatment after hepatocyte ablation (from R0h) significantly increased the number of BECs at R30h, as assessed by Alcam and Tp1:H2B-mCherry expression (Fig. 1E). The Tg(Tp1:H2B-mCherry) line, which expresses stable H2B-mCherry fusion proteins under the Tp1 promoter containing the Notch-responsive element, reveals BECs in the liver (19). Collectively, these data indicate the essential roles of Bmp signaling in BEC-driven liver regeneration.
Figure 1. Bmp signaling is required for BEC-driven liver regeneration.
(A, B) RT-PCR (A) and qPCR (B) data showing the expression levels of smad5, id2a and tbx2b between control and regenerating livers at R6h. (C) WISH images showing the expression of smad5, id2a and tbx2b in control (dashed lines) and regenerating (arrows) livers. Numbers indicate the proportion of larvae exhibiting the expression shown. (D) Single-optical section images showing the expression of Hnf4a and Prox1 or Alcam in regenerating livers. (E) Confocal projection images showing Tp1:H2B-mCherry and Alcam expression in regenerating livers. Quantification of the number of H2B-mCherry/Alcam double-positive cells (i.e., BECs). Scale bars: 150 (C), 20 (D,E) μm; error bars: ±SEM.
Inhibition of Bmp signaling blocks HB-LC differentiation into hepatocytes
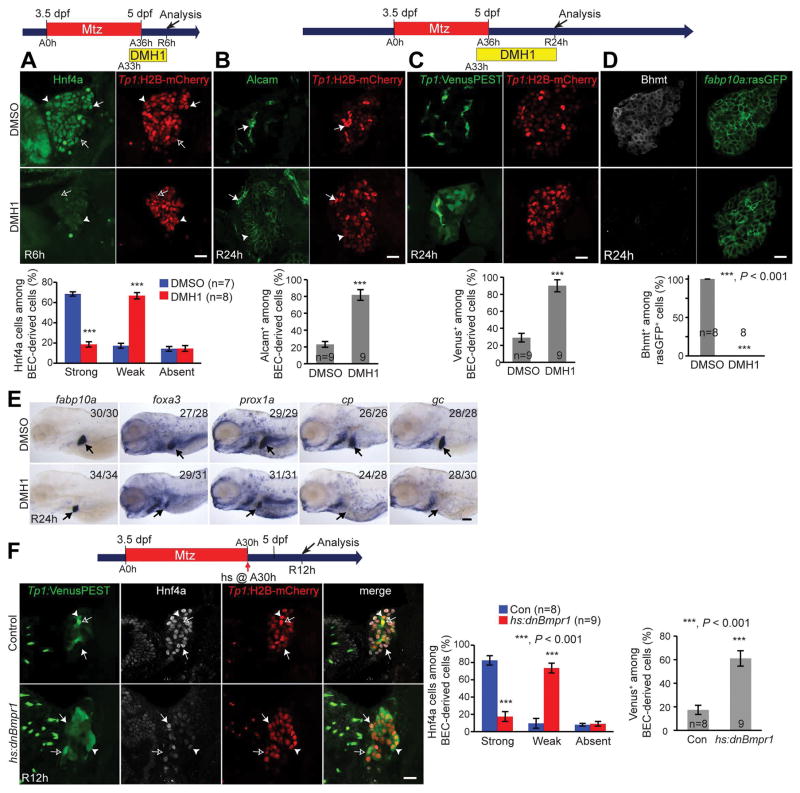
Next, we investigated in detail the effect of Bmp inhibition on earlier stages of BEC-driven liver regeneration. We first determined if Bmp inhibition blocked the dedifferentiation of BECs into HB-LCs. By examining Hnf4a expression at various time points between A24h and A36h, we found that Hnf4a induction in BECs occurred around A33h and that its expression became stronger at A36h (Fig. S2A, arrows). As Hnf4a induction in BECs is indicative of BEC dedifferentiation (4), these data suggest that BECs dedifferentiate into HB-LCs around A33h. This observation was further supported by the expression of fabp10a:rasGFP (Fig. S2B), which is not expressed in hepatoblasts during liver development but induced in BECs during BEC-driven liver regeneration (10). At A33h, there were fabp10a:rasGFP+ cells negative for Hnf4a (Fig. S2C, arrows), but not vice versa, indicating that fabp10a induction in BECs precedes Hnf4a induction. Although Hnf4a expression in BEC-derived cells was greatly reduced at A36h in DMH1-treated regenerating livers compared with their controls (Fig. 1D), the initial induction of Hnf4a and fabp10a:rasGFP in BECs at A33h appeared unaffected in DMH1-treated regenerating livers (Fig. S2A and S2B). These data together with normal Prox1 expression in HB-LCs indicate that Bmp inhibition does not block the dedifferentiation of BECs.
Given that Hnf4a is the master regulator of hepatocyte differentiation (20), the reduced Hnf4a expression upon Bmp inhibition (Fig. 1D) suggests a role for Bmp signaling in HB-LC differentiation into hepatocytes. To address this possibility, we sought to examine DMH1-treated regenerating larvae at later stages, such as R24h. However, the continuous DMH1 treatment from A0h killed most regenerating larvae before R24h, preventing the examination of the regenerating livers at later stages. By treating ablating larvae with DMH1 from different time points, we found that DMH1 treatment from A33h also reduced Hnf4a expression at R6h (Fig. 2A) but did not kill regenerating larvae as late as R24h. At R24h, A33h DMH1-treated regenerating livers exhibited the following phenotypes: (1) sustained Alcam expression in most BEC-derived Tp1:H2B-mCherry+ cells (Fig. 2B); (2) sustained Notch activity, as revealed by Tp1:VenusPEST expression (21), throughout regenerating livers, (Fig. 2C); and (3) no expression of hepatocyte markers, Bhmt, cp, and gc (Fig. 2D and 2E), indicating a defect in HB-LC differentiation into hepatocytes. Despite a lack of hepatocyte marker expression, the expression of foxa3 and prox1a, which are expressed in hepatoblasts, was unaffected in DMH1-treated regenerating livers (Fig. 2E). fabp10a, which is expressed in HB-LCs (Fig. S2B), expression was also unaffected (Fig. 2D and 2E). In addition to the pharmacologic inhibition, we blocked Bmp signaling using the Tg(hs:dnBmpr1) line that expresses dominant-negative Bmpr1 under the hsp70l heat-shock promoter. The overexpression of dnBmpr1 via a single heat-shock at A30h resulted in sustained Notch activity and reduced Hnf4a expression in regenerating livers at R12h (Fig. 2F). All these data indicate the role of Bmp signaling in HB-LC differentiation into hepatocytes.
Figure 2. Bmp inhibition blocks HB-LC differentiation into hepatocytes.
(A) Single-optical section images showing Hnf4a and Tp1:H2B-mCherry expression in regenerating livers. To quantify Hnf4a expression, BEC-derived H2B-mCherry+ cells were divided into three cases: Hnf4astrong (arrows), Hnf4aweak (arrowheads), and Hnf4aabsent (open arrows). Quantification of the percentage of these three cell types among H2B-mCherry+ cells. (B) Single-optical section images showing Alcam and Tp1:H2B-mCherry expression in regenerating livers. In DMH1-treated regenerating livers, Alcam was expressed in both H2B-mCherrystrong (arrows) and H2B-mCherryweak cells (arrowheads), whereas in control regenerating livers, Alcam was expressed only in H2B-mCherrystrong cells. Quantification of the percentage of Alcam+ cells among H2B-mCherry+ cells. (C) Single-optical section images showing the expression of Tp1:VenusPEST and Tp1:H2B-mCherry in regenerating livers. Quantification of the percentage of VenusPEST+ cells among H2B-mCherry+ cells. (D) Single-optical section images showing Bhmt and fabp10a:rasGFP expression in regenerating livers. Quantification of the percentage of Bhmt+ cells among BEC-derived fabp10a:rasGFP+ cells. (E) WISH images showing the expression of fabp10a, foxa3, prox1a, cp and gc in regenerating livers (arrows). (F) Single-optical section images showing Hnf4a, Tp1:VenusPEST, and Tp1:H2B-mCherry expression in regenerating livers. The Tg(hs:dnBmpr1-GFP) line was used to block Bmp signaling via a single heat-shock at A30h. Quantification of the percentage of Hnf4astrong (arrows), Hnf4aweak (arrowheads), and Hnf4aabsent (open arrows) cells and of VenusPEST+ cells among H2B-mCherry+ cells. Scale bars: 20 (A–D, F), 150 (E) μm; error bars: ±SEM.
Inhibition of Bmp signaling maintains HB-LCs in their undifferentiated state
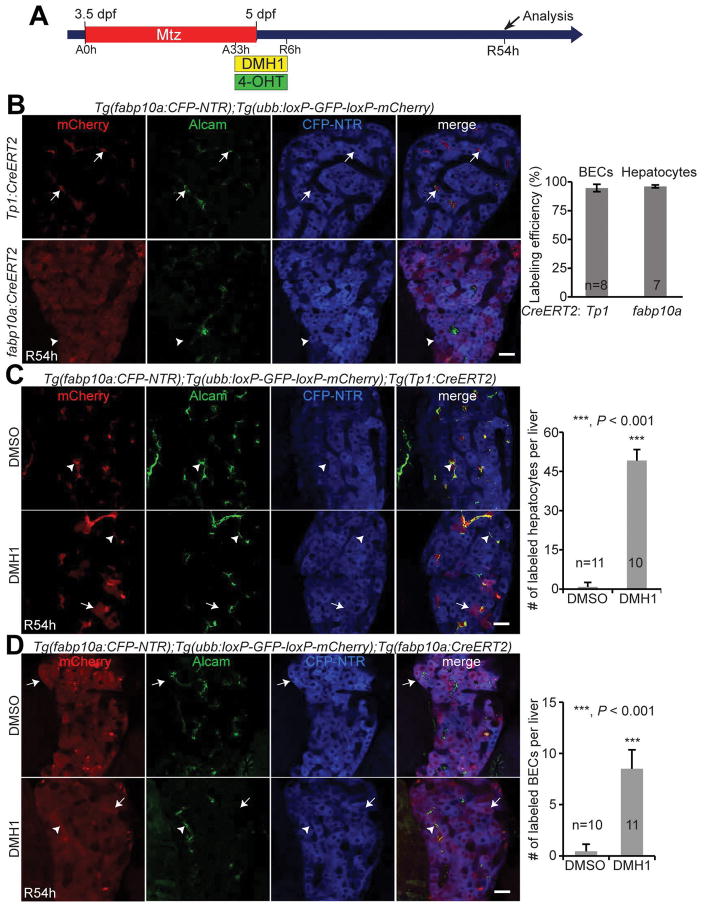
Based on sustained Notch activity and Alcam expression in A33h DMH1-treated regenerating livers, we hypothesized that HB-LCs that failed to differentiate into hepatocytes remained as undifferentiated HB-LCs in DMH1-treated regenerating livers. To test this hypothesis, we used Cre/loxP-mediated lineage tracing and determined the lineages of HB-LCs. To label HB-LCs, we used two Cre lines, Tg(Tp1:CreERT2) that expresses CreERT2 under the Tp1 promoter (22) and Tg(fabp10a:CreERT2) that expresses CreERT2 under the fabp10a promoter (4). Since Notch activity is strong in BECs but weak in HB-LCs, the former line labels most BECs but a few HB-LCs. Likewise, since fabp10a expression is strong in hepatocytes but weak in HB-LCs, the latter line labels most hepatocytes but a few HB-LCs. When Tp1:CreERT2 and fabp10a:CreERT2 were activated by tamoxifen (4-OHT) treatment from A33h to R6h, 95% of BECs and 96% of hepatocytes, respectively, were labeled at R54h (Fig. 3B). However, few hepatocytes were labeled by the Tp1:CreERT2 activation (Fig. 3C) and few BECs were labeled by the fabp10a:CreERT2 activation (Fig. 3D), indicating that few HB-LCs were labeled in these Cre activation settings. This low efficiency of HB-LC labeling can be explained by the weak Notch activity and weak fabp10a expression in HB-LCs and the short duration of HB-LCs in the zebrafish liver injury model. If the HB-LCs fail to differentiate and remain in a progenitor-like state, more HB-LCs will be labeled in the Cre activation settings, thereby more hepatocytes and BECs were labeled by Tp1:CreERT2 and fabp10a:CreERT2, respectively. We observed these phenomena in regenerating livers treated with DMH1 from A33h to R6h (Fig. 3C and 3D), strongly suggesting that Bmp inhibition maintains HB-LCs in their undifferentiated state.
Figure 3. Bmp inhibition maintains HB-LCs as undifferentiated.
(A) Experimental scheme illustrating the stages of Mtz, DMH1, and 4-OHT treatment. (B) Labeling efficiency of the lineage tracing with the Tg(Tp1:CreERT2) and Tg(fabp10a:CreERT2) lines. Arrows point to CFP−/mCherry+/Alcam+ cells (Cre-labeled BECs); arrowheads point to CFP+/mCherry+/Alcam− cells (Cre-labeled hepatocytes). Quantification of the percentage of the labeled BECs and hepatocytes. (C, D) Single-optical section images showing the expression of mCherry, Alcam, and fabp10a:CFP-NTR in regenerating livers at R54h. A Cre reporter line, Tg(ubb:loxP-GFP-loxP-mCherry), was used together with the Tg(Tp1:CreERT2) (C) or the Tg(fabp10a:CreERT2) (D) line. Arrows point to CFP+/mCherry+/Alcam− hepatocytes; arrowheads point to CFP−/mCherry+/Alcam+ BECs. Quantification of the numbers of CFP+/mCherry+/Alcam− hepatocytes and CFP−/mCherry+/Alcam+ BECs per liver. Scale bars: 20 μm; error bars: ±SEM.
Inhibition of Bmp signaling after hepatocyte ablation increases BEC number in regenerating livers via proliferation
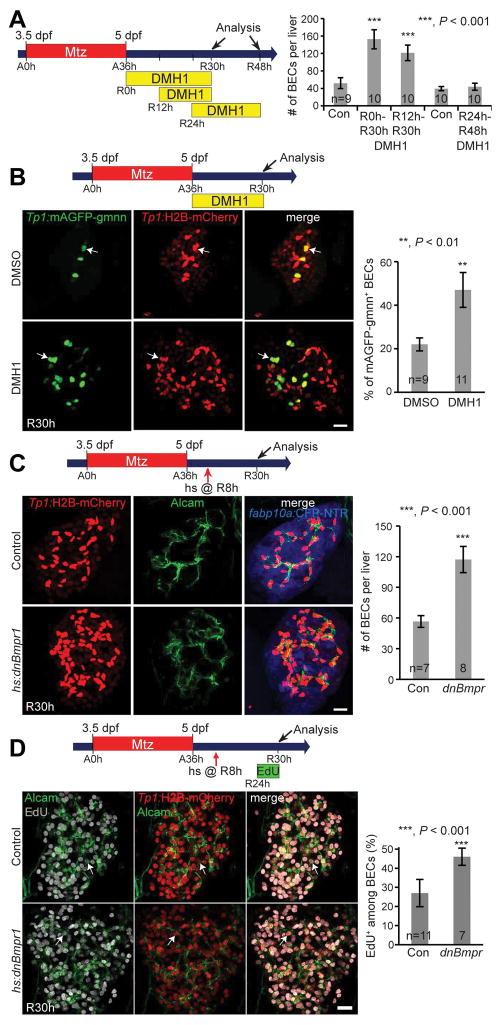
Unlike its treatment from A33h, DMH1 treatment from R0h did not affect HB-LC differentiation into hepatocytes, as assessed by cp and gc expression (Fig. S3). Given the increased number of BECs upon DMH1 treatment from R0h (Fig. 1E), we determined the latest time point from which DMH1 treatment still increased BEC number. We found that DMH1 treatment from R12h, but not R24h, significantly increased BEC number (Fig. 4A). Using a cell-cycle reporter line, Tg(Tp1:mAGFP-gmnn), which reveals BECs in the S/G2/M (but not G0/G1) phases of the cell cycle, we observed that BEC proliferation was significantly increased in DMH1-treated regenerating livers at R30h compared with their controls (Fig. 4B). In addition to the chemical inhibition of Bmp signaling, the overexpression of dnBmpr1 via a single heat-shock at R8h increased BEC proliferation in regenerating livers at R30h (Fig. 4C and 4D). Altogether, these data indicate that Bmp signaling controls the number of BECs in regenerating livers by temporarily suppressing their proliferation.
Figure 4. Bmp inhibition after hepatocyte ablation increases BEC number in regenerating livers via proliferation.
(A) Experimental scheme illustrating the stages of Mtz and DMH1 treatment. Quantification of the total numbers of BECs in regenerating livers treated with DMH1 for three different time-windows. (B) Confocal projection images showing Tp1:H2B-mCherry and Tp1:mAGFP-gmnn expression in regenerating livers. Arrows point to mCherry/mAGFP-gmnn double-positive cells. Quantification of the percentage of mAGFP-gmnn+ cells among H2B-mCherry+ BECs. (C) Confocal projection images showing the expression of Alcam, Tp1:H2B-mCherry, and fabp10a:CFP-NTR in regenerating livers at R30h. Quantification of the total numbers of BECs (H2B-mCherry+/Alcam+) per liver. (D) Confocal projection images showing EdU labeling and Alcam and Tp1:H2B-mCherry expression in regenerating livers. Arrows point to EdU+ BECs. Quantification of the percentage of EdU+ cells among BECs. Scale bars: 20 μm; error bars: ±SEM.
smad5 mutants exhibit a defect in HB-LC differentiation into hepatocytes
Given the upregulation of genes implicated in Bmp signaling, such as smad5, tbx2b, and id2a, in regenerating livers (Fig. 1A–1C), we determined if DMH1 treatment suppressed this upregulation. Indeed, quantitative RT-PCR (qPCR) and WISH showed a reduction of the hepatic expression of smad5, tbx2b, and id2a in DMH1-treated regenerating livers at R6h compared with their controls (Fig. 5A), suggesting a potential role for these genes in mediating Bmp signaling during BEC-driven liver regeneration. To test this possibility, we examined BEC-driven liver regeneration in smad5 mutants. Smad5, together with Smad1 and Smad9, known as receptor-regulated Smads (R-Smads), relays Bmp signaling from the cell surface to the nucleus (23). Zebrafish smad5−/− mutants started to die from 3 dpf but smad5 mRNA injection into one-cell-stage embryos increased their survival time (24), allowing for our liver regeneration assay. Although the mRNA-injected mutants had a smaller liver than their siblings, liver development occurred, as observed by their liver growth (Fig. S4B) and the branching of the intrahepatic biliary structure (Fig. S4C). Since liver size was smaller in the mRNA-injected mutants than in wild-type, we applied Mtz from 5 dpf, when the mutant liver size is similar to that of 3.5-dpf wild-type liver. In this rescue setting, the smad5−/− mutants exhibited sustained Notch activity and Alcam expression throughout regenerating livers (Fig. 5B–5D) and no cp, but faint gc, expression (Fig. 5E) at R24h, recapitulating the HB-LC differentiation defects observed in A33h DMH1-treated regenerating livers. The expression of tbx2b and id2a in regenerating livers also appeared to be reduced in the smad5−/− mutants compared with their siblings (Fig. 5E), further supporting that Bmp signaling regulates tbx2b and id2a expression in regenerating livers. When examined at R48h, continuous DMH1 treatment from A33h was fatal for the zebrafish larvae recovering from severe liver injury, preventing the analysis of BEC-driven liver regeneration at later stages. However, the smad5−/− mutants recovered at R48h from the initial regeneration defects, as displayed by the rapid growth of their regenerating livers (Fig. S4E), normal Notch activity (Fig. S4F), and recovered cp and gc expression (Fig. S4G). Altogether, data from smad5 mutant analyses indicate the role of smad5 in BEC-driven liver regeneration.
Figure 5. smad5 mutants exhibit a defect in HB-LC differentiation into hepatocytes.
(A) qPCR data showing the relative expression levels of smad5, tbx2b, id2a and hnf4a between DMSO- and DMH1-treated regenerating livers at R6h; WISH images showing the expression of smad5, tbx2b and id2a in regenerating livers (arrows). (B) Experimental scheme illustrating the stages of smad5 mRNA injection (red arrow) and Mtz treatment for C–E. (C) Single-optical section images showing Tp1:VenusPEST and Tp1:H2B-mCherry expression in regenerating livers. Quantification of the percentage of VenusPEST+ cells among BEC-derived H2B-mCherry+ cells. (D) Single-optical section images showing Alcam and Tp1:H2B-mCherry expression in regenerating livers. Quantification of the percentage of Alcam+ cells among BEC-derived H2B-mCherry+ cells. (E) WISH images showing the expression of tbx2b, id2a, cp and gc in regenerating livers (arrows). Scale bars: 150 (A, E), 20 (C, D) μm; error bars: ±SEM.
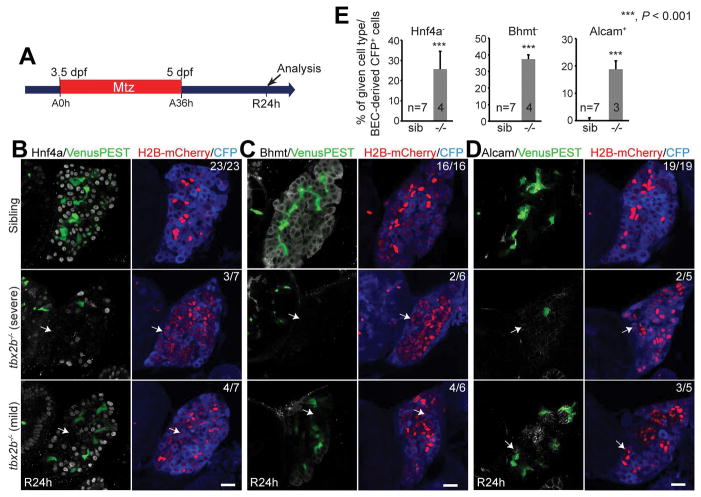
tbx2b mutants exhibit a defect in HB-LC differentiation into hepatocytes
TBX3, a T-box transcription factor, promotes hepatoblast proliferation and maintains the expression of HNF4A and CEBPA, two key transcription factors regulating hepatocyte differentiation, in hepatoblasts (25). TBX2, a close homolog of TBX3, is required for the development of the heart, pharyngeal arch, and optic cup; importantly, its expression in these tissues is regulated by Bmp signaling (26). Thus, to determine if tbx2b was required for BEC-driven liver regeneration, we used zebrafish tbx2b mutants because they develop with normal body morphology (Fig. S5A) and survive long enough for our liver regeneration assay (27). Despite the absence of liver developmental defects (Fig. S5B), tbx2b−/− mutants exhibited a defect in HB-LC differentiation into hepatocytes during BEC-driven liver regeneration. In control regenerating livers, all Tp1:VenusPEST−/Tp1:H2B-mCherry+ cells expressed Hnf4a and Bhmt at R24h, indicating that they are hepatocytes derived from BECs. However, in tbx2b−/− regenerating livers, many Tp1:VenusPEST−/Tp1:H2B-mCherry+ cells not only failed to express Hnf4a and Bhmt (Fig. 6A–6C), but also showed sustained Alcam expression at R24h (Fig. 6D). This phenotype was severe in ~40% of the mutants, having few Bhmt+ hepatocytes, but mild in the rest of them, having a significant number of the hepatocytes (Fig. 6B–6D). To determine whether the observed phenotype at R24h later recovered, we examined regenerating livers at R48h. All mutants at R48h still contained a significant number of Tp1:VenusPEST−/Tp1:H2B-mCherry+ cells negative for Hnf4a (Fig. S5C), suggesting that in the absence of Tbx2b, BEC-derived cells that failed to differentiate into hepatocytes at R24h do not recover at a later time point. We next examined if BEC dedifferentiation was affected in tbx2b−/− mutants. Although Hnf4a expression in regenerating livers at R6h was reduced in the mutants compared with their siblings, fabp10a:rasGFP expression appeared unaffected in the mutants (Fig. S5D), suggesting that BECs dedifferentiate into HB-LCs in the absence of Tbx2b. Altogether, data from tbx2b mutant analyses indicate the crucial role of tbx2b in BEC-driven liver regeneration, in particular, HB-LC differentiation into hepatocytes.
Figure 6. tbx2b mutants exhibit a defect in HB-LC differentiation into hepatocytes.
(A) Experimental scheme illustrating the stage of Mtz treatment and analysis. (B–D) Single-optical section images showing the expression of fabp10a:CFP-NTR, Tp1:VenusPEST, Tp1:H2B-mCherry, and Hnf4a (B), Bhmt (C) or Alcam (D) in regenerating livers. (E) Quantification of the percentage of Hnf4a−, Bhmt−, or Alcam+ cells (arrows) among CFP-NTR+/H2B-mCherry+ cells as shown in B–D. The mild cases were used for quantification. Scale bars: 20 μm; error bars: ±SEM.
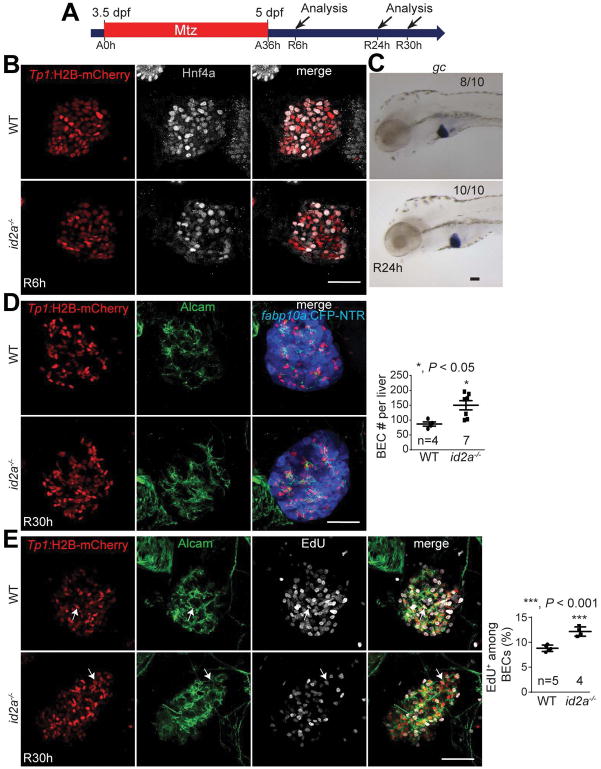
id2a mutants temporarily display an excess of BECs in regenerating livers
Id2 is a well-known downstream target gene of Bmp signaling in many tissues, including pancreatic epithelia (28); the mouse Id2 promoter contains BMP-responsive elements (29). id2a, the zebrafish orthologue of mouse Id2, is also regulated by Bmp signaling in cranial neural crest cells (30). Not only are Id2 and id2a the direct targets of Bmp signaling, but they also mediate the effect of Bmp signaling in these tissues. Given its BEC-specific expression in the developing liver (31) and its regulation by Bmp signaling in regenerating livers (Fig. 5A), we hypothesized that id2a served as a mediator of Bmp signaling in BEC-driven liver regeneration. Using transcription activator-like effector nuclease (TALEN) genome editing technology, we generated id2a mutants containing a 22-bp deletion in its first exon. The mutant larvae did not exhibit any liver developmental defects (Fig. S6A and S6B) and grew normally to adults. BEC-driven liver regeneration appeared unaffected in id2a−/− mutants, as assessed by Hnf4a expression at R6h (Fig. 7B) and gc expression at R24h (Fig. 7C); however, the mutant regenerating livers had significantly more BECs than controls at R30h (Fig. 7D), resembling the excessive BEC phenotype seen in regenerating livers treated with DMH1 from R0h. This increased BEC number was due to increased proliferation, as revealed by EdU labeling (Fig. 7E). However, this BEC phenotype was temporary because there was no difference in BEC number between wild-type and id2a−/− mutant regenerating livers at R72h (Fig. S6C). Altogether, data from id2a mutant analyses suggest that id2a mediates, in part, the effect of Bmp signaling on BEC proliferation during BEC-driven liver regeneration.
Figure 7. id2a mutants have excessive BECs in regenerating livers.
(A) Experimental scheme illustrating the stages of Mtz treatment and analysis. (B) Single-optical section images showing Hnf4a and Tp1:H2B-mCherry expression in regenerating livers. (C) WISH images showing gc expression in regenerating livers. (D) Confocal projection images showing the expression of Alcam, Tp1:H2B-mCherry, and fabp10a:CFP-NTR in regenerating livers. Quantification of the total numbers of BECs (Alcam+/H2B-mCherry+) per liver. (E) Confocal projection images showing EdU labeling and Alcam and Tp1:H2B-mCherry expression in regenerating livers. EdU was treated from R24h for 6 hours. Arrows point to EdU+ BECs. Quantification of the percentage of EdU+ cells among BECs. Scale bars: 50 μm; error bars: ±SEM.
BMP2 addition promotes the differentiation of a murine liver progenitor cell line into hepatocytes in vitro
To explore whether the findings from the zebrafish liver injury model can be translated to mammals, we used a murine liver progenitor cell line that was established from DDC diet-fed mice (32). These cells can efficiently differentiate into hepatocytes or BECs depending on culture conditions (32). In the hepatocyte differentiation condition, addition of BMP2 significantly increased the expression of three hepatocyte markers, G6pc, Tat, and Tdo2 (Fig. 8), consistent with the findings in zebrafish.
Figure 8. BMP2 treatment promotes the differentiation of a murine LPC cell line into hepatocytes in vitro.
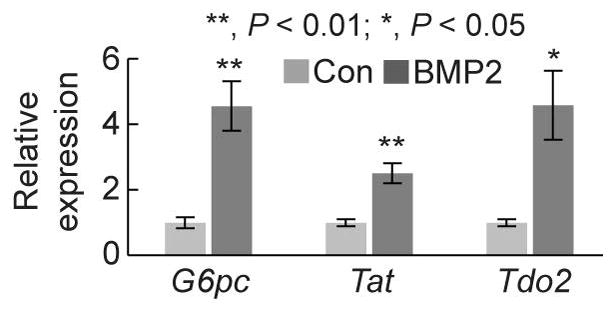
qPCR data showing the relative expression levels of G6pc, Tat, and Tdo2 between control and BMP2 treatment (n=4). Error bars: ±SEM.
Discussion
Using the zebrafish hepatocyte ablation model, we elucidate two distinct roles of Bmp signaling in BEC-driven liver regeneration: (1) initially, Bmp signaling regulates HB-LC differentiation into hepatocytes; (2) later, Bmp signaling controls the proliferation of newly-generated BECs. By analyzing the mutants of smad5, tbx2b, and id2a, genes involved in the Bmp signaling pathway, we discovered that Smad5 is the main receptor-regulated Smad during BEC-driven liver regeneration and that Tbx2b and Id2a mediate its effect on HB-LC differentiation into hepatocytes and on BEC proliferation, respectively.
Bmp (11, 12), Fgf (12, 33), and Wnt/β-catenin signaling (34) are implicated in hepatoblast specification and proliferation during liver development. Given that regeneration often recapitulates development and key developmental factors are re-utilized during regeneration, these signaling pathways may also regulate liver regeneration. During hepatocyte-driven liver regeneration, both Wnt/β-catenin (35) and Fgf signaling (36) promote hepatocyte proliferation. During BEC-driven liver regeneration, Wnt/β-catenin signaling promotes oval cell proliferation (37) and Fgf signaling induces oval cell activation (8). In contrast to Wnt/β-catenin and Fgf signaling, the influence of Bmp signaling on hepatocyte proliferation during hepatocyte-driven liver regeneration is ligand-dependent: BMP4 signaling through Alk3 represses hepatocyte proliferation, whereas BMP7 signaling through Alk2 promotes hepatocyte proliferation (14). Previous in vitro studies have reported on the role of BMP signaling in the differentiation of LPCs into hepatocytes: BMP4 treatment induced the differentiation of rat hepatic progenitor cells (38) and CD133+ hepatic cancer stem cells (39) into hepatocytes. Our in vitro and in vivo data support these findings and further reveal the crucial role of Bmp signaling in BEC-driven liver regeneration, specifically during the differentiation phase of LPCs into hepatocytes.
To further elucidate the mechanisms by which Bmp signaling controls BEC-driven liver regeneration, we analyzed tbx2b and id2a mutants. Our RNAseq data revealed that at R6h, both tbx2b (among the T-box transcription factor genes) and id2a (among the Id family genes) were highly upregulated in regenerating livers compared with the other tbx and id genes (Fig. S7A and S7B), suggesting their importance in BEC-driven liver regeneration. As expected, both tbx2b and id2a expression decreased in both DMH1-treated and smad5−/− regenerating livers. Importantly, tbx2b−/− mutants exhibited a defect in HB-LC differentiation into hepatocytes and id2a mutants exhibited excessive BECs in regenerating livers at R30h, revealing tbx2b and id2a as the key downstream mediators of Bmp signaling that regulates BEC-driven liver regeneration.
Using a morpholino-mediated knockdown approach, we previously reported that during liver development, Id2a positively regulates hepatic outgrowth through hepatoblast proliferation and survival (31). However, we found that during BEC-driven liver regeneration, Id2a negatively regulates the proliferation of newly-generated BECs. Although we did not detect any developmental defects in the id2a−/− mutants, inconsistent with the morpholino results, this discrepancy might be explained by the genetic compensations that can occur in mutants but not in morphants (40). Previous rodent studies have highlighted the role of Id1 and Id2 in hepatocyte-driven liver regeneration. Following partial hepatectomy, not only Id1 was upregulated in liver sinusoidal endothelial cells (LSECs), but Id1−/− mice displayed abnormal liver function and impaired liver regeneration (41). In fact, following acute liver injury, Id1 was induced downstream of LSEC-specific upregulation of CXCR4/CXCR7 (42). As a result, ID1 promotes the secretion of pro-regenerative angiocrine factors, such as Wnt2 and HGF, to aid in hepatocyte proliferation after liver injury (41, 42). Similar to Id1, Id2 levels also increased after partial hepatectomy and bile duct ligation in rats (43), but its role in liver regeneration has not yet been reported.
While Wnt/β-catenin signaling promotes LPC differentiation into hepatocytes, Notch signaling represses the process (44, 45). Here, we presented Bmp signaling as another regulator of this differentiation process. Since we did not observe any changes in Wnt activity between DMSO- and DMH1-treated regenerating livers (Fig. S8), it appears that Bmp signaling does not affect Wnt/β-catenin signaling. However, sustained, weak Notch activity observed in DMH1-treated regenerating livers suggests that Bmp signaling may suppress Notch signaling. Although it was reported that repression of Notch signaling promotes LPC differentiation into hepatocytes (44, 45), the hepatocyte differentiation defects observed in DMH1-treated livers were not rescued by inhibiting Notch signaling globally (data not shown). Thus, it is unlikely that Bmp signaling controls LPC differentiation through Wnt/β-catenin or Notch signaling. However, we do not exclude the possibility that Wnt/β-catenin and Notch signaling control LPC differentiation through Bmp signaling.
The zebrafish hepatocyte ablation model has contributed significantly to a better understanding of BEC- or LPC-driven liver regeneration. We previously reported that Wnt/β-catenin signaling via Wnt2bb regulates the proliferation of newly-generated hepatocytes (4) and that BET proteins regulate BEC dedifferentiation and the proliferation and maturation of newly-generated hepatocytes (10). Using the same model, others also reported on the involvement of Notch signaling in BEC-driven liver regeneration (46). Here, we provide the gene expression profiles of regenerating livers at four distinct stages, A18h, R6h, R12h, and R24h. Although we focus on Bmp signaling and its downstream target genes in this study, these expression profiles will be useful for identifying other crucial genes involved in regulating BEC-driven liver regeneration.
In summary, we discovered that Bmp signaling regulates HB-LC differentiation into hepatocytes, partly, via Tbx2b and controls the proliferation of newly-generated BECs via Id2a. Liver biopsies of human patients suffering from chronic liver diseases display an abundance of oval cells, also known as HB-LCs. To promote the differentiation of these oval cells into hepatocytes in these patients, one possible therapy may involve enhancing hepatic Bmp signaling.
Supplementary Material
Acknowledgments
Financial support: The work was supported in part by grants from the NIH to D.S. (DK101426), M.K. (DK105714) and D.Y.R.S. (DK60322), from the Packard foundation to D.Y.R.S., and from the Japan Society for the Promotion of Science to M.T. (2611007).
We thank the Joung and Peterson labs for id2a TALEN constructs, which were created as part of an effort funded by an NIH grant (GM088040). We also thank Joshua Gamse for tbx2b and Mary Mullins for smad5 mutant fish, and Neil Hukriede and Michael Tsang for discussions.
List of abbreviations
- BEC
biliary epithelial cell
- LPCs
liver progenitor cells
- HB-LCs
hepatoblast-like cells
- BET
bromodomain extraterminal
- Mtz
metronidazole
- dpf
days post-fertilization
- NTR
nitroreductase
- WISH
whole-mount in situ hybridization
- 4-OHT
tamoxifen
- qPCR
quantitative RT-PCR
- TALEN
transcription activator-like effector nuclease
- LSECs
liver sinusoidal endothelial cells
References
Author names in bold designate shared co-first authorship.
- 1.Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miyajima A, Tanaka M, Itoh T. Stem/Progenitor Cells in Liver Development, Homeostasis, Regeneration, and Reprogramming. Cell Stem Cell. 2014;14:561–574. doi: 10.1016/j.stem.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Greenbaum LE. The Ductal Plate: A Source of Progenitors and Hepatocytes in the Adult Liver. Gastroenterology. 2011;141:1152–1155. doi: 10.1053/j.gastro.2011.08.023. [DOI] [PubMed] [Google Scholar]
- 4.Choi TY, Ninov N, Stainier DY, Shin D. Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology. 2014;146:776–788. doi: 10.1053/j.gastro.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He J, Lu H, Zou Q, Luo L. Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology. 2014;146:789–800. e788. doi: 10.1053/j.gastro.2013.11.045. [DOI] [PubMed] [Google Scholar]
- 6.Lu W-Y, Bird TG, Boulter L, Tsuchiya A, Cole AM, Hay T, Guest RV, et al. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nature Cell Biology. 2015;17:971–983. doi: 10.1038/ncb3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowes KN, Brennan BA, Yeoh GC, Olynyk JK. Oval cell numbers in human chronic liver diseases are directly related to disease severity. American Journal of Pathology. 1999;154:537–541. doi: 10.1016/S0002-9440(10)65299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takase HM, Itoh T, Ino S, Wang T, Koji T, Akira S, Takikawa Y, et al. FGF7 is a functional niche signal required for stimulation of adult liver progenitor cells that support liver regeneration. Genes Dev. 2013;27:169–181. doi: 10.1101/gad.204776.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bird TGT, Lu WW-Y, Boulter L, Gordon-Keylock S, Ridgway Ra, Williams MJ, Taube J, et al. Bone marrow injection stimulates hepatic ductular reactions in the absence of injury via macrophage-mediated TWEAK signaling. Proc Natl Acad Sci U S A. 2013;110:6542–6547. doi: 10.1073/pnas.1302168110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ko S, Choi TY, Russell JO, So J, Monga SPS, Shin D. Bromodomain and extraterminal (BET) proteins regulate biliary-driven liver regeneration. J Hepatol. 2016;64:316–325. doi: 10.1016/j.jhep.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossi JM, Dunn NR, Hogan BL, Zaret KS. Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev. 2001;15:1998–2009. doi: 10.1101/gad.904601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin D, Shin CH, Tucker J, Ober EA, Rentzsch F, Poss KD, Hammerschmidt M, et al. Bmp and Fgf signaling are essential for liver specification in zebrafish. Development. 2007;134:2041–2050. doi: 10.1242/dev.000281. [DOI] [PubMed] [Google Scholar]
- 13.Xu CP, Ji WM, van den Brink GR, Peppelenbosch MP. Bone morphogenetic protein-2 is a negative regulator of hepatocyte proliferation downregulated in the regenerating liver. World Journal of Gastroenterology. 2006;12:7621–7625. doi: 10.3748/wjg.v12.i47.7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Do N, Zhao R, Ray K, Ho K, Dib M, Ren X, Kuzontkoski P, et al. BMP4 is a novel paracrine inhibitor of liver regeneration. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1220–1227. doi: 10.1152/ajpgi.00105.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugimoto H, Yang CQ, LeBleu VS, Soubasakos MA, Giraldo M, Zeisberg M, Kalluri R. BMP-7 functions as a novel hormone to facilitate liver regeneration. Faseb Journal. 2007;21:256–264. doi: 10.1096/fj.06-6837com. [DOI] [PubMed] [Google Scholar]
- 16.Wu CC, Kruse F, Vasudevarao MD, Junker JP, Zebrowski DC, Fischer K, Noel ES, et al. Spatially Resolved Genome-wide Transcriptional Profiling Identifies BMP Signaling as Essential Regulator of Zebrafish Cardiomyocyte Regeneration. Dev Cell. 2016;36:36–49. doi: 10.1016/j.devcel.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 17.Choi TY, Khaliq M, Ko S, So J, Shin D. Hepatocyte-specific ablation in zebrafish to study biliary-driven liver regeneration. J Vis Exp. 2015:e52785. doi: 10.3791/52785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakaguchi TF, Sadler KC, Crosnier C, Stainier DY. Endothelial signals modulate hepatocyte apicobasal polarization in zebrafish. Curr Biol. 2008;18:1565–1571. doi: 10.1016/j.cub.2008.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delous M, Yin C, Shin D, Ninov N, Debrito Carten J, Pan L, Ma TP, et al. Sox9b is a key regulator of pancreaticobiliary ductal system development. PLoS Genet. 2012;8:e1002754. doi: 10.1371/journal.pgen.1002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Dev Cell. 2010;18:175–189. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Ninov N, Borius M, Stainier DYR. Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development. 2012;139:1557–1567. doi: 10.1242/dev.076000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ninov N, Hesselson D, Gut P, Zhou A, Fidelin K, Stainier DYR. Metabolic Regulation of Cellular Plasticity in the Pancreas. Current Biology. 2013;23:1242–1250. doi: 10.1016/j.cub.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massague J, Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer C, Mayr T, Nowak M, Schumacher J, Runke G, Bauer H, Wagner DS, et al. Maternally supplied Smad5 is required for ventral specification in zebrafish embryos prior to zygotic Bmp signaling. Dev Biol. 2002;250:263–279. [PubMed] [Google Scholar]
- 25.Ludtke TH, Christoffels VM, Petry M, Kispert A. Tbx3 promotes liver bud expansion during mouse development by suppression of cholangiocyte differentiation. Hepatology. 2009;49:969–978. doi: 10.1002/hep.22700. [DOI] [PubMed] [Google Scholar]
- 26.Shirai M, Imanaka-Yoshida K, Schneider MD, Schwartz RJ, Morisaki T. T-box 2, a mediator of Bmp-Smad signaling, induced hyaluronan synthase 2 and Tgfbeta2 expression and endocardial cushion formation. Proc Natl Acad Sci U S A. 2009;106:18604–18609. doi: 10.1073/pnas.0900635106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snelson CD, Santhakumar K, Halpern ME, Gamse JT. Tbx2b is required for the development of the parapineal organ. Development. 2008;135:1693–1702. doi: 10.1242/dev.016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hua H, Zhang YQ, Dabernat S, Kritzik M, Dietz D, Sterling L, Sarvetnick N. BMP4 regulates pancreatic progenitor cell expansion through Id2. J Biol Chem. 2006;281:13574–13580. doi: 10.1074/jbc.M600526200. [DOI] [PubMed] [Google Scholar]
- 29.Nakahiro T, Kurooka H, Mori K, Sano K, Yokota Y. Identification of BMP-responsive elements in the mouse Id2 gene. Biochem Biophys Res Commun. 2010;399:416–421. doi: 10.1016/j.bbrc.2010.07.090. [DOI] [PubMed] [Google Scholar]
- 30.Das A, Crump JG. Bmps and Id2a Act Upstream of Twist1 To Restrict Ectomesenchyme Potential of the Cranial Neural Crest. PLoS Genet. 2012:8. doi: 10.1371/journal.pgen.1002710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khaliq M, Choi TY, So J, Shin D. Id2a is required for hepatic outgrowth during liver development in zebrafish. Mech Dev. 2015;138(Pt 3):399–414. doi: 10.1016/j.mod.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okabe M, Tsukahara Y, Tanaka M, Suzuki K, Saito S, Kamiya Y, Tsujimura T, et al. Potential hepatic stem cells reside in EpCAM(+) cells of normal and injured mouse liver. Development. 2009;136:1951–1960. doi: 10.1242/dev.031369. [DOI] [PubMed] [Google Scholar]
- 33.Jung J, Zheng M, Goldfarb M, Zaret KS. Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science. 1999;284:1998–2003. doi: 10.1126/science.284.5422.1998. [DOI] [PubMed] [Google Scholar]
- 34.Ober EA, Verkade H, Field HA, Stainier DY. Mesodermal Wnt2b signalling positively regulates liver specification. Nature. 2006;442:688–691. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- 35.Nejak-Bowen KN, Thompson MD, Singh S, Bowen WC, Jr, Dar MJ, Khillan J, Dai C, et al. Accelerated liver regeneration and hepatocarcinogenesis in mice overexpressing serine-45 mutant beta-catenin. Hepatology. 2010;51:1603–1613. doi: 10.1002/hep.23538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Padrissa-Altes S, Bachofner M, Bogorad RL, Pohlmeier L, Rossolini T, Bohm F, Liebisch G, et al. Control of hepatocyte proliferation and survival by Fgf receptors is essential for liver regeneration in mice. Gut. 2015;64:1444–1453. doi: 10.1136/gutjnl-2014-307874. [DOI] [PubMed] [Google Scholar]
- 37.Apte U, Thompson MD, Cui SS, Liu B, Cieply B, Monga SPS. Wnt/beta-catenin signaling mediates oval cell response in rodents. Hepatology. 2008;47:288–295. doi: 10.1002/hep.21973. [DOI] [PubMed] [Google Scholar]
- 38.Fan J, Shen H, Dai Q, Minuk GY, Burzynski FJ, Gong Y. Bone morphogenetic protein-4 induced rat hepatic progenitor cell (WB-F344 cell) differentiation toward hepatocyte lineage. J Cell Physiol. 2009;220:72–81. doi: 10.1002/jcp.21731. [DOI] [PubMed] [Google Scholar]
- 39.Zhang L, Sun H, Zhao F, Lu P, Ge C, Li H, Hou H, et al. BMP4 administration induces differentiation of CD133+ hepatic cancer stem cells, blocking their contributions to hepatocellular carcinoma. Cancer Res. 2012;72:4276–4285. doi: 10.1158/0008-5472.CAN-12-1013. [DOI] [PubMed] [Google Scholar]
- 40.Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, Stainier DY. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- 41.Ding BS, Nolan DJ, Butler JM, James D, Babazadeh AO, Rosenwaks Z, Mittal V, et al. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature. 2010;468:310–315. doi: 10.1038/nature09493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding BS, Cao Z, Lis R, Nolan DJ, Guo P, Simons M, Penfold ME, et al. Divergent angiocrine signals from vascular niche balance liver regeneration and fibrosis. Nature. 2014;505:97–102. doi: 10.1038/nature12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez JL, Sandoval J, Serviddio G, Sastre J, Morante M, Perrelli MG, Martinez-Chantar ML, et al. Id2 leaves the chromatin of the E2F4-p130-controlled c-myc promoter during hepatocyte priming for liver regeneration. Biochem J. 2006;398:431–437. doi: 10.1042/BJ20060380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kitade M, Factor VM, Andersen JB, Tomokuni A, Kaji K, Akita H, Holczbauer A, et al. Specific fate decisions in adult hepatic progenitor cells driven by MET and EGFR signaling. Genes Dev. 2013;27:1706–1717. doi: 10.1101/gad.214601.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boulter L, Govaere O, Bird TG, Radulescu S, Ramachandran P, Pellicoro A, Ridgway RA, et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nature Medicine. 2012;18:572–579. doi: 10.1038/nm.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang MB, Chang A, Choi M, Zhou D, Anania FA, Shin CH. Antagonistic Interaction Between Wnt and Notch Activity Modulates the Regenerative Capacity of a Zebrafish Fibrotic Liver Model. Hepatology. 2014;60:1753–1766. doi: 10.1002/hep.27285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.