Abstract
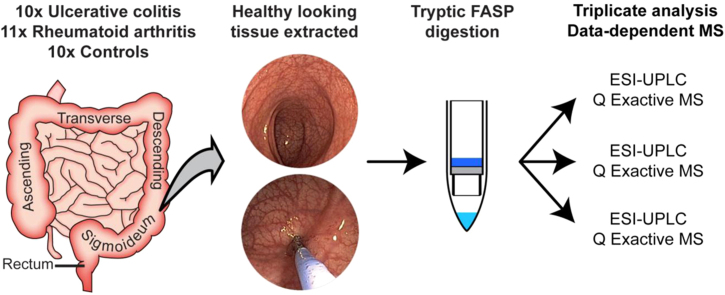
The datasets presented in this article are related to the research articles entitled “Neutrophil Extracellular Traps in Ulcerative Colitis: A Proteome Analysis of Intestinal Biopsies” (Bennike et al., 2015 [1]), and “Proteome Analysis of Rheumatoid Arthritis Gut Mucosa” (Bennike et al., 2017 [2]). The colon mucosa represents the main interacting surface of the gut microbiota and the immune system. Studies have found an altered composition of the gut microbiota in rheumatoid arthritis patients (Zhang et al., 2015; Vaahtovuo et al., 2008; Hazenberg et al., 1992) [5], [6], [7] and inflammatory bowel disease patients (Morgan et al., 2012; Abraham and Medzhitov, 2011; Bennike, 2014) [8], [9], [10]. Therefore, we characterized the proteome of colon mucosa biopsies from 10 inflammatory bowel disease ulcerative colitis (UC) patients, 11 gastrointestinal healthy rheumatoid arthritis (RA) patients, and 10 controls. We conducted the sample preparation and liquid chromatography mass spectrometry (LC-MS/MS) analysis of all samples in one batch, enabling label-free comparison between all biopsies. The datasets are made publicly available to enable critical or extended analyses. The proteomics data and search results, have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD001608 for ulcerative colitis and control samples, and PXD003082 for rheumatoid arthritis samples.
Keywords: Colon mucosa, Proteomics, Rheumatoid arthritis, Ulcerative colitis, Inflammatory bowel diseases, Neutrophil extracellular traps, Dataset, Sigmoidoscopy, Colonoscopy
Graphical abstract
Specifications Table
| Subject area | Biology |
| More specific subject area | Characterization of the proteome of the colon mucosa of ulcerative colitis patients, gastrointestinal healthy rheumatoid arthritis patients, and controls. |
| Type of data | Raw- mass spectrometry files and text/excel files |
| How data was acquired | Mass Spectrometry Liquid Chromatography |
| Data was acquired using a high-resolution/high-accuracy Q Exactive plus (Thermo Scientific) mass spectrometer. | |
| Data format | Raw- and analyzed data. |
| Experimental factors | Human colon mucosal biopsies from ulcerative colitis patients, gastrointestinal healthy rheumatoid arthritis patients, and controls. |
| Experimental features | Biopsies were extracted by colonoscopy and immediately snap-frozen with liquid nitrogen. The biopsies were tryptic digested and analyzed by electrospray ionization liquid chromatography mass spectrometry. |
| Data source location | The Laboratory for Medical Mass Spectrometry, Department of Health Science and Technology, Aalborg University, Fredrik Bajers Vej 7E, 9220 Aalborg East, Denmark |
| Data accessibility | The proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository, [13], [14], [15], [16] with dataset identifiers: |
| PXD001608 – Ulcerative colitis patients, and controls. | |
| PXD003082 – Gastrointestinal healthy rheumatoid arthritis patients. | |
| Direct download links: http://www.ebi.ac.uk/pride/archive/projects/PXD001608 | |
| http://www.ebi.ac.uk/pride/archive/projects/PXD003082 |
Value of the data
-
•
The dataset contains the largest number of identified human proteins from colon mucosa biopsies as of 2017.
-
•
The dataset was obtained in one batch, allowing for label-free comparison of the colon mucosa of ulcerative colitis patients, rheumatoid arthritis patients, and controls.
-
•
The first dataset of the colon mucosa of gastrointestinal healthy RA patients.
-
•
The datasets can be analyzed for novel proteome effects of disease and treatments.
-
•
The datasets allow for extended statistical analysis, and we encourage such collaborations.
1. Data
The datasets in this article provides information on the proteome of the colon mucosa of inflammatory bowel disease patients with ulcerative colitis [1], gastrointestinal healthy rheumatoid arthritis patients [2], and controls. The study was motivated by the finding of an altered composition of the gut microbiota in rheumatoid arthritis patients [5], [6], [7] and inflammatory bowel disease patients [8], [9], [10]. All biopsies were handled on-site by the project group to limit technical variance. The biopsies were randomized, digested using a modified filter-aided sample preparation protein digestion protocol, and analyzed in technical triplicates by high-throughput proteomics on a Q Exactive mass spectrometer. All experimental factors were kept constant, allowing for a label-free quantitative analysis between all samples. The unprocessed proteomics data files (Table 1) and processed search result files (Table 2), have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository, with the dataset identifier PXD001608 for ulcerative colitis and control samples, and PXD003082 for rheumatoid arthritis samples [13], [14].
Table 1.
Raw-datafiles in PXD001608 and PXD003082. All samples were analyzed in technical triplicates, and all raw-files are named accordingly (e.g. Ctrl_10_3 is the third repeat of control 10). “Poor R” signifies a Pearson's correlation coefficient R<0.95 between the technical repeats, and additional data validation is recommended for studies including these datafiles. The number of identified proteins in each replicate is given, wo/w the MaxQuant match between runs feature which transfer MS/MS information between different LC-MS/MS analysis. RA: Rheumatoid arthritis, UC: ulcerative colitis, NA: Not available.
| Filename | Sample | #Proteins matching OFF | #Proteins matching ON | Dataset ID | Comment |
|---|---|---|---|---|---|
| Ctrl_1 | Control | 4362, 4279, 4241 | 5967, 5903, 5897 | PXD001608 | |
| Ctrl_2 | Control | 3613, 3607, 3595 | 5657, 5664, 5723 | PXD001608 | |
| Ctrl_3 | Control | 4203, 4203, 4188 | 5936, 5952, 5939 | PXD001608 | |
| Ctrl_4 | Control | 4245, 4268, 4191 | 5863, 5865, 5849 | PXD001608 | |
| Ctrl_5 | Control | 3961, 3881, 3903 | 5694, 5683, 5632 | PXD001608 | |
| Ctrl_6 | Control | 4290, 4281, 4242 | 5966, 5959, 5932 | PXD001608 | |
| Ctrl_7 | Control | 4080, 4099, 4097 | 5856, 5822, 5817 | PXD001608 | |
| Ctrl_8 | Control | 4269, 4325, 4336 | 5968, 5986, 5966 | PXD001608 | |
| Ctrl_9 | Control | 4560, 4549, 3974 | 6103, 6126, 6103 | PXD001608 | |
| Ctrl_10 | Control | 3974, 3993, 3972 | 5781, 5817, 5779 | PXD001608 | |
| UC_1 | UC | 4290, 4229, 4363 | 5870, 5852, 5907 | PXD001608 | |
| UC_2 | UC | 3328, 3285, 3328 | 5148, 5101, 5139 | PXD001608 | |
| UC_3 | UC | 4455, 4472, 4482 | 6068, 6082, 6041 | PXD001608 | |
| UC_4 | UC | 4236, 4005, NA | 5685, 5773, NA | PXD001608 | UC_4_3 poor R |
| UC_5 | UC | 3097, 3174, NA | 5051, 5017, NA | PXD001608 | UC_5_3 poor R |
| UC_6 | UC | 4458, 4424, 4482 | 6118, 6108, 6115 | PXD001608 | |
| UC_7 | UC | 3657, 3686, 3693 | 5647, 5626, 5575 | PXD001608 | |
| UC_8 | UC | 3356, 3288, 3303 | 5237, 5207, 5164 | PXD001608 | |
| UC_9 | UC | 4681, 4700, 4703 | 6220, 6233, 6223 | PXD001608 | |
| UC_10 | UC | 3762, 3688, 3674 | 5587, 5557, 5562 | PXD001608 | |
| RA_1 | RA | 3900, 3789, 3755 | 5658, 5644, 5632 | PXD003082 | |
| RA_2 | RA | 4347, 4141, 4285 | 6023, 5997, 5973 | PXD003082 | |
| RA_3 | RA | 4654, 4678, 4629 | 6204, 6179, 6180 | PXD003082 | |
| RA_4 | RA | 4298, 4287, 4267 | 5933, 5925, 5899 | PXD003082 | |
| RA_5 | RA | 3472, 3521, 3491 | 5402, 5390, 5344 | PXD003082 | |
| RA_6 | RA | 3545, 3485, 3526 | 5735, 5742, 5728 | PXD003082 | |
| RA_7 | RA | 4538, 4476, 4459 | 6057, 6062, 6071 | PXD003082 | |
| RA_8 | RA | 3619, 3439 NA | 5317, 5310, NA | PXD003082 | RA_8_3 poor R |
| RA_9 | RA | 3361, NA, 3427, | 5256, NA, 5187 | PXD003082 | RA_9_2 poor R |
| RA_10 | RA | 4417, 4354, 4361 | 5991, 6010, 6023 | PXD003082 | |
| RA_11 | RA | 3094, 3055, 3051 | 5005, 5050, 5006 | PXD003082 |
Table 2.
Additional submitted Search and FASTA files in PXD001608 and PXD003082. RA: Rheumatoid arthritis, UC: ulcerative colitis.
| Filename | Dataset ID | Content | Description |
|---|---|---|---|
| CombinedTxtFiles.zip | PXD001608 | Zipped MaxQuant combined txt folder. | Result of the label-free quantitative analysis of UC and controls in MaxQuant. The content of each file is described in “tables.pdf”. |
| 131008_Swissprot_Human_Ref_ proteome.fasta | PXD001608 | Protein FASTA database file. | Protein database used for the UC and controls analysis. |
| MaxQuantOutput.zip | PXD003082 | Zipped MaxQuant combined txt folder. | Result of the label-free quantitative analysis of UC, RA, and controls in MaxQuant. The content of each file is described in “tables.pdf”. |
| UniprotHumanProteome P000005640Isoforms.fasta | PXD003082 | Protein FASTA database file. | Protein database used for the UC, RA, and controls analysis. |
| FASTA file parameters.txt | PXD003082 | FASTA info. | Information regarding the database. |
A cumulated 6768 proteins (FDR<1%) were identified, representing the largest proteome dataset of the colon mucosa so far. Additionally, the dataset represents the first analysis of the colon mucosa of gastrointestinal healthy rheumatoid arthritis patients. The data-analysis result from the analysis with MaxQuant can be downloaded as zipped txt-files, the context of which are described in the tables.pdf also in the zipped file. The result-file proteinGroups.txt, contains all identified proteins at <1% FDR, and information regarding each protein, e.g. the corresponding label-free relative quantitation value (LFQ). Additional information regarding the participants can be found in the publications.
2. Experimental design, materials and methods
2.1. Study cohort and sample collection
The sample material was extracted and processed as described in [1] and 2].
Colon mucosal biopsies (roughly 1 mm3) were sampled 40 cm from the anus by sigmoidoscopy, at the Regional Hospital Silkeborg Denmark, from 10 ulcerative colitis patients, 11 rheumatoid arthritis patients and 10 controls in the period from 2012 to 2013. The biopsies were immediately transferred to cryotubes and snap-frozen in liquid nitrogen followed by storage at minus 80 °C until proteomics sample preparation. All participants had given a written informed consent prior to participation in the study, and the project was approved by The Regional Scientific Ethical Committee (S-20120204) and the Danish Data Protection Agency (2008-58-035).
2.2. Proteomic sample preparation
The biopsies were randomized, and enzymatic digested using a modified filter-aided sample preparation protein [17], [18], [19], [20], [21], [22]. Briefly explained, the biopsies were homogenized in 0.5 mL cold sample buffer (5% sodium deoxycholate, 50 mM triethylammonium bicarbonate, pH 8.5). The lysate protein concentration was estimated by absorbance at 280 nm measured using a NanoDrop 1000 UV–vis spectrophotometer (Thermo Scientific, Waltham, MA, USA). Additionally, the concentration of four biopsy lysates was determined using a bicinchoninic acid assay (BCA) with bovine serum albumin as standard, measured using an Infinite microplate reader (Tecan, Männedorf, Switzerland). The nanodrop measurements were calibrated using the BCA results. 100 µg protein was transferred to 30 kDa molecular weight cutoff spin-filters (Millipore, Billerica, MA, USA) to facilitate buffer exchanges by centrifugation at 15,000g for 15 min between all steps. Protein disulfide bonds were reduced by addition of 100 µL 10 mM tris(2-carboxyethyl)phosphine (Thermo Scientific, Waltham, MA, USA) and alkylated by addition of 100 µL 50 mM 2-iodoacetamide (Sigma-Aldrich, St. Louis, MO, USA) in sample buffer. Two µg sequencing grade modified trypsin (Promega, Madison, WI, USA) diluted in lysis buffer with 0.5% sodium deoxycholate was added to the spin-filter, and the proteins were digested to peptides overnight at 37 °C. The peptide material was eluted from the spin-filter and purified by phase inversions with 1:1 (v/v) ethyl acetate with 1% formic acid, and dried down in a vacuum centrifuge overnight, and stored at −80 °C for a maximum of one week prior to analysis.
2.3. Proteomic analysis
The peptides were analyzed by LC-MS/MS using an UltiMate 3000 UPLC system (Thermo Scientific, Waltham, MA, USA) coupled online to a Q Exactive plus mass spectrometer (Thermo Scientific). Five µg peptide material was loaded onto a 2 cm reverse phase C18-material trapping column and separated on a 50 cm analytical column, both from Acclaim PepMap100 (Thermo Scientific). The liquid phase consisted of 96% solvent A (0.1% formic acid) and 4% solvent B (0.1% formic acid in acetonitrile), at a flow rate of 300 nL/min. The peptides were eluted from the column by increasing to 8% solvent B and subsequently to 30% solvent B on a 225 min ramp gradient, and introduced into the mass spectrometer by a picotip emitter for electrospray ionization (New objective, Woburn, MA, USA). The mass spectrometer was operated in positive mode with data-dependent acquisition, alternating between survey spectra and isolation/fragmentation spectra using a top12 method. Selected eluting peptides were excluded from re-analysis for 30 s. All biopsies were analyzed in triplicates in a random order.
2.4. Data processing
The generated RAW-files were searched with MaxQuant 1.5.2.8 software against the Uniprot Homo sapiens reference proteome database with isoforms (UP000005640, last modified 2015-01-16, entry count 90,434) [23], [24]. Standard settings were employed, with the following abundant peptide modifications included in the search: Carbamidomethylated(C) (fixed), N-terminal protein acetylation (variable), oxidation(M) (variable), and deamidation (N or Q) (variable) [11], [12]. The match between runs feature in MaxQuant was enabled to allow the transfer of confident peptides identifications across LC-MS/MS runs, based on accurate mass-to-charge and retention time. Identified proteins and peptides were filtered to <1% false discovery rate [25]. Label-free quantitation was enabled in MaxQuant to report protein and peptide relative quantities using standard parameters.
Funding sources
The Lundbeck Foundation Denmark (R181-2014-3372), and the Carlsberg Foundation (CF14-0561) are acknowledged for grants enabling the project (TBB grants). Knud and Edith Eriksens Memorial Foundation (“Knudog Edith EriksensMindefond”) and Ferring are acknowledge for grants, enabling the collection of the biological sample material (VA grant). The Obelske Family Foundation and the Svend Andersen Foundation are acknowledged for grants supporting the analytical platform being part of the Danish National Platform for Proteomics (PRO-MS) (AS grants).
Acknowledgements
The authors would like to thank Kasper B. Lauridsen for help establishing the patient cohort, Ditte B. Kristensen for help in the laboratory, and the PRIDE team for making the proteomics data publically available.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2017.09.059.
Transparency document. Supplementary material
Supplementary material
.
References
- 1.Bennike T.B. Neutrophil extracellular traps in ulcerative colitis: a proteome analysis of intestinal biopsies. Inflamm. Bowel Dis. 2015;21:2052–2067. doi: 10.1097/MIB.0000000000000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennike T.B. Proteome analysis of rheumatoid arthritis gut mucosa. J. Proteome Res. 2017;16:346–354. doi: 10.1021/acs.jproteome.6b00598. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015;21:895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 6.Vaahtovuo J., Munukka E., Korkeamäki M., Luukkainen R., Toivanen P. Fecal microbiota in early rheumatoid arthritis. J. Rheumatol. 2008;35:1500–1505. [PubMed] [Google Scholar]
- 7.Hazenberg M.P., Klasen I.S., Kool J., Ruseler-van Embden J.G., Severijnen A.J. Are intestinal bacteria involved in the etiology of rheumatoid arthritis? Review article. Acta Pathol. Microbiol. Immunol. Scand. 1992;100:1–9. doi: 10.1111/j.1699-0463.1992.tb00833.x. [DOI] [PubMed] [Google Scholar]
- 8.Morgan X.C. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abraham C., Medzhitov R. Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology. 2011;140:1729–1737. doi: 10.1053/j.gastro.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennike T. Biomarkers in inflammatory bowel diseases: current status and proteomics identification strategies. World J. Gastroenterol. 2014;20:3231–3244. doi: 10.3748/wjg.v20.i12.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bennike T.B. Comparing the proteome of snap frozen, RNAlater preserved, and formalin-fixed paraffin-embedded human tissue samples. EuPA Open Proteom. 2016;10:9–18. doi: 10.1016/j.euprot.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennike T.B. Proteome stability analysis of snap frozen, RNAlater preserved, and formalin-fixed paraffin-embedded human colon mucosal biopsies. Data Brief. 2016;6:942–947. doi: 10.1016/j.dib.2016.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vizcaíno J.A. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vizcaíno J.A. The PRoteomics IDEntifications PRIDE database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:1063–1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang R. PRIDE Inspector: a tool to visualize and validate MS proteomics data. Nat. Biotechnol. 2012;30:135–137. doi: 10.1038/nbt.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Côté R.G. The PRoteomics IDEntification PRIDE Converter 2 framework: an improved suite of tools to facilitate data submission to the PRIDE database and the ProteomeXchange consortium. Mol. Cell. Proteom. 2012;11:1682–1689. doi: 10.1074/mcp.O112.021543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leon I.R., Schwammle V., Jensen O.N., Sprenger R.R. Quantitative assessment of in-solution digestion efficiency identifies optimal protocols for unbiased protein analysis. Mol. Cell. Proteom. 2013;12:2992–3005. doi: 10.1074/mcp.M112.025585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wisniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 19.Masuda T., Tomita M., Ishihama Y. Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J. Proteome Res. 2008;7:731–740. doi: 10.1021/pr700658q. [DOI] [PubMed] [Google Scholar]
- 20.Manza L.L., Stamer S.L., Ham A.-J.L., Codreanu S.G., Liebler D.C. Sample preparation and digestion for proteomic analyses using spin filters. Proteomics. 2005;5:1742–1745. doi: 10.1002/pmic.200401063. [DOI] [PubMed] [Google Scholar]
- 21.Bennike T. A normative study of the synovial fluid proteome from healthy porcine knee joints. J. Proteome Res. 2014;13:4377–4387. doi: 10.1021/pr500587x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennike T., Lauridsen K.B., Olesen M.K., Andersen V., Birkelund S., Stensballe A. Optimizing the identification of citrullinated peptides by mass spectrometry: utilizing the inability of trypsin to cleave after citrullinated amino acids. J. Proteom. Bioinform. 2013;6:288–295. [Google Scholar]
- 23.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteom. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 25.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material