Abstract
Autophagy is essential in lens organelle degradation. This study aimed to seek potential autophagy-associated long noncoding RNAs (lncRNAs) and their relative mechanisms in human lens development using the “fried egg” lentoid body (LB) generation system. The expression pattern of LC3B in differentiating LBs was similar to that in developing a mouse lens in vivo. Among the massive lncRNAs expressed with a significant difference between induced pluripotent stem cells (iPSCs) and LBs, lncRNA affecting LC3B (ALB), which was predicted to have a co-relationship with the autophagy marker LC3B, was highly expressed in differentiating lens fibers in LBs. This result was consistent with its high expression in human embryonic lenses compared to those in embryonic stem cells (ESCs). Furthermore, lncRNA ALB knockdown resulted in the downregulation of LC3BII at the protein level, therefore inhibiting the autophagy process in human lens epithelial cells (HLECs). Our results identify lncRNA ALB, a potential autophagy regulator in organelle degradation during human lens development, highlighting the importance of lncRNAs in lens development.
Keywords: long noncoding RNA, lentoid bodies, induced pluripotent stem cells, lens
Graphical Abstract
Introduction
Autophagy is a natural, destructive mechanism that disassembles unnecessary or dysfunctional cellular components through a regulated process that allows for the orderly degradation and recycling of cellular components. It has been reported in many biological processes, such as development, aging, aging-related diseases, intracellular homeostasis, cancer, and immunological stress.1, 2 The involvement of autophagy in the lens was first reported by Walton and Menko,3, 4 who discovered autophagosome in rat and chick lens epithelial cells. It is well acknowledged that lens fiber cells gradually lose their organelles and nuclei during development to produce the organelle free zone (OFZ), which contributes to lens transparency.5, 6, 7 Autophagy is implicated to play a key role in lens organelle degradation, as evidenced by the following studies. One group found that the suppression of mTOR signaling leads to the premature loss of organelles and nuclei,8 and another found that the deletion of Pik3c3 in mice leads to congenital cataracts.9 Human congenital cataract families with mutations in the autophagy-related genes FYCO1 and CHMP4B were also reported.10, 11 In addition to its function in lens organelle degradation during development, autophagy was suggested to be essential in maintaining lens epithelial cell homeostasis, as reported by Morishita,9 and that the deletion of autophagy-related 5 (Atg5) produced age-related cataracts, which may be due to the interruption of autophagy in cellular homeostasis. Thus, autophagy is crucial in both lens development and cataract pathology. However, molecular mechanisms in regulating autophagy in the lens are far from being understood.
Long noncoding RNA (lncRNA) is defined as a class of transcripts longer than 200 nt that has limited coding potential.12 lncRNAs have been reported to function in a wide range of biological processes and can regulate gene expression in cis or trans by diverse mechanisms.13 Studies have suggested that lncRNAs play an essential role in cell differentiation, development, metabolism, aging, cancer, et al.12 For example, Xian14 found a novel lncRNA adipogenic differentiation induced noncoding RNA (ADNIR) regulated adipogenesis by transcriptionally activating C/EBPα. Some studies have shown that lncRNAs may regulate autophagy in several physiological and pathological processes,15, 16, 17 indicating the involvement of autophagy-associated lncRNA in lens development and cataracts. Although the expressions of lncRNAs either in a mouse lens during development or in human lens capsules have been analyzed through next-generation RNA sequencing or microarrays,18, 19, 20, 21 their biological functions remain elusive.
Mechanisms in lens development have been well studied in both animal and cellular models.22, 23 However, situations in human lens development are still unclear due to the lack of appropriate cellular models. Our previous study established an efficient system—a “fried egg” method for human lens regeneration in vitro from both embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs).24 The lens generated in the dish (also known as the lentoid body [LB]), which showed a lens-like transparent structure, not only expressed lens-specific markers but also exhibited basic optical characteristics. More importantly, in addition to lens-specific markers, including α, β, γ-crystallins, and MIP, the expression of placodal markers, such as SIX1 and PAX6, and early lens-specific markers, such as SOX1 and PROX1, were also observed at certain time points of LB differentiation. This implicates that LBs generated with the fried egg method at least partially recurred human lens development in a dish. Additionally, the expression of autophagy-associated marker LC3B was also observed during LB differentiation, suggesting the existence of autophagy in LB generation. However, whether the autophagic process during LB differentiation represents its situation in vivo remains to be determined.
In the present study, we verified the similarity of the autophagic process between LB differentiation and lens development in vivo, therefore providing a unique opportunity to investigate lncRNA-associated autophagy in human lens development. Moreover, we report several novel lncRNAs having functions in lens organelle degradation, among which lncRNA ALB (short for affecting LC3B) (ENST00000426012.1 in ENSEMBL) and lncRNA p19101 (TCONS_00021520 in HumanLincRNACatalog), which are co-related with LC3B and SH3GLB1, proved to have similar expression patterns between human embryonic lens and LBs. Finally, the knockdown of lncRNA ALB partially blocked autophagy activity in human lens epithelial cells (HLECs) through decreasing LC3BII expression, verifying its authentic role in autophagy in the lens.
Results
Autophagic Activity during Lens Differentiation in Mice
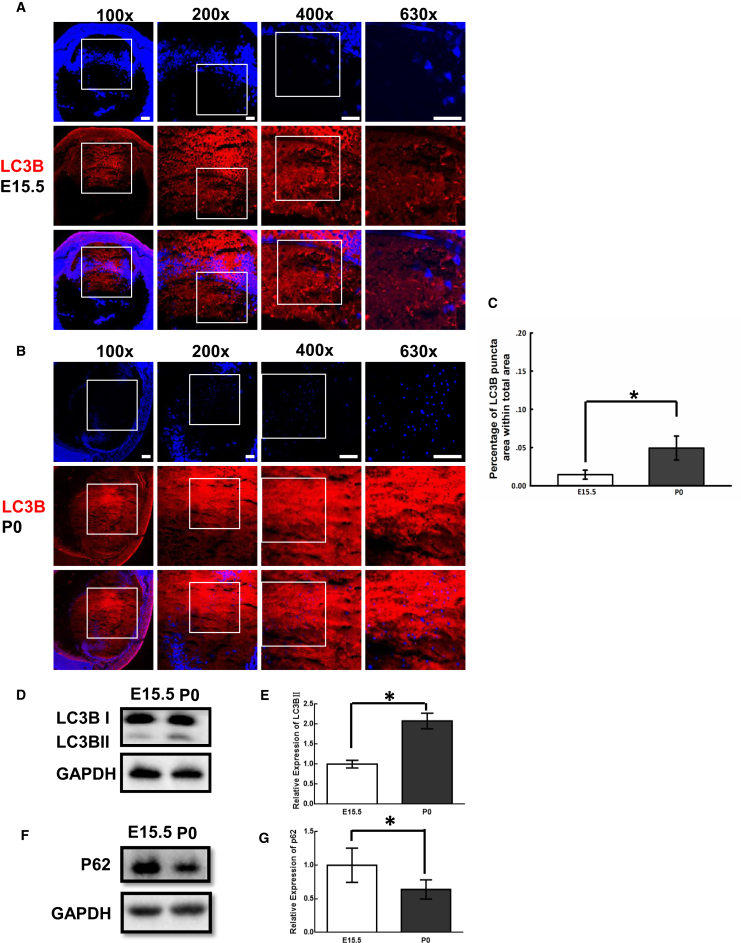
Autophagy is implicated in OFZ formation during lens development,5 so we first analyzed the expression of the autophagy markers LC3B and P62 in a mouse-developing lens by immunofluorescence detection and western blotting. Confocal microscope examination revealed the high expression of LC3B in embryonic and postnatal mouse lenses (Figures 1A and 1B). Furthermore, more LC3B puncta were found in the postnatal (P0) lens than in the embryonic mouse lens (E15.5) (Figures 1A–1C), which was verified by western blot analysis (Figures 1D and 1E). Moreover, western blot analysis revealed that the expression of P62 was lower in P0 than in E15.5 of the mouse lens (Figures 1F and 1G), indicated the potential role of autophagy in the developing lens. These results are consistent with previous studies in which autophagy was suggested to participate in ocular lens organelle degradation.5
Figure 1.
LC3B Expression Pattern in a Mouse Lens
(A and B) Immunofluorescence examination of LC3B (red) in a mouse lens at E15.5 and P0. High-magnification images at 200x, 400x, and 630x are highlighted in the preceding low-magnification images (white square frame) and show the positions of magnified images at equatorial regions of the lens. Nuclei were labeled with DAPI (blue). (C) Quantitative measurements of LC3B puncta area achieved by ImageJ software at the magnification of 630X. Five random fields were measured in one lens (n = 3). (D and F) Western blot assay showed the expression patterns of LC3B and P62 in a mouse lens at E15.5 and P0. GAPDH was used as a reference gene. (E and G) Quantitative measurement of western blotting showed the higher expression of LC3B and lower expression of P62 in a mouse lens at P0 (n = 3). *p < 0.05. Scale bars, 100 μm (100×) and 50 μm (200×, 400×, and 630×).
Autophagic Activity during UiPSC-Derived LB Differentiation
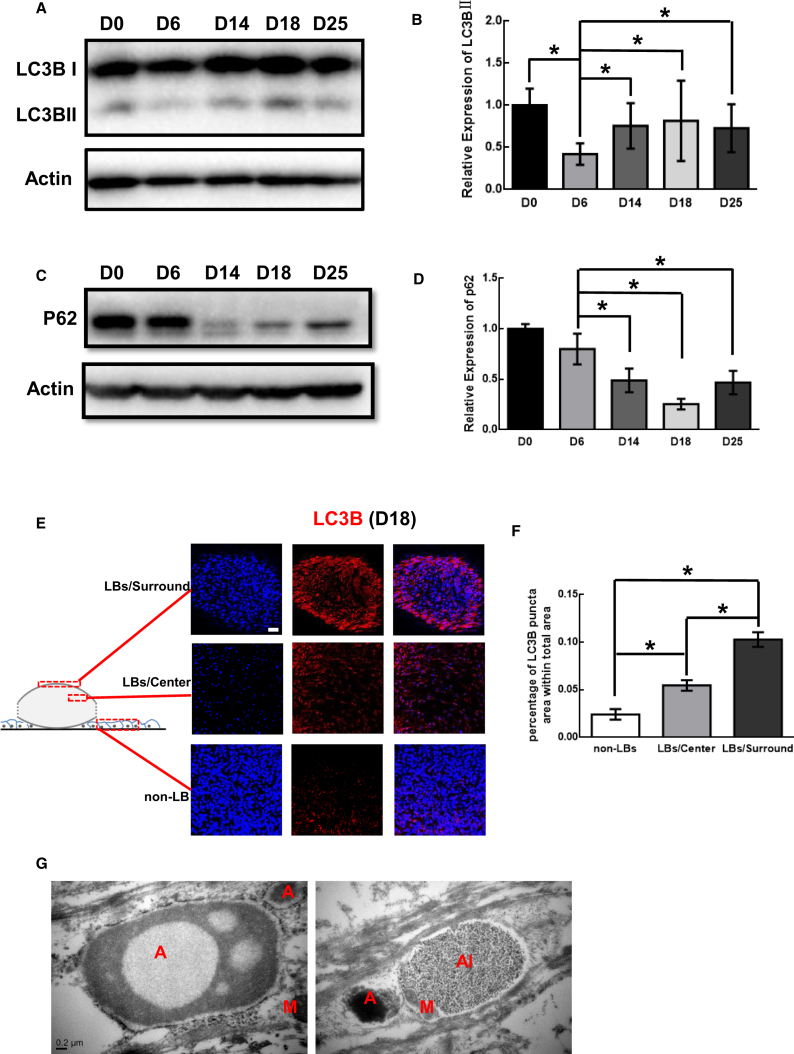
Western blot analysis revealed the expression of LC3BII in mature LBs in our previous study.24 Thus, the expression patterns of LC3B and P62 at different time points during LB differentiation as well as the location of LC3B were examined. Our results showed that more LC3B and less P62 were expressed in differentiating LBs after D14 when massive lens epithelial cells differentiated to fiber cells with organelle degradation (Figures 2A–2D). Furthermore, a confocal microscope examination revealed that more LC3B puncta were in LBs when compared to those in non-LB cells on D18 (Figures 2E and 2F). Furthermore, more LC3B puncta were in the surrounding region of LBs when compared to those in the center region of LBs (Figures 2E and 2F). Additionally, typical autophagosome was observed in differentiating lens fiber cells, which occurred more frequently in immature LBs than in mature LBs (Figure 2G). Our results suggest that the autophagic activity during urinary derived iPSC (UiPSC)-derived LB differentiation using the fried egg method mimicked its role in lens development in vivo to a certain extent. Therefore, this system could be used to study autophagy-mediated organelle degradation during human lens development.
Figure 2.
Autophagic Activity in UiPSC-Induced LB Differentiation
(A and C) Western blotting shows the expression patterns of LC3B and P62 during LB differentiation at multiple time points. Actin was used as a reference gene. (B and D) Quantitative measurements of western blot (n = 3). (E) Immunofluorescence examination of LC3B (red) in the surrounding region of LBs, center region of LBs, and non-LB regions. (F) Quantitative measurements of the LC3B puncta area showed the highest expression of LC3B in the surrounding region of LBs and the lowest expression of LC3B in the non-LBs region (n = 3). (G) The TEM analysis showed the existence of autophagosomes (red A), autophagy lysosome (red AL), and mitochondria (red M) in differentiating LBs. *p < 0.05. Scale bars, 50 μm (C) and 0.2 μm (E).
Transcriptome Analysis of LBs versus UiPSCs
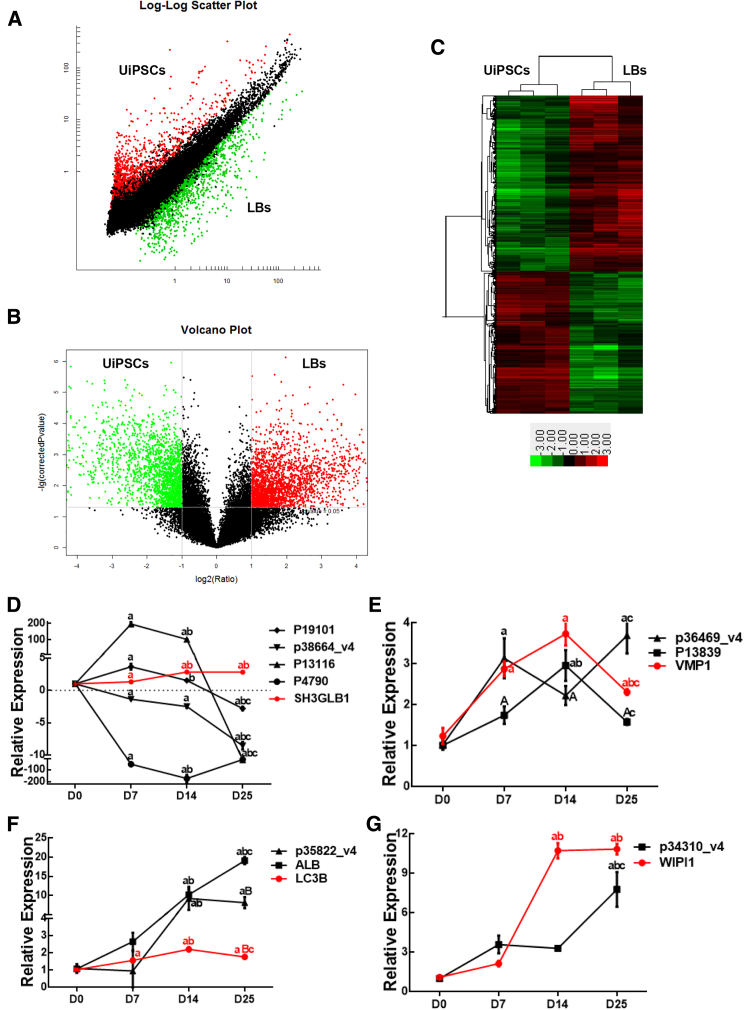
Although the functions of lncRNA and their mechanisms in various tissues were well explored, understanding of lncRNA in the lens has just begun. To start deciphering the molecular mechanisms of the autophagy response and revealing the potential role of lncRNA during the LB differentiation process, we determined the LB (D25) transcriptome (mRNA and lncRNA) to compare it with that of UiPSCs through microarray analysis. A total of 390 lncRNAs were significantly upregulated (fold ≥2.0, p ≤ 0.05) in LBs, and 1,356 were expressed at higher levels in UiPSCs. Figures 3A–3C show a scatterplot, volcano plot, and heatmap of the microarray data. As a start, we performed a focused analysis for (a selection of) lncRNAs involved in the autophagy process during LB differentiation. All raw and processed lncRNA microarray data reported in this manuscript have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO: GES97683.
Figure 3.
Expression Pattern of Autophagy-Associated mRNAs and Their Corresponding lncRNAs during the LB Differentiation Process
(A and B) Log-log scatterplot and volcano plot show the differentially expressed lncRNAs between LBs and UiPSCs. (C) Heatmap was generated from a hierarchical cluster analysis to show the different expression profiling between LBs and UiPSCs. (D) Quantitative real-time PCR analysis of SH3GLB1 and its corresponding lncRNAs, including lncRNA p19101, lncRNA p38664_v4, lncRNA p13116, and lncRNA p4790. (E) Quantitative real-time PCR analysis of VMP1 and its corresponding lncRNAs, including lncRNA p13839 and lncRNA p36469_v4. (F) Quantitative real-time PCR analysis of LC3B and its corresponding lncRNAs, including lncRNA p35822_v4 and lncRNA ALB. (G) Quantitative real-time PCR analysis of WIPI1 and its corresponding lncRNA p34310_v4. The bar represents mean ± SEM (n = 3 independent experiments). a, p < 0.01 versus D0; A, p < 0.05 versus D0; b, p < 0.01 versus D7; B, p < 0.05 versus D7; c, p < 0.01 versus D14.
The lncRNAs for which the expressions are at higher levels in the LBs and genes with which they co-related and autophagy-related genes, including SH3GLB1, WIPI1, MAP1LC3B, VMP7, VMP1,BNIP3L, PTEN, ATG5, and PINK1, are listed in Tables 1 and S1. From this analysis, 10 lncRNAs, which were predicted to correlate with multiple autophagy-related genes (autophagy-associated lncRNA for short), including SH3GLB1, WIPI1, MAP1LC3B, and VMP1, were expressed with significant differences in LBs compared to those in UiPSCs. These results were further verified by performing quantitative real-time PCR assays (Table 1). Furthermore, the expression of other autophagy-associated genes, including ATG13, Beclin1, ATG12, ATG4A, SQSTM, UVRAG, and FYCO1, as mentioned in Brennan’s study,25 which did not show a significant difference between UiPSCs and LBs from microarray analysis, were also analyzed by quantitative real-time PCR assay. The results showed that all genes except FYCO1 increased, along with LB differentiation (Figure S1). Figure S2 shows a scatterplot, volcano plot, and heatmap of the microarray data for the mRNA expression profile between UiPSCs and LBs.
Table 1.
Differentially Expressed Autophagy-Related lncRNA and Their Correlated Genes in LBs versus UiPSCs
| LncRNA ProbeName | LncRNA ID | Database | FC (abs) in Microarray | FC (abs) in Real-Time qPCR | Regulation | Correlated mRNA |
|---|---|---|---|---|---|---|
| p4790 | ENST00000555655.1 | ENSEMBL | 33.81 | 15.17 | down | SH3GLB1 |
| p13116 | ENST00000507926.1 | ENSEMBL | 20.73 | 18.12 | down | |
| p38664_v4 | ENST00000551631.2 | ENSEMBL | 9.39 | 8.54 | down | |
| p19101 | TCONS_00021520 | HumanLincRNACatalog | 8.82 | 2.79 | down | |
| p13515 | ENST00000503797.2 | ENSEMBL | 6.07 | 135.84 | down | |
| p34310_v4 | ENST00000521507.1 | ENSEMBL | 5.19 | 7.79 | up | WIPI1 |
| ALB | ENST00000426012.1 | ENSEMBL | 5.71 | 19.13 | up | MAP1LC3B |
| p35822_v4 | TCONS_00025230 | HumanLincRNACatalog | 5.64 | 8.15 | up | |
| p13839 | ENST00000515570.1 | ENSEMBL | 2.09 | 1.58 | up | VMP1 |
| p36469_v4 | uc010kvv.3 | UCSC | 2.09 | 3.69 | up |
Down means the expression of lncRNA is lower in LBs both in microarray and quantitative real-time PCR analysis. Up means the expression of lncRNA is higher in LBs both in microarray and quantitative real-time PCR analysis.
In addition, lncRNAs correlated with genes involved in lens development (lens-development-associated lncRNAs for short) were analyzed in our study. Among all of the differentially expressed lncRNAs from the microarray data, eight lncRNAs, which were predicted to have co-relationships with several lens-development-related genes, including MAF, MAB21L2, BMP7, CDH2, MEIS1, and CTNNB1, were differentially expressed in LBs compared to UiPSCs (Tables S2 and S3). Quantitative real-time PCR was performed to confirm the above microarray data (Tables S2 and S3).
Finally, a global bioinformatic comparison of the gene expression patterns identified in LBs with that of the normal lens published by others using second-generation sequencing20 was also performed to investigate how complete the LB differentiation is. About 1,000 differentially expressed genes were shared between our microarray analysis for LBs and RNA-Seq results for the mouse lens at 6 different developing stages (Figure S3), indicating the similarity of LBs and the lens in vivo. The top 3,000 expressed genes in LBs or in the mouse lens at different stages are listed in Data S1.
Expression Analysis of lncRNAs during the LB Differentiation Process
Next, the expressions of selected autophagy-associated lncRNAs on D0, D7, D14, and D25 were examined by quantitative real-time PCR assay. Our results revealed that SH3GLB1 increased during LB differentiation, with the highest level on D14. Among four SH3GLB1-corresponding lncRNAs, lncRNA p19101 and lncRNA p13116 increased on D7 and D14 but decreased on D25, whereas lncRNA p4790 and lncRNA p38664_v4 were expressed at lower levels during the entire LB differentiation process when compared to those on D0 (UiPSCs) (Figure 3D). Speaking of the expression of VMP1 and its relative lncRNAs, the highest level of VMP1 and lncRNA p13839 appeared on D14, whereas the expression of lncRNA p36469_v4 showed a trend of fluctuation (Figure 3E). Both LC3B and its relative lncRNAs (lncRNA ALB and lncRNA p35822_v4) upregulated during fiber cell differentiation in LB generation (Figure 3F). The expression of WIPI1 and its relative lncRNA p34310_v4 significantly increased in differentiating LBs (Figure 3G). In general, the results showed that the expression of each lncRNA and its relative gene changed with time during LB differentiation.
Additionally, the expressions of lens-development-associated lncRNAs were also examined at different time points of LB formation. The results are shown in Figure S4. It is worth mentioning that most of the lncRNAs that were either upregulated or downregulated showed significant differences after D14 of LB differentiation when massive cells differentiated to form LB. The expression of lens-development-relative genes fluctuated but was always kept at a high level at a certain point of differentiation (Figure S4).
Expressions of lncRNA ALB and lncRNA p19101 in Human Embryonic Lens
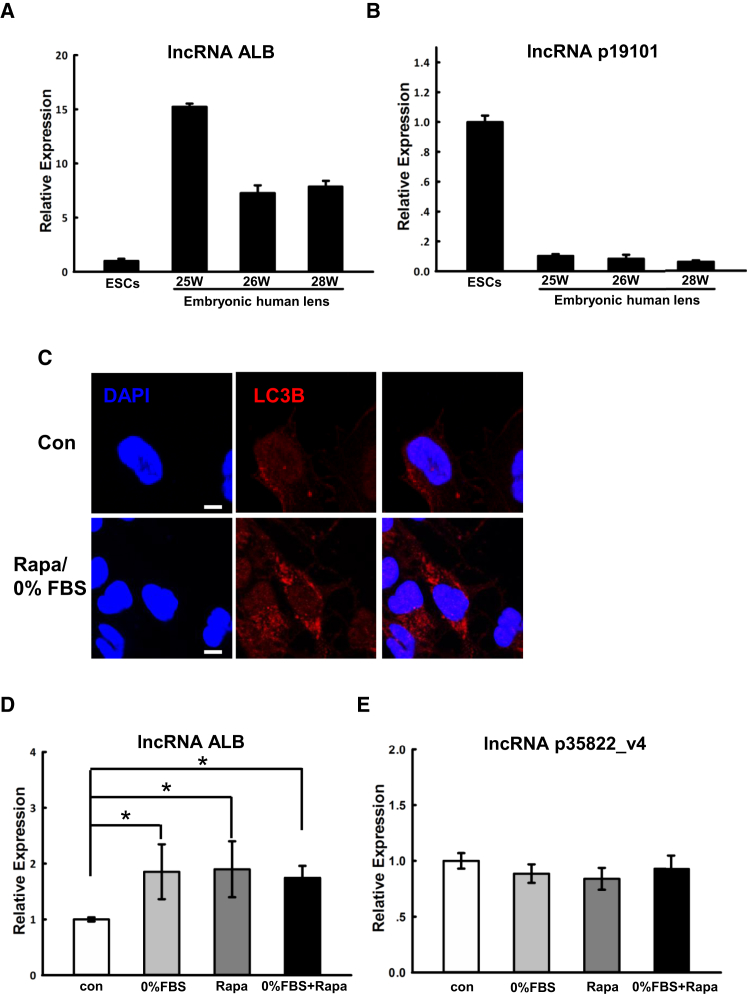
Among the lncRNAs selected above, lncRNA ALB and lncRNA p19101 were next analyzed in a human embryonic lens. Quantitative real-time PCR showed that lncRNA ALB was highly expressed in an embryonic human lens at 25 weeks, 26 weeks, and 28 weeks (Figure 4A), whereas the expression of lncRNA p19101 decreased in an embryonic human lens (Figure 4B) when compared to those in human ESCs. These results are consistent with their expression levels in LBs at relative time points.
Figure 4.
Expression of lncRNA ALB in Human Embryonic Lens and Autophagy-Activated HLECs
(A and B) Quantitative real-time PCR analysis of lncRNA ALB and lncRNA p19101 in human ESCs and an embryonic human lens at 25 weeks, 26 weeks, and 28 weeks. (C) Immunofluorescence examination of LC3B (red) in HLECs with or without rapamycin treatment. Nuclei were labeled with DAPI (blue). (D and E) Quantitative real-time PCR analysis of lncRNA ALB and lncRNA p35822_v4 under four conditions, including normal culture medium (control), 0% FBS, rapamycin, and 0% FBS plus rapamycin. The bar represents mean ± SEM (n = 3 independent experiments). *p < 0.05 versus control. Scale bar, 20 μm (C).
Autophagy Suppression by lncRNA ALB Small Interfering RNA in HLECs
Due to the key function of LC3B in autophagy, we focused on the function of its associated lncRNA ALB and lncRNA p35822_v4 in the following experiments. First, autophagy was triggered by 0% FBS and/or rapamycin, as evidenced by the increasing number of LC3B puncta in the HLECs undergoing autophagy (Figure 4C). Then, the expression of lncRNA ALB and lncRNA p35822_v4 was analyzed by quantitative real-time PCR. The expression of lncRNA ALB stably increased in 0% FBS, rapamycin, and 0% FBS + rapamycin conditions, in which HLECs underwent autophagy when compared to the control (Figure 4D). Alternatively, no significant difference was observed for the expression of lncRNA p35822_v4 between the autophagy conditions and control condition (Figure 4E). Thus, we then explored the co-relationship between lncRNA ALB and LC3B.
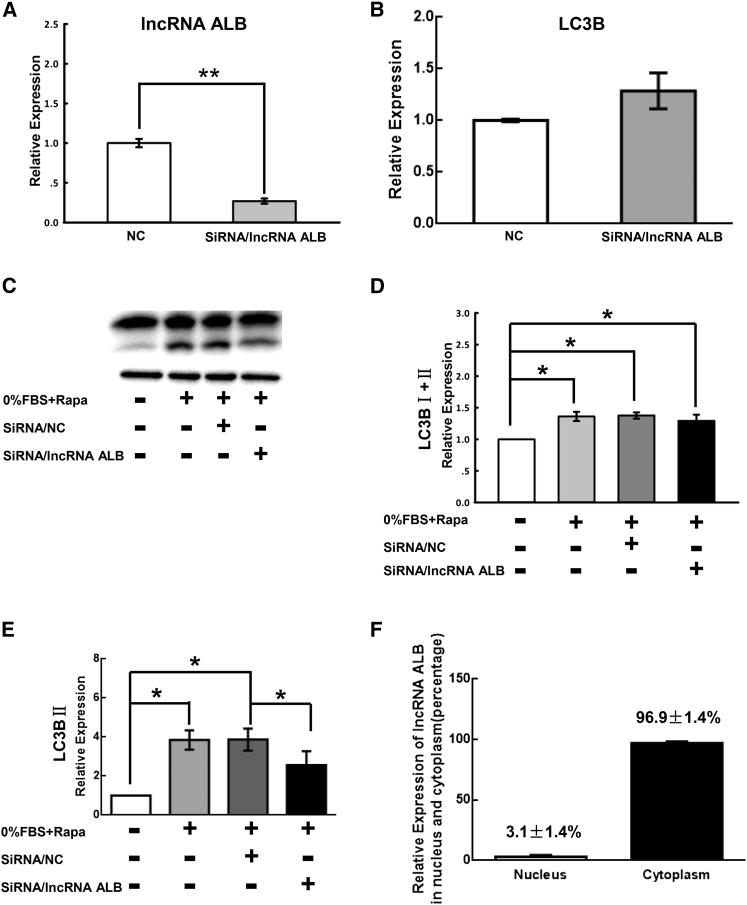
In lncRNA ALB knockdown HLECs, LC3B was analyzed. The results showed that lncRNA ALB knockdown did not reduce the expression of LC3B at the RNA level (Figures 5A and 5B) and total LC3B (I+II) at the protein level (Figures 5C and 5D). However, it significantly reduced the expression of LC3BII, the active form of LC3B in 0% FBS plus rapamycin-treated HLECs (Figures 5C and 5E), thereby partially inhibiting autophagic activity in HLECs. Because the transformation of LC3BI to LC3BII occurred in the cytoplasm, we examined the location of lncRNA ALB. Most of the lncRNA ALB (96.9% ± 1.4%) was expressed in the cytoplasm when compared to its expression in nuclei (3.1% ± 1.4%) (Figure 5F), which provides at least side evidence for our hypothesis that lncRNA ALB may work as a regulator in the transformation of LC3BI to LC3BII.
Figure 5.
Autophagy Suppression by lncRNA ALB siRNA in HLECs
(A and B) Quantitative real-time PCR analysis of lncRNA ALB and LC3B in HLECs with siRNA (NC) or lncRNA ALB siRNA. (C) Western blot analysis of LC3B expression under siRNA (NC) or lncRNA ALB siRNA treatment. Culture mediums without or with 0% FBS plus rapamycin were used as NC and positive control. (D) Quantitative measurements of western blot assay showed the relative expression of total LC3B (I+II) under four conditions. (E) Quantitative measurements of the expression of LC3BII. (F) Quantitative real-time PCR analysis of lncRNA ALB expression in the nucleus and cytoplasm of HLECs. The bar represent mean ± SEM (n = 3 independent experiments). **p < 0.01; *p < 0.05.
Discussion
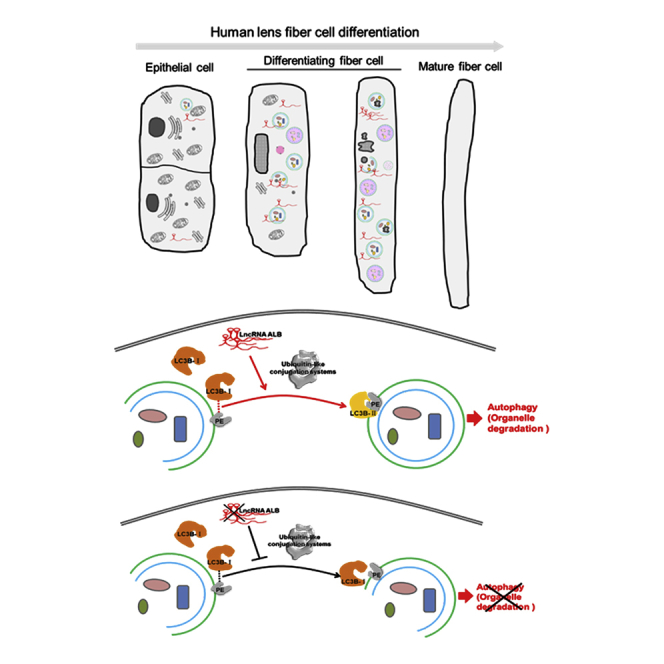
In the present study, we found that the human iPSCs-derived LB differentiation system, through the fried egg method, could partially represent the situation of autophagic activity in lens development in vivo, thereby providing an ideal model for studying autophagy-associated lncRNAs in human lens development. During the LB induction process, lncRNA ALB was found to highly express in differentiating LBs, which is consistent with its expression pattern in a human embryonic lens. Furthermore, lncRNA ALB knockdown decreased the transformation of LC3BI to LC3BII, thereby reducing autophagic activity in HLECs and indicating its potential role in organelle degradation during lens development.
Lens transparency is dependent on enormous factors, during which autophagy-mediated organelle degradation in fiber cell differentiation is widely accepted as the essential process for OFZ formation. Enormous progress has been made in delineating how this phenomenon is induced and regulated in cellular and animal models.5, 8, 9 However, the situations in humans remain elusive due to the lack of appropriate models. In our study, autophagy was observed to be activated during human iPSC-derived LB differentiation, as evidenced by the increase of LC3BII and the frequently observed autophagosome in differentiating fiber cells. This is consistent with the results in a mouse embryonic and postnatal lens, indicating that the LB differentiation system could mimic the lens development in vivo for autophagy activity. This thus provides an ideal opportunity for studying autophagy-associated OFZ formation in humans.
A broad transcriptional program is involved to collectively coordinate lens spatio-temporally differentiation. Recently, regulating RNAs that control gene expression has raised increasing attention in various situations. Although the role of microRNAs (miRNAs) in lenses is well established,26, 27 the role of lncRNAs in lens development and cataracts is still poorly understood. With previous studies reporting the existence of lncRNAs in lens development,18, 19, 20 novel lncRNA ALB is the first, to our knowledge, to potentially play a role in human lens development. Our study reported that lncRNA ALB promotes autophagy by enhancing the transformation of LC3BI to LC3BII in iPSC-derived LB, which could partially represent the situation in lens development in vivo. Therefore, lncRNA ALB was determined to potentially play a role in lens organelle degradation. The results that lncRNA ALB knockdown did not reduce the expression of LC3B at the RNA level and total LC3B (I+II) protein level but significantly reduced the expression of LC3BII may suggest the involvement of lncRNA ALB in conjugation with LC3BI with phosphatidylethanolamine (PE) present in the membrane to form PE-conjugated LC3B proteins (called LC3BII), which is critical in regulating the formation of autophagosomes.28 Experiments investigating the differentiation process of LBs derived from lncRNA ALB knockdown iPSCs/ESCs are needed to investigate lncRNA ALB’s target and relative mechanisms in OFZ formation in human lens development. Furthermore, if lncRNA ALB is conserved in different species, its knockout animal models will be useful in future studies.
The particularity of its location determines the function of lncRNA. In our study, 96.9% of lncRNA ALB was found to be expressed in cytoplasm compared to its expression in nuclei in HLECs, indicating its cytoplasmic function in the posttranscriptional regulation of mRNA. Previous research has shown that TINCR, which is localized to the cytoplasm, interacts with the target protein STAU1 to promote the stability of mRNAs, and therefore functions as the diametric opposite of posttranscriptional silencing by small regulatory RNAs like small interfering RNA (siRNA) or miRNAs.29 Conversely, another group showed that lincRNA-p21 can repress the translation of targeted mRNAs by interacting with the mRNAs of CTNNB1 and JUNB and the translational repressor Rck.30 The mechanism of lncRNA ALB remains elusive; thus, experiments investigating mRNAs and/or proteins with which lncRNA ALB directly interacts are needed in the future for further understanding its potential role during OFZ formation during lens development.
Taken together, our research demonstrated that autophagic activities in LBs derived from human iPSCs/ESCs represented their situations in lens development in vivo. It therefore provides an ideal model for studying OFZ formation during human lens development, during which lncRNA ALB potentially plays a role in regulating LC3BI to LC3BII transformation.
Materials and Methods
Animals
Pregnant C57BL/6 mice were purchased from Silaike Experimental Animal (Shanghai, China) and kept in the Animal Housing Facility at Zhejiang University. All animal experiments were approved by the Institutional Animal Care and Use Committee at Zhejiang University.
Cell Culture
Urinary derived iPSCs (UiPSCs) were generated and characterized in our previous study.24 H9 human ESCs (Cell Bank, Shanghai, China) and UiPSCs were cultured in a feeder-free system. In brief, cells were seeded in matrigel-coated (B&D) dishes, cultured with an mTesR medium (STEMCELL), and passaged with 0.5 mM EDTA in PBS every 4 days.31
HLEC SRA 01-04 was purchased from the Riken cell bank (Tsukuba, Japan). Cells were incubated in DMEM (Gibco) containing 10% fetal bovine serum (FBS) (GIBCO) and passaged with 0.25% trypsin in PBS.
LB Differentiation from UiPSCs
The fried egg method for LB differentiation from UiPSCs/ESCs was described in detail in our previous study.24 In brief, cells were triggered with the 100 ng/mL BMP inhibitor Noggin for 6 days until the wanted cell clusters were mechanically isolated and re-seeded in new dishes. Then, a composition of BMP4 (20 ng/mL), BMP7 (20 ng/mL), and bFGF (100 ng/mL) was added into the medium for 9 days.32 A typical fried-egg-like structure appeared around day 11 of differentiation, when no fried-egg-like cell clusters were mechanically discarded to avoid negative effects on LB formation. Finally, BMP4 and BMP7 were replaced with Wnt3a (20 ng/mL) to accomplish the final differentiation. Mature LBs were obtained around day 25 (D25).
Transmission Electron Microscopy
Autophagosomes in LBs were observed through transmission electron microscopy (TEM) analysis, which was previously described.24 In brief, after fixation with 2.5% glutaraldehyde in phosphate buffer (0.1M, pH 7.0) for 4 hr and postfixation with 1% OsO4 in phosphate buffer (0.1 M, pH 7.0) for 2 hr, the mature LBs (D25) were dehydrated, infiltrated, and embedded in Spurr’s resin. Pictures of the sectioned LBs that were stained with uranyl acetate and alkaline lead citrate were taken using a Hitachi Model H-7650 TEM.
Western Blot
Cell samples during the LB differentiation process at D0 (n = 3), D6 (n = 3), D14 (n = 3), D18 (n = 3), and D25 (n = 3) as well as mouse lenses on embryonic day 15.5 (E15.5) (n = 3) and postnatal day 0 (P0) (n = 3) were lysed and extracted. The protein concentration was qualified by a spectrophotometer (Bio-Rad iMarK Microplate Reader). Then, the expressions of LC3B and P62 were analyzed according to the standard protocol. In brief, all proteins were first separated by running gel and transferred onto a polyvinylidene fluoride (PVDF) blotting membrane. Then, the targeted proteins were incubated overnight with the LC3B (Sigma-Aldrich) primary antibody (1/1,000) or P62 (Cell Signaling Technology) primary antibody (1/1,000), followed by the horseradish peroxidase (HRP)-conjugated second antibody (Cell Signaling Technology) (1:5,000). The images were captured with the ECL detection system (Millipore) and the ChemiDoc MP imaging system (Bio-Rad). Finally, quantitative analysis was performed through the ImageJ software.
Immunofluorescence Examination
LBs
Cell samples were fixed with paraformaldehyde (PFA) (Sigma-Aldrich) (4% in PBS), permeabilized with 0.4% Triton X-100 (Sigma-Aldrich), and incubated overnight with the rabbit anti-LC3B antibody (1/100) (Sigma Aldrich). After incubation with the Alexa Fluor 555 labeled second antibody (1/1,000; Invitrogen) for 2 hr, the nuclei were labeled with DAPI (0.5 μg/mL; Sigma-Aldrich). The images were finally captured using a Leica TCS SP8 confocal microscope (Leica).
Mouse Lens
After being dissected from pregnant mice (E15.5) and pups (P0), mice lenses were immediately embedded in an optimal cutting temperature compound (OCT) and sectioned to 7 μm using a Leica CM1950. Then, the LC3B analysis in sample slices was performed as was described for LBs.
Algorism to Identify LC3B Fluorescence in Differentiating LBs and in Mouse Lens
Representative images of LC3B staining in differentiating LBs and a mouse lens were selected for analysis. An example of the calculation performed is shown in Figure S5 and is described in detail below. The representative images were first turned into 8-bit type and inverted (Figures S5A and S5B). Then, the area of LC3B (area 1) and the area of interest (area 2) were measured after the image’s threshold was adjusted to manifest the LC3B dots (Figure S5C). The value was normalized by the number of pixels in the region. The ratio of area 1/area 2 shows the relative expression of LC3B in a mouse lens or LBs. All image analyses were conducted by ImageJ software.
Microarray Analysis
Total RNAs were extracted from UiPSCs (D0, n = 3) and LBs (D25, n = 3) using the Trizol reagent (Invitrogen) according to the manufacturer’s protocol. The purity and concentration of RNA were determined with a spectrophotometer (NanoDrop ND-1000). Microarray analysis was performed using the Agilent Human lncRNA+mRNA Array V3.0 (CapitalBio Technology, China) containing probes interrogating about 37,000 human lncRNAs and about 34,000 human mRNAs from multiple databases. The lncRNA + mRNA array data were analyzed for data summarization, normalization, and quality control using the GeneSpring software V12.0 (Agilent). The differentially expressed genes were selected as the threshold values of ≥2-fold and ≤2-fold, and a Benjamini-Hochberg-corrected p value of < 0.05. The correlation analysis was performed between the differentially expressed lncRNA and mRNA, and only the pairs with Pearson correlation coefficients not less than 0.99 were selected and considered meaningful. Furthermore, on the basis of the correlation analysis results, the meaningful pairs involving the differentially expressed genes that are involved in lens development and/or autophagy were selected. Therefore, lncRNAs involved in lens development and/or autophagy and investigated in the present study were selected.
Global Bioinformatic Comparison of the Gene Expression Patterns between LBs and Mouse Embryonic and Postnatal Lens
Global bioinformatic comparison of the top 3,000 expressed genes identified in LBs with that of a mouse embryonic and postnatal lens published by others using second-generation sequencing was performed.20 The comparison was achieved on VENNY 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/).
Collection of Human Embryonic Lenses
The human embryonic lenses were collected during abortion at Women’s Hospital, School of Medicine Zhejiang University. The human embryonic lenses were dissected from aborted embryos by an obstetrician and immediately stored in liquid nitrogen for further use. Human embryonic lens collection in this study was approved by the ethics committee of Zhejiang University. Written informed consent was obtained from the participants. The study protocol adhered to the principles of the Declaration of Helsinki.
Quantitative Real-Time PCR
Total RNAs were extracted from cell samples during LB differentiation at D0 (n = 7), D7 (n = 7), D14 (n = 7), and D25 (n = 7) using the Trizol reagent (Invitrogen) according to the manufacturer’s guidelines. RNA samples were stored at −80°C for later use after quality and concentration determination using spectrophotometers (NanoDrop 2000c, Thermo Scientific). RNA samples at different time points from one experiment were then examined via quantitative real-time PCR to validate the expression of selected lncRNA and mRNA selected from the microarray analysis using SYBR Premix Ex Tag (RR420A) on an ABI Fast 7500 RT-PCR system according to the manufacturer’s protocols (TaKaRa).
Total RNAs were also extracted from a human embryonic lens at 25 weeks of gestation (n = 1), 26 weeks of gestation (n = 1), and 28 weeks of gestation (n = 1), and from HLEC samples incubated with or without rapamycin (an autophagy activator) and/or 0% FBS. RNAs from cytoplasm or nuclei of HLECs were extracted according to the manufacturer’s protocols (PARIS). The expressions of genes and lncRNAs in interests were analyzed as described above. Normalization of data was as follows. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous reference. The quantification cycle (Ct) was obtained, and the ΔCt value was calculated with Ct (target gene) − Ct (GAPDH). ΔΔCt (log2ratio) was calculated with ΔCt (control group) − ΔCt (target group).
All primers are listed in Table S4.
Transcript Sequence of lncRNA ALB
lncRNA ALB was collected by Ensembl (Code: ENST00000426012.1) and Havana (Code: OTTHUMT00000078206). The sequence of lncRNA ALB was reported as below: 5′-GTTTTTAATATACATACAAAACCCTGAATGGTCAATTTTTGTGTTGTAGATGGAGACGAGAGATCTGGACTTCTGAACATCTACCGGAAAAGATAAAATGTGCAAGCCCTTCAGAACGCTGGGGAGGTTGGGCTGAACTGCAGTGGGAGAGGAGAAGAGATGCTGGCGTGAAGGGCTGGGGCAGAAGATCAGGGGAAAATTGCGGATGATCTGAATGATAGCAGTGACCACCAGCAACCTACAGAGCTGCCCAACCAGGAAATTGTCAGATGGAAGCAACGCTTGCAGGCAAAGAAGGCTGAAGCAGGCCAAGGCAGGAATGAAACAGTCATAAATGTTGGAGTGAAGCGACTGACCTCCCCGGTCCACACCCCGGCCCAGCGTCCTGCATCTGCTGGAGGATGGTGGAGACCATGTGGAGGATGGGGACTTGGCAGTGGAGGAGAACATGGAAGCAGGATATGGCAGTCCCCTAGCGTCCCAGCCACAAGTCCTCTGAAGAGTGTGGATGACGATGGTGACTTAGGTGCATCCTTCGAATGTCCTGAGGGGGAGACTGGAGAAGCAGACACGCAATTGTGGGAAGAAATCAAAGGCATAAAGAACTCACAGCACAAGTCCAAAGTCTGATCAGCGAGCAGGCCTCTCTGCAGGGTGCAAGGGCACAGCTGGAGAGG-3′.
RNA Knockdown
Ribo lncRNA smart silencer for lncRNA ALB was purchased from Guangzhou RIBOBIO (Guangzhou, China). This product contains a mixture of six target sequences for lncRNA ALB, including 5′-GAACATGGAAGCAGGATAT-3′, 5′-GGAAGAAATCAAAGGCATA-3′, 5′-CAGGAAATTGTCAGATGGA-3′, 5′-TGAAGCAGGCCAAGGCAGGA-3′, 5′-AGTCCTCTGAAGAGTGTGGA-3′, and 5′-CAAAGTCTGATCAGCGAGCA-3′. The siRNA of lncRNA ALB was transfected into HLECs using the ribo FECT Transfection Kit according to the manufacturer’s protocol. In brief, HLECs were seeded in appropriate plates or dishes in a culture medium. Until they reached 30% confluence, cells were transfected with the negative control (NC) of siRNA or lncRNA ALB siRNA. The expressions of lncRNA and LC3B at the RNA level were examined using quantitative real-time PCR. To analyze autophagic activity in lncRNA ALB knockdown HLECs, LC3B protein expression was determined in 0% FBS and rapamycin-treated cell samples that were incubated with the NC of siRNA or lncRNA ALB siRNA using western blotting.
Statistical Analysis
In the present study, a one-way ANOVA, which was followed by least significant difference (LSD) post hoc multiple comparison when more than two groups required comparison, was performed on all experiments with SPSS software (version 17.0, SPSS). Statistical significance was defined as p < 0.05.
Author Contributions
K.Y. and Q.F. developed the concept and designed the study. Z.Q. and L.Z. performed the experiments. D.L., Q.T., H.Y., and Z.C. contributed reagents and materials. Q.F. and Z.Q. analyzed the data and wrote the paper. K.Y. provided administrative support and final approval of the manuscript.
Conflicts of Interest
The authors have declared that no competing interests exist.
Acknowledgments
This work has been supported by the National Natural Science Foundation of China (81371001, 81300641, 81570822, and 81670833), Project of National Clinical Key Discipline of Chinese Ministry of Health, Zhejiang Key Laboratory Fund of China (2011E10006), Program of Zhejiang Medical Technology (2015KYA109), and Zhejiang Province Key Research and Development Program (2015C03042).
Footnotes
Supplemental Information includes five figures, four tables, and one data file and can be found with this article online at https://doi.org/10.1016/j.omtn.2017.09.011.
Supplemental Information
References
- 1.Hurley J.H., Schulman B.A. Atomistic autophagy: the structures of cellular self-digestion. Cell. 2014;157:300–311. doi: 10.1016/j.cell.2014.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rubinsztein D.C., Mariño G., Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 3.Walton J., McAvoy J. Sequential structural response of lens epithelium to retina-conditioned medium. Exp. Eye Res. 1984;39:217–229. doi: 10.1016/0014-4835(84)90010-1. [DOI] [PubMed] [Google Scholar]
- 4.Menko A.S., Klukas K.A., Johnson R.G. Chicken embryo lens cultures mimic differentiation in the lens. Dev. Biol. 1984;103:129–141. doi: 10.1016/0012-1606(84)90014-9. [DOI] [PubMed] [Google Scholar]
- 5.Costello M.J., Brennan L.A., Basu S., Chauss D., Mohamed A., Gilliland K.O., Johnsen S., Menko S., Kantorow M. Autophagy and mitophagy participate in ocular lens organelle degradation. Exp. Eye Res. 2013;116:141–150. doi: 10.1016/j.exer.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wride M.A. Lens fibre cell differentiation and organelle loss: many paths lead to clarity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011;366:1219–1233. doi: 10.1098/rstb.2010.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chauss D., Basu S., Rajakaruna S., Ma Z., Gau V., Anastas S., Brennan L.A., Hejtmancik J.F., Menko A.S., Kantorow M. Differentiation state-specific mitochondrial dynamic regulatory networks are revealed by global transcriptional analysis of the developing chicken lens. G3 (Bethesda) 2014;4:1515–1527. doi: 10.1534/g3.114.012120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basu S., Rajakaruna S., Reyes B., Van Bockstaele E., Menko A.S. Suppression of MAPK/JNK-MTORC1 signaling leads to premature loss of organelles and nuclei by autophagy during terminal differentiation of lens fiber cells. Autophagy. 2014;10:1193–1211. doi: 10.4161/auto.28768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morishita H., Eguchi S., Kimura H., Sasaki J., Sakamaki Y., Robinson M.L., Sasaki T., Mizushima N. Deletion of autophagy-related 5 (Atg5) and Pik3c3 genes in the lens causes cataract independent of programmed organelle degradation. J. Biol. Chem. 2013;288:11436–11447. doi: 10.1074/jbc.M112.437103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J., Ma Z., Jiao X., Fariss R., Kantorow W.L., Kantorow M., Pras E., Frydman M., Pras E., Riazuddin S. Mutations in FYCO1 cause autosomal-recessive congenital cataracts. Am. J. Hum. Genet. 2011;88:827–838. doi: 10.1016/j.ajhg.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sagona A.P., Nezis I.P., Stenmark H. Association of CHMP4B and autophagy with micronuclei: implications for cataract formation. BioMed Res. Int. 2014;2014:974393. doi: 10.1155/2014/974393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 14.Xiao T., Liu L., Li H., Sun Y., Luo H., Li T., Wang S., Dalton S., Zhao R.C., Chen R. Long noncoding RNA ADINR regulates adipogenesis by transcriptionally activating C/EBPα. Stem Cell Reports. 2015;5:856–865. doi: 10.1016/j.stemcr.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K., Liu C.Y., Zhou L.Y., Wang J.X., Wang M., Zhao B., Zhao W.K., Xu S.J., Fan L.H., Zhang X.J. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat. Commun. 2015;6:6779. doi: 10.1038/ncomms7779. [DOI] [PubMed] [Google Scholar]
- 16.Huang S., Lu W., Ge D., Meng N., Li Y., Su L., Zhang S., Zhang Y., Zhao B., Miao J. A new microRNA signal pathway regulated by long noncoding RNA TGFB2-OT1 in autophagy and inflammation of vascular endothelial cells. Autophagy. 2015;11:2172–2183. doi: 10.1080/15548627.2015.1106663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge D., Han L., Huang S., Peng N., Wang P., Jiang Z., Zhao J., Su L., Zhang S., Zhang Y. Identification of a novel MTOR activator and discovery of a competing endogenous RNA regulating autophagy in vascular endothelial cells. Autophagy. 2014;10:957–971. doi: 10.4161/auto.28363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan S.Y., Hackett S.F., Riazuddin S.A. Non-coding RNA profiling of the developing murine lens. Exp. Eye Res. 2016;145:347–351. doi: 10.1016/j.exer.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoang T.V., Kumar P.K., Sutharzan S., Tsonis P.A., Liang C., Robinson M.L. Comparative transcriptome analysis of epithelial and fiber cells in newborn mouse lenses with RNA sequencing. Mol. Vis. 2014;20:1491–1517. [PMC free article] [PubMed] [Google Scholar]
- 20.Khan S.Y., Hackett S.F., Lee M.C., Pourmand N., Talbot C.C., Jr., Riazuddin S.A. Transcriptome profiling of developing murine lens through RNA sequencing. Invest. Ophthalmol. Vis. Sci. 2015;56:4919–4926. doi: 10.1167/iovs.14-16253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen Y., Dong L.F., Zhou R.M., Yao J., Song Y.C., Yang H., Jiang Q., Yan B. Role of long non-coding RNA MIAT in proliferation, apoptosis and migration of lens epithelial cells: a clinical and in vitro study. J. Cell. Mol. Med. 2016;20:537–548. doi: 10.1111/jcmm.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cvekl A., Duncan M.K. Genetic and epigenetic mechanisms of gene regulation during lens development. Prog. Retin. Eye Res. 2007;26:555–597. doi: 10.1016/j.preteyeres.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cvekl A., Ashery-Padan R. The cellular and molecular mechanisms of vertebrate lens development. Development. 2014;141:4432–4447. doi: 10.1242/dev.107953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Q., Qin Z., Jin X., Zhang L., Chen Z., He J., Ji J., Yao K. Generation of functional lentoid bodies from human induced pluripotent stem cells derived from urinary cells. Invest. Ophthalmol. Vis. Sci. 2017;58:517–527. doi: 10.1167/iovs.16-20504. [DOI] [PubMed] [Google Scholar]
- 25.Brennan L.A., Kantorow W.L., Chauss D., McGreal R., He S., Mattucci L., Wei J., Riazuddin S.A., Cvekl A., Hejtmancik J.F. Spatial expression patterns of autophagy genes in the eye lens and induction of autophagy in lens cells. Mol. Vis. 2012;18:1773–1786. [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q.L., Zhang H.Y., Qin Y.J., Meng Q.L., Yao X.L., Guo H.K. MicroRNA-34a promoting apoptosis of human lens epithelial cells through down-regulation of B-cell lymphoma-2 and silent information regulator. Int. J. Ophthalmol. 2016;9:1555–1560. doi: 10.18240/ijo.2016.11.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiang W., Lin H., Wang Q., Chen W., Liu Z., Chen H., Zhang H., Chen W. miR-34a suppresses proliferation and induces apoptosis of human lens epithelial cells by targeting E2F3. Mol. Med. Rep. 2016;14:5049–5056. doi: 10.3892/mmr.2016.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noh H.S., Hah Y.S., Zada S., Ha J.H., Sim G., Hwang J.S., Lai T.H., Nguyen H.Q., Park J.Y., Kim H.J. PEBP1, a RAF kinase inhibitory protein, negatively regulates starvation-induced autophagy by direct interaction with LC3. Autophagy. 2016;12:2183–2196. doi: 10.1080/15548627.2016.1219013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kretz M., Siprashvili Z., Chu C., Webster D.E., Zehnder A., Qu K., Lee C.S., Flockhart R.J., Groff A.F., Chow J. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon J.H., Abdelmohsen K., Srikantan S., Yang X., Martindale J.L., De S., Huarte M., Zhan M., Becker K.G., Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol. Cell. 2012;47:648–655. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beers J., Gulbranson D.R., George N., Siniscalchi L.I., Jones J., Thomson J.A., Chen G. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat. Protoc. 2012;7:2029–2040. doi: 10.1038/nprot.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang C., Yang Y., Brennan L., Bouhassira E.E., Kantorow M., Cvekl A. Efficient generation of lens progenitor cells and lentoid bodies from human embryonic stem cells in chemically defined conditions. FASEB J. 2010;24:3274–3283. doi: 10.1096/fj.10-157255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.