Abstract
Putrescine is widely used in industrial production of bioplastics, pharmaceuticals, agrochemicals, and surfactants. Although engineered Corynebacterium glutamicum has been successfully used to produce high levels of putrescine, the overall cellular physiological and metabolic changes caused by overproduction of putrescine remains unclear. To reveal the transcriptional changes that occur in response to putrescine production in an engineered C. glutamicum strain, a comparative transcriptomic analysis was carried out. Overproduction of putrescine resulted in transcriptional downregulation of genes involved in glycolysis; the TCA cycle, pyruvate degradation, biosynthesis of some amino acids, oxidative phosphorylation; vitamin biosynthesis (thiamine and vitamin 6), metabolism of purine, pyrimidine and sulfur, and ATP-, NAD-, and NADPH-consuming enzymes. The transcriptional levels of genes involved in ornithine biosynthesis and NADPH-forming related enzymes were significantly upregulated in the putrescine producing C. glutamicum strain PUT-ALE. Comparative transcriptomic analysis provided some genetic modification strategies to further improve putrescine production. Repressing ATP- and NADPH-consuming enzyme coding gene expression via CRISPRi enhanced putrescine production.
Keywords: Corynebacterium glutamicum, putrescine, comparative transcriptomic analysis, physiological change, differentially expressed genes
Introduction
Putrescine (1,4-diaminobutane) is widely used as a building block for the industrial production of bioplastics, pharmaceuticals, agrochemicals, and surfactants. For example, putrescine is a raw material used in the production of the bioplastic polyamide nylon-4,6 via polycondensation with adipic acid. Nylon-4,6 is a superior engineering plastic due to its high melting point, high mechanical strength, and excellent solvent resistance. The demand for putrescine is approximately 10,000 tons per year in Europe and is expected to grow (Scott et al., 2007).
The potential commercial demands mean that the efficient biotechnological production of putrescine has become increasingly necessary. After introducing an ornithine decarboxylase gene, putrescine has been produced using engineered Escherichia coli (Qian et al., 2009) and Corynebacterium glutamicum (Schneider and Wendisch, 2010). An engineered E. coli XQ52 (p15SpeC) strain was constructed for putrescine production by a combination of deleting endogenous degradation pathways and replacing the native promoters of the ornithine biosynthetic genes. The strain produced 1.68 g/L of putrescine with a yield of 0.166 g/g glucose in a shake-flask fermentation and 24.2 g/L with a productivity of 0.75 g/L.h in a 6.6-L fed-batch fermentation (Qian et al., 2009). The Wendisch group constructed a series of engineered C. glutamicum strains for putrescine production (Schneider and Wendisch, 2010; Schneider et al., 2012; Choi et al., 2014; Nguyen et al., 2015a,b). Their strategies included: (1) lowering the ornithine carbamoyltransferase gene (argF) expression by modifications of the argF promoter, translational start codon, and ribosome-binding site (Choi et al., 2014); (2) reducing α-ketoglutarate decarboxylase (Kgd) activity by replacing the kgd native start codon GTG with TTG and the native odhI gene with the odhIT15A gene; (3) deleting the snaA gene to eliminate putrescine acetylation (Nguyen et al., 2015b); (4) overexpression of the putrescine transporter gene (cgmA), the glyceraldehyde 3-phosphate dehydrogenase gene (gap), the pyruvate carboxylase gene (pyc) and the feedback-resistant N-acetylglutamate kinase variant gene (argBA49V/M54V). The final engineered C. glutamicum strain NA6 produced 58.1 mM (5.1 g/L) of putrescine with a yield on glucose of 0.26 g/g in a flask culture (Nguyen et al., 2015a), representing the highest values yet seen. The titer and yield of C. glutamicum NA6 were 1.99- and 2-fold higher than that of the parent strain C. glutamicum PUT21 (Nguyen et al., 2015a), respectively. The parent strain C. glutamicum PUT21 produced 19 g/L putrescine with a productivity of 0.55 g/L/h and a yield 0.166 g/g glucose in a fed-batch fermentation (Schneider et al., 2012).
Although engineered C. glutamicum has been successfully employed for the high-level production of putrescine, the overall cellular physiological and metabolic changes caused by the overproduction of putrescine remain unclear. Transcriptome analysis has become an effective approach for monitoring cellular physiological and metabolic changes (Yu et al., 2016). Detailed information on cellular physiological changes cannot only allow for a much better understanding of the underlying regulatory mechanisms but also provide new genetic modification strategies for the further improvement in the production of metabolites. Thus, to understand the cellular physiological and metabolic changes occurring in response to the overproduction of putrescine, we carried out a comparative transcriptomic analysis between the putrescine-producer C. glutamicum PUT-ALE and the wild-type strain C. glutamicum ATCC 13032.
Materials and Methods
Strains, Plasmids, and Primers
The bacterial strains used in this study are listed in Table 1. Plasmids and primers used in this study are presented in Supplementary Table 1.
Table 1.
Strains used in this study.
| Name | Description | Reference/Sources |
|---|---|---|
| Strains | ||
| Corynebacterium glutamicum ATCC 13032 | Wild-type | ATCC |
| C. glutamicum ΔAPE6937R42 | Ornithine producing strain, the evolved strain of C. glutamicum ATCC 13032 (ΔargFΔproBΔspeE), ΔargR | Jiang et al., 2013a |
| C. glutamicum PUT-ALE | Putrescine producer, the metabolically evolved strain of C. glutamicum ΔAPE6937R42 with Δpuo, ΔfabG:: PH36-speC1ECL, ΔbutA and ΔsnaA | Lab storage |
| C. glutamicum PUT-ALE-KT | Putrescine producer, the kgd native GTG start codon in C. glutamicum PUT-ALE was replaced with TTG. | This study |
Plasmid Construction
Genes were amplified from genomes using the responding primers (Supplementary Table 1) and cloned into pEC-XK99E (Kirchner and Tauch, 2003). Gene disruption was performed via two-step homologous recombination using the non-replicable integration vector pK-JL as described by Jiang et al. (2013a,b)).
To enhance specificity and reduce off-target effects, the dcas9 on pCRISPathBrick (Cress et al., 2015) was site-directed mutated into dcas9 (K848A/K1003A/R1060A) as described as Slaymaker et al. (2016) to obtain pCRISPathBrick∗. The p15A ori was amplified from pBAD33 (Guzman et al., 1995) using P15AF and P15AR. The vector backbone was amplified from pEC-XK99E (Kirchner and Tauch, 2003) using primer PEC-AF and PEC-AR. The two PCR products were recombined using ClonExpress II One Step Cloning Kit (Vazyme Biotech Co., Ltd., Nanjing, China) to obtain pEC-XK-p15A. The dcas9∗ gene was amplified from pCRISPathBrick∗ using primers dcas9∗F and dcas9∗R and then cloned into the XmaI/XbaI sites of pEC-XK-p15A to generate pEC-dcas9∗ (Supplementary Figure 1A). The sgRNA sequence was amplified from pTargetF (Jiang et al., 2015) using primers sgRNAF and sgRNAR. The vector backbone was amplified from pXMJPsod using primers psodGF and psodGR. The two PCR products were recombined using ClonExpress II One Step Cloning Kit to obtain the sgRNA plasmid pXMJPsod-sgRNA.
The pXMJPsod-X-sgRNA series (Supplementary Figure 1B), used in target single-gene repression with a targeting N20 sequence of gene loci of interest, was obtained by inverse PCR using primes the target N20F and PsodG-R from pXMJPsod-sgRNA, and followed by self-ligation.
Putrescine Production in Shake Flasks
A single colony was inoculated into 5 mL of seed medium in a test tube, which was aerobically cultured overnight at 200 rpm and 30°C. The overnight seed culture was used to inoculate 50 mL of fermentation medium with an initial OD600 of 0.2. The primary cultures were incubated at 30°C for 72 h in a rotary shaking incubator at 200 rpm. Each liter of seed medium contained 25 g of glucose, 10 g of yeast extract, 10 g of corn steep liquor, 15 g of (NH4)2SO4, 2.5 g of MgSO4 7H2O, 1 g of KH2PO4, 0.5 g of K2HPO4, 0.5 g of Na2HPO4, and 10 g of CaCO3. Each liter of fermentation medium contained 100 g of glucose, 20 g of corn steep liquor, 50 g of (NH4)2SO4, 2.5 g of MgSO4 7H2O, 1 g of KH2PO4, 0.5 g of K2HPO4, 0.5 g of Na2HPO4, 20 mg of FeSO4 7H2O, 20 mg of MnSO4 4H2O, 2 g of molasses, 1 mL of Tween-80, and 10 g of CaCO3. The initial pH of both media described above was adjusted to 7.0.
Analysis of Growth and Metabolite Concentration
Growth was monitored by measuring the optical density of the culture at 600 nm after adding 0.2 M HCl to dissolve CaCO3. The glucose concentration was determined using glucose oxidase and a glucose assay kit (Shanghai Rongsheng Biotech Corporation, Shanghai, China). The putrescine concentration was determined using a Shimadzu HPLC system (LC-20A HPLC, Shimadzu, Japan) equipped with an Inertsil ODS-SP column (5 μm, 4.6 mm × 150 mm, GL Sciences Inc., Tokyo, Japan) as described by Schneider and Wendisch (2010). Putrescine was first derivatized using 9-fluorenylmethyl chloroformate (FMOC). The fluorescent derivatives were detected by excitation at 263 nm (emission at 310 nm). The mobile phase consisted of solvent A (0.05 M sodium acetate, pH 4.2) and solvent B (acetonitrile) with a flow rate of 1.3 mL/min. The following gradient was used: 0 min, 38% B; 5 min, 38% B; 12 min, 57% B; 14 min, 57% B; 20 min, 65% B; 25 min, 76% B; and 35 min, 76% B. A standard curve was constructed from serial dilutions of a standard stock solution of 1,4-diaminobutane.
Transcriptome Analysis
RNA-Seq was performed by GENWIZ (Shuzhou, China) using an Illumina HiSeq sequencer (Illumina, San Diego, CA, United States). Each sample was analyzed in duplicate. Cells cultured for 48 h were harvested by centrifugation at 300 rpm for 2 min to remove CaCO3 and then at 5,000 × g for 15 min and washed twice with PBS. Total RNA was extracted using TRIzol Reagent (Invitrogen) and an RNeasy Mini Kit (Qiagen). Total RNA from each sample was quantified and qualified by an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, United States), NanoDrop (Thermo Fisher Scientific Inc.) and a 1% agarose gel. One μg of total RNA with RIN values above 7 was used for following library preparation. Next generation sequencing library preparations were performed according to the manufacturer’s protocol (NEBNext® UltraTM Directional RNA Library Prep Kit for Illumina®). The rRNA was depleted from the total RNA using a Ribo-Zero rRNA Removal Kit (Bacteria) (Illumina). The rRNA-depleted mRNA was then fragmented and reverse-transcribed. First-strand cDNA was synthesized using ProtoScript II Reverse Transcriptase with random primers and Actinomycin D. The second-strand cDNA was synthesized using Second Strand Synthesis Enzyme Mix (including dACG-TP/dUTP). The double-stranded cDNA was purified using an AxyPrep Mag PCR Clean-up kit (Axygen) and was then treated with End Prep Enzyme Mix to repair both ends and perform dA-tailing of cDNA in one reaction, followed by a T-A ligation to add adaptors to both ends. Size selection of adaptor-ligated DNA was then performed using an AxyPrep Mag PCR Clean-up kit (Axygen)to recover ∼360 bp fragments (with approximate insert sizes of 300 bp). The dUTP-marked second strand was digested with Uracil-Specific Excision Reagent (USER) enzyme (New England Biolabs). Each sample was then amplified by PCR for 11 cycles using P5 and P7 primers, with both primers carrying sequences that can anneal with the flow cell to perform bridge PCR and the P7 primer carrying a six-base index allowing for multiplexing. The PCR products were purified using an AxyPrep Mag PCR Clean-up kit (Axygen), validated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, United States), and quantified with a Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, United States). Next, libraries with different indices were multiplexed and loaded onto an Illumina HiSeq instrument according to the manufacturer’s instructions (Illumina, San Diego, CA, United States). Sequencing was carried out using a 2x150 paired-end (PE) configuration; image analysis and base calling were conducted using the HiSeq Control Software (HCS) + OLB + GAPipeline-1.6 (Illumina) on the HiSeq instrument. The sequences were processed and analyzed by GENEWIZ.
Raw data were processed to generate the pass filter data by Bcl2fastq (v 2.17.1.14), quality checked using the FastQC (v 0.10.1) tool and finally filtered to prepare the clean reads using Cutadapt (v 1.9.1). The clean data were aligned to the reference genome of C. glutamicum ATCC 13032 (Uniprot: UP000000582) using Bowtie 2 (v 2.1.01). The gene transcript expression levels were calculated using HTSeq (v 0.6.1p1) (Anders et al., 2015) and then normalized based on the FPKM (fragments per kilobase of exon per million fragments mapped) method. A false-discovery rate of ≤0.05 (Benjamini and Yekutieli, 2001) and the absolute value of the log2 ratio ≥ 1 were applied as threshold values to define a significant difference in gene expression levels using the DESeg2 (v1.6.3) in Bioconductor package (Anders and Huber, 2010). GO-TermFinder (v0.86) (Boyle et al., 2004) was used in identifying Gene Ontology (GO) terms to annotate a list of enriched genes with a significant p-value of less than 0.05.
Raw sequence data were deposited in the NCBI Sequence Read Archive database (SRA) under the accession number of SRP117800.
Results and Discussion
Culturing of C. glutamicum PUT-ALE and Its Parent Strain C. glutamicum ΔAPE6937R42
In our previous study, we constructed the C. glutamicum PUT-ALE strain from the ornithine producer C. glutamicum ΔAPE6937R42 (Jiang et al., 2013a) for putrescine production using a metabolic evolution strategy (Supplementary Text). We first compared the growth, glucose consumption, and putrescine production of the C. glutamicum PUT-ALE and the wild-type strain C. glutamicum ATCC 13032 strains. As shown in Figure 1, C. glutamicum PUT-ALE produced a higher level of putrescine than the wild-type strain C. glutamicum ATCC 13032. Interestingly, C. glutamicum PUT-ALE (μ = 0.38 h-1) showed a lower growth rate than C. glutamicum ATCC 13032 (μ = 0.43 h-1). This may be because C. glutamicum PUT-ALE and its parent strain C. glutamicum ΔAPE6937R42 are L-arginine and L-proline auxotrophs, resulting from knockouts of the argF and proB genes (Jiang et al., 2013a,b). The rate of glucose consumption by C. glutamicum PUT-ALE was similar to that of the wild-type strain ATCC 13032.
FIGURE 1.
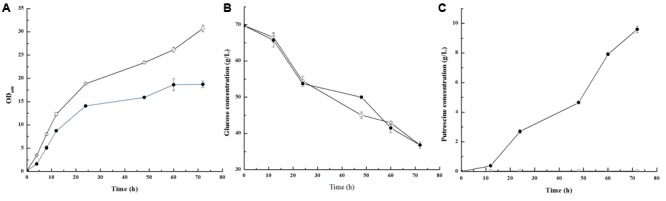
The growth (A), glucose consumption (B) and putrescine production (C) in Corynebacterium glutamicum PUT-ALE (•) and ATCC 13032 (∘). Data represent the average of three replicates and error bars represent standard deviation.
Transcriptomic Changes
To identify the cellular physiological and metabolic changes occurring in response to the overproduction of putrescine, we analyzed the transcriptomic changes between the putrescine-producer C. glutamicum PUT-ALE and the wild-type strain C. glutamicum ATCC13032. The putrescine production resulted in the differential expression of 607 genes, of which 283 were upregulated and 324 were downregulated (Supplementary Table 2). We also analyzed the transcriptomic changes between the putrescine-producer C. glutamicum PUT-ALE and its parent strain C. glutamicum ΔAPE6937R42. A total of 269 genes showed significantly different expression patterns (Supplementary Table 3). Of them, only 29 genes were related to metabolism. Thus, we focused on differentially expressed genes (DEGs) between C. glutamicum PUT-ALE and the wild-type strain C. glutamicum ATCC13032 in this study.
The GO project provides a controlled vocabulary to describe gene products within three categories: biological process, molecular function and cellular component (Boyle et al., 2004). GO enrichment analysis has become a commonly used approach for functional studies, and the GO analysis of DEGs can help biologists better understand the functional relevance of DEGs. In Figure 2, the results of a GO analysis of DEGs for C. glutamicum PUT-ALE vs. ATCC 13032 is presented.
FIGURE 2.
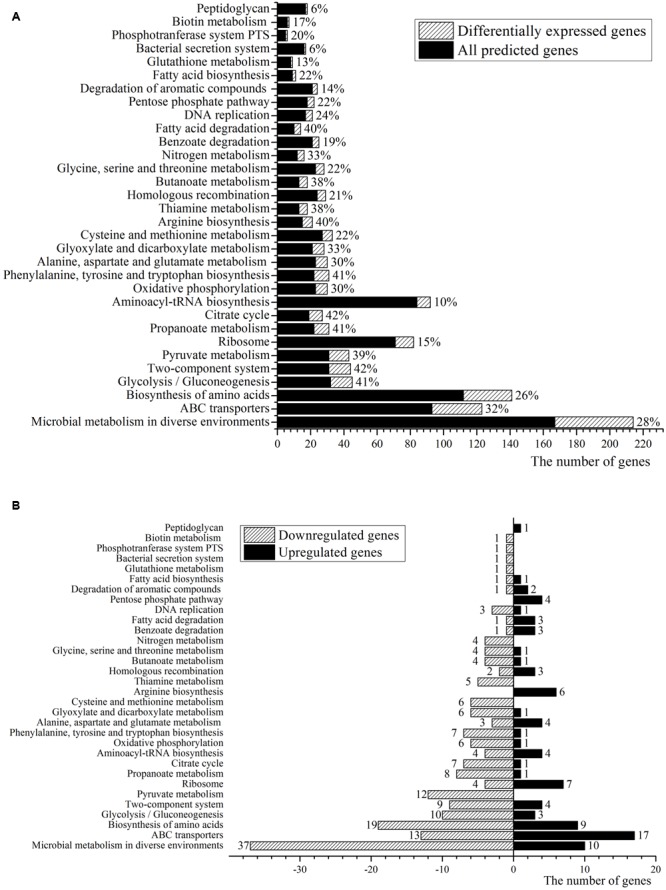
Pathway gene ontology enrichment analysis. (A) The ratio of the DEGs in the total number of genes detected. (B) The numbers of the DEGs.
DEGs involved in metabolic pathways are presented in Figures 3 and 4. As shown in Figure 3, most of the genes (glpX, fda, gpmB, eno, pyk, aceE, prpC1, acn, kgd, sdhAB, mdh, aceAB) involved in the glycolysis and tricarboxylic acid (TCA) cycle were significantly downregulated in C. glutamicum PUT-ALE compared to C. glutamicum ATCC13032. The low rate of growth of C. glutamicum PUT-ALE is consistent with the observed downregulated data. The pyc gene in C. glutamicum PUT-ALE was also downregulated. The pyruvate carboxylase encoded by pyc is one of the most important anaplerotic enzymes in C. glutamicum. Overexpression of the pyc gene can drive greater EMP flux into the TCA cycle to strengthen it. It has been demonstrated that overexpression of the pyc gene increased L-glutamate (Shirai et al., 2007; Hasegawa et al., 2008), L-arginine (Man et al., 2016b) and putrescine (Nguyen et al., 2015a) production in C. glutamicum. Thus, we expressed pyc or its mutant pyc458 from a plasmid in C. glutamicum PUT-ALE. As shown in Table 2, overexpression of the native pyc gene slightly increased putrescine production, while overexpression of the mutated pyc458 gene markedly increased putrescine production by 16% to 133.51 ± 7.20 mM. It has been reported that pyc458 is a beneficial mutation for L-lysine production (Ohnishi et al., 2002).
FIGURE 3.
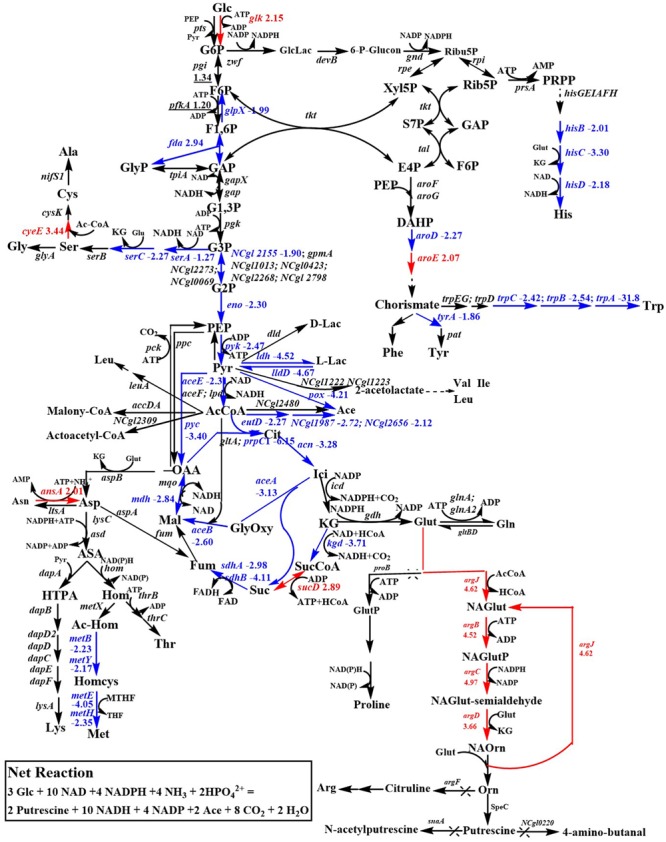
Differentially expressed genes involved in glycolysis, the TCA cycle, pyruvate metabolism, amino acid biosynthesis and the putrescine biosynthetic pathway. The numbers indicate the values of the log2 ratios of the expression levels in C. glutamicum PUT-ALE vs. C. glutamicum ATCC 13032. Red indicates upregulation. Blue indicates downregulation. Glc, glucose; G6P, glucose 6-phosphate; F6P, fructose 6-phosphate; F1,6P, fructose 1,6-bisphosphate; GAP, D-Glyceraldehyde 3-phosphate; GlyP, glycerone phosphate; G1,3P, 1,3-bisphospho-D-glycerate; G3P, 3-phosphoglycerate; G2P, 2-phospho-(R)-glycerate; PEP, phosphoenolpyruvate; Pyr, pyruvate; AcCoA, acetyl-CoA; GlcLac, D-glucono-1,5-lactone 6-phosphate; 6-P-glucon, 6-phospho-D-gluconate; Ribu5P, D-Ribulose 5-phosphate; Rib5P, D-ribose 5-phosphate; Xyl5P, D-Xylulose 5-phosphate; S7P, D-sedoheptulose 7-phosphate; E4P, D-erythrose 4-phosphate; PRPP, 5-phosphoribosyl diphosphate; His, L-histidine; DAHP, 3-deoxy-arabino-heptulonate 7-phosphate; Trp, L-tryptophan; Phe, L-phenylalanine; Tyr: L-tyrosine; D-Lac, D-Lactate; L-Lac, L-lactate; Ace, acetate; Val, L-valine; Ile, L-isoleucine; Leu, L-leucine; Ser, L-serine; Gly, L-glycine; Cys, L-cysteine; Ala, L-alanine; Cit, citrate; Ici, isocitrate; KG, 2-oxoglutarate; SucCoA, succinyl-CoA; Suc, succinate; Fum, fumarate; Mal, malate; OAA, oxaloacetate; Asp, L-aspartate; Asn, L-asparagine; ASA, L-aspartate 4-semialdehyde; HTPA, (2S,4S)-4-hydroxy-2,3,4,5-tetrahydrodipicolinate; Lys, L-lysine; Hom, homoserine; Thr, L-threonine; Ac-Hom, O-acetylhomoserine; Homcys, L-homocysteine; Met, L-methionine. Glut, L-glutamate; Gln, L-glutamine; GlutP, L-glutamate 5-phosphate; NAGlut, N-acetylglutamate; NAGlutP, N-acetyl-glutamyl 5-phosphate; NAGlut-semialdehyde, N-acetylglutamate semialdehyde; NAOrn, N-acetyl-ornithine; Orn, ornithine; Arg, L-arginine.
FIGURE 4.
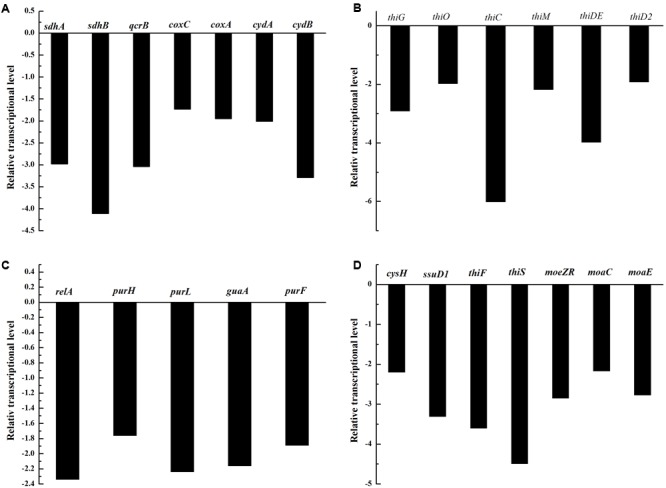
The relative transcriptional levels of genes involved in oxidative phosphorylation (A), vitamin biosynthesis (B), the metabolism of purine and pyrimidine (C), and sulfur metabolism (D).
Table 2.
Effect of the pyc and kgd gene on putrescine production in C. glutamicum PUT-ALE.
| Strain | OD600 | Putrescine (mM) | Yield (%, g/g) |
|---|---|---|---|
| C. glutamicum PUT-ALE (pEC-XK99E) | 19.41 ± 0.75 | 115.12 ± 2.42 | 27.0 ± 0.1 |
| C. glutamicum PUT-ALE (pEC-pyc) | 15.93 ± 0.35 | 123.18 ± 2.71 | 27.3 ± 0.6 |
| C. glutamicum PUT-ALE (pEC-pyc458) | 16.98 ± 0.44 | 133.51 ± 7.20 | 28.1 ± 1.5 |
| C. glutamicum PUT-ALE | 17.64 ± 0.27 | 107.95 ± 2.31 | 27.6 ± 1.1 |
| C. glutamicum PUT-ALE-KT | 20.04 ± 0.78 | 114.39 ± 2.14 | 28.4 ± 1.5 |
Data represent the average of three replicates and error bars represent standard deviation.
The transcription level of the kgd gene was also downregulated in C. glutamicum PUT-ALE. Alpha-ketoglutarate (KG) is a key node of the TCA cycle, and α-ketoglutarate decarboxylase (encoded by kgd) catalyzes the oxidative decarboxylation of KG to synthesize succinyl coenzyme A. The downregulation of kgd transcription can channel increased carbon flux into the glutamate biosynthetic pathway, enhancing putrescine production. Many groups have reported that decreasing the Kgd activity in Corynebacterium, or even deleting kgd, increased the production of glutamate (Asakura et al., 2007; Kim et al., 2009), the glutamate-derived compound putrescine (Nguyen et al., 2015a), gamma-aminobutyric acid (Jorge et al., 2017) and L-arginine (Chen et al., 2015; Man et al., 2016b). It has been demonstrated that the exchanging the translational start codon of the kgd gene from GTG to TTG reduced the Kgd activity from 11 to 7 mU/mg (Nguyen et al., 2015a). Thus, we replaced the native GTG start codon of the C. glutamicum PUT-ALE kgd gene with TTG to obtain C. glutamicum PUT-ALE-KT. The resulting strain (C. glutamicum PUT-ALE-KT) produced a higher level of putrescine (114.39 ± 2.14 mM) than C. glutamicum PUT-ALE (107.95 ± 2.31, Table 2), indicating that decreasing the activity of Kgd may be a strategy for further improving putrescine production.
In Figure 3, it is observed that may genes that are involved in pyruvate metabolism were significantly down-regulated in C. glutamicum PUT-ALE, such as ldh, lldD, pox, eutD, acyP, and ackA. The downregulation of pyruvate metabolism can drive carbon flux toward glycolysis for putrescine biosynthesis. Genes involved in the putrescine biosynthetic pathway, such as argJ, argB, argC, and argD were significantly upregulated in C. glutamicum PUT-ALE (Figure 3).
We also observed that some genes involved in the serine, methionine, histidine, tryptophan, and tyrosine biosynthetic pathway were significantly downregulated (Figure 3). These genes include serA, serC, metB, metY, metE, metH, hisB, hisC, hisD, aroD, trpC, trpB, trpA, and tyrA. The enzyme encoded by serC or hisC catalyzes the glutamate-consuming reaction. The downregulation of serC and hisC transcription may provide more glutamate for putrescine biosynthesis.
As shown in Figure 4A, the transcriptional levels of genes involved in oxidative phosphorylation were down-regulated, such as sdhA, sdhB, qcrB, coxC, coxA, cydA, and cydB. Genes involved in thiamine and vitamin B6 biosynthesis, such as thiG, thiO, thiC, thiM, thiDE, and thiD2, were also down-regulated (Figure 4B). The transcriptional levels of genes involved in purine and pyrimidine metabolism, such as relA, purH, purL, guaA, and purF were down-regulated (Figure 4C), as were genes involved in sulfur metabolism, such as cysH, ssuD1, thiF, thiS, moeZR, moaC, and moaE (Figure 4D). Of the above genes, thiM, thiDE, thiD2, relA, purl, guaA, and moeZR encode adenosine triphosphate (ATP)-consuming enzymes. The transcriptional downregulation of these genes could result in more ATP being available for putrescine production.
ATP is the most important energy source for metabolic reaction and pathways, playing an important role in cell growth and the production of target metabolites. Many ATP-consuming enzyme encoding genes, such as rbsK, cysD, cysN, pknG, pknB, bioD, iolC, mthfs, coaE, chlI, glgC, and moeZR, were downregulated in C. glutamicum PUT-ALE (Supplementary Table 2). It has been reported that increasing the ATP supply enhanced L-arginine production in C. glutamicum (Man et al., 2016a). The protein kinases encoded by pknG and pknB phosphorylate the α-ketoglutarate decarboxylase inhibitor OdhI, and unphosphorylated OdhI inhibits α-ketoglutarate decarboxylase activity (Niebisch et al., 2006; Schultz et al., 2009; Raasch et al., 2014). Thus, the decreased transcription of pknG and pknB in C. glutamicum PUT-ALE may increase the ability of OdhI to inhibit α-ketoglutarate decarboxylase. The regulation of OdhI phosphorylation by the deletion of the protein kinase encoding gene pknG has been previously shown to increase glutamate production (Schultz et al., 2007).
In Figure 3, it is observed that synthesizing one mole of putrescine requires 2 moles of NADPH and 5 moles of NAD. Thus, NADPH availability and transhydrogenation between NAD and NADP are important for putrescine production. The transcriptional levels of the NADPH-consuming enzyme encoding genes [rhcM2 and NAD (FAD)-dependent dehydrogenase gene NCgl2615] and the NAD-consuming enzyme encoding genes (gabD3, iolG, and fdhF) were significantly downregulated. The transcriptional levels of NADPH-forming enzyme encoding genes, such as proA, aldH, and mdhB, were significantly upregulated in C. glutamicum PUT-ALE (Supplementary Table 2). The expression patterns can increase NADPH or NAD availability for putrescine production. It has been demonstrated that increasing NADPH availability enhances L-ornithine production (Jiang et al., 2013b; Hwang and Cho, 2014; Kim et al., 2015).
CRISPRi system is a powerful tool to repress expression of targeted genes (Qi et al., 2013). It has successfully applied to repress genes for improving L-lysine and L-glutamate production in C. glutamicum (Cleto et al., 2016). Thus, we established a CRISPRi system, which contains the dcas9 (K848A/K1003A/R1060A) plasmid pEC-dcas9∗ (Supplementary Figure 1A) and the sgRNA plasmid pXMJPsod-X-sgRNA (Supplementary Figure 1B). To validate the effects of ATP- and NADPH-consuming enzyme genes, we applied the CRISPRi system to repress expression of ATP- and NADPH-consuming enzyme encoding genes in C. glutamicum PUT-ALE. The results were presented in Table 3. Repressing ATP-consuming enzyme encoding genes, such as carB, xylB, accDA, purL, coaA, pknG, and panC2 resulted in increasing putrescine production of 5–10%. Repressing the dxr, aroE, or trxB expression enhanced putrescine production by 13, 19, or 20%, respectively. The dxr encodes 1-deoxy-D-xylulose 5-phosphate reductoisomerase which catalyzes the reduction of 1-deoxy-D-xylulose 5-phosphate to 2-C-methyl-D-erythritol 4-phosphate in the presence of NADPH. The aroE encodes shikimate dehydrogenase which catalyzes NAD+-dependent oxidation of shikimate to 3-dehydroshikimate. The trxB encodes thioredoxin reductase which catalyzes the reduction of thioredoxin disulfide to thioredoxin in the presence of NADPH. Repressing the dxr, trxB, or aroE expression can provide more NADPH or NAD for putrescine production.
Table 3.
Effect of perturbations of ATP- and NADPH-consuming enzyme encoding genes on putrescine production in C. glutamicum PUT-ALE.
| Gene targeted | Gene encoding enzyme/protein | OD600 | Putrescine (mM) | Ratio |
|---|---|---|---|---|
| ATP-consuming | ||||
| Control | 16.83 ± 0.18 | 103.66 ± 3.29 | 1.00 | |
| carB | Carbamoyl-phosphate synthase large subunit [EC6.3.5.5] | 17.27 ± 0.20 | 109.00 ± 4.36 | 1.05 |
| thrB1 | Homoserine kinase [EC 2.7.1.39] | 17.02 ± 0.20 | 105.05 ± 0.45 | 1.01 |
| coaA | Pantothenate kinase [EC 2.7.1.33], | 16.56 ± 0.51 | 110.18 ± 1.32 | 1.06 |
| glnA | Glutamine synthetase [EC 6.3.1.2] | 15.63 ± 0.06 | 86.44 ± 5.17 | 0.86 |
| nadD | Nicotinate-nucleotide adenylyltransferase [EC 2.7.7.18] | 16.71 ± 0.18 | 95.68 ± 0.93 | 0.92 |
| hemH | Phosphoribosylaminoimidazole-succinocarboxamide synthase [EC 6.3.2.6], | 16.55 ± 0.35 | 86.74 ± 8.33 | 0.84 |
| xylB | Xylulokinase [EC 2.7.1.17] | 16.62 ± 0.42 | 108.88 ± 0.07 | 1.05 |
| guaA | GMP synthase (glutamine-hydrolysing) [EC:6.3.5.2], | 17.25 ± 0.24 | 82.71 ± 1.40 | 0.80 |
| accBC | Acyl-CoA carboxylase | 18.26 ± 0.42 | 80.20 ± 0.55 | 0.77 |
| accDA | Acetyl-CoA carboxylase beta subunit [EC 6.4.1.2] | 17.36 ± 0.10 | 109.24 ± 0.04 | 1.05 |
| purL | Phosphoribosylformylglycinamidine synthase [EC 6.3.5.3] | 16.60 ± 0.54 | 108.86 ± 0.59 | 1.05 |
| purQ | Phosphoribosylformylglycinamidine synthase [EC 6.3.5.3] | 17.91 ± 0.38 | 103.78 ± 5.31 | 1.00 |
| panC1 | Pantoate-beta-alanine ligase [EC 6.3.2.1] | 18.98 ± 1.34 | 107.24 ± 0.98 | 1.03 |
| panC2 | Pantoate-beta-alanine ligase [EC 6.3.2.1] | 17.98 ± 0.58 | 113.94 ± 2.98 | 1.10 |
| pknG | Serine/threonine protein kinases [EC 2.7.11.1] | 17.59 ± 0.35 | 109.57 ± 1.21 | 1.06 |
| NADPH-consuming | ||||
| Control | 19.80 ± 0.30 | 108.99 ± 2.51 | 1.00 | |
| pobA | p-hydroxybenzoate 3-monooxygenase [EC 1.14.13.2] | 15.90 ± 0.24 | 105.36 ± 2.53 | 0.93 |
| aldH | 2,5-dioxopentanoate dehydrogenase [EC 1.2.1.26] | 16.07 ± 0.15 | 92.65 ± 3.83 | 0.88 |
| fabG1 | 3-oxoacyl-[acyl-carrier protein] reductase [EC 1.1.1.100], | 16.08 ± 0.26 | 108.96 ± 0.79 | 0.96 |
| adhC | Maleylacetate reductase [EC 1.3.1.32] | 15.62 ± 0.16 | 101.45 ± 2.38 | 0.93 |
| gor | Dihydrolipoamide dehydrogenase/glutathione oxidoreductase and related enzymes, [EC 1.8.1.7] | 15.52 ± 0.35 | 94.92 ± 3.05 | 0.87 |
| dxr | 1-deoxy-D-xylulose-5-phosphate reductoisomerase [EC 1.1.1.267] | 15.36 ± 0.50 | 123.18 ± 0.55 | 1.13 |
| asd | aspartate-semialdehyde dehydrogenase [EC 1.2.1.11] | 15.54 ± 0.18 | 99.7 ± 1.48 | 0.91 |
| proA | Glutamate-5-semialdehyde dehydrogenase [EC 1.2.1.41] | 16.21 ± 0.19 | 98.51 ± 0.87 | 0.90 |
| NCgl2558 | Transcriptional regulators | 15.87 ± 0.12 | 99.06 ± 1.92 | 0.87 |
| thyX | Thymidylate synthase (FAD) [EC 2.1.1.148] | 19.77 ± 0.48 | 103.71 ± 3.55 | 0.95 |
| aroE | Shikimate dehydrogenase [EC 1.1.1.25], | 16.74 ± 0.06 | 129.29 ± 1.76 | 1.19 |
| sir | Sulfite reductase (ferredoxin) [EC 1.8.7.1] | 17.82 ± 0.30 | 106.87 ± 2.02 | 0.98 |
| NCgl0503 | Aldo/keto reductases | 15.18 ± 2.58 | 110.53 ± 3.30 | 1.01 |
| ddh | diaminopimelate dehydrogenase [EC 1.4.1.16] | 15.93 ± 0.09 | 98.82 ± 2.68 | 0.91 |
| ilvC | Ketol-acid reductoisomerase [EC 1.1.1.86] | 16.32 ± 0.30 | 102.49 ± 3.71 | 0.94 |
| qor | NADPH:quinone reductase and related Zn-dependent oxidoreductases [EC 1.6.5.5] | 16.83 ± 0.27 | 108.78 ± 0.34 | 0.98 |
| trxB | Thioredoxin reductase (NADPH) [EC 1.8.1.9] | 17.16 ± 0.36 | 131.12 ± 0.88 | 1.20 |
| NCgl0200 | NADPH:quinone reductase and related Zn-dependent oxidoreductases | 16.26 ± 0.24 | 109.30 ± 0.76 | 0.99 |
Data represent the average of three replicates and error bars represent standard deviation.
A total of 76 secretion and membrane transport protein encoding genes were significantly differentially expressed in C. glutamicum PUT-ALE (Supplementary Table 2). Of these genes, 30 were downregulated and 46 were upregulated. The differential expression may affect the metabolite transport. It has been previously shown that CgmA is a putrescine export permease and that overexpression of the cgmA gene increased putrescine production in C. glutamicum (Nguyen et al., 2015a,b). We also observed that the transcriptional of the cgmA gene in C. glutamicum PUT-ALE was significantly upregulated (Supplementary Table 2). A total of 30 transcription factors were significantly differentially expressed in C. glutamicum PUT-ALE (Supplementary Table 2). Of these genes, 13 were downregulated and 17 were upregulated. In addition, 378 other genes, such as unknown, transposase and ribosomal RNA genes, were significantly differentially expressed in C. glutamicum PUT-ALE (Supplementary Table 2). Of these genes, 189 were downregulated and 189 were upregulated.
Conclusion
We comparatively analyzed the transcriptomic changes in response to putrescine production in the strain C. glutamicum PUT-ALE. The overproduction of putrescine resulted in the transcriptional downregulation of genes involved in: glycolysis, the TCA cycle, pyruvate degradation, the biosynthesis of some amino acids, oxidative phosphorylation, vitamin biosynthesis (thiamine and vitamin 6), the metabolism of purine, pyrimidine and sulfur; and ATP-, NAD- and NADPH-consuming enzymes. The transcriptional levels of genes involved in ornithine biosynthesis and those encoding NADPH-forming enzymes were upregulated in the putrescine producer C. glutamicum PUT-ALE. The comparative transcriptomic analysis provided some genetic modification strategies for further improving putrescine production. Overexpression of pyc or its mutant pyc458, and replacing the kgd native start codon GTG with TTG further improved putrescine production. Repressing ATP- and NADPH-consuming enzyme coding gene expression via CRISPRi also enhanced putrescine production. To the best of our knowledge, this is the first report on increasing putrescine production via repressing ATP- and NADPH-consuming enzyme coding gene expression.
Author Contributions
ZL performed the experiments. J-ZL directed the project and wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the National Natural Science Foundation of China (grant no. 21276289), the Natural Science Foundation of Guangdong Province (no. 2015A030311036), the Project of the Scientific and Technical Program of Guangdong Province (no. 2015A010107004) and the Project of the Scientific and Technical Program of Guangzhou (no. 201607010028).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.01987/full#supplementary-material
References
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W. (2015). HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y., Kimura E., Usuda Y., Kawahara Y., Matsui K., Osumi T., et al. (2007). Altered metabolic flux due to deletion of odhA causes L-glutamate overproduction in Corynebacterium glutamicum. Appl. Environ. Microbiol. 73 1308–1319. 10.1128/AEM.01867-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Yekutieli D. (2001). The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29 1165–1188. 10.1186/1471-2105-9-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle E. I., Weng S. A., Gollub J., Jin H., Botstein D., Cherry J. M., et al. (2004). GO::termfinder – open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics 20 3710–3715. 10.1093/bioinformatics/bth456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. L., Chen X. L., Wan F., Zhang B., Chen J. C., Xiong Y. H. (2015). Effect of Tween 40 and DtsR1 on L-arginine overproduction in Corynebacterium crenatum. Microb. Cell Fact. 14 119. 10.1186/s12934-015-0310-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Kyeong H. H., Choi J. M., Kim H. S. (2014). Rational design of ornithine decarboxylase with high catalytic activity for the production of putrescine. Appl. Microbiol. Biotechnol. 98 7483–7490. 10.1007/s00253-014-5669-8 [DOI] [PubMed] [Google Scholar]
- Cleto S., Jensen J. V. K., Wendisch V. F., Lu T. K. (2016). Corynebacterium glutamicum metabolic engineering with CRISPR interference (CRISPRi). ACS Synth. Biol. 5 375–385. 10.1021/acssynbio.5b00216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cress B. F., Toparlak O. D., Guleria S., Lebovich M., Stieglitz J. T., Englaender J. A., et al. (2015). CRISPathBrick: modular combinatorial assembly of type II-A CRISPR arrays for dCas9-mediated multiplex transcriptional repression in E. coli. ACS Synth. Biol. 4 987–1000. 10.1021/acssynbio.5b00012 [DOI] [PubMed] [Google Scholar]
- Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177 4121–4130. 10.1128/jb.177.14.4121-4130.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa T., Hashimoto K. I., Kawasaki H., Nakamatsu T. (2008). Changes in enzyme activities at the pyruvate node in glutamate-overproducing Corynebacterium glutamicum. J. Biosci. Bioeng. 105 12–19. 10.1263/jbb.105.12 [DOI] [PubMed] [Google Scholar]
- Hwang G. H., Cho J. Y. (2014). Enhancement of L-ornithine production by disruption of three genes encoding putative oxidoreductases in Corynebacterium glutamicum. J. Ind. Microbiol. Biotechnol. 41 573–578. 10.1007/s10295-013-1398-8 [DOI] [PubMed] [Google Scholar]
- Jiang L. Y., Chen S. G., Zhang Y. Y., Liu J. Z. (2013a). Metabolic evolution of Corynebacterium glutamicum for increased production of L-ornithine. BMC Biotechnol. 13:47. 10.1186/1472-6750-13-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L. Y., Zhang Y. Y., Li Z., Liu J. Z. (2013b). Metabolic engineering of Corynebacterium glutamicum for increasing the production of L-ornithine by increasing NADPH availability. J. Ind. Microbiol. Biotechnol. 40 1143–1151. 10.1007/s10295-013-1306-2 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Chen B., Duan C., Sun B., Yang J., Yang S. (2015). Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 81 2506–2514. 10.1128/AEM.04023-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorge J. M. P., Nguyen A. Q. D., Perez-Garcia F., Kind S., Wendisch V. F. (2017). Improved fermentative production of gamma-aminobutyric acid via the putrescine route: systems metabolic engineering for production from glucose, amino sugars, and xylose. Biotechnol. Bioeng. 114 862–873. 10.1002/bit.26211 [DOI] [PubMed] [Google Scholar]
- Kim J., Hirasawa T., Sato Y., Nagahisa K., Furusawa C., Shimizu H. (2009). Effect of odhA overexpression and odhA antisense RNA expression on Tween-40-triggered glutamate production by Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 81 1097–1106. 10.1007/s00253-008-1743-4 [DOI] [PubMed] [Google Scholar]
- Kim S. Y., Lee J., Lee S. Y. (2015). Metabolic engineering of Corynebacterium glutamicum for the production of L-ornithine. Biotechnol. Bioeng. 112 416–421. 10.1002/bit.25440 [DOI] [PubMed] [Google Scholar]
- Kirchner O., Tauch A. (2003). Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J. Biotechnol. 104 287–299. 10.1016/S0168-1656(03)00148-2 [DOI] [PubMed] [Google Scholar]
- Man Z. W., Rao Z. M., Xu M. J., Guo J., Yang T. W., Zhang X., et al. (2016a). Improvement of the intracellular environment for enhancing L-arginine production of Corynebacterium glutamicum by inactivation of H2O2-forming flavin reductases and optimization of ATP supply. Metab. Eng. 38 310–321. 10.1016/j.ymben.2016.07.009 [DOI] [PubMed] [Google Scholar]
- Man Z. W., Xu M. J., Rao Z. M., Guo J., Yang T. W., Zhang X., et al. (2016b). Systems pathway engineering of Corynebacterium crenatum for improved L-arginine production. Sci. Rep. 6:28629. 10.1038/srep28629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A. Q. D., Schneider J., Reddy G. K., Wendisch V. F. (2015a). Fermentative production of the diamine putrescine: system metabolic engineering of Corynebacterium glutamicum. Metabolites 5 211–231. 10.3390/metabo5020211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A. Q. D., Schneider J., Wendisch V. F. (2015b). Elimination of polyamine N-acetylation and regulatory engineering improved putrescine production by Corynebacterium glutamicum. J. Biotechnol. 201 75–85. 10.1016/j.jbiotec.2014.10.035 [DOI] [PubMed] [Google Scholar]
- Niebisch A., Kabus A., Schultz C., Weil B., Bott M. (2006). Corynebacterial protein kinase G controls 2-oxoglutarate dehydrogenase activity via the phosphorylation status of the OdhI protein. J. Biol. Chem. 281 12300–12307. 10.1074/jbc.M512515200 [DOI] [PubMed] [Google Scholar]
- Ohnishi J., Mitsuhashi S., Hayashi M., Ando S., Yokoi H., Ochiai K., et al. (2002). A novel methodology employing Corynebacterium glutamicum genome information to generate a new L-lysine-producing mutant. Appl. Microbiol. Biotechnol. 58 217–223. 10.1007/s00253-001-0883-6 [DOI] [PubMed] [Google Scholar]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., Arkin A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152 1173–1183. 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z. G., Xia X. X., Lee S. Y. (2009). Metabolic engineering of Escherichia coli for the production of putrescine: a four carbon diamine. Biotechnol. Bioeng. 104 651–662. 10.1002/bit.22502 [DOI] [PubMed] [Google Scholar]
- Raasch K., Bocola M., Labahn J., Leitner A., Eggeling L., Bott M. (2014). Interaction of 2-oxoglutarate dehydrogenase OdhA with its inhibitor OdhI in Corynebacterium glutamicum: mutants and a model. J. Biotechnol. 191 99–105. 10.1016/j.jbiotec.2014.05.023 [DOI] [PubMed] [Google Scholar]
- Schneider J., Eberhardt D., Wendisch V. F. (2012). Improving putrescine production by Corynebacterium glutamicum by fine-tuning ornithine transcarbamoylase activity using a plasmid addiction system. Appl. Microbiol. Biotechnol. 95 169–178. 10.1007/s00253-012-3956-9 [DOI] [PubMed] [Google Scholar]
- Schneider J., Wendisch V. F. (2010). Putrescine production by engineered Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 88 859–868. 10.1007/s00253-010-2778-x [DOI] [PubMed] [Google Scholar]
- Schultz C., Niebisch A., Gebel L., Bott M. (2007). Glutamate production by Corynebacterium glutamicum: dependence on the oxoglutarate dehydrogenase inhibitor protein OdhI and protein kinase PknG. Appl. Microbiol. Biotechnol. 76 691–700. 10.1007/s00253-007-0933-9 [DOI] [PubMed] [Google Scholar]
- Schultz C., Niebisch A., Schwaiger A., Viets U., Metzger S., Bramkamp M., et al. (2009). Genetic and biochemical analysis of the serine/threonine protein kinases PknA, PknB, PknG and PknL of Corynebacterium glutamicum: evidence for non-essentiality and for phosphorylation of OdhI and FtsZ by multiple kinases. Mol. Microbiol. 74 724–741. 10.1111/j.1365-2958.2009.06897.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott E., Peter F., Sanders J. (2007). Biomass in the manufacture of industrial products – the use of proteins and amino acids. Appl. Microbiol. Biotechnol. 75 751–762. 10.1007/s00253-007-0932-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai T., Fujimura K., Furusawa C., Nagahisa K., Shioya S., Shimizu H. (2007). Study on roles of anaplerotic pathways in glutamate overproduction of Corynebacterium glutamicum by metabolic flux analysis. Microb. Cell Fact. 6 19. 10.1186/1475-2859-6-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker I. M., Gao L. Y., Zetsche B., Scott D. A., Yan W. X., Zhang F. (2016). Rationally engineered Cas9 nucleases with improved specificity. Science 351 84–88. 10.1126/science.aad5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. L., Jin H. Y., Cheng X. L., Wang Q., Qi Q. S. (2016). Transcriptomic analysis for elucidating the physiological effects of 5-aminolevulinic acid accumulation on Corynebacterium glutamicum. Microbiol. Res. 192 292–299. 10.1016/j.micres.2016.08.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


