Abstract
The Rcs phosphorelay system, a non-orthodox two-component regulatory system, integrates environmental signals, regulates gene expression, and alters the physiological behavior of members of the Enterobacteriaceae family of Gram-negative bacteria. Recent studies of Rcs system focused on protein interactions, functions, and the evolution of Rcs system components and its auxiliary regulatory proteins. Herein we review the latest advances on the Rcs system proteins, and discuss the roles that the Rcs system plays in the environmental adaptation of various Enterobacteriaceae species.
Keywords: Rcs phosphorelay system, envelope stress, acetylation, auxiliary regulators, molecular evolution
Introduction
The Regulator of Capsule Synthesis phosphorelay system was originally identified as a positive regulator of colanic acid capsule in Escherichia coli (Gottesman et al., 1985). Colanic acid capsule is composed of glucose, galactose, and glucuronic acid. Overproduction of colanic acid results in a distinctive ‘mucoid’ colony phenotype (Gottesman et al., 1985). Subsequently studies have shown that Rcs phosphorelay system is a non-orthodox two-component signal transduction system (TCS) present in members of the Enterobacteriaceae family of Gram-negative bacteria (Gottesman et al., 1985; Allen et al., 1987; Brill et al., 1988; Majdalani and Gottesman, 2005; Huang et al., 2006). The Rcs system is composed of three core proteins, the transmembrane sensor kinase RcsC, the transmembrane protein RcsD, and the response regulator RcsB (Majdalani and Gottesman, 2005). The response regulator RcsB and the membrane-localized hybrid sensor kinase RcsC represent the classical members of bacterial TCS (Stout and Gottesman, 1990), whereas the membrane-bound sensor RcsD lacks kinase activity (Majdalani and Gottesman, 2005; Clarke, 2010). Several accessory components have also been described, such as auxiliary regulators RcsA, BglJ, GadE, MatA (EcpR), DctR, TviA, RflM, and RmpA (Table 1; McCallum and Whitfield, 1991; Virlogeux et al., 1996; Winter et al., 2009; Castanié-Cornet et al., 2010; Cheng et al., 2010; Venkatesh et al., 2010; Kühne et al., 2016; Pannen et al., 2016), and lipoprotein RcsF that senses envelope stress (Gervais and Drapeau, 1992; Majdalani et al., 2005; Figure 1A). As a non-orthodox TCS, the Rcs system engages in complex multistep phosphorelay signal transduction (Figure 1; Takeda et al., 2001; Majdalani and Gottesman, 2005). Upon receiving an extracytoplasmic stimulus, likely via RcsF, the hybrid sensor RcsC autophosphorylates at a conserved histidine residue (His479 in RcsC from E. coli) on its histidine kinase (HK) domain in an ATP-dependent manner. The phosphoryl group is then transferred to an aspartate residue (Asp875 in RcsC from E. coli) on the phosphoryl receiver (PR) domain of RcsC, and subsequently to a histidine residue (His842 in RcsD from E. coli) on the unique C-terminal histidine-containing phosphotransmitter (HPt) domain of RcsD. Finally, the phosphoryl group is transmitted to an aspartate residue (Asp56 in RcsB from E. coli) on the PR domain of RcsB (Chen et al., 2001; Takeda et al., 2001; Clarke et al., 2002; Majdalani and Gottesman, 2005). In addition, in the absence of extracytoplasmic stress or in response to certain metabolic stress, RcsB can also be phosphorylated by the central metabolite acetyl phosphate (AcP), an intracellular low-molecular-weight phosphoryl group donor (Fredericks et al., 2006; Hu et al., 2013).
Table 1.
Homodimer and heterodimers of RcsB.
| Dimer | Species | Function(s) of the dimer | Reference |
|---|---|---|---|
| RcsB–RcsB | E. coli and others | Activation of rprA which positively regulates RpoS in E. coli, repression of glutamate decarboxylase gadA in E. coli and etc. | Ferrières et al., 2009;Castanié-Cornet et al., 2010 |
| RcsA–RcsB | E. coli and others | Activation of cps operon involved in capsular polysaccharide biosynthesis in E. coli, activation of yjbEFGH involved in EPS synthesis in E. coli and etc. | Stout et al., 1991;Majdalani and Gottesman, 2005;Ferrières et al., 2007 |
| BglJ–RcsB | E. coli | Activation of bgl loci involved in aryl-β, D-glucoside synthesis, activation of transcription factor leuO and etc. | Venkatesh et al., 2010;Stratmann et al., 2012 |
| GadE–RcsB | E. coli | Activation of gad genes involved in acid resistance | Castanié-Cornet et al., 2010 |
| MatA–RcsB | E. coli | Activation of mat operon involved in Mat fimbria synthesis | Pannen et al., 2016 |
| DctR–RcsB | E. coli | Protection against organic acids while none of direct target genes were identified | Pannen et al., 2016 |
| RmpA–RcsB | K. pneumoniae | Activation of cps operon involved in capsular polysaccharide biosynthesis | Cheng et al., 2010 |
| RflM–RcsB | S. enterica | Repression of flhDC which is involved in flagellar synthesis | Kühne et al., 2016 |
| TviA–RcsB | S. typhi and S. enterica | Activation of viaA and viaB involved in Vi antigen synthesis | Virlogeux et al., 1996;Winter et al., 2009 |
FIGURE 1.
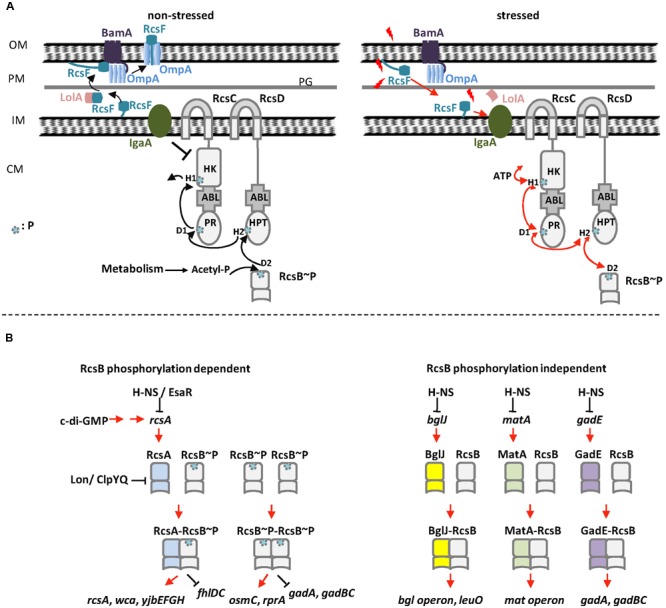
The Rcs phosphorelay system in E. coli. (A) Regulation of RcsB phosphorylation in E. coli. Left: in the absence of stress, RcsF is funneled by BamA and sequestered by OmpA as a surface-exposed protein. YrfF (IgaA in S. enterica serovar Typhimurium) inhibits the kinase activity of RcsC, resulting in dephosphorylation of RcsB. In this situation, AcP might act as a phosphoryl group donor to keep a low level of RcsB phosphorylation. Right: in the presence of stress, RcsF does not interact with BamA and OmpA but interacts with IgaA that release the inhibition of YrfF (IgaA in S. enterica serovar Typhimurium). RcsC autophosphorylates at the H1 position of its HK domain in an ATP-dependent manner. The phosphoryl group is then transferred to D1 on the PR domain of RcsC, then to H2 on the HPt domain of RcsD and finally to RcsB. (B) The homo- and heterodimers of RcsB in E. coli. Left: RcsB forms homodimer or heterodimer with RcsA to regulate target genes in a phosphorylation dependent manner. Right: RcsB forms heterodimer with BglJ, GadE, or MatA to regulate target genes in a phosphorylation independent manner. OM, outer membrane; PM, periplasm; IM, inner membrane; CM, cytoplasm; PG, peptidoglycan; P, phosphoryl group.
Highlights.
Regulator of Capsule Synthesis phosphorelay system: a non-orthodox two-component signal transduction (TCS) system with multistep cascade, His–Asp–His–Asp, by three core proteins RcsC, RcsD, and RcsB.
Envelope stress: stress by alteration of bacterial envelope that is a complex extracytoplasmic compartment.
Lysine acetylation: a reversible posttranslational modification of proteins and plays a key role in regulating gene expression, with an addition of an acetyl moiety to the 𝜀-amino group of a lysine residue.
Molecular evolution: the process of change in the sequence composition of cellular molecules such as DNA, RNA, and proteins across generation.
RcsB can form homodimer or heterodimers with the auxiliary proteins, which then interact with a conserved motif in target genes to modulate their transcription. The involvement of a hybrid sensor kinase, additional functional domains, multiple phosphodonors, several acceptor sites, and a variety of auxiliary proteins results in a high complex signaling system, which likely increases the flexibility of the Rcs system and provides multiple checkpoints to facilitate precise regulation of gene expression. Recent studies on the Rcs system focused on protein interactions, functions, modifications, and on the evolution of the Rcs components. Herein we review the latest advances of the complex Rcs system and discuss the mechanisms by which Rcs system senses environmental changes and regulates gene expression to allow Enterobacteriaceae species to adapt to different environments.
Inputs Regulating RCS Regulon
The first environmental signal reported to activate the Rcs system was osmotic upshift in E. coli, and several other regulatory inputs have been discovered since then (Sledjeski and Gottesman, 1996). The activation or repression of Rcs system, in turn, regulates its target genes to relieve the damages caused by the input signals. For example, lysozyme induces Rcs system, which upregulates the transcription of lysozyme inhibitors in E. coli (Callewaert et al., 2009). Similarly, the Rcs system is activated upon oxidative stress in Salmonella, which regulates the transcription of dps gene that protects the bacterial DNA against damage resulting from host reactive oxygen species (ROS) production during infection (Farizano et al., 2014). In the following section, we will discuss several examples of inputs that regulate the Rcs system.
Perturbation of the Envelope Leads to Activation of Rcs System
The envelope protects the bacterial cell from toxic substances or environmental stress. Extracellular or intracellular signals may cause perturbation of the cell envelope, which activates the Rcs system (Majdalani et al., 2005). In E. coli, these signals include the damage of periplasmic peptidoglycan caused by lysozyme (Callewaert et al., 2009), the transient cell wall-deficient (or L-form) state caused by exposure to β-lactam antibiotics (Hirakawa et al., 2003; Glover et al., 2009), defective lipoprotein sorting by lolA mutation (Tao et al., 2012), loss of the periplasm-spanning protein TolA (Vianney et al., 2005; Morgan et al., 2014), and modification of the peptidoglycan in the absence of certain penicillin-binding proteins PBP4, PBP5, PBP7, or AmpH (Table 2; Evans et al., 2013). Rcs system is also induced by perturbation of the cell envelope in other bacteria. For example, a change in the concentration of osmoregulated periplasmic glucans (OPGs) in Dickeya dadantii (Bontemps-Gallo et al., 2013), the absence of UDP-glucose dehydrogenase in Edwardsiella tarda (Lv et al., 2012), alteration in the enterobacterial common antigen (ECA) structure in Serratia marcescens (Castelli and Vescovi, 2011), and the outer membrane damage caused by oxidative stress in Salmonella can modulate Rcs phosphorelay system (Table 2; Farizano et al., 2014). In summary, the perturbation of the envelope caused by these extracellular or intracellular signals can be sensed by Rcs system, which in turn regulates its target genes to allow cells to adapt to the environmental or genetic changes.
Table 2.
Extracellular or intracellular signals that cause envelope stress and induce Rcs system.
| Signal | Description | Species | Function | Reference |
|---|---|---|---|---|
| Lysozyme | N-acetylmuramide glycanohydrolase | E. coli | Destruction of the periplasmic peptidoglycan | Callewaert et al., 2009 |
| β-lactam antibiotics | Chemicals repressing penicillin binding protein (PBPs) | E. coli | Inhibition of peptidoglycan formation and transition to L-form state | Hirakawa et al., 2003; Glover et al., 2009 |
| lolA mutation Ile93-Cys/Phe140Cys | Lipoprotein-specific chaperone lolA | E. coli | Defect in lipoprotein sorting | Tao et al., 2012 |
| pbp4, pbp5, pbp7, or ampH deletion | Penicillin-binding protein pbp4, etc. | E. coli | Modification of the peptidoglycan | Evans et al., 2013 |
| tolA deletion | Periplasm spanning protein tolA | E. coli | Outer membrane perturbation | Morgan et al., 2014 |
| mdoH deletion | mdoH, glucosyltransferase for the synthesis of periplasmic membrane-derived oligosaccharides | E. coli | Defect in membrane-derived oligosaccharides synthesis | Shiba et al., 2012 |
| The concentration of OPGs | OPGs, osmoregulated periplasmic glucans | D. dadantii | Sensing the osmolarity of medium | Bontemps-Gallo et al., 2013 |
| ugd deletion | UDP-glucose dehydrogenase ugd | E. tarda | Defect in LPS, a truncated core with no O-antigen attached | Lv et al., 2012 |
| wecG mutation | ManNAcA transferases wecG | S. marcescens | Alteration in the enterobacterial common antigen (ECA) structure | Castelli and Vescovi, 2011 |
Detecting Envelope Stress via RcsF
RcsF, a small lipoprotein (14 kDa), is membrane-associated via a lipidated N-terminal membrane-anchored helix at the outer membrane, with a loop and a central four-stranded β-sheet in the periplasmic space (Castanié-Cornet et al., 2006). Acting as a mediator between environmental stimuli and Rcs phosphorelay system, RcsF activates Rcs system signaling in response to different types of envelope stress (Shiba et al., 2012; Laloux and Collet, 2017). The mechanism by which envelope stress induces the Rcs system via RcsF has been recently investigated by Silhavy’s lab and Collet’s lab (Cho et al., 2014; Konovalova et al., 2014). Both of them found that Rcs system interacts with BamA and three β-barrels (OmpA, OmpF, and OmpC) to detect envelope stress. Cho et al. (2014) proposed that the interaction between RcsF and BamA plays a major role in RcsF sensing (Figure 1A). In the absence of envelope stress, RcsF is trapped by the outer membrane β-barrel assembly machinery (Bam) and funneled to β-barrels (Cho et al., 2014; Konovalova et al., 2014). In this situation, RcsF cannot interact with YrfF (IgaA in Salmonella enterica serovar Typhimurium) to activate Rcs system (Figure 1A). In the presence of envelope stress, the interaction between RcsF and BamA is impaired and therefore RcsF cannot be funneled to OmpA/C/F. This allows RcsF to be exposed to the periplasm and free to activate Rcs system (Figure 1A). This model proposed that RcsF senses envelope stress by monitoring the Bam complex (Cho et al., 2014). Konovalova et al. (2014, 2016) proposed another model to explain how RcsF senses the lipopolysaccharide (LPS) defects. In their model, RcsF forms a complex with β-barrels and uses its positively charged, surface-exposed N-terminal domain to directly monitor lateral interactions between LPS molecules, which in turn regulate Rcs system (Konovalova et al., 2014, 2016). These two models might represent two different ways for RcsF to sense different types of stress (as reviewed in Laloux and Collet, 2017). Their findings elucidate the mechanism by which RcsF senses and transmits signals as a sentinel in the transduction signal pathway and are important to dissect the regulatory mechanism of Rcs system.
The functional N-terminal domain of RcsF contains five basic amino acid residues and six prolines (Umekawa et al., 2013). The positively charged residues are important for RcsF to sense alterations in LPS (Konovalova et al., 2016). The prolines might also be important for RcsF to sense environmental signals but their role has not been investigated. RcsF has two non-consecutive disulfide bridges, Cys74–Cys118 and Cys109–Cys124 in the E. coli protein, whose formation is catalyzed by the periplasmic disulfide isomerase DsbC (Leverrier et al., 2011; Rogov et al., 2011). Both of the disulfide bridges Cys74–Cys118 and Cys109–Cys124 are important for the activity of RcsF (Rogov et al., 2011). However, whether the formation of disulfide bridges is involved in sensing the environmental signal also needs to be investigated. Future work should continue to reveal the detailed mechanism of RcsF sensing at the molecular level.
Factors Regulating the Expression and Stability of Rcs System Proteins
Rcs system can be regulated by the expression level or stability of Rcs system proteins (Majdalani and Gottesman, 2005). For example, expression of rcsA is autoregulated by the RcsA–RcsB heterodimer in E. coli, S. enterica, and Klebsiella pneumoniae (Wang and Harshey, 2009). rcsB is transcribed from two promoters, PrcsDB and PrcsB in S. enterica serovar Typhimurium (Wang and Harshey, 2009; Pescaretti Mde et al., 2010). RcsB represses rcsD gene expression by binding directly to the PrcsDB promoter, negatively autoregulating rcsB in S. enterica serovar Typhimurium (Pescaretti Mde et al., 2010). Furthermore, the rcsA gene is indirectly upregulated by c-di-GMP in E. coli (Sledjeski and Gottesman, 1995; Mendez-Ortiz et al., 2006) and negatively regulated by the quorum-sensing regulator EsaR in Pantoea stewartii (Carlier and von Bodman, 2006).
All auxiliary regulators seem to be repressed by the nucleoid-associated global repressor protein H-NS (Sledjeski and Gottesman, 1995; Krin et al., 2010; Venkatesh et al., 2010; Martinez-Santos et al., 2012; Pannen et al., 2016). In addition, transcription of dctR is activated by the regulator YdeO in response to extracellular acid stress (Yamanaka et al., 2014). The expression of matA in the mat (ecp) operon encoding fimbrial adhesin is induced at acidic pH, low temperature, or high acetate concentration (Lehti et al., 2013). Finally, RcsA is unstable at 37°C and can be degraded by the ClpYQ and Lon proteases (Allen et al., 1987; Stout et al., 1991; Chang et al., 2016). The regulatory roles of RcsB heterodimers are largely controlled by the availability and regulation of the auxiliary regulators.
Functions and Interactions of RCS System Components
As mentioned above, Rcs system components include RcsC, RcsD, RcsB, and auxiliary regulators such as RcsA. Compared to other TCS proteins, Rcs system components exhibit high sequence and structural specificity among homologs (Majdalani and Gottesman, 2005; Clarke, 2010). In this section, we summarize the latest advances in our knowledge of the functions and interactions of Rcs system proteins.
RcsC Performs Dual Functions
RcsC is located at the inner membrane, with most of the protein located in the cytoplasm and a small periplasmic domain in the periplasmic space (Löhr et al., 2005). The atomic structure of the C-terminus of E. coli RcsC was determined by NMR, revealing a novel α–β-loop (ABL) domain and a PR domain (Löhr et al., 2005; Rogov et al., 2006). Upon phosphorylation of the RcsC-PR domain, it is recognized by RcsD-HPt and phosphoryl transmission takes place (Löhr et al., 2005). Meanwhile, RcsC-ABL facilitates phosphoryl transfer between RcsC-HK and RcsC-PR, or between RcsC-PR and RcsD-HPt (Rogov et al., 2006). These interactions mediate the signal transduction function of RcsC, while the first identified ABL domain of RcsC revealed a coordination module. Thus, RcsC contributes to the complexity of prokaryotic signaling systems.
RcsC was initially identified as a kinase in the Rcs phosphorelay system. As we know, many kinases are also phosphatases, and subsequent studies showed that RcsC also possesses phosphatase activity (Garcia-Calderon et al., 2005). RcsC functions together with RcsD as a phosphatase to precisely control the low levels of phosphorylated RcsB in the cell in the absence of environmental signal. Residues Ala904, Asp875, and Thr903 are required for the putative phosphatase activity of RcsC-PR (Garcia-Calderon et al., 2005). Recently, Ancona et al. (2015) reported that RcsC also exhibits dual functionality in Erwinia amylovora. Mutation of Thr904 (corresponding to Thr903 in E. coli RcsC) and Ala905 (corresponding to Ala904 in E. coli RcsC) constitutively activates the Rcs system (Ancona et al., 2015). The dual functions of RcsC are essential for its regulatory role to prevent the constitutive activation of the Rcs system. However, the molecular mechanism of its phosphatase activity is not yet clear.
Interactions between RcsD and Other Rcs System Proteins
RcsD is an inner membrane protein with a substantial periplasmic domain. Unlike RcsC, RcsD does not show any autophosphorylation activity, due to the lack of a canonical active site (Majdalani and Gottesman, 2005). The structure of the HPt domain of E. coli RcsD has been determined and shown to be similar to ArcB-HPt (Rogov et al., 2004). RcsD-HPt has a recognition area in close vicinity to the phosphoryl site (His842) that is recognized by two cognate PR domains, RcsB-PR and RcsC-PR. The interaction interfaces of the two domains with RcsD-HPt largely overlap. Meanwhile, Schmoe et al. (2011) describe a complex between RcsD-ABL and RcsB. Earlier Rogov et al. (2006) describe a complex between RcsC-ABL and RcsD. These complexes probably enhance the interaction between membrane-bound proteins and soluble components to increase the rate and efficiency of the phosphorelay signaling cascade (Schmoe et al., 2011).
Post-translational Modification of RcsB
The C-terminal DNA-binding domain of RcsB, belonging to the FixJ/NarL family, forms a helix-turn-helix (HTH) DNA-binding motif (Pristovsek et al., 2003). The N-terminal domain of RcsB, containing the phosphoacceptor site, belongs to the family of two-component receiver domains (Filippova et al., 2016). This receiver domain participates in RcsB dimer formation and also contributes to dimer formation with other transcription factor partners. In response to environmental stimuli, RcsB is phosphorylated by RcsD, and phosphorylation modulates its own DNA-binding affinity at the promoter region of target genes (Majdalani and Gottesman, 2005; Ancona et al., 2015). The phosphorylation status of RcsB is critical for biofilm development in S. enterica serovar Typhimurium (Latasa et al., 2012). Phosphorylated RcsB stimulates the transcription of sRNA rprA, and the accumulation of rprA leads to repression of the transcriptional regulator CsgD, resulting in inhibition of biofilm development (Latasa et al., 2012). In the absence of environmental signals, RcsC and RcsD might act as a phosphatase to dephosphorylate RcsB to maintain a low level of rcsB expression (Clarke, 2010; Hu et al., 2013). In this situation, AcP, the intermediate of the Pta-AckA pathway, might provide the phosphoryl group for RcsB, ensuring a low level of RcsB phosphorylation (Hu et al., 2013). Recently, Filippova et al. (2016) solved the crystal structure of non-phosphorylated receiver domain of RcsB. They found that the RcsB receiver domain can exist as a dimer in solution (Filippova et al., 2016). They suggested the non-phosphorylated and phosphorylated states of RcsB use a similar dimerization interface in the homodimer or heterodimers (Filippova et al., 2016). How phosphorylation affects the DNA binding ability and regulatory roles of RcsB is still not clear.
Another RcsB modification is lysine acetylation. RcsB can be acetylated by AcP and deacetylated by CobB (Thao et al., 2010; Hu et al., 2013). Thao et al. (2010) found that Lys180 of RcsB can be acetylated in vitro, while Hu et al. (2013) reported that several lysines in RcsB can be acetylated in vivo and acetylation of Lys154 negatively regulates RcsB activity. Castano-Cerezo et al. (2014) reported that acetylation of RcsB prevents DNA binding, activates flagella biosynthesis and motility, and compromises acid stress survival. Their finding indicates that acetylation might play an important role as same as phosphorylation in regulation of the activity of RcsB. Although Rcs system is important for enterobacteria, constitutive activation of the Rcs system could reduce the fitness and virulence (Ancona et al., 2015). Thus phosphorylation and acetylation might act together to tightly control the activity of RcsB, which in turn fine-tunes the function of Rcs system to adapt to the differential environment. The inter-conversion between phosphorylation and acetylation of RcsB remains largely unclear and needs to be investigated.
Homodimer and Heterodimers of RcsB
RcsB binds DNA either as a homodimer or heterodimer with auxiliary regulators (Table 1). The auxiliary proteins cooperate with RcsB to modulate transcription by binding to specific DNA motifs. The motifs binding by different dimers have a similarity in one half that is probably bound by RcsB, whereas the other half varies. The interaction of RcsB with auxiliary proteins might alter the specificity or strengthen the ability of DNA binding (Salscheider et al., 2014).
RcsB homodimer regulates targets once RcsB is phosphorylated. The binding sites are usually located upstream of the -35 region of the RNA polymerase-binding site (Majdalani and Gottesman, 2005). RcsB homodimer positively regulates sRNA rprA and the osmoregulated gene osmC in E. coli, as well as other targets in an RcsB phosphorylation-dependent manner (Majdalani and Gottesman, 2005; Clarke, 2010). RcsA stabilizes RcsA–RcsB complex to the RcsAB box and enhances the regulation activity of RcsB (Majdalani and Gottesman, 2005; Clarke, 2010). It is worth noting that RcsA’s function is dependent of phosphorylation of RcsB, whereas the other auxiliary regulators such as MatA, BglJ, and GadE act independently of phosphorylation of RcsB (Castanié-Cornet et al., 2010; Venkatesh et al., 2010; Pannen et al., 2016). For phosphorylation-independent heterodimers, it is not the level of RcsB phosphorylation in the cytoplasm but rather the presence of different auxiliary regulators that is important for its functions (Pannen et al., 2016). In addition, GadE–RcsB heterodimer activates the transcription of glutamate-dependent (Gad) acid resistant genes gadA/BC, but acetylation of RcsB negatively regulates gadA/BC (Castanié-Cornet et al., 2010; Castano-Cerezo et al., 2014). This indicates the function of phosphorylation-independent heterodimers might be affected by the acetylation of RcsB.
Castanié-Cornet et al. (2010) and Venkatesh et al. (2010) reported that BglJ–RcsB and GadE–RcsB heterodimers are active independent of RcsB phosphorylation. Pannen et al. (2016) found that MatA–RcsB heterodimer is also active independent of RcsB phosphorylation. In addition, they investigated conserved or surface-exposed residues and their roles in the activity of RcsB in E. coli. Their results revealed that mutation of residues in the vicinity of the phosphorylation site such as Asp56, Asp11, Thr87, and Lys109 impairs the activity of phosphorylation-dependent dimers such as RcsA–RcsB and RcsB–RcsB. Meanwhile, residue Lys109 also affects the activity of phosphorylation-independent heterodimers such as BglJ–RcsB and MatA–RcsB. One possible explanation is that a phosphorylation-induced structural change releases the DNA-binding domain that is otherwise buried or inhibited by the non-phosphorylated form of the PR domain. For both phosphorylation-dependent and -independent dimers, mutation of surface-exposed residue Ile14 impairs the activity of the dimers, indicating that RcsB and its auxiliary proteins interact via a surface similar to that with which NarL and related proteins interact (Pannen et al., 2016). Their findings suggest that phosphorylation might cause a structure change to stabilize the active form of RcsB–RcsB and RcsA–RcsB, while the heterodimers of BglJ–RcsB and MatA–RcsB are intrinsically active.
The heterodimers formed by the transcriptional regulator RcsB with auxiliary regulatory proteins modulate the DNA binding specificity and expands the regulatory role of Rcs system. In addition, the flexibility of the Rcs system regulatory role is further increased because some of the RcsB heterodimers depend on RcsB phosphorylation but some of them do not depend on RcsB phosphorylation. Multiple auxiliary regulators with varying dependence on the phosphorylation of RcsB endow flexibility to the Rcs system that may allow the bacteria to adapt to diverse environments.
Targets of the RCS Regulon
Rcs system was first reported to regulate the expression in E. coli (Gottesman et al., 1985). Later many Rcs system regulated genes were identified in enterobacteria using transcriptome or proteome analysis (Hinchliffe et al., 2008; Bury-Mone et al., 2009; Mariscotti and Garcia-del Portillo, 2009; Wang et al., 2012). Rcs system predominantly regulates genes involved in the production of important cell surface-associated structures (e.g., flagella, LPS, and fimbriae), exopolysaccharide, and environmental stress-related genes (Fredericks et al., 2006; McMahon et al., 2012; Tao et al., 2012; Farizano et al., 2014; Fang et al., 2015). Rcs phosphorelay system regulates these genes to remodeling the cell surface or metabolism to adapt to the stressed environment. In this section, we discuss the Rcs-regulated target genes involved in biofilm formation, acid resistance, and virulence.
Role of Rcs System in Biofilm Formation in Y. pestis
The plague pathogen Y. pestis forms biofilms in the midgut of its flea host to enhance transmission, and cyclic-di-GMP positively regulates biofilm development. In Y. pestis, c-di-GMP is synthesized by diguanylate cyclases HmsT and HmsD, and degraded by HmsP (Sun et al., 2011). We showed that the transcription of hmsT is directly repressed by RcsB in Y. pestis (Sun et al., 2012). RcsB binds to a conserved Rcs-binding motif which overlaps with the transcriptional start site of hmsT, leading to the repression of the transcription of hmsT (Sun et al., 2012). In addition, we also found that RcsB positively and directly regulates the hmsCDE operon (Guo et al., 2015). Furthermore, Fang et al. (2015) found that RcsB also directly represses the transcription of hmsHFRS, which is responsible for the biosynthesis and translocation of biofilm matrix exopolysaccharide. In summary, the Rcs system acts as a master repressor of biofilm formation by regulating the production of cyclic-di-GMP and biofilm matrix exopolysaccharide synthesis to facilitate environmental adaptation in Y. pestis.
Other Functions of Rcs System
Hundreds of genes were found to be regulated by the Rcs system using genome-wide approaches in E. amylovora, E. coli, Y. pestis, and S. enterica (Hinchliffe et al., 2008; Bury-Mone et al., 2009; Mariscotti and Garcia-del Portillo, 2009; Wang et al., 2012). Numerous Rcs-associated phenotypes have been characterized including quorum sensing in E. coli (Shimada et al., 2013), cell division in Proteus mirabilis (Howery et al., 2015), lipoprotein sorting in E. coli (Tao et al., 2012), and biogenesis of outer membrane vesicles in S. marcescens (McMahon et al., 2012). Recent advances on the targets of the Rcs system have focused on acid resistance and virulence.
The Gad system is the most efficient acid resistant system in E. coli (Foster, 2004; De Biase and Pennacchietti, 2012; Lund et al., 2014). RcsB controls the expression of gad genes and is required for acid resistance in E. coli (Castanié-Cornet et al., 2007). Castanié-Cornet et al. (2007, 2010) reported that the basal activity of RcsB is necessary and sufficient for GadE dependent regulation of gad genes, whereas increased RcsB activity through activation of the Rcs phosphorelay system or through overproduction of the protein leads to general repression of the gad genes. This result is consistent with the later findings that the regulatory role of RcsB/GadE is independent of RcsB phosphorylation (Castanié-Cornet et al., 2010), while the regulatory role of RcsB homodimer is dependent of RcsB phosphorylation. Further studies showed that activation of transcription gadA, encoding a glutamate decarboxylase, requires binding of RcsB/GadE to a GAD box, while repression of gadA transcription is caused by binding of RcsB to an RcsB box which lies in the -10 and -35 regions of gadA promoter (Castanié-Cornet et al., 2010). In response to acid stress, RcsB might function together with GadE to precisely regulate the expression of acid resistance-related genes (Castanié-Cornet et al., 2007; Johnson et al., 2011). However, a number of acid resistance-related genes including safA, slp, and gadW are regulated by RcsB but not GadE (Johnson et al., 2011). Johnson et al. (2011) speculated that RcsB might work with the response regulator EvgA as a heterodimer to regulate acid resistance-related genes.
The Rcs system also contributes to bacterial virulence. LcrF, a master transcriptional regulator of virulence genes in Yersinia pseudotuberculosis, is regulated by Rcs system (Li et al., 2015). Rcs system can also modulate the interaction between pathogen and host. For example, RcsB regulates the colonization of E. coli on the mouse intestine (Lasaro et al., 2014). In addition, deletion of cytoskeletal element mreC induces RcsC, resulted in downregulation of flagella systems and the SPI-1 type 3 secretion system (T3SS) and attenuated virulence in Salmonella (Bulmer et al., 2012). In S. marcescens, RcsB also represses the pore-forming toxin ShlA that is responsible for early induction of autophagy in host cells (Di Venanzio et al., 2014). Therefore, the Rcs system can affect the interaction between pathogen and host by regulating the assembly of the secretion system, colonization of the host, and early induction of autophagy in host cells.
Adaptation of the RCS Regulon Genes
Mutation of genes, especially those encoding global regulators such as the Rcs system, can alter cellular signaling circuits, expression of target genes, and physiological behavior, which in turn affect the ability of an organism to adapt to its environment. In the following section, we will discuss the examples of the molecular evolution of Rcs system genes as far as we know.
RcsA and RcsD in Y. pseudotuberculosis and Y. pestis
The rcsA gene in Y. pestis contains a 30 bp internal duplication, and is therefore nonfunctional, whereas rcsA in the ancestral Y. pseudotuberculosis species is functional and can repress biofilm formation. When Y. pestis is supplemented with the functional Y. pseudotuberculosis RcsA, biofilm formation is repressed and even abolished in fleas. Therefore, the conversion of rcsA to a pseudogene during the evolution of Y. pestis represents a case of positive selection (Sun et al., 2008). Furthermore, the loss of rcsA in Y. pestis following the gain of phospholipase D encoded by ymt, together with the loss of two minor genes in the bacterial progenitor, facilitated the transmission of fleas by stimulating c-di-GMP-mediated biofilm formation in the flea foregut (Sun et al., 2014). The evolutionary history of rcsA in Y. pestis is one example that a few genetic changes such as gene losses can alter the regulation of targets and consequently the infection phenotype, leading to arthropod–borne transmission. Despite an apparent frameshift, RcsD remains functional and acts in concert with RcsB to promote biofilm formation in Y. pestis, but it functions in a way that is opposite to RcsD in Y. pseudotuberculosis (Sun et al., 2008). The effect of mutating rcsD on Rcs system signal transduction and its biological significance requires further experimental investigation.
RcsB in E. coli O157: H7
Escherichia coli O157: H7 (O157) is a frequent cause of foodborne disease in humans, and is transmitted to human through food vehicles. To cause infection, O157 needs to adapt to and survive in diverse conditions. Curli fimbriae play an important role in surface attachment, biofilm formation, and confer cell protection from toxic compounds. The production of curli in E. coli O157: H7 is intrastrain and interstrain variant (Carter et al., 2012). The curli producing (C+) O157 variants are much more acid sensitive, but grow better in nutrient-limited environment than curli-deficient (C-) variants. Deletion of rcsB gene is responsible for the C+ phenotypes in two O157 strains, isolated during the 1993 U.S. hamburger-associated outbreak (Carter et al., 2012). In addition, the C+ O157 variants can also be generated by insertion or single nucleotide change in rcsB gene (Chen et al., 2016; Sharma et al., 2017). RcsB affects bacterial survival in growth-restrictive environments through regulation of genes related to biofilm formation and certain metabolic functions. Inactivation of rcsB in O157 might enhance the persistence and survival ability in nutrient limited environment, but decrease the resistance to heat shock and acid stress. Thus, mutation of rcsB generates a distinct O157 subpopulation that may confer this pathogen with survival advantages in the ever changing environment.
Conclusion and Future Perspectives
Recent advances on the Rcs system have helped to unravel the interactions, functions, and evolution of Rcs system proteins. Moreover, knowledge on the auxiliary regulatory proteins, regulation of the Rcs regulon, targets, and physiological behavior has considerably extended. Modifications of RcsB, dual functions of RcsC, involvement of multiple auxiliary proteins, and adaptation of Rcs system genes contributed together to increase the complexity and flexibility of the Rcs system that leads to the precise control of its target genes. However, many questions concerning Rcs system structure and function remain unclear. For example, the Rcs system appears to be functional in the absence of RcsF, and it remains unknown how Rcs system senses signals that are independent of RcsF. In addition, similar to RcsC, RcsD also has a periplasmic domain. Whether the periplasmic domain of RcsD is also involved in sensing environmental signals and regulating Rcs system also needs to be clarified. Finally, many auxiliary proteins have now been identified, but how they are involved in the response to differential environmental signals and how they regulate physiological behavior in a coordinated manner to facilitate adaptation requires further investigation. In summary, the Rcs phosphorelay system provides a unique model for investigating the complexity of genetic regulation, environmental adaptation, and evolution in bacteria.
Author Contributions
X-PG and Y-CS wrote the manuscript. Both authors have approved the final version for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank Dr. Lin Lin for the critical reading of the manuscript and helpful discussion.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (31700072 and 31670139), the Special Key of Biosafety Technologies for the National Major Research and Development Program of China (2016YFC1202600), and the CAMS Innovation Fund for Medical Sciences (2016-I2M-1-013).
References
- Allen P., Hart C. A., Saunders J. R. (1987). Isolation from Klebsiella and characterization of two rcs genes that activate colanic acid capsular biosynthesis in Escherichia coli. J. Gen. Microbiol. 133 331–340. 10.1099/00221287-133-2-331 [DOI] [PubMed] [Google Scholar]
- Ancona V., Chatnaparat T., Zhao Y. (2015). Conserved aspartate and lysine residues of RcsB are required for amylovoran biosynthesis, virulence, and DNA binding in Erwinia amylovora. Mol. Genet. Genomics 290 1265–1276. 10.1007/s00438-015-0988-8 [DOI] [PubMed] [Google Scholar]
- Bontemps-Gallo S., Madec E., Dondeyne J., Delrue B., Robbe-Masselot C., Vidal O., et al. (2013). Concentration of osmoregulated periplasmic glucans (OPGs) modulates the activation level of the RcsCD RcsB phosphorelay in the phytopathogen bacteria Dickeya dadantii. Environ. Microbiol. 15 881–894. 10.1111/1462-2920.12054 [DOI] [PubMed] [Google Scholar]
- Brill J. A., Quinlan-Walshe C., Gottesman S. (1988). Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 170 2599–2611. 10.1128/jb.170.6.2599-2611.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer D. M., Kharraz L., Grant A. J., Dean P., Morgan F. J., Karavolos M. H., et al. (2012). The bacterial cytoskeleton modulates motility, type 3 secretion, and colonization in Salmonella. PLOS Pathog. 8:e1002500. 10.1371/journal.ppat.1002500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bury-Mone S., Nomane Y., Reymond N., Barbet R., Jacquet E., Imbeaud S., et al. (2009). Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLOS Genet. 5:e1000651. 10.1371/journal.pgen.1000651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callewaert L., Vanoirbeek K. G. A., Lurquin I., Michiels C. W., Aertsen A. (2009). The Rcs system two-component system regulates expression of lysozyme inhibitors and is induced by exposure to lysozyme. J. Bacteriol. 191 1979–1981. 10.1128/JB.01549-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier A. L., von Bodman S. B. (2006). The rcsA promoter of Pantoea stewartii subsp. stewartii features a low-level constitutive promoter and an EsaR quorum-sensing-regulated promoter. J. Bacteriol. 188 4581–4584. 10.1128/JB.00211-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M. Q., Parker C. T., Louie J. W., Huynh S., Fagerquist C. K., Mandrell R. E. (2012). RcsB contributes to the distinct stress fitness among Escherichia coli O157:H7 curli variants of the 1993 hamburger-associated outbreak strains. Appl. Environ. Microbiol. 78 7706-7719. 10.1128/AEM.02157-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanié-Cornet M. P., Cam K., Bastiat B., Cros A., Bordes P., Gutierrez C. (2010). Acid stress response in Escherichia coli: mechanism of regulation of gadA transcription by RcsB and GadE. Nucleic Acids Res. 38 3546–3554. 10.1093/nar/gkq097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanié-Cornet M. P., Cam K., Jacq A. (2006). RcsF is an outer membrane lipoprotein involved in the RcsCDB phosphorelay signaling pathway in Escherichia coli. J. Bacteriol. 188 4264–4270. 10.1128/JB.00004-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanié-Cornet M. P., Treffandier H., Francez-Charlot A., Gutierrez C., Cam K. (2007). The glutamate-dependent acid resistance system in Escherichia coli: essential and dual role of the His-Asp phosphorelay RcsCDB/AF. Microbiology 153 238–246. 10.1099/mic.0.29278-0 [DOI] [PubMed] [Google Scholar]
- Castano-Cerezo S., Bernal V., Post H., Fuhrer T., Cappadona S., Sanchez-Diaz N. C., et al. (2014). Protein acetylation affects acetate metabolism, motility and acid stress response in Escherichia coli. Mol. Syst. Biol. 10 762. 10.15252/msb.20145227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelli M. E., Vescovi E. G. (2011). The Rcs signal transduction pathway is triggered by enterobacterial common antigen structure alterations in Serratia marcescens. J. Bacteriol. 193 63–74. 10.1128/JB.00839-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. Y., Hu H. T., Tsai C. H., Wu W. F. (2016). The degradation of RcsA by ClpYQ (HslUV) protease in Escherichia coli. Microbiol. Res. 184 42–50. 10.1016/j.micres.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Nguyen L. H., Cottrell B. J., Irwin P. L., Uhlich G. A. (2016). Multiple mechanisms responsible for strong Congo-red-binding variants of Escherichia coli O157: H7 strains. Pathog. Dis. 74:ftv123. 10.1093/femspd/ftv123 [DOI] [PubMed] [Google Scholar]
- Chen M. H., Takeda S., Yamada H., Ishii Y., Yamashino T., Mizuno T. (2001). Characterization of the RcsC–> YojN–> RcsB phosphorelay signaling pathway involved in capsular synthesis in Escherichia coli. Biosci. Biotechnol. Biochem. 65 2364–2367. 10.1271/bbb.65.2364 [DOI] [PubMed] [Google Scholar]
- Cheng H. Y., Chen Y. S., Wu C. Y., Chang H. Y., Lai Y. C., Peng H. L. (2010). RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J. Bacteriol. 192 3144–3158. 10.1128/JB.00031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. H., Szewczyk J., Pesavento C., Zietek M., Banzhaf M., Roszczenko P., et al. (2014). Detecting envelope stress by monitoring beta-barrel assembly. Cell 159 1652–1664. 10.1016/j.cell.2014.11.045 [DOI] [PubMed] [Google Scholar]
- Clarke D. J. (2010). The Rcs phosphorelay: more than just a two-component pathway. Future Microbiol. 5 1173–1184. 10.2217/fmb.10.83 [DOI] [PubMed] [Google Scholar]
- Clarke D. J., Joyce S. A., Toutain C. M., Jacq A., Holland I. B. (2002). Genetic analysis of the RcsC sensor kinase from Escherichia coli K-12. J. Bacteriol. 184 1204–1208. 10.1128/jb.184.4.1204-1208.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biase D., Pennacchietti E. (2012). Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: function, distribution and biomedical implications of the gadBC operon. Mol. Microbiol. 86 770–786. 10.1111/mmi.12020 [DOI] [PubMed] [Google Scholar]
- Di Venanzio G., Stepanenko T. M., Garcia Vescovi E. (2014). Serratia marcescens ShlA pore-forming toxin is responsible for early induction of autophagy in host cells and is transcriptionally regulated by RcsB. Infect. Immun. 82 3542–3554. 10.1128/IAI.01682-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans K. L., Kannan S., Li G., de Pedro M. A., Young K. D. (2013). Eliminating a set of four penicillin binding proteins triggers the Rcs phosphorelay and Cpx stress responses in Escherichia coli. J. Bacteriol. 195 4415–4424. 10.1128/JB.00596-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang N., Yang H., Fang H., Liu L., Zhang Y., Wang L., et al. (2015). RcsAB is a major repressor of Yersinia biofilm development through directly acting on hmsCDE, hmsT, and hmsHFRS. Sci. Rep. 5:9566. 10.1038/srep09566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farizano J. V., Torres M. A., Pescaretti Mde L., Delgado M. A. (2014). The RcsCDB regulatory system plays a crucial role in the protection of Salmonella enterica serovar Typhimurium against oxidative stress. Microbiology 160(Pt 10) 2190–2199. 10.1099/mic.0.081133-0 [DOI] [PubMed] [Google Scholar]
- Ferrières L., Aslam S. N., Cooper R. M., Clarke D. J. (2007). The yjbEFGH locus in Escherichia coli K-12 is an operon encoding proteins involved in exopolysaccharide production. Microbiology 153 1070–1080. 10.1099/mic.0.2006/002907-0 [DOI] [PubMed] [Google Scholar]
- Ferrières L., Thompson A., Clarke D. J. (2009). Elevated levels of sigma S inhibit biofilm formation in Escherichia coli: a role for the Rcs phosphorelay. Microbiology 155 3544–3553. 10.1099/mic.0.032722-0 [DOI] [PubMed] [Google Scholar]
- Filippova E. V., Wawrzak Z., Ruan J., Pshenychnyi S., Schultz R. M., Wolfe A. J., et al. (2016). Crystal structure of nonphosphorylated receiver domain of the stress response regulator RcsB from Escherichia coli. Protein Sci. 25 2216–2224. 10.1002/pro.3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J. W. (2004). Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2 898–907. 10.1038/nrmicro1021 [DOI] [PubMed] [Google Scholar]
- Fredericks C. E., Shibata S., Aizawa S., Reimann S. A., Wolfe A. J. (2006). Acetyl phosphate-sensitive regulation of flagellar biogenesis and capsular biosynthesis depends on the Rcs phosphorelay. Mol. Microbiol. 61 734–747. 10.1111/j.1365-2958.2006.05260.x [DOI] [PubMed] [Google Scholar]
- Garcia-Calderon C. B., Garcia-Quintanilla M., Casadesus J., Ramos-Morales F. (2005). Virulence attenuation in Salmonella enterica rcsC mutants with constitutive activation of the Rcs system. Microbiology 151 579–588. 10.1099/mic.0.27520-0 [DOI] [PubMed] [Google Scholar]
- Gervais F. G., Drapeau G. R. (1992). Identification, cloning, and characterization of rcsF, a new regulator gene for exopolysaccharide synthesis that suppresses the division mutation ftsZ84 in Escherichia coli K-12. J. Bacteriol. 174 8016–8022. 10.1128/jb.174.24.8016-8022.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover W. A., Yang Y., Zhang Y. (2009). Insights into the molecular basis of L-form formation and survival in Escherichia coli. PLOS ONE 4:e7316. 10.1371/journal.pone.0007316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S., Trisler P., Torres-Cabassa A. (1985). Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J. Bacteriol. 162 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X. P., Ren G. X., Zhu H., Mao X. J., Sun Y. C. (2015). Differential regulation of the hmsCDE operon in Yersinia pestis and Yersinia pseudotuberculosis by the Rcs phosphorelay system. Sci. Rep. 5:8412. 10.1038/srep08412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchliffe S. J., Howard S. L., Huang Y. H., Clarke D. J., Wren B. W. (2008). The importance of the Rcs phosphorelay in the survival and pathogenesis of the enteropathogenic yersiniae. Microbiology 154 1117–1131. 10.1099/mic.0.2007/012534-0 [DOI] [PubMed] [Google Scholar]
- Hirakawa H., Nishino K., Yamada J., Hirata T., Yamaguchi A. (2003). Beta-lactam resistance modulated by the overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J. Antimicrob. Chemother. 52 576–582. 10.1093/jac/dkg406 [DOI] [PubMed] [Google Scholar]
- Howery K. E., Clemmer K. M., Simsek E., Kim M., Rather P. N. (2015). Regulation of the min cell division inhibition complex by the Rcs phosphorelay in Proteus mirabilis. J. Bacteriol. 197 2499–2507. 10.1128/JB.00094-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L. I., Chi B. K., Kuhn M. L., Filippova E. V., Walker-Peddakotla A. J., Basell K., et al. (2013). Acetylation of the response regulator RcsB controls transcription from a small RNA promoter. J. Bacteriol. 195 4174–4186. 10.1128/JB.00383-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. H., Ferrières L., Clarke D. J. (2006). The role of the Rcs phosphorelay in Enterobacteriaceae. Res. Microbiol. 157 206–212. 10.1016/j.resmic.2005.11.005 [DOI] [PubMed] [Google Scholar]
- Johnson M. D., Burton N. A., Gutierrez B., Painter K., Lund P. A. (2011). RcsB is required for inducible acid resistance in Escherichia coli and acts at gadE-dependent and -independent promoters. J. Bacteriol. 193 3653–3656. 10.1128/JB.05040-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konovalova A., Mitchell A. M., Silhavy T. J. (2016). A lipoprotein/β-barrel complex monitors lipopolysaccharide integrity transducing information across the outer membrane. eLife 5:e15276. 10.7554/eLife.15276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konovalova A., Perlman D. H., Cowles C. E., Silhavy T. J. (2014). Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of beta-barrel proteins. Proc. Natl. Acad. Sci. U.S.A. 111 E4350–E4358. 10.1073/pnas.1417138111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krin E., Danchin A., Soutourina O. (2010). RcsB plays a central role in H-NS-dependent regulation of motility and acid stress resistance in Escherichia coli. Res. Microbiol. 161 363–371. 10.1016/j.resmic.2010.04.002 [DOI] [PubMed] [Google Scholar]
- Kühne C., Singer H. M., Grabisch E., Codutti L., Carlomagno T., Scrima A., et al. (2016). RflM mediates target specificity of the RcsCDB phosphorelay system for transcriptional repression of flagellar synthesis in Salmonella enterica. Mol. Microbiol. 101 841–855. 10.1111/mmi.13427 [DOI] [PubMed] [Google Scholar]
- Laloux G., Collet J. F. (2017). Major Tom to ground control: how lipoproteins communicate extra-cytoplasmic stress to the decision center of the cell. J. Bacteriol. 10.1128/JB.00216-17 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasaro M., Liu Z., Bishar R., Kelly K., Chattopadhyay S., Paul S., et al. (2014). Escherichia coli isolate for studying colonization of the mouse intestine and its application to two-component signaling knockouts. J. Bacteriol. 196 1723–1732. 10.1128/JB.01296-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latasa C., Garcia B., Echeverz M., Toledo-Arana A., Valle J., Campoy S., et al. (2012). Salmonella biofilm development depends on the phosphorylation status of RcsB. J. Bacteriol. 194 3708–3722. 10.1128/JB.00361-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti T. A., Bauchart P., Kukkonen M., Dobrindt U., Korhonen T. K., Westerlund-Wikström B. (2013). Phylogenetic group-associated differences in regulation of the common colonization factor Mat fimbria in Escherichia coli. Mol. Microbiol. 87 1200–1222. 10.1111/mmi.12161 [DOI] [PubMed] [Google Scholar]
- Leverrier P., Declercq J. P., Denoncin K., Vertommen D., Hiniker A., Cho S. H., et al. (2011). Crystal structure of the outer membrane protein RcsF, a new substrate for the periplasmic protein-disulfide isomerase DsbC. J. Biol. Chem. 286 16734–16742. 10.1074/jbc.M111.224865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Hu Y., Francis M. S., Chen S. (2015). RcsB positively regulates the Yersinia Ysc-Yop type III secretion system by activating expression of the master transcriptional regulator LcrF. Environ. Microbiol. 17 1219–1233. 10.1111/1462-2920.12556 [DOI] [PubMed] [Google Scholar]
- Löhr F., Rogov V. V., Shi M., Bernhard F., Dötsch V. (2005). Triple-resonance methods for complete resonance assignment of aromatic protons and directly bound heteronuclei in histidine and tryptophan residues. J. Biomol. NMR 32 309–328. 10.1007/s10858-005-1195-4 [DOI] [PubMed] [Google Scholar]
- Lund P., Tramonti A., De Biase D. (2014). Coping with low pH: molecular strategies in neutralophilic bacteria. FEMS. Microbiol. Rev. 38 1091–1125. 10.1111/1574-6976.12076 [DOI] [PubMed] [Google Scholar]
- Lv Y., Zheng J., Yang M., Wang Q., Zhang Y. (2012). An Edwardsiella tarda mutant lacking UDP-glucose dehydrogenase shows pleiotropic phenotypes, attenuated virulence, and potential as a vaccine candidate. Vet. Microbiol. 160 506–512. 10.1016/j.vetmic.2012.06.006 [DOI] [PubMed] [Google Scholar]
- Majdalani N., Gottesman S. (2005). The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59 379–405. 10.1146/annurev.micro.59.050405.101230 [DOI] [PubMed] [Google Scholar]
- Majdalani N., Heck M., Stout V., Gottesman S. (2005). Role of RcsF in signaling to the Rcs phosphorelay pathway in Escherichia coli. J. Bacteriol. 187 6770–6778. 10.1128/JB.187.19.6770-6778.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariscotti J. F., Garcia-del Portillo F. (2009). Genome expression analyses revealing the modulation of the Salmonella Rcs regulon by the attenuator IgaA. J. Bacteriol. 191 1855–1867. 10.1128/JB.01604-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Santos V. I., Medrano-Lopez A., Saldana Z., Giron J. A., Puente J. L. (2012). Transcriptional regulation of the ecp operon by EcpR, IHF, and H-NS in attaching and effacing Escherichia coli. J. Bacteriol. 194 5020–5033. 10.1128/JB.00915-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum K. L., Whitfield C. (1991). The rcsA Gene of Klebsiella pneumoniae O1: K20 is involved in expression of the serotype-specific K (capsular) antigen. Infect. Immun. 59 494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon K. J., Castelli M. E., Garcia Vescovi E., Feldman M. F. (2012). Biogenesis of outer membrane vesicles in Serratia marcescens is thermoregulated and can be induced by activation of the Rcs phosphorelay system. J. Bacteriol. 194 3241–3249. 10.1128/JB.00016-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Ortiz M. M., Hyodo M., Hayakawa Y., Membrillo-Hernandez J. (2006). Genome-wide transcriptional profile of Escherichia coli in response to high levels of the second messenger 3′, 5′-cyclic diguanylic acid. J. Biol. Chem. 281 8090–8099. 10.1074/jbc.M510701200 [DOI] [PubMed] [Google Scholar]
- Morgan J. K., Ortiz J. A., Riordan J. T. (2014). The role for TolA in enterohemorrhagic Escherichia coli pathogenesis and virulence gene transcription. Microb. Pathog. 77 42–52. 10.1016/j.micpath.2014.10.010 [DOI] [PubMed] [Google Scholar]
- Pannen D., Fabisch M., Gausling L., Schnetz K. (2016). Interaction of the RcsB response regulator with auxiliary transcription regulators in Escherichia coli. J. Biol. Chem. 291 2357–2370. 10.1074/jbc.M115.696815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pescaretti Mde L., Lopez F. E., Morero R. D., Delgado M. A. (2010). Transcriptional autoregulation of the RcsCDB phosphorelay system in Salmonella enterica serovar Typhimurium. Microbiology 156 3513–3521. 10.1099/mic.0.041319-0 [DOI] [PubMed] [Google Scholar]
- Pristovsek P., Sengupta K., Löhr F., Schafer B., von Trebra M. W., Ruterjans H., et al. (2003). Structural analysis of the DNA-binding domain of the Erwinia amylovora RcsB protein and its interaction with the RcsAB box. J. Biol. Chem. 278 17752–17759. 10.1074/jbc.M301328200 [DOI] [PubMed] [Google Scholar]
- Rogov V. V., Bernhard F., Löhr F., Dötsch V. (2004). Solution structure of the Escherichia coli YojN histidine-phosphotransferase domain and its interaction with cognate phosphoryl receiver domains. J. Mol. Biol. 343 1035–1048. 10.1016/j.jmb.2004.08.096 [DOI] [PubMed] [Google Scholar]
- Rogov V. V., Rogova N. Y., Bernhard F., Koglin A., Löhr F., Dötsch V. (2006). A new structural domain in the Escherichia coli RcsC hybrid sensor kinase connects histidine kinase and phosphoreceiver domains. J. Mol. Biol. 364 68–79. 10.1016/j.jmb.2006.07.052 [DOI] [PubMed] [Google Scholar]
- Rogov V. V., Rogova N. Y., Bernhard F., Löhr F., Dötsch V. (2011). A disulfide bridge network within the soluble periplasmic domain determines structure and function of the outer membrane protein RCSF. J. Biol. Chem. 286 18775–18783. 10.1074/jbc.M111.230185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salscheider S. L., Jahn A., Schnetz K. (2014). Transcriptional regulation by BglJ-RcsB, a pleiotropic heteromeric activator in Escherichia coli. Nucleic Acids Res. 42 2999–3008. 10.1093/nar/gkt1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoe K., Rogov V. V., Rogova N. Y., Löhr F., Guntert P., Bernhard F., et al. (2011). Structural insights into Rcs phosphotransfer: the newly identified RcsD-ABL domain enhances interaction with the response regulator RcsB. Structure 19 577–587. 10.1016/j.str.2011.01.012 [DOI] [PubMed] [Google Scholar]
- Sharma V. K., Bayles D. O., Alt D. P., Looft T., Brunelle B. W., Stasko J. A. (2017). Disruption of rcsB by a duplicated sequence in a curli-producing Escherichia coli O157:H7 results in differential gene expression in relation to biofilm formation, stress responses and metabolism. BMC Microbiol. 17:56. 10.1186/s12866-017-0966-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba Y., Miyagawa H., Nagahama H., Matsumoto K., Kondo D., Matsuoka S., et al. (2012). Exploring the relationship between lipoprotein mislocalization and activation of the Rcs signal transduction system in Escherichia coli. Microbiology 158 1238–1248. 10.1099/mic.0.056945-0 [DOI] [PubMed] [Google Scholar]
- Shimada K., Ogasawara H., Yamada K., Shimura M., Kori A., Shimada T., et al. (2013). Screening of promoter-specific transcription factors: multiple regulators for the sdiA gene involved in cell division control and quorum sensing. Microbiology 159 2501–2512. 10.1099/mic.0.067538-0 [DOI] [PubMed] [Google Scholar]
- Sledjeski D., Gottesman S. (1995). A small RNA acts as an antisilencer of H-NS-silenced rcsA gene of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 92 2003–2007. 10.1073/pnas.92.6.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski D. D., Gottesman S. (1996). Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 178 1204–1206. 10.1128/jb.178.4.1204-1206.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout V., Gottesman S. (1990). RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J. Bacteriol. 172 659–669. 10.1128/jb.172.2.659-669.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout V., Torres-Cabassa A., Maurizi M. R., Gutnick D., Gottesman S. (1991). RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J. Bacteriol. 173 1738–1747. 10.1128/jb.173.5.1738-1747.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann T., Pul Ü., Wurm R., Wagner R., Schnetz K. (2012). RcsB-BglJ activates the Escherichia coli leuO gene, encoding an H-NS antagonist and pleiotropic regulator of virulence determinants. Mol. Microbiol. 83 1109–1123. 10.1111/j.1365-2958.2012.07993.x [DOI] [PubMed] [Google Scholar]
- Sun Y. C., Guo X. P., Hinnebusch B. J., Darby C. (2012). The Yersinia pestis Rcs phosphorelay inhibits biofilm formation by repressing transcription of the diguanylate cyclase gene hmsT. J. Bacteriol. 194 2020–2026. 10.1128/JB.06243-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. C., Hinnebusch B. J., Darby C. (2008). Experimental evidence for negative selection in the evolution of a Yersinia pestis pseudogene. Proc. Natl. Acad. Sci. U.S.A. 105 8097–8101. 10.1073/pnas.0803525105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. C., Jarrett C. O., Bosio C. F., Hinnebusch B. J. (2014). Retracing the evolutionary path that led to flea-borne transmission of Yersinia pestis. Cell Host Microbe 15 578–586. 10.1016/j.chom.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. C., Koumoutsi A., Jarrett C., Lawrence K., Gherardini F. C., Darby C., et al. (2011). Differential control of Yersinia pestis biofilm formation in vitro and in the flea vector by two c-di-GMP diguanylate cyclases. PLOS ONE 6:e19267. 10.1371/journal.pone.0019267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S., Fujisawa Y., Matsubara M., Aiba H., Mizuno T. (2001). A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC – > YojN – > RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol. Microbiol. 40 440–450. 10.1046/j.1365-2958.2001.02393.x [DOI] [PubMed] [Google Scholar]
- Tao K., Narita S., Tokuda H. (2012). Defective lipoprotein sorting induces lolA expression through the Rcs stress response phosphorelay system. J. Bacteriol. 194 3643–3650. 10.1128/JB.00553-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao S., Chen C. S., Zhu H., Escalante-Semerena J. C. (2010). Nepsilon-lysine acetylation of a bacterial transcription factor inhibits Its DNA-binding activity. PLOS ONE 5:e15123. 10.1371/journal.pone.0015123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umekawa M., Miyagawa H., Kondo D., Matsuoka S., Matsumoto K., Hara H. (2013). Importance of the proline-rich region for the regulatory function of RcsF, an outer membrane lipoprotein component of the Escherichia coli Rcs signal transduction system. Microbiology 159 1818–1827. 10.1099/mic.0.069328-0 [DOI] [PubMed] [Google Scholar]
- Venkatesh G. R., Kembou Koungni F. C., Paukner A., Stratmann T., Blissenbach B., Schnetz K. (2010). BglJ-RcsB Heterodimers Relieve Repression of the Escherichia coli bgl Operon by H-NS. J. Bacteriol. 192 6456–6464. 10.1128/JB.00807-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vianney A., Jubelin G., Renault S., Dorel C., Lejeune P., Lazzaroni J. C. (2005). Escherichia coli tol and rcs genes participate in the complex network affecting curli synthesis. Microbiology 151 2487–2497. 10.1099/mic.0.27913-0 [DOI] [PubMed] [Google Scholar]
- Virlogeux I., Waxin H., Ecobichon C., Lee J. O., Popoff M. Y. (1996). Characterization of the rcsA and rcsB genes from Salmonella typhi: rcsB through tviA is involved in regulation of Vi antigen synthesis. J. Bacteriol. 178 1691–1698. 10.1128/jb.178.6.1691-1698.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Qi M., Calla B., Korban S. S., Clough S. J., Cock P. J., et al. (2012). Genome-wide identification of genes regulated by the Rcs phosphorelay system in Erwinia amylovora. Mol. Plant Microbe Interact. 25 6–17. 10.1094/MPMI-08-11-0207 [DOI] [PubMed] [Google Scholar]
- Wang Q., Harshey R. M. (2009). Rcs signalling-activated transcription of rcsA induces strong anti-sense transcription of upstream fliPQR flagellar genes from a weak intergenic promoter: regulatory roles for the anti-sense transcript in virulence and motility. Mol. Microbiol. 74 71–84. 10.1111/j.1365-2958.2009.06851.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter S. E., Winter M. G., Thiennimitr P., Gerriets V. A., Nuccio S. P., Russmann H., et al. (2009). The TviA auxiliary protein renders the Salmonella enterica serotype Typhi RcsB regulon responsive to changes in osmolarity. Mol. Microbiol. 74 175–193. 10.1111/j.1365-2958.2009.06859.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka Y., Oshima T., Ishihama A., Yamamoto K. (2014). Characterization of the YdeO regulon in Escherichia coli. PLOS ONE 9:e111962. 10.1371/journal.pone.0111962 [DOI] [PMC free article] [PubMed] [Google Scholar]


