Abstract
Magnetic resonance imaging (MRI) examinations provide high-resolution information about the anatomic structure of the kidneys and are used to measure total kidney volume (TKV) in patients with Autosomal Dominant Polycystic Kidney Disease (ADPKD). Height adjusted TKV (HtTKV) has become the gold-standard imaging biomarker for ADPKD progression at early stages of the disease when estimated glomerular filtration rate (eGFR) is still normal. However, HtTKV does not take advantage of the wealth of information provided by MRI. Here we tested whether image texture features provide additional insights into the ADPKD kidney that may be used as complementary information to existing biomarkers. A retrospective cohort of 122 patients from The Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease (CRISP study) was identified who had T2-weighted MRIs, and eGFR values over 70 mL/min/1.73m2 at the time of their baseline scan. We computed nine distinct image texture features for each patient. The ability of each feature to predict subsequent progression to CKD stage 3A, 3B, and 30% reduction in eGFR at eight-year follow up was assessed. A multiple linear regression model was developed incorporating age, baseline eGFR, HtTKV, and three image texture features identified by stability feature selection (Entropy, Correlation, and Energy). Including texture in a multiple linear regression model (predicting percent change in eGFR) improved Pearson correlation coefficient from -0.51 (using age, eGFR, and HtTKV) to -0.70 (adding texture). Thus, texture analysis offers an approach to refine ADPKD prognosis and should be further explored for its utility in individualized clinical decision making and outcome prediction.
Keywords: Gray level co-occurrence matrix, magnetic resonance imaging, multiple linear regression, polycystic kidney disease, total kidney volume
Introduction
Radiological imaging plays an important role in diagnosis and management of patients with autosomal dominant polycystic kidney disease (ADPKD) 1. Total kidney volume (TKV), measured by magnetic resonance imaging (MRI), computed tomography (CT), or ultrasound (US), predicts the decline of renal function 2-4 and is used to ascertain the effectiveness of treatment interventions 5-7. Recently, the U.S. Food and Drug Administration and the European Medicines Agency qualified TKV, in combination with patient age and estimated glomerular filtration rate (eGFR), as a prognostic imaging biomarker for ADPKD research and clinical trials. Particularly, height adjusted TKV (HtTKV) is the strongest prognostic biomarker of renal function worsening and progression to end-stage renal disease (ESRD) in patients at an early stage of ADPKD when eGFR is within the normal range. This has led to the development of a number of automated approaches to accurately and reproducibly measure HtTKV 8-11. Although HtTKV provides important information regarding kidney size, which is linked to overall disease progression, it does not take advantage of the wealth of information available within an MRI or CT image related to renal tissue structure.
There are a number of possible imaging biomarkers beyond HtTKV. One would be to measure the cystic load, including quantification of the number, size, and composition of renal cysts. This could lead to a deeper understanding of a patient's particular phenotype in terms of whether, for example, their kidneys are composed of a few large simple exophitic cysts, or whether they have numerous complex (e.g., proteinaceous) cysts. However, performing segmentation of renal cysts from radiological scans of ADPKD patients is extremely time consuming and challenging, particularly in patients with a moderate to severe cyst burden 12. Therefore, automated approaches are being actively pursued. A number of studies have published cyst segmentation techniques. These techniques have included threshold-based approaches 13, 14, as well as more advanced, semi-automated techniques including shape- or boundary-detection 15. In a majority of these studies, T2-weighted MRI scans are used due to the difference in signal intensity of fluid-filled cysts (high signal intensity) compared to renal parenchyma (low signal intensity) 16. However, these techniques typically rely on the assumption that all cysts have similar signal intensities. However, complex cysts often are darker, and therefore, most often only simple fluid-filled cysts are delineated. Techniques that can differentiate not only cysts from parenchyma, but also classify different types of cysts (simple, proteinaceous, infected) will be necessary to accurately characterize renal cystic burden in more detail.
Another opportunity is through the use of quantitative MRI techniques 17. These techniques could provide a deeper understanding of the composition of an ADPKD patient's renal tissue. For example, magnetization transfer imaging (MTI) has been shown to identify fibrotic tissue within kidneys 18-20, perfusion imaging (e.g., arterial spin labeling) can identify reduced perfusion territories 21, and blood oxygen level dependent (BOLD) imaging reflects regional renal oxygenation status 22-24. Other techniques such as measurement of renal blood flow 25-27, magnetic resonance elastography (to measure tissue stiffness) 28, sodium imaging 29, diffusion weighted imaging (DWI) 30 with intra-voxel incoherent motion (IVIM) processing 31, and chemical exchange saturation transfer imaging 32 are additional imaging techniques currently being explored which could show utility in ADPKD management. However, the information regarding how these imaging findings relate to progression of the disease in humans may not be available for quite some time.
Advanced image processing techniques offer a unique opportunity to retrospectively explore the large number of imaging examinations available and correlate this information with known progression of the disease. Texture analysis is one technique which performs an ensemble of mathematical computations to greatly increase the information provided by conventional radiological images 33. Image texture refers to the appearance, structure, and arrangement of different intensity levels within an image and has been used in organ segmentation tasks (e.g., segmentation of kidneys in US images 34, and CT images 35), as well as clinical decision making (e.g., classifying breast tumors 36, early diagnosis of multiple sclerosis 37, and differentiating liver lesions 38).
In this manuscript, we examined whether image textures can serve as imaging biomarkers in ADPKD. We hypothesize that texture based imaging biomarkers provide quantifiable parameters of renal tissue structure that will better classify patients and improve individual prognosis compared to HtTKV.
A retrospective cohort of 122 patients from The Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease (CRISP study) was identified who had T2-weighted MRIs, eGFR values >70 mL/min/1.73m2 at the time of their baseline scan, and typical presentations of the disease, according to the definition provided by Irazabal et al. 39 Based on their eGFR measurements performed 8-years later, the patients were split into two groups according to whether they had or had not reached CKD stage 3A, or CKD stage 3B, or had a 30% reduction in eGFR (a potentially acceptable surrogate endpoint for the development of ESRD in some circumstances). 40
Results
Patient Cohort
In order to explore the potential of texture features as candidate imaging biomarkers, a retrospective cohort of patients from the CRISP study was identified with T2-weighted MRIs, eGFR values >70 mL/min/1.73m2 at the time of their baseline scan, and typical presentations of the disease (N=122) based on the description provided by Irazabal et al. 39 Based on eGFR measurements performed 8-years later, the patients were split into two groups according to whether they had (N=44) or had not (N=78) reached CKD stage 3A (eGFR <60 mL/min/1.73m2), whether they had (N=22) or had not (N=100) reached CKD stage 3B (eGFR <45 mL/min/1.73m2), or whether a 30% decrease of eGFR had (N=47) or had not (N=75) occurred.
A wide range of disease phenotypes were observed in this study's cohort. Shown in Figure 1 are examples of 6 patients with different renal cyst appearances along with age, sex, HtTKV, and genetic mutation information. Although some of these differences are obvious and one might suspect clinically that the disease severity will be different, no quantifiable parameters (i.e. imaging biomarkers) have yet been developed to characterize these differences.
Figure 1.
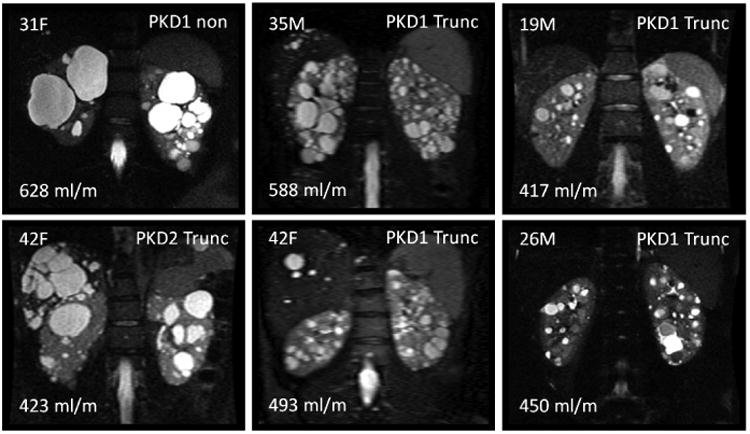
Examples highlighting phenotypic differences of 6 different ADPKD patients from this study. Shown in each panel is the age and sex of the patient in the upper left, the genetic mutation in the upper right, and the HtTKV measurement in the bottom left. Although visually many differences are apparent, no quantifiable image-based morphological parameters have yet been utilized beyond TKV and cystic burden of the kidneys to characterize ADPKD phenotypes.
Texture Analysis
Image texture features offer a promising approach to provide additional insights into PKD phenotypes. Figure 2 depicts how image texture features convey differences in tissue heterogeneity. In particular, how similar signal intensity regions in the MR image have drastically different textural depictions. Figure 3 shows the nine image texture features calculated in this study, based on a single middle coronal slice for the same patient. Each image texture feature calculates distinct properties, reflecting differences in kidney phenotype.
Figure 2.
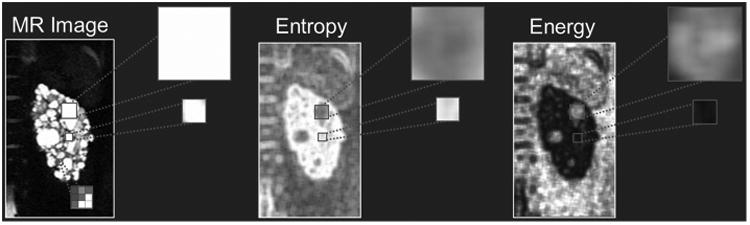
Texture feature analysis is an image processing technique that can inform on phenotypic differences of the kidneys. Shown in the left panel is a T2-weighted MRI of the left kidney of one of the patients in this study. If we consider two different cystic regions (zoomed-in panels), the information (in terms of image intensity) is similar. However, in the two example texture images of Entropy (middle panel) and Energy (right panel), information regarding size and position is ingrained in the image. Notice how the two cyst regions look very different in the texture images, and thus quantifiable values can be extracted which convey tissue structural information such as cyst size and number. Other structural tissue changes (e.g., fibrotic tissue or cyst classification) could likely be conveyed within the texture images.
Figure 3.
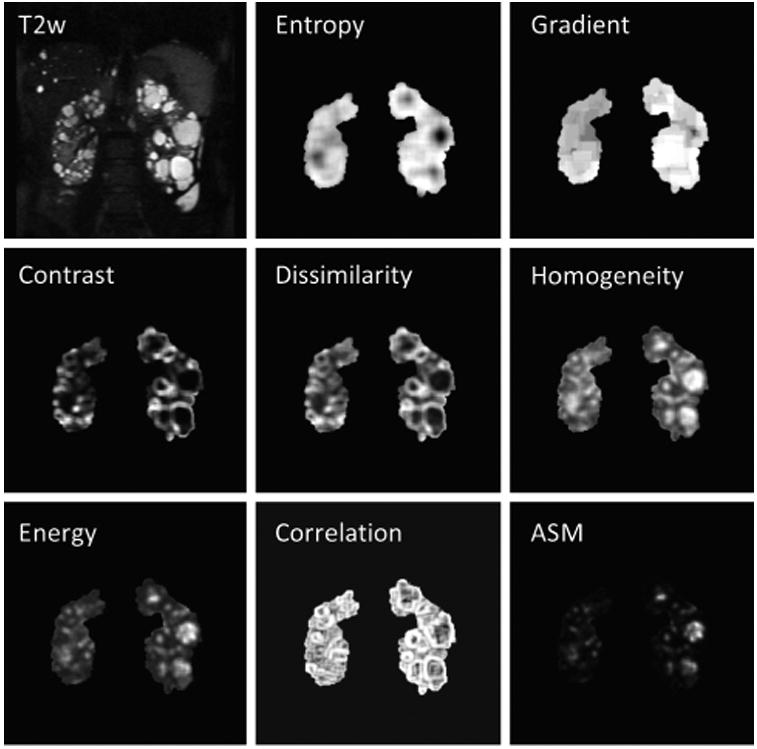
Examples of the nine derived imaging texture features for a single patient. From top left, by row then column: T2-weighted MR image from which gray scale values are extracted and texture analysis is performed, 1st order Entropy, 1st order Gradient, 2nd order GLCM Contrast, 2nd order GLCM Dissimilarity, 2nd order Homogeneity, 2nd order GLCM Energy, 2nd order GLCM Correlation, and 2nd order GLCM angular second moment (ASM). Entropy - Measures the degree of disorder within the kidney. Kidneys with seemingly random cyst distributions will have a higher Entropy measurement. Note how large cystic regions appear dark, whereas in regions of many small cysts a high Entropy value is calculated. Gradient – is a measure of gray scale changes. Kidneys with many small cysts will have more detectable edges and larger local changes in gray scale. Contrast - Measures the local variations in the gray-level cooccurrence matrix. Low values mean that the gray levels are similar throughout the kidneys. Dissimilarity – Measures differences in GLCM elements, which relates to a measure of renal tissue heterogeneity. Homogeneity – Measures the closeness of the distribution of elements in the GLCM to the GLCM diagonal. Note how both large cystic regions as well as regions with no cystic burden appear bright. Energy – Provides the sum of squared elements in the GLCM and is a measure of tissue uniformity. It is closely related to the inverse of Entropy. Correlation - Measures gray scale value dependence of kidney voxels (the joint probability occurrence of specified pixel pairs). Angular Second Moment (ASM) – Is also a measure of tissue homogeneity.
Predictive Power of Biomarkers
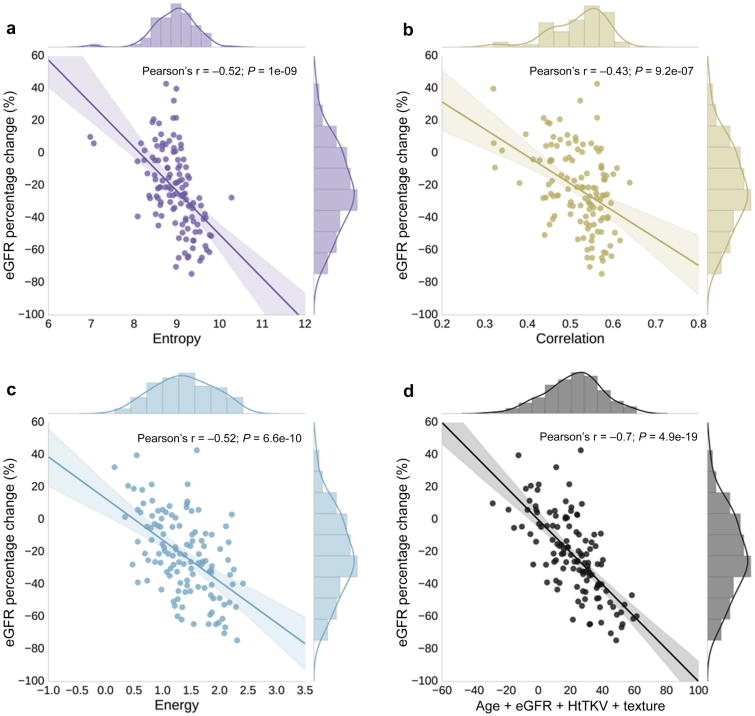
At baseline, age and eGFR had relatively little predictive value of subsequent renal function decline, whereas HtTKV had a much better predictive value as presented in Table 1 and shown in the ROC curves of Figure 4. For the three comparisons (progression to CKD stage 3A, progression to CKD stage 3B, and 30% change in eGFR) HtTKV had strong predictive power for progression to CKD stage 3A and 3B (Az = 0.81 and 0.84, respectively), while it was less informative for prediction of a 30% or more change in eGFR (Az = 0.73). Shown in Figure 5 are the regression analyses results, comparing baseline age, eGFR, and HtTKV to percent change in eGFR at 8 year follow up. Pearson's r was -0.07 (p = 0.45) for age, -0.03 (p=0.74) for baseline eGFR, and -0.48 (p < 0.01) for HtTKV. There exist a number of patients whose prognosis was not accurately determined using these more traditional biomarkers by themselves. A multiple linear regression model incorporating age, baseline eGFR, and HtTKV performed relatively well at distinguishing between the patient groupings (Figure 4, model labelled ‘Traditional’), and obtained an r = -0.51 (p < 0.01), as shown in the bottom right panel of Figure 5.
Table 1.
Summary statistics for age, eGFR, HtTKV, and the three image texture features identified by the stability feature selection approach. Statistics are given for progression to CKD stage 3A, stage 3B, and 30% cut-off threshold for percent change in eGFR. Each group has the median and median absolute deviation (m ± mad) listed between groups, p-value calculated from Wilcoxon rank-sum test, sensitivity, specificity, area under ROC curve (Az) with 95% confidence interval, and the cutoff threshold used for ROC analysis. A p-value < 0.01 was deemed significant (highlighted with an asterisk).
| Progression to CKD Stage 3A | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Biomarker | >60 | <60 | p-value | Sensitivity | Specificity | Az | Threshold |
| Age | 30.0 ± 9.0 | 34.6 ± 5.5 | 0.013 | 0.89 | 0.41 | 0.64 [0.58-0.72] | 26.5 |
| eGFR | 100.0 ± 16.0 | 81.3 ± 5.6 | < 0.01* | 0.68 | 0.83 | 0.77 [0.62-0.91] | 94.2 |
| HtTKV | 403 ± 118 | 724 ± 239 | < 0.01* | 0.82 | 0.67 | 0.81 [0.75-0.87] | 457 |
| Entropy | 8.80 ± 0.23 | 9.36 ± 0.17 | < 0.01* | 0.84 | 0.96 | 0.93 [0.88-0.97] | 9.2 |
| Correlation | 0.51 ± 0.05 | 0.55 ± 0.02 | < 0.01* | 0.80 | 0.63 | 0.72 [0.64-0.79] | 0.53 |
| Energy | 1.17 ± 0.35 | 1.82 ± 0.34 | < 0.01* | 0.57 | 0.90 | 0.80 [0.74-0.87] | 1.74 |
|
| |||||||
| Progression to CKD Stage 3B | |||||||
|
| |||||||
| Biomarker | >45 | <45 | p-value | Sensitivity | Specificity | Az | Threshold |
|
| |||||||
| Age | 31.7 ± 7.5 | 34.6 ± 5.9 | 0.17 | 0.96 | 0.28 | 0.59 [0.50-0.70] | 24.6 |
| eGFR | 96.3 ± 14.2 | 80.8 ± 4.4 | < 0.01* | 0.52 | 1.00 | 0.75 [0.70-0.81] | 94.2 |
| HtTKV | 428 ± 143 | 896 ± 324 | < 0.01* | 0.73 | 0.83 | 0.82 [0.73-0.90] | 700 |
| Entropy | 8.92 ± 0.28 | 9.38 ± 0.17 | < 0.01* | 0.87 | 0.76 | 0.86 [0.78-0.91] | 9.1 |
| Correlation | 0.52 ± 0.05 | 0.57 ± 0.02 | < 0.01* | 1.00 | 0.58 | 0.79 [0.71-0.85] | 0.53 |
| Energy | 1.25 ± 0.34 | 1.86 ± 0.23 | < 0.01* | 1.00 | 0.51 | 0.80 [0.74-0.87] | 1.26 |
|
| |||||||
| Percent Change in eGFR | |||||||
|
| |||||||
| Biomarker | <30% | >30% | p-value | Sensitivity | Specificity | Az | Threshold |
|
| |||||||
| Age | 31.8 ± 7.4 | 32.1 ± 7.3 | 0.469 | 0.77 | 0.35 | 0.54 [0.47-0.62] | 26.5 |
| eGFR | 94 2 ± 12.7 | 87.4 ± 9.3 | 0.350 | 0.45 | 0.77 | 0.55 [0.45-0.63] | 97.7 |
| HtTKV | 415 ± 129 | 610 ± 215 | < 0.01* | 0.81 | 0.53 | 0.73 [0.64-0.80] | 423 |
| Entropy | 8.83 ± 0.24 | 9.28 ± 0.24 | < 0.01* | 0.70 | 0.85 | 0.82 [0.73-0.88] | 9.1 |
| Correlation | 0.51 ± 0.05 | 0.55 ± 0.02 | < 0.01* | 0.77 | 0.63 | 0.69 [0.62-0.78] | 0.53 |
| Energy | 1.18 ± 0.34 | 1.74 ± 0.35 | < 0.01* | 0.49 | 0.91 | 0.75 [0.69-0.83] | 1.81 |
Figure 4.
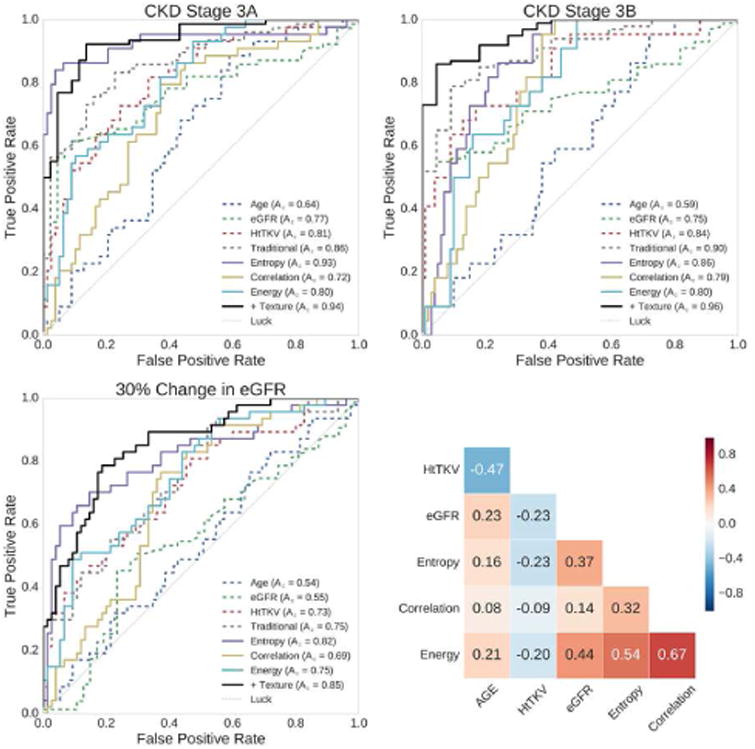
Individual biomarkers show strong predictive power for progression to CKD stage 3A, CKD stage 3B, and 30% change in eGFR in 8-year follow-up exams. Shown in the top left panel are the ROC curves for prediction to progression of CKD stage 3A, in the top right are the ROC curves for prediction of progression to CKD stage 3B, and in the bottom left are the ROC curves for prediction of a 30% or more decrease in eGFR. The individual biomarkers include age, eGFR, HtTKV, Entropy, Correlation, and Energy. In the bottom right is the correlation matrix showing how the individual features relate to one another. These biomarkers were included in a multiple linear regression model in order to ascertain the added value over traditional image derived modeling. The traditional model (‘Traditional’) used age, eGFR, and HtTKV, while the texture model (‘+ Texture’) also included Entropy, Correlation, and Energy. Compared with the traditional model, the addition of texture improved the predictive power to CKD stage 3A from an Az of 0.86 to an Az of 0.94, to CKD stage 3B from an Az of 0.90 to an Az of 0.96, and for a 30% or more change in eGFR from an Az of 0.75 to an Az of 0.85. Coloring in ROC curves corresponds to colors in Figures 5 and 6, and dashed biomarkers are ‘Traditional’, whereas solid lines are texture based.
Figure 5.
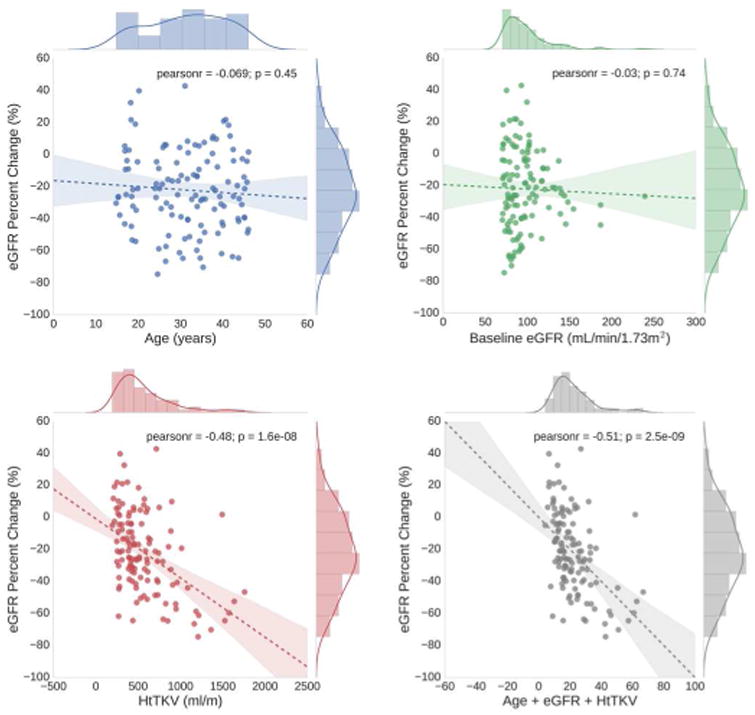
Shown here are the regression analyses results, comparing baseline traditional biomarkers (age, baseline eGFR, and HtTKV) to percent change in eGFR at 8 year follow up. Age (top left) and baseline eGFR (top right) have very low correlation, whereas HtTKV (bottom left) has a fairly high correlation, with subsequent percent change in eGFR. A multiple linear regression model incorporating these biomarkers obtained a Pearson's r = -0.51 (bottom right panel). Coloring corresponds to those used for each feature in Figure 4.
We used stability of texture features as the basis for selecting three texture metrics (median of Entropy with window = 31, 25th percentile of Correlation with window = 7, and skewness of Energy with window = 7). Focusing on stability rather than just the best performing for this cohort should better generalize to other patient cohorts. The correlation between the three traditional biomarkers (age, eGFR, and HtTKV) and the three image texture features is shown in the bottom right panel of Figure 4. We found that these three texture features correlated well with a 30% or more decrease in eGFR, and differentiated the patients who progressed to either CKD stage 3A or 3B (Table 1 and Figure 6). Entropy had the strongest predictive power of the three, with an Az = 0.93 for progression to CKD stage 3A, an Az = 0.86 for progression to CKD stage 3B, and an Az = 0.82 for predicting a 30% or more reduction in eGFR.
Figure 6.
Individual texture biomarkers showed strong correlation with subsequent percent change in eGFR. Shown here are the regression analyses results, comparing texture biomarkers (Entropy, Correlation, and Energy) to percent change in eGFR at 8 year follow up. Shown in the bottom right panel is the multiple linear regression model incorporating traditional biomarkers and texture features. This multiple linear regression model incorporating image texture features helped further refine prediction of subsequent renal function decline improving Pearson's r from -0.51 (traditional biomarkers alone, bottom right panel of Figure 5) to -0.70 (addition of image texture features). Coloring corresponds to those used for each feature in Figure 4.
Texture biomarkers also had a strong correlation with percent change in eGFR as shown in Figure 6. The Pearson's r for Entropy texture was -0.52 (p < 0.01), for Correlation texture was -0.43 (p < 0.01), and for Energy texture was -0.52 (p < 0.01). The three image texture features were included in a multiple linear regression model in order to ascertain the added value over traditional image derived modeling (age + eGFR + HtTKV). Incorporating the texture features improved prediction based on correlation coefficient, obtaining an r = -0.70 (p < 0.01) as shown in the bottom right panel of Figure 6. Also, summary statistics for the two models are shown in Table 2.
Table 2.
Statistics comparing the traditional prediction model (Age, eGFR, HtTKV) with the texture model. Pearson's r is defined as the square root of the adjusted r-square for the prediction models. SE is the standard error, and Sres is the residual standard deviation. Also shown are the standard errors and confidence intervals for the regression coefficients (slope and bias). The multiple linear regression models have a slope of -1, and bias of 0.
| Statistic | Traditional Model | +Texture Model |
|---|---|---|
|
| ||
| Pearson's r | -0.51 | -0.70 |
| SE | 3.9 | 2.6 |
| Sres | 21.6 | 17.9 |
| Slope SE | 0.155 | 0.094 |
| Slope CI | [-1.307 -0.693] | [-1.186 -0.814] |
| Bias SE | 3.95 | 2.64 |
| Bias CI | [-7.81 7.81] | [-5.22 5.22] |
Reproducibility of Texture Features
Texture derived imaging biomarkers were highly reproducible. In order to determine the reproducibility of the image derived texture features, 30 cases were reanalyzed by a second observer. The two major sources for variability were segmentation of the kidneys and the measurement of CSF which is used to linearly normalize the MR scans. For the mean±std comparison of the two segmentations: Jaccard similarity metric was 0.97±0.01, precision was 0.98±0.01, and percent volume error was 0.95±1.14%. For measurement of CSF, the percent difference was 3.2±5.0%. More importantly, the impact on texture measures was small: the percent difference for Entropy was 0.10±0.23%, for Correlation was 0.91±3.2%, and for Energy was 0.13±0.35%.
Discussion
This current study presents the first report of texture analysis for ADPKD assessment. A cohort of patients (from the CRISP study) with normal renal function at baseline and typical presentations of the disease was identified. We assessed the value of various biomarkers to predict progression to CKD stage 3A and 3B 8-years from baseline, as well as percent change in eGFR. MRIs were pre-processed, standardized, and kidneys were segmented. Texture features were extracted from the images and evaluated for their individual performance of distinguishing rapid vs slow progressing groups. Lastly, a multiple linear regression model was developed that demonstrated the added value of texture image analysis over current models.
Individual texture features were found to predict subsequent renal function decline. These features characterized factors such as the degree of disorder within the kidney, differences in cyst size and number, as well as whether the kidneys were composed of many similar appearing regions to differentiate the patients. The combination of the features identified by stability selection utilizing the multiple linear regression model system allowed for the greatest prediction ability for subsequent renal function decline. It is likely that the combination of texture feature analysis with other non-image based biomarkers will improve personalized clinical decision making of ADPKD. We believe that these texture metrics can function as a complementary tool to existing classification methods, such as the HtTKV-based prognostic model developed by Irazabal et al. 39 and the proPKD score developed by Cornec-Le Gall et al 41. In particular, based on these results (Table 2), there is a 23% narrowing of the prediction intervals when including the MR-derived texture features. As an example, using the traditional model, a prediction of a 30% decrease in eGFR has a 95% confidence interval with a fairly wide range (from 16.5 to 43.5%), whereas including texture has a 95% confidence interval which is much more precise (from 26.5 to 33.5%). Thus, incorporating image texture into current prediction models greatly improves disease prognosis precision.
In addition, this approach of texture analysis may be preferable to other imaging methods including new quantitative MRI techniques, as they may be more challenging to put into practice. Texture analysis can be easily applied to routinely acquired MRI examinations. Therefore, retrospective studies (such as the current study) can be pursued to search for new imaging biomarkers of the disease when known disease progression data is available. Quantitative MRI techniques are more difficult to acquire and may be harder to standardize. So although newer imaging techniques hold a great deal of promise in terms of tissue characterization and functional evaluation, their association with disease severity and disease progression will take many years to assess.
The phenotypical description provided by image texture features as they relate to characterizing the PKD kidney is important to understand in order to best apply these techniques in future studies. Briefly, we highlight these details for the three image texture features identified by Stability Selection and used in the regression model. Entropy - Measures the degree of disorder within the kidney. Kidneys with seemingly random cyst distributions will have a higher Entropy measurement. Correlation - Measures gray scale value dependence of kidney voxels (the joint probability occurrence of specified pixel pairs). High values mean the kidneys are composed of many similar appearing regions. Energy – Provides the sum of squared elements in the GLCM and is a measure of tissue uniformity. The power of texture analysis comes from the fact that we can derive a number of measureable parameters regarding kidney structure which can be used to further refine an individual's prognosis based on phenotype.
The approach of texture feature analysis may also be preferable to other image processing techniques such as cyst segmentation. If cyst segmentation is performed manually, significant time is required, making a study of even moderate size nearly impossible. In addition, variability in the measurements will likely be fairly high as discerning what are, and what are not cysts, as well as defining cyst edges in scans of ADPKD patients is extremely difficult. On the other hand, if performed automatically, questions regarding the accuracy are difficult to ascertain. Fortunately, texture feature analysis provides information regarding cystic phenotype without requiring cysts to be individually segmented and classified.
CKD, which is characterized by reduced renal function, is a common outcome for ADPKD patients. However, the time course of disease progression to various stages of CKD is far from fully understood. Therefore, the development of new tools to better characterize the evolution of the disease is needed. We believe that the new imaging biomarkers provided by image texture feature analysis, along with automated TKV, will significantly improve the assessment of patient prognosis, and will facilitate the ability to more quickly judge the effectiveness of interventions.
There exist a large number of challenges in terms of applying texture analysis to clinical practice as well as research studies. For example, image acquisition protocols, as well as image quality should be kept as similar as possible since MRI shows a greater non-linear influence on signal intensity and on quantification of heterogeneity (compared with CT). However, non-contrast enhanced CT will not provide sufficient soft tissue contrast to make texture analysis effective. To be most effectively utilized, standardization of the image acquisition protocols is needed.
Future studies should be performed to evaluate texture analysis in larger and more diverse patient populations to fully understand how these new imaging biomarkers improve our understanding of ADPKD phenotypes for disease prognosis and clinical decision making. Adding other clinical data in non-biased populations needs to be explored, as well as analysis of texture feature analysis on other imaging modalities (e.g., CT) and different sequences (e.g., T1-weighted MRI).
Methods
Approval from Mayo Clinic's Institutional Review Board was obtained for this retrospective study.
MRI Data
The MRIs were coronal single shot fast spin echo (SSFSE) T2 sequences, acquired with a GE (GE Medical Systems, Waukesha, WI) scanner, with matrix size 256×256×Z (with Z large enough to cover the full extent of the kidneys within the imaged volume). Specifically, B0: 1.5T, TE: 190ms, TR: minimum, pixel size: 1.5mm, slice thickness: 3.0mm, slice spacing: 3.0mm. Images were typically acquired over several breath holds.
Pre-Processing
T2-weighted abdominal MRIs often have signal intensity non-uniformities, including both intra-42 and inter-slice 43 artifacts. Intra-slice intensity artifacts come from several sources (e.g., choice of radio-frequency coil, sample geometry, etc.) which degrade image quality and introduce low frequency intensity changes. Inter-slice intensity variations are the result of gradient eddy currents and cross-talk between slices. These cause interleaved ‘dark’ and ‘bright’ slices, which can be drastically different. To correct for these issues, we used an approach developed in-house which sequentially corrects inter-slice intensity, and intra-slice intensity variations 44, 45.
Finally, although texture analysis is rather robust to differences in absolute image signal ranges, the images were normalized based on the cerebral spinal fluid (CSF) in the adjacent spinal canal to have standardized tissue signal. Measurement of CSF intensity was performed for each scan by drawing a region-of-interest (ROI) containing purely CSF. The mean value of this ROI was then normalized to have a value of 1,000 (i.e., CSF was normalized to have the same value in each MRI scan).
Kidney Segmentation
A trained medical image analyst (M.E.E.) performed kidney segmentation semi-automatically utilizing the MIROS software package (https://github.com/TLKline/PyCysticImage) 46. The software package has an interactive viewer that allows visualization of the image data in coronal, sagittal, and axial planes. Segmentations can be overlaid and edited with a range of interactive tools. The MIROS algorithm was implemented in the Python programming language and has a push-button that starts the interactive tool for defining crude polygon contours of the cystic organ. We performed the MIROS method obtaining user input every third slice (every 9mm). After segmentation of each kidney by MIROS, the interactive tools were used to perform quality assurance and finalize the segmentation.
Texture Features
Gray level (1st order)
First order texture features capture basic characteristics of the intensities within an image. Global intensity features were computed for each MRI. In addition, Gradient and Entropy local intensity features were computed at five different window sizes (7, 15, 23, 31, and 41). In order to account for the anisotropic nature of the voxel size, texture feature filters were calculated in a slice-by-slice fashion, for every slice within the volume.
Gray level co-occurrence matrix (2nd order)
Gray level co-occurrence matrix (GLCM) characterizes the second order statistics of the spatial distribution of gray levels in an image 47. For each image, four GLCM (corresponding to four directions) were calculated at three different window sizes (7, 15, and 31), and 6 features (Contrast, Dissimilarity, Homogeneity, Energy, Correlation, and Angular Second Moment) were derived from the GLCMs. The GLCM based features were implemented in the Python programming language.
Texture-filtered features
Texture-filtered versions of the images were created for each patient. In the case of the 2nd order features, averaging over the four directions was performed (making the derived textures direction invariant). Finally, for each of the kidney ROIs, the mean, median, standard deviation, skewness, and the 25th and 75th percentiles were calculated for the filtered textures. These values were then used as features to search for notable textural differences between the two groups of patients. Calculation of all texture features was performed in a matter of minutes for each patient scan.
Reproducibility
In order to determine the reproducibility of the image derived texture features, 30 cases were reanalyzed by a second observer. Similarity metrics were calculated between the segmentations derived by the two observers, as well as comparison of the measured CSF values. Finally, the difference between the computed texture values was characterized.
Classification System
Feature selection
A robust feature selection method known as Stability Selection was used to identify candidate features for developing the multiple linear regression model 48. The feature selection algorithm is applied on different subsets of data with different subsets of features. The process is repeated and the number of times a particular feature is selected as important is evaluated. The most stable features get a score of 1 while weak features get a score of 0.
Multiple linear regression
The multiple linear regression modelling approach was implemented in Python with the sklearn toolkit which uses a least squares approach (function ‘sklearn.linear_model.LinearRegression’). The approach minimizes the sum of the squared residuals in order to adjust the variable weights to best fit the data.5/10/2017
Statistical Analysis
Analysis based on Wilcoxon rank sum tests were performed to calculate statistical significance of each imaging biomarker, as well as the area-under-ROCs (Az), and the sensitivity and specificity of the features was also computed. Bootstrap resampling was used to estimate confidence intervals for Az. Regression analysis was performed to characterize the relationship between individual biomarkers and the percent change in eGFR.
Acknowledgments
Sources of support: This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases under NIH Grant/Award Number P30 DK090728 to the “Mayo Clinic Robert M. and Billie Kelley Pirnie Translational Polycystic Kidney Disease Center”, the PKD Foundation under grant 206g16a, and the National Cancer Institute (NCI) under grant/award CA160045. The CRISP study is supported by cooperative agreements from the NIDDK of the NIH (DK056943, DK056956, DK056957, DK056961) and by the National Center for Research Resources General Clinical Research Centers at each institution (RR000039, Emory University; RR00585, Mayo College of Medicine; RR23940, Kansas University Medical Center; RR000032, University of Alabama at Birmingham) and the National Center for Research Resources Clinical and Translational Science Awards at each institution (RR025008, Emory University; RR024150, Mayo College of Medicine; RR033179, Kansas University Medical Center; RR025777 and UL1 TR000165, University of Alabama at Birmingham; RR024153, University of Pittsburgh School of Medicine).
Footnotes
Disclosure: All the authors declared no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chapman AB, Devuyst O, Eckardt KU, et al. Autosomal-dominant polycystic kidney disease (ADPKD): executive summary from a Kidney Disease: Improving Global Outcomes (KDIGO) Controversies Conference. Kidney Int. 2015;88:17–27. doi: 10.1038/ki.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grantham JJ, Torres VE, Chapman AB, et al. Volume progression in polycystic kidney disease. N Engl J Med. 2016;354:2122–2130. doi: 10.1056/NEJMoa054341. [DOI] [PubMed] [Google Scholar]
- 3.Chapman AB, Bost JE, Torres VE, et al. Kidney volume and functional outcomes in autosomal dominant polycystic kidney disease. Clin J Am Soc Nephrol. 2012;7:479–486. doi: 10.2215/CJN.09500911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhutani H, Smith V, Rahbari-Oskoui F, et al. A comparison of ultrasound and magnetic resonance imaging shows that kidney length predicts chronic kidney disease in autosomal dominant polycystic kidney disease. Kidney Int. 2015;88:146–151. doi: 10.1038/ki.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torres VE, Chapman AB, Devuyst O, et al. Tolvaptan in patients with autosomal dominant polycystic kidney disease. N Engl J Med. 2012;367:2407–2418. doi: 10.1056/NEJMoa1205511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caroli A, Perico N, Perna A, et al. Effect of longacting somatostatin analogue on kidney and cyst growth in autosomal dominant polycystic kidney disease (ALADIN): a randomised, placebo-controlled, multicentre trial. Lancet. 2013;382:1485–1495. doi: 10.1016/S0140-6736(13)61407-5. [DOI] [PubMed] [Google Scholar]
- 7.Schrier RW, Abebe KZ, Perrone RD, et al. Blood pressure in early autosomal dominant polycystic kidney disease. N Engl J Med. 2014;371:2255–2266. doi: 10.1056/NEJMoa1402685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mignani R, Corsi C, De Marco M, et al. Assessment of kidney volume in polycystic kidney disease using magnetic resonance imaging without contrast medium. Am J Nephrol. 2011;33:176–184. doi: 10.1159/000324039. [DOI] [PubMed] [Google Scholar]
- 9.Cohen BA, Barash I, Kim DC, et al. Intraobserver and interobserver variability of renal volume measurements in polycystic kidney disease using a semiautomated MR segmentation algorithm. AJR Am J Roentgenol. 2012;199:387–393. doi: 10.2214/AJR.11.8043. [DOI] [PubMed] [Google Scholar]
- 10.Kline TL, Korfiatis P, Edwards ME, et al. Automatic total kidney volume measurement on follow-up MRIs to faciliate monitoring of ADPKD progression. Nephrol Dial Transplant. 2016;31:241–248. doi: 10.1093/ndt/gfv314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turco D, Severi S, Mignani R, et al. Reliability of Total Renal Volume Computation in Polycystic Kidney Disease From Magnetic Resonance Imaging. Acad Radiol. 2015;22:1376–1384. doi: 10.1016/j.acra.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 12.Kistler AD, Poster D, Krauer F, et al. Increases in kidney volume in autosomal dominant polycystic kidney disease can be detected within 6 months. Kidney Int. 2009;75:235–241. doi: 10.1038/ki.2008.558. [DOI] [PubMed] [Google Scholar]
- 13.King BF, Reed JE, Bergstralh EJ, et al. Quantification and longitudinal trends of kidney, renal cyst, and renal parenchyma volumes in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2000;11:1505–1511. doi: 10.1681/ASN.V1181505. [DOI] [PubMed] [Google Scholar]
- 14.Bae KT, Zhu F, Chapman AB, et al. Magnetic resonance imaging evaluation of hepatic cysts in early autosomal-dominant polycystic kidney disease: the Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease cohort. Clin J Am Soc Nephrol. 2006;1:64–69. doi: 10.2215/CJN.00080605. [DOI] [PubMed] [Google Scholar]
- 15.Bae K, Park B, Sun H, et al. Segmentation of individual renal cysts from MR images in patients with autosomal dominant polycystic kidney disease. Clin J Am Soc Nephrol. 2013;8:1089–1097. doi: 10.2215/CJN.10561012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman AB, Guay-Woodford LM, Grantham JJ, et al. Renal structure in early autosomal-dominant polycystic kidney disease (ADPKD): The Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease (CRISP) cohort. Kidney Int. 2003;64:1035–1045. doi: 10.1046/j.1523-1755.2003.00185.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang JL, Morrell G, Rusinek H, et al. New magnetic resonance imaging methods in nephrology. Kidney Int. 2014;85:768–778. doi: 10.1038/ki.2013.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kajander S, Kallio T, Alanen A, et al. Imaging end-stage kidney disease in adults. Low-field MR imaging with magnetization transfer vs. ultrasonography. Acta Radiol. 2000;41:357–360. doi: 10.1080/028418500127345460. [DOI] [PubMed] [Google Scholar]
- 19.Ebrahimi B, Macura SI, Knudsen BE, et al. Fibrosis detection in renal artery stenosis mouse model using magnetization transfer MRI. Proc SPIE 8672, Medical Imaging 2013: Biomedical Applications in Molecular, Structural, and Functional Imaging, 867205. 2013;8672:867. [Google Scholar]
- 20.Kline TL, Irazabal MV, Ebrahimi B, et al. Utilizing magnetization transfer imaging to investigate tissue remodeling in a murine model of autosomal dominant polycystic kidney disease. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2015 doi: 10.1002/mrm.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sourbron SP, Michaely HJ, Reiser MF, et al. MRI-measurement of perfusion and glomerular filtration in the human kidney with a separable compartment model. Invest Radiol. 2008;43:40–48. doi: 10.1097/RLI.0b013e31815597c5. [DOI] [PubMed] [Google Scholar]
- 22.Li LP, Halter S, Prasad PV. Blood oxygen level-dependent MR imaging of the kidneys. Magn Reson Imaging Clin N Am. 2008;16:613–625. viii. doi: 10.1016/j.mric.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pedersen M, Dissing TH, Morkenborg J, et al. Validation of quantitative BOLD MRI measurements in kidney: application to unilateral ureteral obstruction. Kidney Int. 2005;67:2305–2312. doi: 10.1111/j.1523-1755.2005.00334.x. [DOI] [PubMed] [Google Scholar]
- 24.Prasad PV, Edelman RR, Epstein FH. Noninvasive evaluation of intrarenal oxygenation with BOLD MRI. Circulation. 1996;94:3271–3275. doi: 10.1161/01.cir.94.12.3271. [DOI] [PubMed] [Google Scholar]
- 25.Khatir DS, Pedersen M, Jespersen B, et al. Reproducibility of MRI renal artery blood flow and BOLD measurements in patients with chronic kidney disease and healthy controls. J Magn Reson Imaging. 2014;40:1091–1098. doi: 10.1002/jmri.24446. [DOI] [PubMed] [Google Scholar]
- 26.King BF, Torres VE, Brummer ME, et al. Magnetic resonance measurements of renal blood flow as a marker of disease severity in autosomal-dominant polycystic kidney disease. Kidney Int. 2003;64:2214–2221. doi: 10.1046/j.1523-1755.2003.00326.x. [DOI] [PubMed] [Google Scholar]
- 27.Torres VE, King BF, Chapman AB, et al. Magnetic resonance measurements of renal blood flow and disease progression in autosomal dominant polycystic kidney disease. Clin J Am Soc Nephrol. 2007;2:112–120. doi: 10.2215/CJN.00910306. [DOI] [PubMed] [Google Scholar]
- 28.Warner L, Yin M, Glaser KJ, et al. Noninvasive In vivo assessment of renal tissue elasticity during graded renal ischemia using MR elastography. Invest Radiol. 2011;46:509–514. doi: 10.1097/RLI.0b013e3182183a95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maril N, Margalit R, Mispelter J, et al. Functional sodium magnetic resonance imaging of the intact rat kidney. Kidney Int. 2004;65:927–935. doi: 10.1111/j.1523-1755.2004.00475.x. [DOI] [PubMed] [Google Scholar]
- 30.Thoeny HC, De Keyzer F, Oyen RH, et al. Diffusion-weighted MR imaging of kidneys in healthy volunteers and patients with parenchymal diseases: initial experience. Radiology. 2005;235:911–917. doi: 10.1148/radiol.2353040554. [DOI] [PubMed] [Google Scholar]
- 31.Chandarana H, Lee VS, Hecht E, et al. Comparison of biexponential and monoexponential model of diffusion weighted imaging in evaluation of renal lesions: preliminary experience. Invest Radiol. 2011;46:285–291. doi: 10.1097/RLI.0b013e3181ffc485. [DOI] [PubMed] [Google Scholar]
- 32.Wang F, Kopylov D, Zu Z, et al. Mapping murine diabetic kidney disease using chemical exchange saturation transfer MRI. Magn Reson Med. 2015 doi: 10.1002/mrm.26045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castellano G, Bonilha L, Li LM, et al. Texture analysis of medical images. Clin Radiol. 2004;59:1061–1069. doi: 10.1016/j.crad.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 34.Xie J, Jiang Y, Tsui HT. Segmentation of kidney from ultrasound images based on texture and shape priors. IEEE Trans Med Imaging. 2005;24:45–57. doi: 10.1109/tmi.2004.837792. [DOI] [PubMed] [Google Scholar]
- 35.Lin DT, Lei CC, Hung SW. Computer-aided kidney segmentation on abdominal CT images. IEEE Trans Inf Technol Biomed. 2006;10:59–65. doi: 10.1109/titb.2005.855561. [DOI] [PubMed] [Google Scholar]
- 36.Chen W, Giger ML, Li H, et al. Volumetric texture analysis of breast lesions on contrast-enhanced magnetic resonance images. Magn Reson Med. 2007;58:562–571. doi: 10.1002/mrm.21347. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Tong L, Wang L, et al. Texture analysis of multiple sclerosis: a comparative study. Magn Reson Imaging. 2008;26:1160–1166. doi: 10.1016/j.mri.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida H, Casalino DD, Keserci B, et al. Wavelet-packet-based texture analysis for differentiation between benign and malignant liver tumours in ultrasound images. Phys Med Biol. 2003;48:3735–3753. doi: 10.1088/0031-9155/48/22/008. [DOI] [PubMed] [Google Scholar]
- 39.Irazabal MV, Rangel LJ, Bergstralh EJ, et al. Imaging classification of autosomal dominant polycystic kidney disease: a simple model for selecting patients for clinical trials. J Am Soc Nephrol. 2015;26:160–172. doi: 10.1681/ASN.2013101138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levey AS, Inker LA, Matsushita K, et al. GFR decline as an end point for clinical trials in CKD: a scientific workshop sponsored by the National Kidney Foundation and the US Food and Drug Administration. Am J Kidney Dis. 2014;64:821–835. doi: 10.1053/j.ajkd.2014.07.030. [DOI] [PubMed] [Google Scholar]
- 41.Cornec-Le Gall E, Audrezet MP, Rousseau A, et al. The PROPKD Score: A New Algorithm to Predict Renal Survival in Autosomal Dominant Polycystic Kidney Disease. J Am Soc Nephrol. 2015 doi: 10.1681/ASN.2015010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Belaroussi B, Milles J, Carme S, et al. Intensity non-uniformity correction in MRI: existing methods and their validation. Med Image Anal. 2006;10:234–246. doi: 10.1016/j.media.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt MA. Department of Computing Science. Edmonton: University of Albert; 2005. Method for Standardizing MR Intensities between Slices and Volumes. [Google Scholar]
- 44.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 45.Tustison NJ, Avants BB, Gee JC. Directly manipulated free-form deformation image registration. IEEE Trans Image Process. 2009;18:624–635. doi: 10.1109/TIP.2008.2010072. [DOI] [PubMed] [Google Scholar]
- 46.Kline TL, Edwards ME, Korfiatis P, et al. Semi-automated segmentation of polycystic kidneys in T2-weighted magnetic resonance images. Am J Roentgenol. 2016;207:605–613. doi: 10.2214/AJR.15.15875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haralick RM, Shanmugam K, Dinstein IH. Textural Features for Image Classification. Systems, Man and Cybernetics, IEEE Transactions on. 1973;SMC-3:610–621. [Google Scholar]
- 48.Meinshausen N, Buhlmann P. Stability selection. J R Stat Soc B. 2010;72:417–473. [Google Scholar]