Highlights
-
•
We tried to establish the epigenetics of malignant transformation in PCC/PGLs.
-
•
Benign and malignant PCC/PGLs were examined using whole genomic methylation analysis.
-
•
Selected candidate CpGs were narrowed down by analysis of individual genomic regions.
-
•
Two were left as the final candidates, related to ACSBG1 and MAST1 respectively.
-
•
Epigenetics in these genes might be involved in malignant transformation of PCC/PGLs.
Keywords: Pheochromocytoma, Paraganglioma, Methylation, ACSBG1, MAST1
Abstract
Aims
In recent years, aberrant DNA methylation of specific CpG sites has been detected in many types of malignant tumors, and the epigenetic regulation of promoter CpG sites is considered an important mechanism underlying carcinogenesis. This study aimed to establish the epigenetics of the malignant transformation of malignant pheochromocytoma (PCC) and paraganglioma (PGL) by performing a methylation analysis.
Materials and methods
Based on the results of the Infinium HumanMethylation450 BeadChip array using DNA samples of PCC/PGL patients, candidate CpG sites that were hyper/hypo-methylated in metastatic tumors relative to those in the primary tumors of 2 patients with malignant PCC/PGL were selected. The methylation levels of the chosen candidate CpG sites were evaluated quantitatively.
Results
Twelve CpG sites were selected as hypermethylated candidates, and 16 CpG sites were selected as hypomethylated candidates. Using two quantitative methylation analysis methods, one hypermethylated site (cg02119938) and one hypomethylated site (cg26870725) remained as candidates. These sites were related to ACSBG1 (acyl-CoA synthetase bubblegum family member 1) and MAST1 (microtubule-associated serine-threonine kinase 1), respectively. Immunohistochemical studies of ACSBG1 and MAST1 revealed that epigenetic changes in the malignant transformation of PCC/PGL might be associated with ACSBG1 silencing or MAST1 overexpression.
Conclusions
Here, we report two noteworthy genes, ACSBG1 and MAST1; the aberrant promoter methylation/demethylation of these genes might be involved in their silencing/expression in malignant PCC/PGL. Further investigations are necessary to determine the role of ACSBG1 and/or MAST1 expression in malignant transformation and to establish pathological markers that can evaluate the malignant potential of PCC/PGL.
Introduction
Pheochromocytoma (PCC) and paraganglioma (PGL) are rare tumors derived from the chromaffin tissue of the adrenal medulla or sympathetic/parasympathetic ganglia. These tumors occur in 0.3 or fewer people per million people per year [1], and most of them are benign. The prevalence of malignant PCC/PGL is reported to be 3% to 36% of all PCC/PGL cases [2], [3], [4].
Malignant PCC/PGL is unique because it is very difficult to diagnose early. Unlike many other malignancies, neither reliable histological markers in pathological specimens nor biochemical markers in peripheral samples are available to distinguish between benign and malignant tumors, and the diagnosis of malignancy cannot be made until metastasis to a non-chromaffin organ(s) has been identified. Furthermore, the time required for metastasis to develop, with a mean duration of 8.5 ± 6.0 years after primary surgery, presents an additional challenge in the diagnosis [5].
Because no effective therapeutic methods for progressive malignant PCC/PGL have been reported, malignant PCC/PGL has a very poor prognosis. To establish a useful clinical marker for early diagnosis and an effective treatment for this disease, it is important to clarify the mechanisms of the malignant transformation of PCC/PGL.
In recent years, DNA methylation of specific CpG islands (CpGi) has been detected in many types of malignant tumors, and the epigenetic regulation of promoter CpGis of tumor-related genes is considered an important mechanism for cancerous change. In this study, we hypothesized that PCC/PGL tumors, which are benign tumors originally, undergo metastasis progressively through epigenetic events. To test this hypothesis, we investigated the methylation of malignant PCC/PGL tumors and tried to establish the epigenetics of their malignant transformation.
Materials and methods
Study design
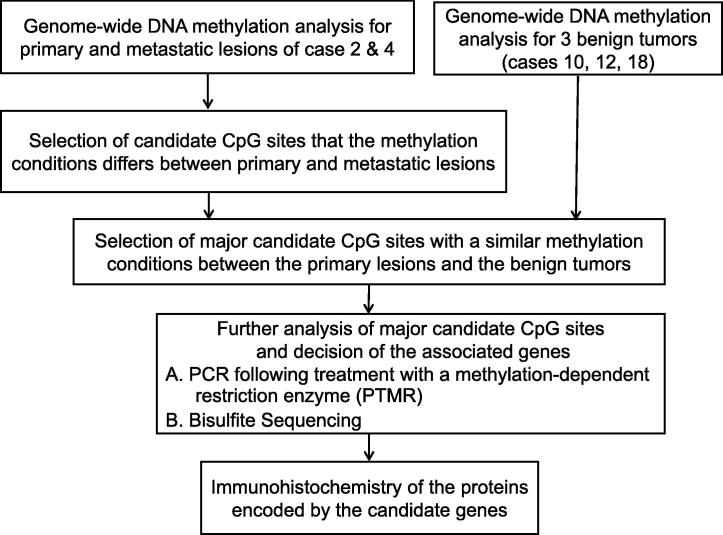
A genome-wide methylation analysis of primary and metastatic tumors in a malignant PCC/PGL patient was performed. Based on the results of this analysis, we selected several candidate CpG sites. To elucidate the pathological role of each candidate CpG site, we evaluated the quantitative methylation states of these sites using multiple procedures, including PCR following treatment with a methylation-dependent restriction enzyme (PTMR) and bisulfite sequencing in a larger number of patients (Fig. 1).
Fig. 1.
Outline of the study design.
Patients and tissue samples
We used the fresh frozen (FF) or formalin-fixed paraffin-embedded (FFPE) tissues of 19 human PCCs and PGLs (12 females and 7 males/15 PCC and 4 PGL specimens). The tissue specimens were obtained during surgery conducted at Hamamatsu University Hospital and the affiliated hospitals from 1991 to 2015. Table 1 presents a summary of the attended 19 cases. This study was conducted with the approval of these hospitals' institutional review boards. We have obtained written informed consent from all of the patients or their families.
Table 1.
Clinical details of the patients in this study.
| Case | Age (y.) | Gender | DM | HT | FH | Primary | Size (cm) | Metastatic lesion | Metastatasis-free survival (mo.) | Germline mutation | Follow-up (mo.) | MIB1 LI(%) | Sample | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Malignant | ||||||||||||||
| 1 | 29 | F | − | − | − | Bladder | Metastasis | 3.5 | Bone | 36 | DOD | 8.9 | FFPE | |
| 2 | 58 | M | + | + | − | Adrenal | Primary | 10 | Bone, lung, | 93 | DOD | 2.2 | FFPE | |
| Metastasis | 0.5 | Anterior mediastinum | 18.3 | FFPE | ||||||||||
| 3 | 62 | F | + | − | − | Retroperitoneal | Metastasis | 6 | Liver | 3 | DOD | 7.8 | FFPE | |
| 4 | 12 | F | − | + | − | Bladder | Primary | 5.7 | Bone, lung | 133 | SDHB | 240 | <1 | FFPE |
| Metastasis | 3.5 | 15.4 | FFPE | |||||||||||
| 5 | 54 | M | + | − | − | Retroperitoneal | Metastasis | 12 | Lung, bone | a.d | DOD | 12.6 | FF | |
| 6 | 34 | M | − | + | − | Bladder | Primary | 6.1 | Lung, bone | 27 | DOD | 7.5 | FFPE | |
| Metastasis | 4.5 | 9.9 | FF | |||||||||||
| 7 | 37 | F | + | + | − | Adrenal | Primary | 3.5 | Lung, liver, bone | 37 | DOD | <1 | FFPE | |
| Metastasis | 4 | 13.6 | FFPE | |||||||||||
| 8 | 58 | F | + | + | − | Adrenal | Metastasis | 8 | Lung, liver, bone | 12 | 17 | 11.7 | FF | |
| 9 | 55 | F | − | − | − | Adrenal | Metastasis | 3.8 | Liver | a.d. | RET | 12 | 5.0 | FFPE |
| Benign | ||||||||||||||
| 10 | 59 | M | + | + | − | Adrenal | 4.2 | 260 | 1.1 | FF | ||||
| 11 | 19 | F | + | + | − | Adrenal | 5.5 | VHL | 233 | <1 | FF | |||
| 12 | 48 | F | + | + | − | Adrenal | 10 | 215 | 1.7 | FF | ||||
| 13 | 55 | F | + | + | + | Adrenal | 3 | RET | 208 | <1 | FFPE | |||
| 14 | 51 | M | + | + | − | Adrenal | 3 | 198 | <1 | FFPE | ||||
| 15 | 23 | M | − | + | − | Adrenal | 8.5 | RET | 171 | <1 | FFPE | |||
| 16 | 47 | F | + | + | − | Adrenal | 12 | 171 | <1 | FFPE | ||||
| 17 | 56 | M | + | + | − | Retroperitoneul | 5.5 | 160 | <1 | FFPE | ||||
| 18 | 58 | F | + | + | − | Adrenal | 3.2 | 143 | <1 | FF | ||||
| 19 | 25 | F | − | + | − | Adrenal | 6.5 | RET | 142 | <1 | FFPE | |||
DM: diabetes mellitus, HT: hypertension, FH: family history of pheochromocytomas or paragangliomas, Metastatasis-free survival: how long the metastatic lesion was found, MIB1 LI: MIB-1 labeling index, SDHB: succinate dehydrogenase complex subunit B, VHL: von Hippel-Limdau disease tumor suppressor, RET: rearranged during transfection, a.d.: at diagnosis, DOD: dead of disease, Primary: primary tumor, Meta: metastatic lesion, n.d.: not done, FFPE: Formalin-fixed paraffin-embedded, FF: Fresh frozen.
Based on information concerning the tumor recurrence and survival of each patient as of the date of entry into this study, diagnoses of benign or malignant PCC/PGL were made. According to a report, the average time to diagnosis of malignant PCC/PGL from primary surgery based on the existence of metastatic lesion was 8.5 years (maximum 17 years) in the western region of Sweden [5]. Cohort studies of Japanese PCC/PGL patients that examined metastatic-free survival have not yet been published. In our study, the mean metastatic-free survival was 48.7 (range from 3 to 133; SD 47.0) months, and the longest was 133 months (11 years) among 9 malignant patients. Based on these observations, we defined benign PCC/PGL here as cases in which the patients were disease free for 133 months or longer. Nine malignant PCC/PGLs and 10 benign tumors were studied. The diagnoses of all 9 patients with malignant PCC/PGL in this study were based on the identification of distant metastasis in non-chromaffin organs, not on invasion to nearby tissue.
We were fortunate to obtain two types of tumors at different stages (both primary and metastatic tumors in a single patient) from two of the 9 patients with malignant PCC/PGLs (Cases 2 and 4). Case 2 was a male patient who was diagnosed with right adrenal PCC without metastatic lesions at 58 years of age and who underwent adrenalectomy; neither adjuvant chemotherapy nor irradiation was administered. Eight years later, metastasis of the plural lymph nodes, including the Virchow lymph node, was diagnosed. The patient died of respiratory failure with tumor growth to the trachea. We considered the adrenal PCC of this patient as the primary tumor and the biopsied Virchow lymph node as the metastatic tumor. Case 4 was a female patient who was diagnosed with PGL in the bladder at 12 years of age and underwent segmental resection of the bladder. At 23 years of age, bone metastases to the rib, lung and sternum were diagnosed. The patient underwent surgery to remove all of the detected metastatic tumors and received adjuvant chemotherapy. No recurrence was observed during the period of enrollment in this study, 5 years after the last course of chemotherapy. We considered the bladder PGL of this patient as the primary tumor and a lung metastatic lesion as the metastatic tumor.
We analyzed the primary and metastatic tumors of Cases 2 and 4 using the Ion Ampliseq Comprehensive Cancer Panel (Life Technologies, Carlsbad, CA, USA) to evaluate somatic mutations. We detected no significant variants in these samples (data not shown).
DNA extraction
Before extraction, the tumor area was delineated macro- and microscopically in each operatively resected specimen and was then carefully macrodissected using a scalpel. DNA was extracted using a DNA/RNA Mini kit (Qiagen, Valencia, CA, USA) for FF tissues and using a Recover All Total Nucleic Acid Isolation kit (Ambion, Foster City, CA, USA) for FFPE tissues. We extracted between 30 and 100 μg of DNA from 20 mg of the FF tumor with an average yield of 64.8 μg. Additionally, we extracted between 5 and 30 μg of DNA from 20 mg of the FFPE tumor with an average yield of 18.7 μg.
Genome-wide DNA methylation analysis
Genome-wide screening of DNA methylation in 7 samples, including the malignant-primary and malignant-metastatic lesions of Cases 2 and 4, respectively, and 3 other benign tumors (from Cases 10, 12, and 18) was performed using the Infinium HumanMethylation450 BeadChip (Illumina 450 K) array, which covers 485,577 CpG sites on the human genome (Illumina, San Diego, CA, USA). The arrays were scanned using the iScan System (Illumina), and the obtained data were analyzed using GenomeStudio Methylation Module Software (Illumina). The methylation level of each CpG site is expressed by the β value, which corresponds to “no methylation” as β = 0 and “full methylation” as β = 1.
Selection of candidate CpG sites
Initially, in order to avoid potential confounding factors, CpG sites assessed with probes targeting several regions of same or different chromosomes and probes associated with sex chromosomes, were excluded from further analysis [6].
Methylated CpG sites that met two or more of the following three criteria were selected: First, the CpG sites in which the β values of the metastatic lesion minus that of the primary tumor is 100 high ranks were defined for Case 2. Second, CpG sites in which the difference is 100 high ranks were defined for Case 4. Third, CpG sites in which the difference is higher than 0.4 were defined for both Cases 2 and 4. Similarly, the unmethylated CpG sites in which the metastatic lesion was more unmethylated than the primary lesion were identified.
Subsequently, the β values of these candidate CpG sites in three benign tumors (Cases 10, 12, and 18) were also assessed. We identified major candidates of methylated CpG sites for which the β values of two or three benign tumors were lower than 0.4, likely indicating that the epigenetic change at these sites had not occurred in the benign tumor. Similarly, we identified major candidates of unmethylated sites for which the β values of two or three benign tumors were higher than 0.4.
Methylation analysis of the candidate CpG sites
PCR following treatment with a methylation-dependent restriction enzyme (PTMR)
PTMR was performed according to the method reported by Shigematsu et al. [7]. In this study, we employed two types of methylation-dependent restriction enzymes: MspJI (New England Biolabs, Beverly, MA, USA), which cleaves DNA 9 bp downstream from the mCNNR (N = A, T, G or C; R = G or C) sequence [8], [9], and FspEI (New England Biolabs), which cleaves DNA 12 bp downstream from the CmC sequence [8]. One microgram of the extracted DNA was incubated with 4 units of the appropriate two enzymes at 37 °C for 8 h in 30 μl of readymade reaction buffer (New England Biolabs) [10]. As fully methylated/fully unmethylated control genome DNAs, EpiScope Methylated HCT116 gDNA/EpiScope Unmethylated HCT116 DKO gDNA (Takara, Tokyo, Japan) were also treated with the enzyme. Subsequently, amplification of the enzyme-treated samples and paired untreated samples was carried out using specific primers that encompassed the target MspJI/FspEI site. The PCR conditions used are given in Supplemental Table 1. Agarose electrophoresis was performed to visualize the PCR products, and we quantified their digitized intensities using the Atto CS Analyzer (Atto, Tokyo, Japan). We defined a sample as methylated when the intensity of the band was less than 50% after enzyme treatment (Supplemental Fig. 1).
Bisulfite sequencing
Two micrograms of the isolated DNA sample were bisulfite converted using the MethylEasy Xceed Kit (Human Genetic Signatures, Randwick NSW, Australia). The bisulfite-treated DNA was amplified by PCR with primers specific for the target CpG site. The PCR conditions used are given in Supplemental Table 2.
The purified PCR products were treated with two enzymes, shrimp alkaline phosphatase and exonuclease I. Following the sequencing reaction using the BigDye Terminator v3.1 cycle sequencing Kit (Applied Biosystems, Foster City, CA, USA), the final samples were sequenced using the ABI 3500xL Genetic Analyzer (Applied Biosystems) as PCR chromatograms. The methylation level was calculated using the peak height of unconverted cytosine (C) and peak height of converted thymine (T) for each CpG site within the DNA amplicon (methylation level = C/(C + T)).
Immunohistochemistry (IHC)
Antibody against Acyl-CoA synthetase, bubblegum family, member 1 (ACSBG1) was purchased from Abcam (Cambridge, UK) (Rabbit polyclonal antibody). Antibody against microtubule associated serine/threonine kinase 1 (MAST1) was purchased from Abnova (Taipei, Taiwan) (Rabbit polyclonal antibody). Antibodies against Ki-67 and chromogranin A (both prediluted mouse monoclonal antibodies) were purchased from DakoCytomation Denmark A/S (Glostrup, Denmark).
Immunostaining of ACSBG1, MAST1, Ki-67 and chromogranin A was performed using Histofine Simplestain immunostaining kit (Nichirei, Tokyo, Japan) on serial tissue sections (3 μm) cut from paraffin-embedded specimens. To retrieve the antigenicity of ACSBG1, MAST1 and Ki-67, the sections were pretreated by hydrated autoclaving in 10 mM sodium citrate buffer (pH 6.0) at 120 °C for 5 min. Each primary antibody was applied to the tissue sections for 18 h at 4 °C. The antigen-antibody complexes on the specimens were visualized by immersion in 3.3′-diaminobenzidine solution in 50 mM Tris/HCl buffer containing 10 mM sodium azide and 0.006% hydrogen peroxide. The human normal pancreas was used as a positive control for ACSBG1. The human normal stomach was used as a positive control for MAST1.
Statistical analysis
Statistical analyses were performed using JMP software, version 9.0.2 (SAS Institute, Cary, NC, USA). The difference between the mean values of the two groups of samples was evaluated using the t-test. Fisher’s exact test was used to statistically analyze the positive rate of IHC in the two groups of samples. P values < 0.05 were considered to indicate significance.
Results
Genome-wide DNA methylation analysis
We determined DNA methylation profiles of 7 samples including malignant-primary tumors (Cases 2 and 4), malignant-metastatic lesions (Cases 2 and 4) and benign tumors (Cases 10, 12 and 18), using the Illumina 450 K array. We made a heatmap of whole genome methylation status with these samples, and assessed them by hierarchical cluster analysis (Supplemental Fig. 2). The primary tumors were close to the benign tumors. The metastatic lesions clustered separately from the primary and benign tumors.
Selection of candidate CpG sites
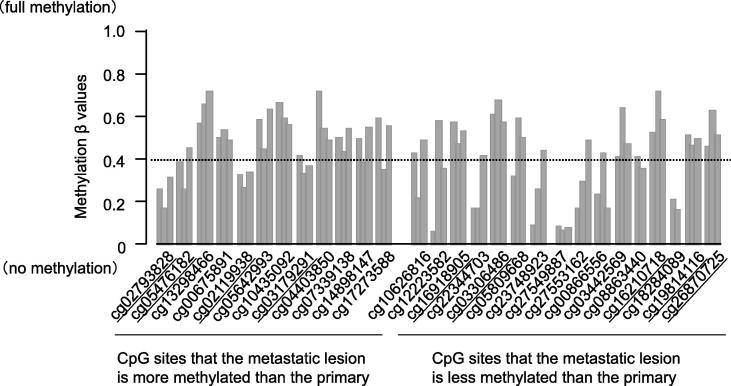
Twelve methylated and 16 unmethylated candidate CpG sites were selected according to the criteria described in the Materials and Methods section (Supplemental Fig. 3). Additionally, based on three benign tumors, we finally selected 4 major methylated candidates and 5 major unmethylated candidates for further analysis (Fig. 2 and Table 2).
Fig. 2.
Methylation β values of the candidate CpG sites in three benign PCCs. Bar graphs represent Cases 10, 12 and 18, from left to right. The dotted line represents β value = 0.4. We identified hyper/hypo-methylated CpG candidates that were in opposite methylation status to those of benign tumors. We selected them with the β values of the benign tumors were lower/higher than 0.4 respectively. Underlining indicates the candidate CpG sites selected for further analysis.
Table 2.
Candidate CpG sites of hyper/hypo-methylation in malignant pheochromocytoma and paraganglioma.
| Target ID | Gene | Benign tumor | CHR | Relation to CpG Island | PTMR |
Bisulfite sequencing |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Enzyme | Product size (bp) | Annealing temp. (°C) | No. of CpG sites in PCR product | Product size (bp) | Annealing temp. (°C) | |||||
| cg02793828 | – | U | 16 | Shore | n.d. | n.d. | n.d. | 5 | 145 | 55 |
| cg05476182 | PHF15 | U | 5 | – | MspJI | 224 | 56 | 1 | 173 | 57 |
| cg02119938 | ACSBG1 | U | 15 | – | FspEI | 234 | 58 | 6 | 165 | 57 |
| cg03179291 | CAMKK1 | U | 17 | Shore | MspJI | 152 | 58 | 8 | 155 | 57 |
| cg16918905 | – | M | 2 | Island | MspJI | 229 | 57 | 14 | 169 | 57 |
| cg03306486 | APC2 | M | 19 | Island | n.d. | n.d. | n.d. | 16 | 162 | 57 |
| cg16210718 | GP1BB;SEPT5 | M | 22 | Island | MspJI | 208 | 58 | 14 | 166 | 57 |
| cg19814116 | KCNAB2 | M | 1 | Island | n.d. | n.d. | n.d. | 8 | 152 | 57 |
| cg26870725 | MAST1 | M | 19 | Shore | MspJI | 214 | 58 | 6 | 179 | 55 |
TargetID: a unique CpG site identifier from the Illumina CG database, CHR: Chromosome, PTMR: PCR following treatment with a methylation-dependent restriction enzyme, U: Unmethylated, M: Methylated, PHF15: PHD finger protein 15, ACSBG1: Acyl-CoA Synthetase, Bubblegum Family, member 1, CAMKK1: Calcium/calmodulin-dependent protein kinase kinase 1, APC2: adenomatosis polyposis coli 2, GP1BB: Glycoprotein Ib (platelet), beta polypeptide, SEPT5: Septin-5, KCNAB2: potassium voltage-gated channel, shaker-related subfamily, beta member 2, MAST1: microtubule associated serine/threonine kinase 1, Island: CpG Island, Shore: within 2000 bp of CpG Island.
n.d.: not done.
PTMR and bisulfite sequencing
Ten benign tumors, 3 malignant-primaries and 8 malignant-metastatic lesions (Cases 1–8) were examined by PTMR and bisulfite sequencing.
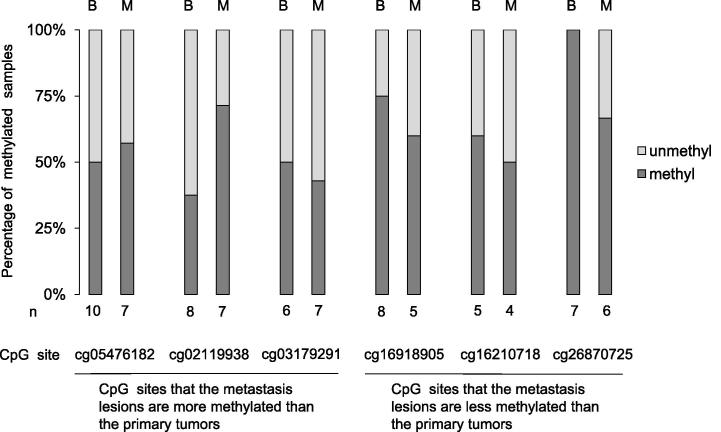
Six of the 9 candidate CpG sites could be cleaved by the enzymes MspJI or FspEI, and 3 sites could not. Among the 6 CpG sites examined, cg26870725, in which all 7 benign tumors were methylated but 2 of the 6 (33.3%) metastatic lesions were unmethylated, seemed to be specific (Fig. 3 and Table 3). In other CpG sites, an evident tendency of the methylation level was not demonstrated in the 7 benign tumors, and we concluded that the likelihood of linking the remaining 5 CpG sites except cg26870725 to malignant transformation was remote.
Fig. 3.
Methylation analysis of candidate CpG sites using PTMR. B: Benign tumor, M: Metastatic lesion. unmethyl: unmethylated, methyl: methylated, PTMR: PCR following treatment with a methylation-dependent restriction enzyme.
Table 3.
Methylation analysis using PTMR.
| Target ID | Benign |
Primary |
Metastatic lesion |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Methyl | Unmethyl | Percentage of methyl | Methyl | Unmethyl | Percentage of methyl | Methyl | Unmethyl | Percentage of methyl | |
| Methylated candidates | |||||||||
| cg05476182 | 5 | 5 | 50.0% | 2 | 0 | 100% | 4 | 3 | 57.1% |
| cg02119938 | 3 | 5 | 37.5% | 1 | 1 | 50.0% | 5 | 2 | 71.4% |
| cg03179291 | 3 | 3 | 50.0% | 0 | 1 | 0.0% | 3 | 4 | 42.8% |
| Unmethylated candidates | |||||||||
| cg16918905 | 6 | 2 | 75.0% | 1 | 0 | 100% | 3 | 2 | 60.0% |
| cg16210718 | 3 | 2 | 60.0% | 1 | 0 | 100% | 2 | 2 | 50.0% |
| cg26870725 | 7 | 0 | 100% | 1 | 0 | 100% | 4 | 2 | 66.7% |
PTMR: PCR following treatment with a methylation-dependent restriction enzyme, methyl: methylated samples, unmethyl: unmethylated samples.
All 9 candidate CpG sites could be examined by bisulfite sequencing. Because not only the target CpG site (Target) but also the peripheral CpG sites (Periphery) existed in the PCR products, we evaluated the methylation level of the average of both Target and Periphery. There was no significant difference between the benign tumors and metastatic lesions in all of the candidate CpG sites examined (Table 4). Among them, we considered cg02119938, in which the methylation level of two of five metastatic lesions was much higher than those of the benign tumors, and cg26870725, in which the methylation level of one of four metastatic lesions was much lower than those of the benign tumors (Supplemental Fig. 4).
Table 4.
Methylation analysis using Bisulfite Sequencing.
| TargetID | Benign tumor |
Primary |
Metastatic lesion |
p-value⁎ | |
|---|---|---|---|---|---|
| Mean ± SE | Mean ± SE | Mean ± SE | |||
| Methylated candidates | |||||
| cg02793828 | Target | 0.595 ± 0.060 | 0.613 ± 0.082 | 0.572 ± 0.071 | 0.380 |
| Periphery | 0.702 ± 0.057 | 0.876 ± 0.018 | 0.758 ± 0.065 | 0.528 | |
| cg05476182 | Target | 0.580 ± 0.122 | 1.000 | 0.790 ± 0.149 | 0.265 |
| Periphery | n.d. | n.d. | n.d. | n.d. | |
| cg02119938 | Target | 0.268 ± 0.097 | n.d. | 0.477 ± 0.146 | 0.523 |
| Periphery | 0.322 ± 0.082 | n.d. | 0.455 ± 0.122 | 0.528 | |
| cg03179291 | Target | 0.715 ± 0.050 | 0.681 ± 0.086 | 0.688 ± 0.054 | 0.721 |
| Periphery | 0.819 ± 0.037 | 0.859 ± 0.069 | 0.812 ± 0.044 | 0.911 | |
| Unmethylated candidates | |||||
| cg16918905 | Target | 0.608 ± 0.093 | 1.000 | 0.794 ± 0.124 | 0.728 |
| Periphery | 0.714 ± 0.067 | 0.907 | 0.775 ± 0.089 | 0.901 | |
| cg03306486 | Target | 0.750 ± 0.094 | 0.941 ± 0.127 | 0.950 ± 0.071 | 0.256 |
| Periphery | 0.808 ± 0.024 | 0.926 ± 0.033 | 0.913 ± 0.018 | 0.591 | |
| cg16210718 | Target | 0.635 ± 0.068 | 0.235 | 0.579 ± 0.083 | 0.620 |
| Periphery | 0.677 ± 0.052 | 0.838 | 0.805 ± 0.064 | 0.158 | |
| cg19814116 | Target | 0.231 ± 0.116 | 0.725 | 0.465 ± 0.150 | 0.236 |
| Periphery | 0.290 ± 0.100 | 0.275 | 0.570 ± 0.115 | 0.085 | |
| cg26870725 | Target | 0.844 ± 0.138 | n.d. | 0.737 ± 0.169 | 0.635 |
| Periphery | 0.747 ± 0.124 | n.d. | 0.728 ± 0.152 | 0.924 | |
Target: Methylation status of target CpG site only.
Periphery: Average of methylation status of CpG sites included in PCR product.
n.d.: not done.
t-test: Benign tumors vs Metastatic lesions.
The results of PTMR and bisulfite sequencing revealed that cg02119938 of the metastatic lesions tended to be more methylated than that of benign tumors and cg26870725 of metastatic lesions tended to be less methylated than that of benign tumors. Cg02119938 and cg26870725 were associated with the ACSBG1 gene and MAST1 gene, respectively.
Immunohistochemistry (IHC)
All of the specimens from the examined 22 tumors (19 patients) were immunohistochemically positive for chromogranin A, supporting the clinical diagnosis of PCC/PGL. Additionally, we detected immunoreactivity for Ki-67 in the nuclei of tumor cells in some of the examined specimens. The labeling index is summarized in Table 1. The index varied from <1.0% to 18.3%, and a significant difference was recognized between benign and malignant-metastatic tumors (p < 0.001), but was not found between benign and malignant-primary tumors (p = 0.32)
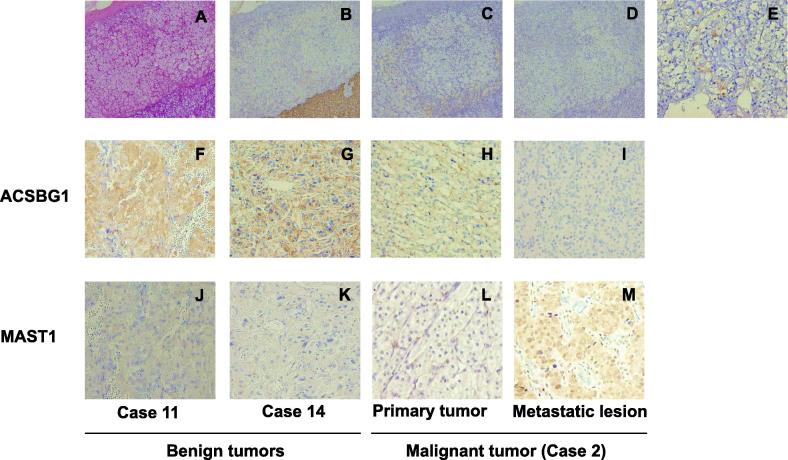
The IHC of three normal adrenal glands revealed that the zona glomerulosa, zona fasciculate and medulla were negative for ACSBG1, whereas the cytoplasm of the zona reticularis was positive. Of the 10 benign cases, nine (90.0%) were positive for ACSBG1, as were all 3 primary lesions from the malignant cases. But 6/9 (66.7%) of the metastatic lesions were negative. A significant difference was recognized between benign and metastatic tumors (p = 0.020).
Of the three normal adrenal glands, the three zones of the adrenal cortex and medulla were all immunohistochemically negative for MAST1. It has been reported that the distribution of the MAST1 included the nucleus and cytoplasm of the heart, brain, spleen, lung, liver, skeletal muscle, kidney, and testis [11]. All 10 benign tumors and all 3 primary lesions from the malignant cases were negative for MAST1; of the 9 metastatic lesions, 6 (66.7%) were positive. A significant difference was recognized between benign and metastatic tumors (p = 0.003) (Fig. 4 and Table 5).
Fig. 4.
Hematoxylin-eosin staining and immunohistochemical analysis of Chromogranin A, ACSBG1 and MAST1. Panel A shows the Hematoxylin-eosin staining of normal adrenal tissue. Panels B-M show the immunohistochemical staining of Chromogranin A (B), ACSBG1 (C, F–I) and MAST1 (D, E, J–M). Panels A–E show normal adrenals, and Panels F–K show benign tumors; Panels H and L show primary lesions of malignant tumors, and panels I and M show metastatic lesions. Normal adrenal medulla is shown at a higher magnification in panel E. The magnification of panels A–D is 200x. The magnification of panels E–M is 400x.
Table 5.
Immunohistochemistry of ACSBG1 and MAST1.
| Case | ACSBG1 | MAST1 | |
|---|---|---|---|
| Normal adrenal | |||
| Cortex | + | − | |
| Medulla | − | + | |
| Benign tumor | |||
| 10 | + | − | |
| 11 | + | − | |
| 12 | + | − | |
| 13 | + | − | |
| 14 | + | − | |
| 15 | + | − | |
| 16 | + | − | |
| 17 | − | − | |
| 18 | + | − | |
| 19 | + | − | |
| 9/10 | 0/10 | ||
| Malignant tumor | |||
| Primary | |||
| 2 | + | − | |
| 4 | + | − | |
| 6 | + | − | |
| 3/3 | 0/3 | ||
| Metastatic lesion | |||
| 1 | + | + | |
| 2 | − | + | |
| 3 | + | + | |
| 4 | + | + | |
| 5 | − | − | |
| 6 | − | + | |
| 7 | − | − | |
| 8 | − | − | |
| 9 | − | + | |
| 3/9 | 6/9 | ||
| p-value⁎ | 0.020 | 0.003 | |
ACSBG1: Acyl-CoA Synthetase, Bubblegum Family, member 1.
MAST1: microtubule associated serine/threonine kinase 1.
Fisher’s exact test: Benign tumors vs Metastatic lesions.
Discussion
Epigenetic changes of tumor-related genes play a major role in carcinogenesis. It is well known that promoter hypermethylation of tumor-suppressor genes causes the development and differentiation of cancer cells by their inactivation [12]. However, global DNA hypomethylation is also detected in plural types of cancer cells [13], [14]. Recently, it was reported that the hypomethylation of specific promoters in colon cancer cells might activate the aberrant expression of oncogenes and induce the loss of imprinting in vitro [15]. Although several genetic variants associated with malignant PCC/PGL have been identified [16], [17], [18], little is known concerning the epigenetic changes in malignant PCC/PGL. There are several reports on the genome-wide methylation analysis of metastatic PCC/PGL. Letouzé et al. reported 191 genes that characterized in significant hypermethylation in their promoter CpG island and were downregulated in succinate dehydrogenase (SDH) B-mutated PCC/PGLs [19]. We evaluated the methylation status of CpG sites associated with these 191 genes in our sample set (Supplemental Fig. 5). The majority of the 191 genes were hypermethylated in the metastatic lesion compared to the primary tumor of case 4 carrying a SDHB mutation. In case 2, which does not have a SDHx mutation, the opposite effect was seen. De Cubas et al. reported significant hypermethylation of cg06351503 related to RDBP (negative elongation factor complex member E) in metastatic tumors and proposed its clinical value for the evaluation of the potential risk of metastasis [20]. In this study, the β values of cg06351503 were 0.080, 0.770 (Case 2; primary and metastatic tumor), 0.584 and 0.892 (Case 4; primary and metastatic tumor), respectively. The metastatic lesion exhibited hypermethylation in Case 2 but not in Case 4. The epigenetic condition of cg06351503 in Case 2 is consistent with the finding of de Cubas et al. The two proposed candidates in this study, cg02119938 and cg26870725, were examined using the Illumina 450 K array but not using the Illumina 27 K array employed by de Cubas et al. Recently, Richter et al. reported hypermethylation of SDHC promoter in a PGLs patient without germline mutation of the gene, and they proposed the possibility that not germline mutations but gene inactivation by epigenetic change in promoter of SDHC may cause PGL tumor development in some instances [21]. Analysis of methylation patterns in our samples demonstrated that SDHC promoter methylation does not play a role in metastatic transformation (Supplemental Fig. 6).
In most previous reports that discussed the epigenetic changes of malignant transformation, the methylation levels of malignant tumors were assessed compared with those of non-neoplastic tissue or the benign tumor of another patient. In this study, we compared two types of tumor at different stages, both primary and metastatic tumors, in a patient. This procedure has an advantage in its correction for individual differences in the DNA methylation state. It was reported that probes targeting CpG loci associated with single nucleotide polymorphisms (SNPs) near or within the probe sequence may influence corresponding methylated probes [19]. Our procedure of comparing two types of tumor in a patient has an advantage in also obviating interference of individual SNPs. Meanwhile, the effect of aging should be considered in the analysis because aging decreases the methylation level of global DNA and increases that of CpG islands in patients [22], [23]. Distant metastasis, which was diagnosed after a long metastatic-free period, was common to Cases 2 and 4, which we subjected to epigenetic analysis comparing primary tumor and metastasis in this study. However, the differences between these two patients, including gender, age of first-diagnosis, and germ-line mutation, might be not negligible. This is a subject of future study in a larger number of patients.
Several reports on large-scale epigenetic studies of plural types of malignancies using the Illumina 450 K array had provided valuable information concerning new tumor-related genes [7], [24]. DNA extracted from FFPE tissue is fragmented. Additionally, the longer the preservation duration is, the more fragmented the DNA becomes. In this study, DNA from the oldest FFPE tissue (Case 2; primary tumor) was preserved for 26 years, and the DNA size was approximately 500 bp in contrast to approximately 20,000 bp from FF tumors (data not shown). Some samples from old FFPE tissue in this study were decreased in the number of analyzable CpG sites, and the possibility exists that some meaningful CpG sites may be overlooked.
Genetic intra-tumor heterogeneity has also been known [25], [26], and the extracted DNA from large-sized tumors such as PCC/PGLs may present heterogeneous results according to the site from which the material is sampled. These factors also might have affected the results of this study.
There are several methods for DNA methylation quantitative analysis of individual genomic regions. These methods are based on the three principles: substitution by bisulfite treatment, DNA digestion with a methylation-dependent restriction enzyme and immunoprecipitation based on the recognition of methylated DNA with a binding protein or antibody. Of the three, bisulfite sequencing is one of popular methods. However, it is known to damage both the quantity and quality of DNA samples for subsequent procedure. It has been reported that analyzable DNA decreases to approximately 10% after the treatment [27]. In this study, we adopted two experimental methods: PTMR and bisulfite sequencing. The MspJI family (MspJI, FspEI and LpnPI) is a recently characterized modification-dependent restriction enzyme that cleaves specific DNA sites according to their methylation state [8], [9]. Shigematsu et al. reported a unique technique, qPTMR by which the methylation level was measured using a combination of digestion with the enzymes MspJI and qPCR [7]. This method is useful because it does not involve steps that damage the quantity of DNA samples, including bisulfite treatment, but has the shortcoming in which there may be no sequences that the enzyme can cleave in some of the target CpG sites.
The ACSBG1 gene is located on q25.1 of chromosome 15, and it encodes ACSBG1 protein, which is a subtype of ACSBG [28]. ACSBG1 is one of the enzymes associated with the metabolism of sphingolipid, which is a long-chain fatty acid [29], and it is localized in the endoplasmic reticulum and mitochondria-associated membranes in the brain, testis, ovary and adrenal gland. It was not evident whether the genetic or epigenetic abnormality of ACSBG1 causes the pathological phenotype of diseases, including malignancies. In this study, the results, great majority of the benign tumors and the malignant-primary tumors were positive for ACSBG1 suggested the possibility that the neoplastic change in chromaffin cells is related to epigenetic changes in the promoter lesion of ACSBG1, resulting in gene activation. Furthermore, the result that some of the metastatic tumors were negative for ACSBG1 suggests that the promoter is hypermethylated again, resulting in the silencing of ACSBG1. Functional analysis (for example, using knockdown with the siRNA for this gene) is required for further clarification of the mechanism underlying the malignant transformation.
The MAST1 gene is located on p13.13 of chromosome 19, and it encodes MAST1 protein, which belongs to the MAST kinase family. The members of the family are MAST1, 2, 3, 4, and MAST-like. MAST kinase family genes are characterized by the presence of a serine/threonine kinase domain. MAST1 is expressed in multiple tissues, particularly the brain [11]. The MAST gene is thought to be required for normal cell division. Gene alterations of MAST have been reported to result in several different mitotic abnormalities. The mitotic defects are commonly observed in carcinomas. Therefore, MAST genes are considered instrumental for carcinogenesis [30]. In addition, according to the report by Robinson et al., MAST1 or MAST2 gene fusions were detected in 3–5% of breast cancer tissues and cell lines. The authors demonstrated that the overexpression of MAST1 or MAST2 gene fusions caused higher rates of cell proliferation in vitro and assessed the potential oncogenic functions of these gene fusions [31]. From the above results, the MAST kinase may be one of the key proteins involved in oncogenesis. In our study, the methylation analysis revealed that the hypomethylation of cg26870725, which is associated with MAST1, is observed in the metastatic lesions of malignant PCC/PGLs. These results implicate the gene activation of MAST1 in malignant PCC/PGLs. Positive rate of MAST1 IHC in the metastatic lesion group of samples was significantly higher than that in the benign tumor group. We conclude that epigenetic change in the malignant transformation of PCC/PGL is linked to MAST1 gene overexpression by hypomethylation.
Cases 2 and 4 were extremely unlike considering baseline characteristics including gender, age of onset, and presence of the SDHB germline mutation. We thought that especially the genetic background is not ignored, because it is well known that the presence of germline mutations is related to the percentage of malignant tumors in them. On the other hand, the relationship between germline mutation and metastatic-free duration has not been known, and we thought that our cases could be representative malignant PCC/PGLs with long metastatic-free duration. Another limitation is the low number of samples. A longer sample collection period and larger-scale field cohort trials would be needed to clarify the relationship between these genes and the malignant transformation involving PCC/PGL.
Our results are inconclusive concerning whether the hypermethylation of ACSBG1 and hypomethylation of MAST1 in malignant PCC/PGL are involved in the malignant transformation of PCC/PGL. However, these effects seem likely because the two genes changed their expression level in tumor cells according to the change in malignant potential. Further investigation into the two noteworthy CpG sites and two genes may provide valuable information for the prediction of malignant prognoses in PCC/PGL patients.
Conclusions
The aberrant promoter methylation/demethylation of the ACSBG1 and MAST1 genes might be involved in their silencing/expression in malignant PCC/PGL. Further investigations are necessary to determine how ACSBG1 and/or MAST1 expression are involved in malignant transformation and to establish pathological markers that can be used to evaluate malignant potential in cases of PCC/PGL.
Conflicts of interest
All the authors declare that no conflicts of interest exist regarding the publication of this paper.
Disclosure summary
The authors have nothing to disclose.
Acknowledgments
The following investigators collaborated on the project and offered samples: Satoshi Baba of the Hamamatsu University School of Medicine; Koji Nagayama, Hiroki Mori and Takachika Ozawa of the Hamamatsu Medical Center; and Rieko Genma and Yoshiro Otsuki of the Seirei Hamamatsu General Hospital.
Footnotes
This work was supported by Japan Society for the Promotion of Science KAKENHI Grants (23590666), Charitable Trust Laboratory Medicine Research Foundation of Japan and Kurozumi Medical Foundation.
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jcte.2016.12.004.
Appendix A. Supplementary data
Electrophoresis of the PTMR (FspEI) products of cg02119938. M/C: methylated control DNA, U/C: unmethylated control DNA, +: Sample DNA treated with a methylation-sensitive restriction enzyme, −: Untreated sample DNA. The size of the PCR product is 235 bp. PTMR: PCR following treatment with a methylation-dependent restriction enzyme.
Heatmap of whole genome methylation profile ordered by similarity assessed by hierarchical cluster analysis. The heatmap was created based on data of all 7 samples analyzed by the Illumina 450K array in this study. Blank indicates unavailable data.
Venn diagram showing the number of candidate CpG sites for which the methylation status differs between primary and metastatic lesions. CpG sites that satisfied two or more of three criteria were selected. a. CpG sites that are methylated more strongly in the metastatic lesion than in the primary tumor. b. CpG sites that are less methylated in the metastatic lesion than in the primary tumor.
The results of methylation analysis using bisulfite sequencing. Open circles represent methylation levels in benign tumors. Closed circles represent methylation levels in metastatic lesions. Methylation levels are calculated as C/(C+T) at each CpG site and are expressed as follows: “no methylation”, 0; “full methylation”, 1. The solid line represents an average value for each CpG site. Target: only the target CpG; Periphery: peripheral CpG sites existed in the PCR product. In the PCR product of cg05476182, only the target CpG site existed; there were no other CpG sites.
Heatmap of the methylation pattern of 186 CpG sites associated with the 191 genes, that Letouze et al. reported [19]. Blank indicates unavailable data.
Heatmap of the methylation pattern of 14 CpG sites associated with the SDHC promoter. The data of cg15152945 in Case18 was not available.
References
- 1.Waguespack S.G., Rich T., Grubbs E., Ying A.K., Perrier N.D., Ayala-Ramirez M. A current review of the etiology, diagnosis, and treatment of pediatric pheochromocytoma and paraganglioma. J Clin Endocrinol Metab. 2010;95:2023–2037. doi: 10.1210/jc.2009-2830. [DOI] [PubMed] [Google Scholar]
- 2.Ayala-Ramirez M., Feng L., Habra M.A., Rich T., Dickson P.V., Perrier N. Clinical benefits of systemic chemotherapy for patients with metastatic pheochromocytomas or sympathetic extra-adrenal paragangliomas: insights from the largest single-institutional experience. Cancer. 2012;118:2804–2812. doi: 10.1002/cncr.26577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edstrom Elder E., Hjelm Skog A.L., Hoog A., Hamberger B. The management of benign and malignant pheochromocytoma and abdominal paraganglioma. Eur J Surg Oncol. 2003;29:278–283. doi: 10.1053/ejso.2002.1413. [DOI] [PubMed] [Google Scholar]
- 4.Huang H., Abraham J., Hung E., Averbuch S., Merino M., Steinberg S.M. Treatment of malignant pheochromocytoma/paraganglioma with cyclophosphamide, vincristine, and dacarbazine: recommendation from a 22-year follow-up of 18 patients. Cancer. 2008;113:2020–2028. doi: 10.1002/cncr.23812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khorram-Manesh A., Ahlman H., Nilsson O., Friberg P., Oden A., Stenstrom G. Long-term outcome of a large series of patients surgically treated for pheochromocytoma. J Intern Med. 2005;258:55–66. doi: 10.1111/j.1365-2796.2005.01504.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y.A., Lemire M., Choufani S., Butcher D.T., Grafodatskaya D., Zanke B.W. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shigematsu Y., Niwa T., Yamashita S., Taniguchi H., Kushima R., Katai H. Identification of a DNA methylation marker that detects the presence of lymph node metastases of gastric cancers. Oncol Lett. 2012;4:268–274. doi: 10.3892/ol.2012.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen-Karni D., Xu D., Apone L., Fomenkov A., Sun Z., Davis P.J. The MspJI family of modification-dependent restriction endonucleases for epigenetic studies. Proc Natl Acad Sci USA. 2011;108:11040–11045. doi: 10.1073/pnas.1018448108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng Y., Cohen-Karni D., Xu D., Chin H.G., Wilson G., Pradhan S. A unique family of Mrr-like modification-dependent restriction endonucleases. Nucleic Acids Res. 2010;38:5527–5534. doi: 10.1093/nar/gkq327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horton J.R., Mabuchi M.Y., Cohen-Karni D., Zhang X., Griggs R.M., Samaranayake M. Structure and cleavage activity of the tetrameric MspJI DNA modification-dependent restriction endonuclease. Nucleic Acids Res. 2012;40:9763–9773. doi: 10.1093/nar/gks719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garland P., Quraishe S., French P., O'Connor V. Expression of the MAST family of serine/threonine kinases. Brain Res. 2008;1195:12–19. doi: 10.1016/j.brainres.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 12.Baylin S.B., Jones P.A. A decade of exploring the cancer epigenome – biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang M., Zhang Y., Fei J., Chang X., Fan W., Qian X. Rapid quantification of DNA methylation by measuring relative peak heights in direct bisulfite-PCR sequencing traces. Lab Invest. 2010;90:282–290. doi: 10.1038/labinvest.2009.132. [DOI] [PubMed] [Google Scholar]
- 14.Kanwal R., Gupta S. Epigenetic modifications in cancer. Clin Genet. 2012;81:303–311. doi: 10.1111/j.1399-0004.2011.01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui H. Loss of imprinting of IGF2 as an epigenetic marker for the risk of human cancer. Dis Markers. 2007;23:105–112. doi: 10.1155/2007/363464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fishbein L., Nathanson K.L. Pheochromocytoma and paraganglioma: understanding the complexities of the genetic background. Cancer Genet. 2012;205:1–11. doi: 10.1016/j.cancergen.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gimm O., DeMicco C., Perren A., Giammarile F., Walz M.K., Brunaud L. Malignant pheochromocytomas and paragangliomas: a diagnostic challenge. Langenbecks Arch Surg. 2012;397:155–177. doi: 10.1007/s00423-011-0880-x. [DOI] [PubMed] [Google Scholar]
- 18.Kolackov K., Tupikowski K., Bednarek-Tupikowska G. Genetic aspects of pheochromocytoma. Adv Clin Exp Med. 2012;21:821–829. [PubMed] [Google Scholar]
- 19.Letouze E., Martinelli C., Loriot C., Burnichon N., Abermil N., Ottolenghi C. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–752. doi: 10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 20.de Cubas A.A., Korpershoek E., Inglada-Perez L., Letouze E., Curras-Freixes M., Fernandez A.F. DNA methylation profiling in pheochromocytoma and paraganglioma reveals diagnostic and prognostic markers. Clin Cancer Res. 2015;21:3020–3030. doi: 10.1158/1078-0432.CCR-14-2804. [DOI] [PubMed] [Google Scholar]
- 21.Richter S., Klink B., Nacke B., de Cubas A.A., Mangelis A., Rapizzi E. Epigenetic mutation of the succinate dehydrogenase C promoter in a patient with two paragangliomas. J Clin Endocrinol Metab. 2016;101:359–363. doi: 10.1210/jc.2015-3856. [DOI] [PubMed] [Google Scholar]
- 22.Bollati V., Schwartz J., Wright R., Litonjua A., Tarantini L., Suh H. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev. 2009;130:234–239. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fraga M.F., Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 24.Conesa-Zamora P., Garcia-Solano J., Turpin Mdel C., Sebastian-Leon P., Torres-Moreno D., Estrada E. Methylome profiling reveals functions and genes which are differentially methylated in serrated compared to conventional colorectal carcinoma. Clin Epigenetics. 2015;7:101. doi: 10.1186/s13148-015-0128-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerlinger M., Rowan A.J., Horswell S., Larkin J., Endesfelder D., Gronroos E. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pribluda A., de la Cruz C.C., Jackson E.L. Intratumoral heterogeneity: from diversity comes resistance. Clin Cancer Res. 2015;21:2916–2923. doi: 10.1158/1078-0432.CCR-14-1213. [DOI] [PubMed] [Google Scholar]
- 27.Grunau C., Clark S.J., Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.13.e65. [E65-5] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mashek D.G., Li L.O., Coleman R.A. Long-chain acyl-CoA synthetases and fatty acid channeling. Future Lipidol. 2007;2:465–476. doi: 10.2217/17460875.2.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohkuni A., Ohno Y., Kihara A. Identification of acyl-CoA synthetases involved in the mammalian sphingosine 1-phosphate metabolic pathway. Biochem Biophys Res Commun. 2013;442:195–201. doi: 10.1016/j.bbrc.2013.11.036. [DOI] [PubMed] [Google Scholar]
- 30.Wang X., Fredericksen Z.S., Vierkant R.A., Kosel M.L., Pankratz V.S., Cerhan J.R. Association of genetic variation in mitotic kinases with breast cancer risk. Breast Cancer Res Treat. 2010;119:453–462. doi: 10.1007/s10549-009-0404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson D.R., Kalyana-Sundaram S., Wu Y.M., Shankar S., Cao X., Ateeq B. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med. 2011;17:1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electrophoresis of the PTMR (FspEI) products of cg02119938. M/C: methylated control DNA, U/C: unmethylated control DNA, +: Sample DNA treated with a methylation-sensitive restriction enzyme, −: Untreated sample DNA. The size of the PCR product is 235 bp. PTMR: PCR following treatment with a methylation-dependent restriction enzyme.
Heatmap of whole genome methylation profile ordered by similarity assessed by hierarchical cluster analysis. The heatmap was created based on data of all 7 samples analyzed by the Illumina 450K array in this study. Blank indicates unavailable data.
Venn diagram showing the number of candidate CpG sites for which the methylation status differs between primary and metastatic lesions. CpG sites that satisfied two or more of three criteria were selected. a. CpG sites that are methylated more strongly in the metastatic lesion than in the primary tumor. b. CpG sites that are less methylated in the metastatic lesion than in the primary tumor.
The results of methylation analysis using bisulfite sequencing. Open circles represent methylation levels in benign tumors. Closed circles represent methylation levels in metastatic lesions. Methylation levels are calculated as C/(C+T) at each CpG site and are expressed as follows: “no methylation”, 0; “full methylation”, 1. The solid line represents an average value for each CpG site. Target: only the target CpG; Periphery: peripheral CpG sites existed in the PCR product. In the PCR product of cg05476182, only the target CpG site existed; there were no other CpG sites.
Heatmap of the methylation pattern of 186 CpG sites associated with the 191 genes, that Letouze et al. reported [19]. Blank indicates unavailable data.
Heatmap of the methylation pattern of 14 CpG sites associated with the SDHC promoter. The data of cg15152945 in Case18 was not available.