Abstract
Aim
4β‐Hydroxycholesterol (4βOHC) is sensitive towards induction or inhibition of CYP3A4, but its potential usefulness as a dosing biomarker remains to be demonstrated. The aim of this study was to investigate the correlation between 4βOHC levels and steady‐state concentrations (Css) of quetiapine, a CYP3A4 substrate with high presystemic metabolism, in psychiatric patients.
Methods
Serum samples from 151 patients treated with quetiapine as immediate release (IR; n = 98) or slow release (XR; n = 53) tablets were included for analysis of 4βOHC. In all patients, Css of quetiapine had been measured at trough level, i.e. 10–14 and 17–25 h post‐dosing for IR and XR tablets, respectively. Correlations between 4βOHC levels and dose‐adjusted Css (C/D ratios) of quetiapine were tested by univariate (Spearman's) and multivariate (multiple linear regression) analyses. Gender, age (≥60 vs. <60 years) and tablet formulation were included as potential covariates in the multivariate analysis.
Results
Correlations between 4βOHC levels and quetiapine C/D ratios were highly significant both for IR‐ and XR‐treated patients (P < 0.0001). Estimated Spearman r values were −0.47 (95% confidence interval −0.62, −0.30) and −0.56 (−0.72, −0.33), respectively. The relationship between 4βOHC level and quetiapine C/D ratio was also significant in the multiple linear regression analysis (P < 0.001), including gender (P = 0.023) and age (P = 0.003) as significant covariates.
Conclusions
The present study shows that 4βOHC level is significantly correlated with steady‐state concentration of quetiapine. This supports the potential usefulness of 4βOHC as a phenotype biomarker for individualized dosing of quetiapine and other drugs where systemic exposure is mainly determined by CYP3A4 metabolism.
Keywords: 4β‐hydroxycholesterol, biomarker, CYP3A4, quetiapine
What is Already Known about this Subject
4β‐Hydroxycholesterol (4βOHC) is a promising endogenous CYP3A(4) biomarker.
4βOHC is sensitive to coadministration of potent inducers and inhibitors of CYP3A4, but the potential usefulness of 4βOHC as dosing biomarker remains to be demonstrated.
Quetiapine is a frequently prescribed, low‐bioavailable antipsychotic drug with extensive pharmacokinetic variability, which is mainly determined by individual differences in CYP3A4 metabolism.
What this Study Adds
The close correlation between 4βOHC level and dose‐adjusted steady‐state serum concentration of orally administered quetiapine shown in the present study provides strong evidence that 4βOHC reflects basal CYP3A4 activity in humans.
The findings of this study support the clinical pharmacological potential of 4βOHC as a biomarker for individualized dosing of quetiapine, and possibly other drugs where CYP3A4 metabolism is a main determinant of systemic exposure.
Introduction
Cytochrome P450 3A4 (CYP3A4) is the most important enzyme in drug metabolism being involved in biotransformation of 30–50% of all clinically used drugs 1. The individual variability in CYP3A4 phenotype is extensive, as reflected by the more than 10‐fold interindividual variability in clearance of midazolam 2, the golden standard CYP3A4 probe agent. Genetic polymorphisms seem to play a secondary role for CYP3A4 phenotype variability 3, 4, in contrast to several other CYP enzymes. Thus, the potential for individualized dosing based on genotyping of drugs primarily metabolized by CYP3A4 is limited. Instead, phenotype biomarkers are more appropriate for capturing the various environmental factors that mainly determine individual variability in CYP3A4‐mediated drug metabolism.
There is great interest in finding simple and practical measures/biomarkers for determination of CYP3A4 phenotype. In drug development, coadministration of microdoses of exogenous CYP3A4 probes, such as midazolam 5 or quinine 6, is used to identify and quantify potential interactions associated to CYP3A4 inhibition or induction of new drug candidates. However, exogenous probes are less suitable as phenotype biomarkers for individualized dosing of CYP3A4‐metabolized drugs in clinical practice. For this latter purpose, endogenous biomarkers would be more appropriate from a practical point of view.
Perhaps the most promising endogenous CYP3A4 biomarker is 4β‐hydroxycholesterol (4βOHC) 7, 8, 9, 10. In humans, 4βOHC is formed from cholesterol mainly by CYP3A4 11. In vitro data suggest that CYP3A5 is of minor importance in 4βOHC formation 12, but in vivo studies reporting elevated levels of 4βOHC in CYP3A5 expressers indicate some involvement of CYP3A5 in 4βOHC formation as well 13, 14, 15, 16.
Several studies have shown that 4βOHC levels change significantly following administration of both CYP3A4 inducers and inhibitors 3, 7, 8, 9, 11, 17, 18, 19. Change in 4βOHC level seems to be more sensitive towards CYP3A4 induction than inhibition, which possibly reflects its long elimination half‐life 7, 12. A claimed limitation of 4βOHC as a potential in vivo CYP3A4 biomarker is the assumption of its exclusive hepatic formation 6, 20. However, we recently published data suggesting that 4βOHC is also formed by intestinal CYP3A4 17, which is crucial to serve as a dosing biomarker for orally administered CYP3A4 substrates. Despite these promising features as a CYP3A4 phenotype biomarker, the potential usefulness of 4βOHC as a dosing biomarker of CYP3A4‐metabolized drugs remains to be demonstrated.
Quetiapine is a commonly used atypical antipsychotic agent with low oral bioavailability (~9%), which is restricted by substantial presystemic metabolism via CYP3A4 21. As 70% of orally administered quetiapine is absorbed from the gut 22, this indicates a relevant contribution of intestinal metabolism in limiting its bioavailability. Quetiapine has a short elimination half life (5–7 h) 22, and an extended release tablet formulation has been marketed to reduce dosing frequencies and fluctuations in plasma concentration. The interindividual pharmacokinetic variability of quetiapine is extensive 23, and its exposure is highly sensitive towards coadministration of both CYP3A4 inducers and inhibitors 24. Thus, to obtain knowledge on the potential usefulness of 4βOHC as dosing biomarker of a drug mainly metabolized by CYP3A4, the aim of this study was to investigate the correlation between 4βOHC levels and dose‐adjusted steady‐state serum concentrations of quetiapine in psychiatric patients treated with immediate‐release (IR) or extended‐release (XR) tablets.
Methods
Study material
During a one‐year prospective study period, residual patient samples submitted for therapeutic drug monitoring (TDM) analysis of quetiapine and the active, CYP3A4‐mediated metabolite N‐desalkylquetiapine (NDQ) at the Center for Psychopharmacology, Diakonhjemmet Hospital, Oslo, Norway, were routinely collected for possible reanalysis of 4βOHC. If the serum samples contained sufficient volumes for 4βOHC reanalysis (>1 ml), and accompanying requisition forms contained information about prescribed quetiapine dose (mg per day), tablet formulation (IR or XR), and sampling time (hours between last dose intake and blood sampling), these were considered for inclusion in the study. Exclusion criteria were (i) blood sampling time outside predefined criteria (10–14 h after last dose intake for IR tablets or 17–25 h for XR tablets), (ii) measured concentrations of quetiapine or NDQ outside the validated concentration ranges (20–2400 nmol l−1 and 50–1500 nmol l−1, respectively), and (iii) dose adjustments or treatment initiation performed less than 3 days before blood sampling.
Requisition forms accompanying samples eligible for inclusion were reviewed to identify information about prescribed quetiapine dose, time interval between last dose intake and blood sampling, potential recent treatment initiation or dose adjustments, and comedication with CYP3A4 inhibitors or inducers. For the latter purpose the Flockhart Cytochrome P450 Drug Interaction Table was applied 25, although this does not provide a complete overview of CYP3A4 inhibitors/inducers. Data on the measured serum concentrations of quetiapine and NDQ were retrieved directly from the TDM files. If multiple samples/measurements were available from the same patient within the study period, the first registered sample compliant with the defined criteria was included. When included patient samples contained sufficient volumes, total cholesterol concentrations were also determined to enable calculation of 4βOHC‐to‐cholesterol (4βOHC/C) ratios. Cholesterol measurements were performed by a standard method at the Department of Medical Biochemistry, Diakonhjemmet Hospital, Oslo, Norway.
The study was approved by the Regional Committee for Medical and Health Research Ethics (case number 2014/1191) and the Hospital Investigational Review board. Ethical approval was given without requirement of patient consent since the study was based on already collected serum samples and existing laboratory data from a routine TDM service.
Analytical assay of quetiapine and N‐desalkylquetiapine
Determination of serum concentrations of quetiapine and NDQ was performed by a validated UPLC‐MS/MS method used in routine TDM practice. Briefly, serum samples (500 μl) were prepared by protein precipitation with 1000 μl cold acetonitrile solution including the internal standard (promazine). After centrifugation, aliquots of 5 μl were injected onto an Aquility UPLC system coupled to a Micromass Quattro Premier tandem MS detector (Waters, Milford, MA, USA). An UPLC BEH C18 column RP shield (1.7 μm, 1 × 100 mm) from Waters was used for chromatographic separation, using a gradient elution with a mix of acetonitrile and ammonium acetate (pH = 4.8) as mobile phase at a flow rate of 0.2 ml min−1. The total run time per sample was 5 min and the retention times were approximately 2.2 min and 2.3 min for NDQ and quetiapine, respectively. MS/MS detection was performed by electrospray ionization (ESI) in positive mode using multiple reaction monitoring (MRM) at the transitions m/z 296 → 210 (NDQ) and m/z 384 → 253 (quetiapine).
At the lowest validated concentration (defined as lower limit of quantification; LLOQ), which was 20 nmol l−1 for quetiapine and 50 nmol l−1 for NDQ, the intra‐ and interday precision and accuracy was <14%. At the highest validated concentration (2400 nmol l−1 for quetiapine and 1500 nmol l−1 for NDQ), the intra‐ and interday precision and accuracy was <8%.
Analytical assay of 4βOHC
4βOHC was determined in the remaining serum volumes of clinical samples submitted for TDM analyses by a previously published UPLC‐MS/MS method 17. Briefly, aliquots of 10 μl purified samples including the added internal standard, i.e. deuterated 4βOHC (4βOHC‐D7; IS), were injected on the same UPLC column as described for analysis of quetiapine and NDQ. A gradient elution with a mix of water and methanol (85–95%) was used as mobile phase for chromatographic separation (flow rate 0.150 ml min−1, column temperature 40°C). MS/MS detection was obtained by an atmospheric pressure chemical ionization (APCI) probe operated in positive mode using MRM at the transitions m/z 385 → 367 (4βOHC) and m/z 392 → 374 (4βOHC‐D7; IS). The total runtime per sample was 10 min.
The intra‐ and interday precision and accuracy of the 4βOHC assay was <15% at 25 nmol l−1 (defined as LLOQ) and <4% at 1600 nmol l−1 (defined as upper limit of quantification; ULOQ). In line with requirements specified in a recently published consensus paper on 4βOHC analysis 26, 4αOHC and 4βOHC were chromatographically separated with retention times of 2.5 and 3 min, respectively.
Statistical analyses
The Spearman's signed rank test was used to investigate univariate correlations between 4βOHC serum levels and C/D ratios of quetiapine and NDQ/quetiapine metabolic ratios. In the multivariate analyses, associations between 4βOHC levels and quetiapine C/D and metabolic ratios were tested in multiple linear regression models including age (≥60 vs. <60 years), gender (females vs. males), and tablet formulation (IR vs. XR), as potential covariates (‘<60 years’, ‘females’ and ‘IR’ encoded as reference). The distribution of measured C/D ratios of quetiapine and metabolic ratios did not pass normality tests, and were therefore log‐transformed (ln) before multiple linear regression analyses. After multivariate analyses, model constants/intercepts and effect estimates of tested variables (β values with 95% confidence intervals; CI) were back‐transformed and presented as nominal values. R 2 values from the multiple linear regression analyses were used as measure of explained variability in quetiapine C/D ratios and metabolic ratios.
In addition to statistical analyses related to quetiapine measurements, measured 4βOHC levels were compared between females and males, patients ≥60 and <60 years, and IR and XR tablet formulations, by Mann–Whitney tests. For patients with sufficient rest volumes of TDM samples available for reanalysis of both 4βOHC and total cholesterol concentration in serum, the Spearman's test was used to investigate the correlation between unadjusted 4βOHC concentrations and 4βOHC/C ratios.
IBM SPSS Statistics 21 (IBM SPSS Statistics, IBM Corp., Armonk, NY) was used for statistical analyses, while GraphPad Prism version 7 (GraphPad Software, Inc., San Diego, CA, USA) was used for graphical illustrations. P < 0.05 was considered statistically significant.
Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY 27, and are permanently archived in the Concise Guide to PHARMACOLOGY 2015/16 28, 29.
Results
Patient inclusion and characteristics
During the prospective one‐year collection period, 392 patients with routinely measured serum concentrations of quetiapine were identified to have (i) sufficient residual sample volumes for 4βOHC reanalysis, and (ii) available information about daily dosing and sampling time. Among these, 241 patients were excluded due to blood sampling time outside predefined criteria, measured drug/metabolite concentrations outside the validated ranges, or absence of steady‐state conditions. The remaining 151 quetiapine‐treated patients were included in the study for reanalysis of 4βOHC. After reviewing the TDM requisition forms from which the respective serum samples of the included patients originated, comedication with known inducers or inhibitors of CYP3A4 was not identified.
The majority of the included patients (n = 98, 65%) received treatment with the IR tablet formulation of quetiapine, while the remaining received XR tablets. The gender distribution within the included patient population was balanced (54% females). Median age in the whole population was 44 years (range 15–92 years) with a higher age in IR‐ than XR‐treated patients, i.e. median 45 vs. 36 years.
Serum concentrations of 4βOHC in the entire study population ranged from 17 to 302 nmol l−1 (median 75 nmol l−1). Measured serum concentrations of 4βOHC were significantly higher in females than males (median 89 vs. 60 nmol l−1, P < 0.0001), but no significant difference in 4βOHC levels was observed between older and younger patients, or between XR‐ and IR‐treated patients. Sufficient serum volumes for supplementary measurements of total cholesterol (C) concentration, in addition to 4βOHC, were available for 86 of the 151 included patients (57%; median C concentration 4.8 nmol l−1, range 2.8–7.5). A tight, positive correlation between absolute 4βOHC (unadjusted) concentration and 4βOHC/C ratio was observed in these cases (Spearman's r = 0.90, 95% CI 0.84, 0.93; P < 0.0001; Figure 1).
Figure 1.
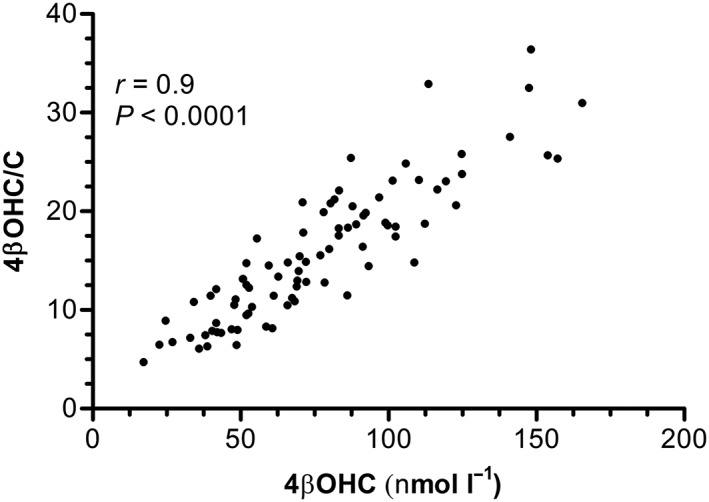
Relationship between unadjusted 4β‐hydroxycholesterol levels (4βOHC, nmol l−1) and 4βOHC‐to‐cholesterol (C) ratios (4βOHC/C) in the 86 patients where residual sample volumes were sufficient for reanalysis of both 4βOHC and C. P‐value estimated by Spearman's rank correlation test (r [rho] = correlation coefficient)
Univariate correlations between 4βOHC level and quetiapine steady‐state concentration and metabolic ratio
Correlation plots between 4βOHC levels and (i) quetiapine dose‐adjusted steady‐state (trough) serum concentrations (C/D) ratios, and (ii) metabolic ratios, are presented in Figure 2. Highly significant, negative correlations were observed between 4βOHC levels and C/D ratios of quetiapine in IR‐treated patients (Spearman's r = −0.47, 95% CI −0.62, −0.30; P < 0.0001), XR‐treated patients (Spearman's r = −0.56, 95% CI −0.72, −0.33; P < 0.0001) and in the pooled population (Spearman's r = −0.51, 95% CI −0.62, −0.38; P < 0.0001) (Figure 2).
Figure 2.
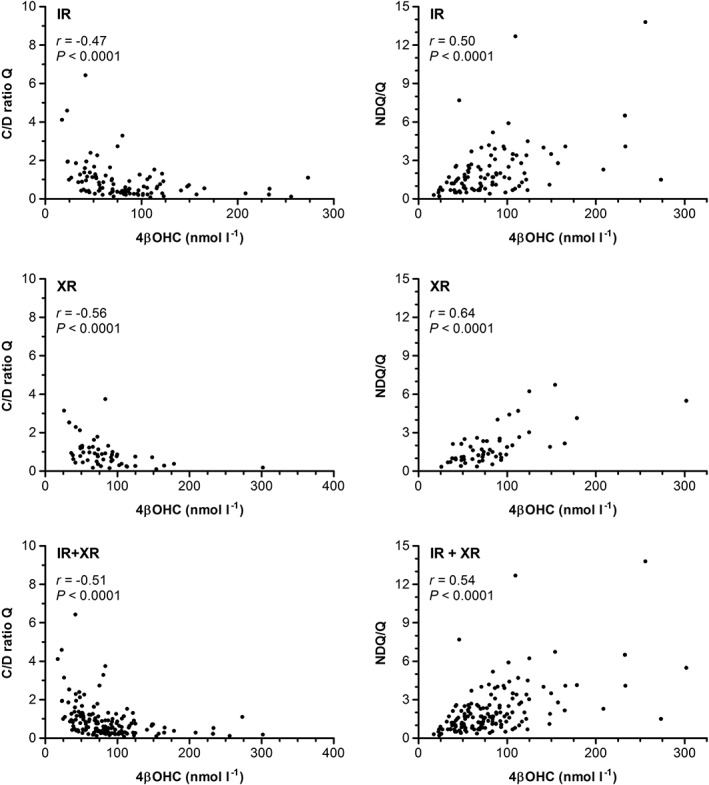
Correlations between 4β‐hydroxycholesterol (4βOHC) levels and (i) dose‐adjusted serum concentration (C/D ratio) of quetiapine (Q) (left panel), and (ii) N‐desalkylquetiapine‐to‐quetiapine (NDQ/Q) metabolic ratios (right panel), for immediate release (IR), extended release (XR) and pooled samples (IR + XR). P‐values estimated by Spearman's rank correlation tests (r [rho] = correlation coefficient)
We also observed statistically significant, positive correlations between 4βOHC levels and metabolic ratios in IR‐treated patients (r = 0.50, 95% CI 0.32, 0.64; P < 0.0001), XR‐treated patients (r = 0.64, 95% CI 0.44, 0.78; P < 0.0001) and in the pooled population (r = 0.54, 95% CI 0.41, 0.64; P < 0.0001) (Figure 2).
4βOHC level and other candidate variables' association with quetiapine steady‐state concentration and metabolic ratio in multivariate analyses
Results of the multiple linear regression analyses are summarized in Table 1 (quetiapine C/D ratio) and Table 2 (metabolic ratio). Briefly, 4βOHC level, gender and age, but not tablet formulation, were significantly associated with C/D ratio and metabolic ratio of quetiapine. 4βOHC level was found to be the strongest explanatory variable for quetiapine C/D ratio in the multivariate analysis (Table 1). After correction for covariate effects, C/D ratio of quetiapine was estimated to be reduced by 0.7% per unit (nmol l−1) 4βOHC increase above the lowest measured value in the dataset, i.e. 17 nmol l−1 (P < 0.001, Table 1). In addition, males were estimated to have approximately 30% higher C/D ratio of quetiapine compared to females (P = 0.023), and patients ≥60 years were estimated to obtain approximately 60% higher C/D ratio of quetiapine compared to younger patients (P = 0.003, Table 1).
Table 1.
Estimated effects of 4βOHC level, gender, age, and tablet formulation, on dose‐adjusted serum concentration (C/D ratio; nmol l−1 mg−1 day−1) of quetiapine in multiple linear regression analysis
| Variable | Estimated fold change (95% CI) [Link] | P |
|---|---|---|
| 4βOHC per unit (nmol/l −1 ) increase a | 0.993 (0.991–0.995) | <0.001 |
| Male vs. female | 1.327 (1.043–1.689) | 0.023 |
| ≥60 vs. <60 years | 1.627 (1.186–2.232) | 0.003 |
| XR vs. IR | 1.242 (0.974–1.584) | N.S. |
| Model fit (R 2 ) = 0.283 | ||
4βOHC, 4β‐hydroxycholesterol; C/D ratio, dose‐adjusted serum concentration; CI, confidence interval; IR, immediate release; N.S., not significant; XR, extended release
Effect estimates (β values) are presented as back transformed ln values from the multiple linear regression analysis (back transformed intercept/constant of the model = 0.739 nmol l−1 mg−1 day−1).
Unit increase from the lowest measured 4βOHC in the dataset, i.e. 17 nmol l−1)
Table 2.
Estimated effects of 4βOHC level, gender, age, and tablet formulation, on quetiapine metabolic ratio (NDQ/quetiapine) in multiple linear regression analysis
| Variable | Estimated fold change (95% CI) a | P |
|---|---|---|
| 4βOHC per unit (nmol/l −1 ) increase b | 1.007 (1.005–1.009) | <0.001 |
| Male vs. female | 0.694 (0.561–0.861) | 0.001 |
| ≥60 vs. <60 years | 0.736 (0.557–0.971) | 0.033 |
| XR vs. IR | 0.843 (0.681–1.045) | N.S. |
| Model fit (R 2 ) = 0.356 | ||
4βOHC, 4β‐hydroxycholesterol; CI, confidence interval; IR, immediate release; NDQ, N‐desalkylquetiapine; N.S., not significant; XR, extended release
Effect estimates (β values) are presented as back transformed ln values from the multivariate model analysis (back transformed intercept/constant of the model = 1.344 nmol l−1 mg−1 day−1).
Unit increase from the lowest measured 4βOHC in the dataset, i.e. 17 nmol l−1)
4βOHC level, gender and age were also significantly associated with individual variability in NDQ/quetiapine metabolic ratios in multivariate analysis (Table 2). While increasing levels of 4βOHC were associated with higher metabolic ratios, significantly lower metabolic ratios were estimated in males than females and in patients ≥60 vs. <60 years (Table 2).
Model fits (R 2) of the multiple linear regression analyses were 0.283 and 0.356 for quetiapine C/D ratio (Table 1) and NDQ/quetiapine metabolic ratio (Table 2), respectively.
Discussion
This study shows that 4βOHC level is closely correlated with dose‐adjusted steady‐state trough serum concentration of orally administered quetiapine, a CYP3A4 substrate with low bioavailability due to extensive presystemic metabolism 21. The relationship between 4βOHC level and quetiapine exposure in the included population of psychiatric patients was highly significant in both univariate and multivariate statistical analyses.
To our knowledge, this is the first study to show a clear correlation between 4βOHC level and exposure of a CYP3A4 substrate. One previous study failed to show significant associations between 4βOHC levels and clearance estimates of the CYP3A4 substrates docetaxel and paclitaxel following intravenous administration 30. This latter might reflect potential relevance of non‐CYP3A4 mechanisms in clearance of docetaxel and paclitaxel, but in light of the possible role of intestinal CYP3A4 in biomarker formation 17, the intravenous administration of these substrates could also be a factor of relevance for the reported lack of associations with 4βOHC levels in that study.
Tacrolimus, a combined substrate of CYP3A4/5 and the efflux transporter P‐glycoprotein (P‐gp), shows great interindividual variability in oral clearance 31. A recent study by Vanhove et al. reported a correlation between 4βOHC level and weight‐adjusted oral tacrolimus clearance in stable renal transplant patients 32. However, this correlation could not be reproduced in the early phase after transplantation in a subsequent study by Størset et al. 33, 34. This indicates that other factors than CYP3A(4) phenotype are more important for the pharmacokinetic variability of tacrolimus in the early phase after transplantation, which limits the potential clinical utility of 4βOHC level as biomarker for estimating tacrolimus dose requirements. Overall, the variable degree of correlation with exposure of different substrates shows that clinical implementation of 4βOHC level and as a generic biomarker for individualized dosing of CYP3A4‐metabolized drug is not realistic, but its potential relevance as dosing biomarker probably needs be evaluated for each drug separately.
The metabolite NDQ is almost exclusively formed by CYP3A4 35, and the close correlation between 4βOHC levels and NDQ/quetiapine metabolic ratios supports that 4βOHC level reflects basal CYP3A4 activity. Gender differences in NDQ/quetiapine ratios and 4βOHC levels were of similar magnitudes (i.e. 30–50% higher in females than males), which provide clear evidence that basal CYP3A4 activity is higher in females than males. The underlying mechanism for the gender difference in CYP3A4 phenotype is uncertain, but we recently observed that females appeared to be more responsive towards induction of CYP3A4 activity during use of carbamazepine 17. The higher basal CYP3A4 activity in females than males might therefore be due to greater responsiveness towards exogenous or endogenous factors increasing enzyme activity.
Previous studies have reported conflicting results regarding the impact of age on interindividual differences in quetiapine exposure 23, 36. In the present study, we observed significantly higher dose‐adjusted serum concentration of quetiapine in older vs. younger patients in the multiple linear regression analysis. This observation was accompanied by a lower NDQ/quetiapine metabolic ratio in older vs. younger patients. However, the slight, nonsignificantly higher 4βOHC level found in older compared to younger patients does not indicate that altered quetiapine pharmacokinetics in the elderly is caused by reduced CYP3A4 activity. Another, more likely mechanism behind higher dose‐adjusted serum concentration of quetiapine, is reduced clearance in the elderly due to an age decline in hepatic blood flow.
In the present study, we observed a close, apparently linear correlation between unadjusted 4βOHC concentrations and 4βOHC/C ratios in those patients with sufficient sample volumes for reanalysis of both 4βOHC and total cholesterol levels. These data therefore support that absolute 4βOHC level generally will work well as phenotype biomarker of CYP3A4, which is in line with a previous publication by Diczfalusy et al. reporting that differences in cholesterol levels only explain <10% of the variability in serum concentration of 4βOHC 15.
Consistent with the extensive individual variability in CYP3A4 phenotype, 4βOHC levels ranged >15‐fold in the study population. In the multivariate analysis, one unit (nmol l−1) increase in 4βOHC level was associated with an estimated 0.7% decrease in steady‐state trough serum concentration of quetiapine. This means that raising 4βOHC level by, for example 30 nmol l−1, will result in approximately 20% reduction in quetiapine exposure. This kind of quantitative estimate is subject to considerable uncertainty, and it should be further investigated to what extent pretreatment 4βOHC level, together with other significant explanatory variables, could predict steady‐state trough serum concentration of quetiapine at an individual level. Moreover, it would be of interest to investigate whether βOHC level predicts trough concentrations, as measured in this study, differently than maximum concentrations (C max) and area under the concentration vs. time curves (AUC) of quetiapine.
The multiple linear regression analysis explained approximately 30% of the total variability in dose‐adjusted serum concentration of quetiapine. Studies have shown that quetiapine is a P‐gp (ABCB1) substrate 37, 38, and individual differences in P‐gp phenotype likely explain some of the remaining variability in quetiapine exposure. Methodological limitations associated with the use of retrospective and naturalistic data in this study have probably also contributed to variability in quetiapine measurements. Details about dose, time between the last dose intake and blood sampling, and comedication, were based on requisition forms filled out by physicians, which could be inaccurate. In addition, degree of patient adherence to quetiapine treatment is another, uncontrolled factor that may have introduced nonpharmacological variability in the data material. Despite these limitations, the large number of included patients, together with the high explanatory degree of the included biological variables in the multivariate analysis, strengthens the study findings.
In conclusion, the present study shows that 4βOHC level is significantly correlated with steady‐state serum concentration of quetiapine in psychiatric patients. This illustrates the clinical pharmacological potential of 4βOHC as an endogenous CYP3A4 biomarker for individualized drug dosing, and future studies should evaluate to what extent pretreatment 4βOHC level can predict dose‐adjusted concentrations of quetiapine and other drugs mainly metabolized by CYP3A4.
Competing Interests
There are no competing interests to declare.
We acknowledge the South‐Eastern Norway Regional Health Authority for PhD funding to author C.G. Moreover, we are grateful to the Department of Medical Biochemistry at Diakonhjemmet Hospital for performing cholesterol measurements.
Contributors
All the authors contributed to the study design. C.G. was responsible for the data collection and performance of laboratory analyses and statistical tests. C.G. and E.M. contributed to the drafting of the manuscript. All authors reviewed the manuscript and approved the final version.
Gjestad, C. , Haslemo, T. , Andreassen, O. A. , and Molden, E. (2017) 4β‐Hydroxycholesterol level significantly correlates with steady‐state serum concentration of the CYP3A4 substrate quetiapine in psychiatric patients. Br J Clin Pharmacol, 83: 2398–2405. doi: 10.1111/bcp.13341.
References
- 1. Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 2013; 138: 103–141. [DOI] [PubMed] [Google Scholar]
- 2. Lin YS, Lockwood GF, Graham MA, Brian WR, Loi CM, Dobrinska MR, et al. In‐vivo phenotyping for CYP3A by a single‐point determination of midazolam plasma concentration. Pharmacogenetics 2001; 11: 781–791. [DOI] [PubMed] [Google Scholar]
- 3. Hole K, Gjestad C, Heitmann KM, Haslemo T, Molden E, Bremer S. Impact of genetic and nongenetic factors on interindividual variability in 4beta‐hydroxycholesterol concentration. Eur J Clin Pharmacol 2017; 73: 317–324. [DOI] [PubMed] [Google Scholar]
- 4. Klein K, Zanger UM. Pharmacogenomics of cytochrome P450 3A4: recent progress toward the ‘missing heritability’ problem. Front Genet 2013; 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fuhr U, Jetter A, Kirchheiner J. Appropriate phenotyping procedures for drug metabolizing enzymes and transporters in humans and their simultaneous use in the ‘cocktail’ approach. Clin Pharmacol Ther 2007; 81: 270–283. [DOI] [PubMed] [Google Scholar]
- 6. Bjorkhem‐Bergman L, Backstrom T, Nylen H, Ronquist‐Nii Y, Bredberg E, Andersson TB, et al. Quinine compared to 4beta‐hydroxycholesterol and midazolam as markers for CYP3A induction by rifampicin. Drug Metab Dispos 2014; 29: 352–355. [DOI] [PubMed] [Google Scholar]
- 7. Diczfalusy U, Kanebratt KP, Bredberg E, Andersson TB, Bottiger Y, Bertilsson L. 4beta‐hydroxycholesterol as an endogenous marker for CYP3A4/5 activity: stability and half‐life of elimination after induction with rifampicin. Br J Clin Pharmacol 2009; 67: 38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wide K, Larsson H, Bertilsson L, Diczfalusy U. Time course of the increase in 4beta‐hydroxycholesterol concentration during carbamazepine treatment of paediatric patients with epilepsy. Br J Clin Pharmacol 2008; 65: 708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kanebratt KP, Diczfalusy U, Backstrom T, Sparve E, Bredberg E, Bottiger Y, et al. Cytochrome P450 induction by rifampicin in healthy subjects: determination using the Karolinska cocktail and the endogenous CYP3A4 marker 4beta‐hydroxycholesterol. Clin Pharmacol Ther 2008; 84: 589–594. [DOI] [PubMed] [Google Scholar]
- 10. Tomalik‐Scharte D, Lutjohann D, Doroshyenko O, Frank D, Jetter A, Fuhr U. Plasma 4beta‐hydroxycholesterol: an endogenous CYP3A metric? Clin Pharmacol Ther 2009; 86: 147–153. [DOI] [PubMed] [Google Scholar]
- 11. Bodin K, Bretillon L, Aden Y, Bertilsson L, Broome U, Einarsson C, et al. Antiepileptic drugs increase plasma levels of 4beta‐hydroxycholesterol in humans: evidence for involvement of cytochrome p450 3A4. J Biol Chem 2001; 276: 38685–38689. [DOI] [PubMed] [Google Scholar]
- 12. Bodin K, Andersson U, Rystedt E, Ellis E, Norlin M, Pikuleva I, et al. Metabolism of 4 beta‐hydroxycholesterol in humans. J Biol Chem 2002; 277: 31534–31540. [DOI] [PubMed] [Google Scholar]
- 13. Diczfalusy U, Miura J, Roh HK, Mirghani RA, Sayi J, Larsson H, et al. 4beta‐Hydroxycholesterol is a new endogenous CYP3A marker: relationship to CYP3A5 genotype, quinine 3‐hydroxylation and sex in Koreans, Swedes and Tanzanians. Pharmacogenet Genomics 2008; 18: 201–208. [DOI] [PubMed] [Google Scholar]
- 14. Suzuki Y, Fujioka T, Sato F, Matsumoto K, Muraya N, Tanaka R, et al. CYP3A5 polymorphism affects the increase in CYP3A activity after living kidney transplantation in patients with end stage renal disease. Br J Clin Pharmacol 2015; 80: 1421–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diczfalusy U, Nylen H, Elander P, Bertilsson L. 4beta‐Hydroxycholesterol, an endogenous marker of CYP3A4/5 activity in humans. Br J Clin Pharmacol 2011; 71: 183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gebeyehu E, Engidawork E, Bijnsdorp A, Aminy A, Diczfalusy U, Aklillu E. Sex and CYP3A5 genotype influence total CYP3A activity: high CYP3A activity and a unique distribution of CYP3A5 variant alleles in Ethiopians. Pharmacogenomics J 2011; 11: 130–137. [DOI] [PubMed] [Google Scholar]
- 17. Gjestad C, Huynh DK, Haslemo T, Molden E. 4beta‐hydroxycholesterol correlates with dose but not steady‐state concentration of carbamazepine: indication of intestinal CYP3A in biomarker formation? Br J Clin Pharmacol 2016; 81: 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Josephson F, Bertilsson L, Bottiger Y, Flamholc L, Gisslen M, Ormaasen V, et al. CYP3A induction and inhibition by different antiretroviral regimens reflected by changes in plasma 4beta‐hydroxycholesterol levels. Eur J Clin Pharmacol 2008; 64: 775–781. [DOI] [PubMed] [Google Scholar]
- 19. Lutjohann D, Marinova M, Schneider B, Oldenburg J, von Bergmann K, Bieber T, et al. 4beta‐Hydroxycholesterol as a marker of CYP3A4 inhibition in vivo – effects of itraconazole in man. Int J Clin Pharmacol Ther 2009; 47: 709–715. [DOI] [PubMed] [Google Scholar]
- 20. Bjorkhem‐Bergman L, Backstrom T, Nylen H, Ronquist‐Nii Y, Bredberg E, Andersson TB, et al. Comparison of endogenous 4beta‐hydroxycholesterol with midazolam as markers for CYP3A4 induction by rifampicin. Drug Metab Dispos 2013; 41: 1488–1493. [DOI] [PubMed] [Google Scholar]
- 21. Narala A, Veerabrahma K. Preparation, characterization and evaluation of quetiapine fumarate solid lipid nanoparticles to improve the oral bioavailability. J Pharm 2013; 2013: 265741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. DeVane CL, Nemeroff CB. Clinical pharmacokinetics of quetiapine: an atypical antipsychotic. Clin Pharmacokinet 2001; 40: 509–522. [DOI] [PubMed] [Google Scholar]
- 23. Bakken GV, Rudberg I, Molden E, Refsum H, Hermann M. Pharmacokinetic variability of quetiapine and the active metabolite N‐desalkylquetiapine in psychiatric patients. Ther Drug Monit 2011; 33: 222–226. [DOI] [PubMed] [Google Scholar]
- 24. Grimm SW, Richtand NM, Winter HR, Stams KR, Reele SB. Effects of cytochrome P450 3A modulators ketoconazole and carbamazepine on quetiapine pharmacokinetics. Br J Clin Pharmacol 2006; 61: 58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Flockhart D. Drug interactions: cytochrome P450 drug interaction table. Indiana University School of Medicine (medicine.iupui.edu/clinpharm/ddis, last accessed 22 October 2016).
- 26. Aubry AF, Dean B, Diczfalusy U, Goodenough A, Iffland A, McLeod J, et al. Recommendations on the development of a bioanalytical assay for 4beta‐hydroxycholesterol, an emerging endogenous biomarker of CYP3A activity. AAPS J 2016; 18: 1056–1066. [DOI] [PubMed] [Google Scholar]
- 27. Southan C, Sharman JL, Benson HE, Faccenda E, Pawson AJ, Alexander SP, et al The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucl Acids Res 2016; 44: D1054–D1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alexander SPH, Fabbro D, Kelly E, Marrion N, Peters JA, Benson HE, et al The Concise Guide to PHARMACOLOGY 2015/16: Enzymes. Br J Pharmacol 2015; 172: 6024–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alexander SPH, Kelly E, Marrion N, Peters JA, Benson HE, Faccenda E, et al The Concise Guide to PHARMACOLOGY 2015/16: Transporters. Br J Pharmacol 2015; 172: 6110–6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Graan AJ, Sparreboom A, de Bruijn P, de Jonge E, van der Holt B, Wiemer EA, et al. 4beta‐hydroxycholesterol as an endogenous CYP3A marker in cancer patients treated with taxanes. Br J Clin Pharmacol 2015; 80: 560–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Staatz CE, Tett SE. Clinical pharmacokinetics and pharmacodynamics of tacrolimus in solid organ transplantation. Clin Pharmacokinet 2004; 43: 623–653. [DOI] [PubMed] [Google Scholar]
- 32. Vanhove T, de Jonge H, de Loor H, Annaert P, Diczfalusy U, Kuypers DR. Comparative performance of oral midazolam clearance and plasma 4beta‐hydroxycholesterol to explain interindividual variability in tacrolimus clearance. Br J Clin Pharmacol 2016; 82: 1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Størset E, Hole K, Midtvedt K, Bergan S, Molden E, Asberg A. Bodyweight adjustments introduce significant correlations between CYP3A metrics and tacrolimus clearance. Br J Clin Pharmacol 2016; 83: 1350–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Størset E, Hole K, Midtvedt K, Bergan S, Molden E, Asberg A. The CYP3A biomarker 4beta‐hydroxycholesterol does not improve tacrolimus dose predictions early after kidney transplantation. Br J Clin Pharmacol 2017; 83: 1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bakken GV, Rudberg I, Christensen H, Molden E, Refsum H, Hermann M. Metabolism of quetiapine by CYP3A4 and CYP3A5 in presence or absence of cytochrome B5. Drug Metab Dispos 2009; 37: 254–258. [DOI] [PubMed] [Google Scholar]
- 36. Bakken GV, Molden E, Hermann M. Impact of genetic variability in CYP2D6, CYP3A5, and ABCB1 on serum concentrations of quetiapine and N‐desalkylquetiapine in psychiatric patients. Ther Drug Monit 2015; 37: 256–261. [DOI] [PubMed] [Google Scholar]
- 37. Nikisch G, Baumann P, Oneda B, Kiessling B, Weisser H, Mathe AA, et al. Cytochrome P450 and ABCB1 genetics: association with quetiapine and norquetiapine plasma and cerebrospinal fluid concentrations and with clinical response in patients suffering from schizophrenia: a pilot study. J Psychopharmacol 2011; 25: 896–907. [DOI] [PubMed] [Google Scholar]
- 38. Boulton DW, DeVane CL, Liston HL, Markowitz JS. In vitro P‐glycoprotein affinity for atypical and conventional antipsychotics. Life Sci 2002; 71: 163–169. [DOI] [PubMed] [Google Scholar]


