Abstract
Objective(s):
Listeria monocytogens, Bacillus cereus and Campylobacter jejuni are three toxin producing bacteria over the world, especially in Iran, and it is essential to find a certain, rapid procedure to identify these microorganisms. In this research, these bacteria were simultaneously detected by multiplex PCR technique in foods.
Materials and Methods:
The primary approval of bacterial strains was performed by biochemical tests. PCR primers were designed based on the nucleotide sequences of the NHEB/NHEC gene of B. cereus, the hly gene of L. monocytogenes and the C gene of C. jejuni. The specificity of Multiplex PCR method was determined using seven food poisoning bacteria including Salmonella typhi, Shigella dysentery, Yersinia pestis, Staphylococcus aureus, Clostridium perfringens, Clostridium botulinum and Vibrio cholerae. To confirm the reaction, DNA extraction was performed from 30 food samples (milk), and gene amplification was performed by PCR. The length of amplified fragments was 300 bp, 210 bp and 160 bp for NHEB/NHEC, hly and C genes, respectively.
Results:
The detection limits of the PCR method were 5, 4 and 3 pg for L. monocytogenes, B. cereus and C. jejuni, respectively. Specifisity test showed that this reaction is spesific to these 3 bacteria.
Conclusion:
In this study, we introduced a new multiplex PCR method for simultsnus detection of L. monocytogens, B. cereus and C. jejuni. These results can be used for detection of other toxin producing bacteria in food.
Keywords: Bacillus cereus, Campylobacter jejuni, Hly, Listeria Monocytogenes, Multiplex PCR, NHEB/NHEC
Introduction
Food poisoning is a serious and ongoing issue in public health over the world. Approximately, 76 million illnesses and 5000 deaths occur annually due to the prevalence of food poisoning (1). On the other hand, food safety of individuals is considered as one of the most important public health problems in the world. Food poisoning is caused by toxins produced by bacterial, virus, or protozoa (2). To date, using the culture-based methods to identify and distinguish microorganisms contaminating foods has been considered as Gold Standard (3), albeit, traditional methods are often time-consuming and expensive (3, 4). Therefore it is necessary to apply new tools for the identification of individuals’ food safety to study the presence of pathogens in this regard. Recent developments in biotechnology are due to the rapid evolution of methods to identify pathogens by which the identification using primarily (manual) methods has been prevented. The detection methods based on
DNA assay including polymerase chain reaction (PCR), DNA hybridization and DNA microarray reduce the cost and the time of reaction. In addition, these methods are not only able to identify the microbial agents, but also are able to track the origin of the contamination, which is based on molecular epidemiology. PCR is a technique that has led to the revolution in molecular biology through many applications in the detection of genetic diseases and infections, as well as identifying the pathogens in foodstuffs (5). Today, the assay based on this technique has led to the rapid and specific detection of intoxicating agents in food, and commercial kits act accordingly for the specific detection of food poisoning. Listeria monocytogenes has several exotoxins, which have a key role in its pathogenesis causing some infections such as septicemia, encephalitis, meningitis, premature infants and premature labor. The presence of L. monocytogenes in food samples is shown by PCR methods (6).
Bacillus cereus is one of the major bacterial in food poisoning, which causes 2 type of poisoning in form of vomiting and diarrhea (7, 8). B. cereus spores can grow at refrigeration temperatures and survive at high temperature conditions (9, 10) and is one of the resistant agents in milk pasteurization process (11). To identify the prevalence of virulence genes of B. cereus in children foods, a prevalence study was performed and the results showed that 42% of infant foods are contaminated with B. cereus (12).
Campylobacter jejuni is a Gram-negative, oxidase positive bacterium, which is one of the health problems in developed European countries (13). In 2008, among 27 European countries, 190,566 Campylobacteriosis cases were reported. This bacterium is often found in poultry meat (14). In several studies, it was shown that C. jejuni is a common poisoning bacteria in food samples (15-19).
Various methods for rapid nucleic acid-based identification of bacteria are used. Some of these techniques include real time PCR and microarray, each in turn has some advantages and disadvantages. Nucleic acid sequence-based amplification (NASABA) and loop-mediated isothermal amplification (LAMP) techniques are used to identify microorganisms with a high sensitivity and specificity and low cost. The main problem in these methods is the complicated primers design and the need for live microorganisms. The other drawback of these methods is handling of RNA due to its short half-life. PCR is a molecular method to detect pathogens in the food samples. This method has high sensitivity and specificity, and is able to provide interpretable results automatically. The simplicity of PCR compared with other methods, introduced it as a preferred method in any situation.
This study is aimed to launch a Multiplex PCR method for simultaneous diagnosis of three common causes of food poisoning (L. monocytogenes, B. cereus and C. jejuni) in short time with a high sensitivity and specificity.
Materials and Methods
Bacteria strains, cultivation and counting
The bacteria for the study were obtained from Academic Centers as follows:
C. jejuni bacterium obtained from Microbiology Department, Tehran University of Medical Sciences.
B. cereus bacterium obtained from Tarbiat Modarres University.
L. monocytogenes bacterium obtained from Iranian Centre of Industrial and Medical Fungi and Bacteria Collection (PTCC 1298)
Following bacteria were prepared from the Iranian Reference Laboratory: Salmonella typhi ATCC 700931, Shigella dysentery ATCC 13313, Yersinia pestis ATCCBAA-1511D-5, Staphylococcus aureus ATCC 25923, Clostridium perfringes ATCC 13124, Clostridium botulinum ATCC 3502, Vibrio cholera ATCC 9459.
Cultivation and DNA extraction
Bacteria were inoculated in Luria Bertani (LB) agar and broth medium and incubated at 37 °C for 18 to 24 hr, and then 1.5 ml of an overnight culture was centrifuged at 5000 rpm for 5 min. To confirm the bacteria, the shape and structure of colonies were studied. Biochemical tests were also used for bacterial identification. Then, 1.5 to 5 ml of the broth medium was used for DNA extraction (20) as follows: 1.5 ml of an overnight culture was centrifuged at 5000 rpm for 3 to 5 min. The supernatant was discarded and the pellet was resuspended in 200 µl sodium chloride-Tris-EDTA (STE) buffer and kept for 10 min at room temperature. Then, 200 µl sodium dodecyl sulfate (SDS) buffer was added followed by severe vortexing. Then 200 µl sodium acetate was added and the tube was inverted 10 times and centrifuged at 10000 rpm for 5 min at 4 °C. The supernatant was transferred to a fresh tube, isopropanol was added as the same volume and the mixture was incubated for 20 min at -20 °C. Subsequently, the tube was centrifuged at 10000 rpm for 15 min at 4 °C to precipitate DNA.
The precipitated DNA was dried and resuspended in 100 µl Tris EDTA (TE) buffer containing 10 microgarm RNase A and incubated for 1 hr at 37 °C.
Then, the supernatant was discarded and the pellet was dried for 20 to 30 min at 60-65 °C, dissolved in 100 µl distilled water, and 1 µl of the solution was used as template for PCR.
Selection of gene fragments and the preparation of appropriate primers
Three pairs of primers specific to hly gene for L. monocytogenes, NHB/C gene for B. cereus, and C gene for C. jejuni were designed. The characteristics of primers such as loops, melting points, etc., were analyzed by DNASIS, BLAST and Oligo molecular software. The primers were synthesized by MWG Co. (Germany) and DNA Technology AISCo. (Denmark). The quality of primers was evaluated using 15% polyacrylamide gel electrophoresis. The sequences of primes are shown in Table 1.
Table 1.
The sequence of primers used in this study
| sSize | Primer sequences | Bacteria |
|---|---|---|
| 210 bp | LF: CGCAACAAACTGAAGCAAAGG | Listeria monocytogenes |
| LR:TTGGCGGCACATTTGTCAC | ||
| 160bp | CF:GGAAAATCAAATAAAGTTAGAGGTAGAA | Campylobacter Jejuni |
| CR:CCATAAGCACTAGCTAGCTGATTATC | ||
| 300bp | N/BC –F:ACATTGCGAAAGATAGCTGGA | Bacillus cereus |
| N/BC-R:TGTTCTGCTGCAAAAAGGATG |
L-f and L-r primers amplify a 210- bp gene fragment, which relates to the hly gene of L. monocytogenes. C-4 and C-1 primers amplify a 160- bp gene fragment, which relates to the oxidoreductase gene of C. jejuni. N/BC-r and, N/BC-f primers amplify a 300- bp gene fragment, which relates to the non hemolysin B/C gene of B. cereus
The polymerase chain reaction
DNA ladder, restriction enzymes and Taq DNA polymerase were purchased from Fermentase Co. (xxx). PCR was performed in final volume of 25 µl. Composition was set 0.5 µM of each primer pairs, 1 unit of Taq DNA polymerase, 0.2 µM of each dNTPs, 2.5 µl of PCR buffer, 2 mM MgCl2 and template DNA. DNA template concentration was optimized for each DNA sample including 2, 4, and 3 µl (concentra-tion) for C. jejuni, L. monocytogenes, and B. cereus, respectively. In addition, PCR settings were as follows: incubation at 94 °C for 5 min; 30 cycles of denaturation at 94 °C for 60 sec, primer annealing at L-f and L-r primers amplify a 210- bp gene fragment, which relates to the hly gene of L. monocytogenes. C-4 and C-1 primers amplify a 160- bp gene fragment, which relates to the oxidoreductase gene of C. jejuni. N/BC-r and, N/BC-f primers amplify a 300- bp gene fragment, which relates to the non hemolysin B/C gene of B. cereus 56 °C for 60 sec, primer extension at 72 °C for 60 sec; and final extension at 72 °C for 3 min.
The amplification products were detected by agarose gel electrophoresis. The electrophoresis was run on 2% agarose gel at a constant voltage of 80 V for 30 min in Tris-borate-EDTA (TBE) buffer.
Determination of the reaction sensitivity in terms of the number of bacteria
To calculate the reaction sensitivity in terms of the number of bacteria, after culturing the bacteria and measuring the optical density (OD), the dilutions of 1010 were prepared from the desired culture, then PCR was performed for all dilutions and colony count was performed to calculate the number of bacteria in each dilution. For sequencing, 50 µl of each amplified product was purified using PCR product purification kit (Fermentase Co.). The purified products were sequenced at Sina genes Co. (USA).
Determination of the specificity of PCR
To determine the specificity of PCR, a PCR reaction was performed using the genomic DNA extracted from L. monocytogenes, B. cereus, C. jejuni, S. typhi, Sh. dysentery, Y. pestis, S. aureus, C. perfringens, C. botulinum, and V. cholerae, as template, and the same designed primers. The amplification products were detected by 2% agarose gel electrophoresis.
Detection of bacteria in food samples
Analysis of experimentally milk samples
Before performing the PCR, conventional culti-vation method was used to investigate the milk contamination. Results showed that none of the samples were contaminated with these bacteria. To confirm the multiplex PCR method for detection of B. cereus, C. jejuni and L. monocytogenes, 30 pasteurized milk samples were prepared from the milk procurement centers of livestock in Tehran.
Each milk sample was diluted with TSBYE at the ratio of 1:10, mixed well and incubated overnight (18 hr) at 37 °C. 1 ml was obtained at the end of incubation time and processed for DNA extraction by boiling method. 1.5 µl of purified DNA was used as template in PCR reaction.
Results
PCR product results with genomic samples in Multiplex PCR
Multiplex PCR method was used for the simultane-ous identification of C. jejuni, L. monocytogenes and B. cereus. As shown in Figure 1, the 210 bp fragment related to the amplification of a part of the hly gene of L. monocytogenes, 160 bp fragment related to the amplification of a part of the oxidoreductase gene (C gene) of C. jejuni and the 300 bp fragment related to the amplification of NHB/C genes of B. cereus.
Figure 1.
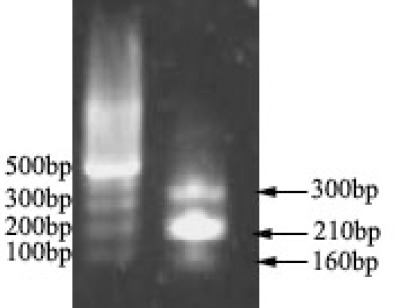
Results obtained from the multiplex PCR product of bacterial genomics.
Lane 1: 100 bp DNA ladder
Lane 2: L. monocytogenes (210 bp),C. jejuni (160 bp), B. cereus (300 bp)
Study of the specificity of PCR
The specific primers designed for the amplified genes of L. monocytogenes, C. jejuni and B. cereus were used for PCR with genomic DNA extracted from S. typhi, Sh. dysentery, Y. pestis, S. aureus, C. perfringens, C. botulinum, and V. cholera. As shown in Figure 2, no PCR product was observed, indicating specificity of the primers.
Figure 2.
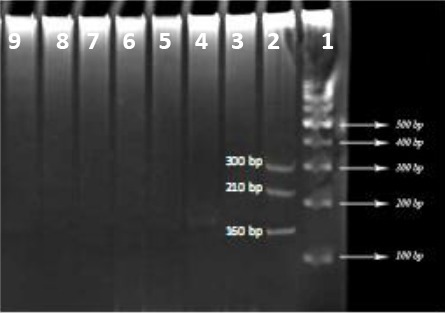
Study of the specificity of PCR with six bacterial strains
Lane 1: 100 bp DNA marker
Lane 2: Triple PCR products
Lane 3-8: PCR products with six bacterial strains to study the specificity of reaction
3Study of the sensitivity of PCR
To determine the sensitivity of reaction with the three bacteria, PCR was performed with different dilutions. The sensitivity of reaction was expressed in terms of the number of bacteria. As shown in Table 2, the number of L.monocytogenes, B. cereus and C. jejuni were 5, 4, and 3 pg, respectively.
Table 2.
Results of PCR for detection of C. jejuni, L. monocytogenes, and B. cereus randomly in food samples
| Organism | Number of sample | Positive in PCR |
|---|---|---|
| Listeria monocytogenes | 30 | 2 |
| Bacillus cereus | 30 | 1 |
| Campylobacter jejuni | 30 | 1 |
Analysis of experimentally milk samples
As stated in the methods section, contaminated food samples were determined using PCR reactions and the results were shown in Table 2.
Discussion
The incidence of food poisoning is considered as a major issue in today’s world. So far, nearly 250 types of food poisoning disease have occurred in the world in which more than two-thirds of them have bacterial origin. Among the bacteria causing food poisoning, L. monocytogenes, C. jejuni and B. cereus are particularly important (21, 22). Simultaneous amplification of greater than one particular locus is essential for a rapid diagnosis of multiple microbes. It is just a technique termed as Multiplex PCR, through which a number of distinct primer packages tend to be blended into a one PCR assay (23). Apparently, the planning on the primers can be a main factor in the progress of multiplex PCR assay. There could be possibly much relationship between the multiple primer packages, and so the primer concentrations of it may have to possibly be adjusted so as to create reputable brings of all of the PCR products.
Figure 3.
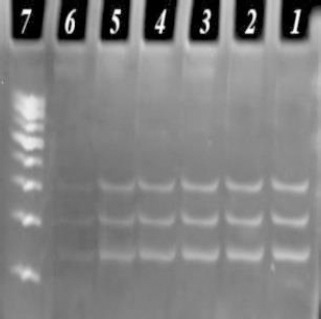
Study of the sensitivity of PCR on 1%5 polyacrylamide gel electrophoresis
Lane 7: 100 bp DNA marker
Lane 1-6: PCR products with the different dilutions of L. monocytogenes, B. cereus and C. jejuni with 101-106
Meanwhile, the actual primer packages need to be designed with the same annealing temperature, though supplying a means to identify the amplicons using thermal microbial online communities and also to assess active design, like in the course of fermentation or within response to environment versions. Finding and detection of the lowest rates of bacterial spoilage in food is necessary because these bacteria cause illness and diseases in human. The Multiplex PCR had suitable high rate of sensibility and could detect 101 to 102 organisms per ml of growth in LB media. This procedure was sufficient to reveal the presence of these bacteria in food samples. However, the detection limit of food samples is higher when compared with pure culture (103 to 104CFU per ml). multiplex PCR gives a faster prognosis compared with straightforward PCR over the simultaneous amplification of a number of genes. Even so, various units of specific primers are widely used within Multiplex PCR assay, while just one pair of specific primers are widely used within regular PCR assay. Primer design and style is essential to the progress of multiplex PCR, because primer units should have similar annealing heat to produce a successful multiplex PCR assay. In addition, the actual focus of primers can be crucial in the multiplex PCR. This is due to conversation may well come about relating to the multiple primer units inside mPCR that will leads to primer dimers, thus, the actual focus of primers may well should be fine-tuned to ensure the manufacturing of trusted PCR products (24). Different indicators such as PCR buffer levels, the balance concerning magnesium chloride in addition to dNTP levels, the actual volumes of DNA, cycling conditions and Taq DNA polymerase may increase the success rate of multiplex PCR assay (25, 26).
In this study, standard bacteria were used and the selected primers were specifically able to identify their virulence genetic agents. The size of primers was selected in a way that the fragments with sizes of 160, 300 and 210 bp could be easily provided on agarose and polyacrylamide gel systems. This method allows a rapid detection of genome, the preparation of reaction mix-ture, the performance of rapid PCR cycles, electropho-resis (containing ethidium bromide) and the investiga-tion of results in less than 1.5 hr.
In this study, the gene encoding Listeriolysin O (hly gene, 210 bp) was used to prepare the strain specific primers and to identify L. monocytogenes, the oxido-reductase gene was also used to identify C. jejuni (160 bp), and the NheB/C gene (300 bp) was applied for B. cereus.
In a study conducted by Gilbert et al (2003), the specificity for the target bacteria (C. jejuni) was examined that was lower than the specificity achieved in this study (27). In another study, the specificity reported for the target bacteria (L. monocytogenes) was lower than the specificity achieved in this study (28). In a similar study performed in 2010, the sensitivity of two bacterial agents in foodstuffs was evaluated, in which the sensitivity was compared to this method (29). In another study on the identification of B. cereus, an acceptable specificity were obtained (30). In another study, a sensitivity of identifying microorganisms with 20 pg was reported that was less than the sensitivity of our study (31). Also, the results of a study showed that detection limit of real time PCR for C. jejuni in food samples was lower than Multiplex PCR in our study (detection limit was 4.3 CFU/ml) (32). In another study, the reported detection limit for L. monocytogenes in food samples was also fewer than this work (33).
Conclusion
Based on the results of this study, Multiplex PCR can be used for the rapid and simultaneous detection of L. monocytogenes, C. jejuni and B. cereus in food samples. Given the fact that these bacteria are world widely common in food poisoning epidemics, this research introduced an effective method for early, accurate and cost-effective detection of these bacteria that are responsible for the prevalence of food poisoning. This method can be considered as a suitable identification method for epidemiologic tests.
Acknowledgment
The authors of this article appreciate the cooperation of Tehran University of Medical Sciences and Comprehensive Imam Hossein University.
References
- 1.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, et al. Food-related illness an d death in the United States. Emerg Infect Dis. 1999;5:607. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tauxe RV. Emerging foodborne pathogens. Int J Food Microbiol. 2002;78:31–41. doi: 10.1016/s0168-1605(02)00232-5. [DOI] [PubMed] [Google Scholar]
- 3.Jasson V, Jacxsens L, Luning P, Rajkovic A, Uyttendaele M, et al. Alternative microbial methods:An overview and selection criteria. Food Microbiol. 2010;27:710–730. doi: 10.1016/j.fm.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Singh A, Poshtiban S, Evoy S. Recent advances in bacteriophage based biosensors for food-borne pathogen detection. Sensors. 2013;13:1763–1786. doi: 10.3390/s130201763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calvo J, Zaragoza P, Osta R. Technical note:A quick and more sensitive method to identify pork in processed and unprocessed food by PCR amplification of a new specific DNA fragment. J Anim Sci. 2001;79:2108–2112. doi: 10.2527/2001.7982108x. [DOI] [PubMed] [Google Scholar]
- 6.Gilmore MS, Cruz-Rodz AL, Leimeister-Wächter M, Kreft J, Goebel W, et al. A Bacillus cereus cytolytic determinant, cereolysin AB, which comprises the phospholipase C and sphingomyelinase genes:nucleotide sequence and genetic linkage. J Bacteriol. 1989;171:744–753. doi: 10.1128/jb.171.2.744-753.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schoeni JL, Lee Wong AC. Bacillus cereus food poisoning and its toxins. J Food Protect. 2005;68:636–648. doi: 10.4315/0362-028x-68.3.636. [DOI] [PubMed] [Google Scholar]
- 8.Dierick K, Van Coillie E, Swiecicka I, Meyfroidt G, Devlieger H, Meulemans A, et al. Fatal family outbreak of Bacillus cereus-associated food poisoning. J Clin Microbiol. 2005;43:4277–4279. doi: 10.1128/JCM.43.8.4277-4279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Te Giffel M, Beumer RR, Granum PE, Rombouts FM. Isolation and characterisation of Bacillus cereus from pasteurised milk in household refrigerators in the Netherlands. Int J Food Microbiol. 1997;34:307–318. doi: 10.1016/s0168-1605(96)01204-4. [DOI] [PubMed] [Google Scholar]
- 10.Valero M, Hernandez-Herrero L, Giner M. Survival, isolation and characterization of a psychrotrophic Bacillus cereus strain from a mayonnaise-based ready-to-eat vegetable salad. Food Microbiol. 2007;24:671–677. doi: 10.1016/j.fm.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Novak JS, Call J, Tomasula P, Luchansky JB. An assessment of pasteurization treatment of water, media, and milk with respect to Bacillus spores. J Food Protect. 2005;68:751–757. doi: 10.4315/0362-028x-68.4.751. [DOI] [PubMed] [Google Scholar]
- 12.Rahimi E, Abdos F, Momtaz H, Baghbadorani ZT, Jalali M. Bacillus cereus in infant foods:prevalence study and distribution of enterotoxigenic virulence factors in Isfahan Province, Iran. ScientificWorldJournal. 2013;2013:292571. doi: 10.1155/2013/292571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trends E. Sources of zoonoses and zoonotic agents and food-borne outbreaks in the European Union in 2008. EFSA J. 2010;8:1496. [Google Scholar]
- 14.Tam CC, Higgins CD, Neal KR, Rodrigues LC, Millership SE, O’Brien SJ, et al. Chicken consumption and use of acid-suppressing medications as risk factors for Campylobacter enteritis England. Emerg Infect Dis. 2009;15:1402. doi: 10.3201/eid1509.080773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassanzadeh P, Motamedifar M. Occurrence of Campylobacter jejuni in Shiraz, southwestIran. Med Princ Pract. 2007;16:59–62. doi: 10.1159/000096142. [DOI] [PubMed] [Google Scholar]
- 16.Khoshbakht R, Tabatabaei M, Hosseinzadeh S, Shekarforoush SS, Aski HS. Distribution of nine virulence-associated genes in Campylobacter jejuni and C. coli isolated from broiler feces in Shiraz, Southern Iran. Foodborne Pathog Dis. 2013;10:764–770. doi: 10.1089/fpd.2013.1489. [DOI] [PubMed] [Google Scholar]
- 17.Ghorbanalizadgan M, Bakhshi B, Kazemnejad Lili A, Najar-Peerayeh S, Nikmanesh B. A molecular survey of Campylobacter jejuni and Campylobacter coli virulence and diversity. Iran Biomed J. 2014;18:158. doi: 10.6091/ibj.1359.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamidian M, Sanaei M, Bolfion M, Dabiri H, Zali MR, Walther-Rasmussen J. Prevalence of putative virulence markers in Campylobacter jejuni and Campylobacter coli isolated from hospitalized children, raw chicken, and raw beef in Tehran Iran. Can J Microbiol. 2011;57:143–148. doi: 10.1139/w10-089. [DOI] [PubMed] [Google Scholar]
- 19.Hosseinzadeh S, Shekarforoush SS, Ansari-Lari M, EsalatPanah-Fard Jahromi M, Berizi E, Abdollahi M. Prevalence and risk factors for listeria monocytogenes in broiler flocks in Shiraz, southern Iran. Foodborne Pathog Dis. 2012;9:568–572. doi: 10.1089/fpd.2011.1080. [DOI] [PubMed] [Google Scholar]
- 20.Burke D, Dawson D, Stearns T. Methods in yeast genetics:a Cold Spring Harbor Laboratory course manual 2000. Cold Spring Harbor Laboratory Course Manual Press [Google Scholar]
- 21.Choma C, Guinebretière MH, Carlin F, Schmitt P, Velge P, Granum PE, et al. Prevalence, characterization and growth of Bacillus cereus in commercial cooked chilled foods containing vegetables. J Appl Microbiol. 2000;88:617–625. doi: 10.1046/j.1365-2672.2000.00998.x. [DOI] [PubMed] [Google Scholar]
- 22.Kathariou S. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J Food Protect. 2002;65:1811–1829. doi: 10.4315/0362-028x-65.11.1811. [DOI] [PubMed] [Google Scholar]
- 23.Chen HM, Lin CW. Hydrogel-coated streptavidin piezoelectric biosensors and applications to selective detection of strep-tag displaying cells. Biotechnol Progress. 2007;23:741–748. doi: 10.1021/bp060328h. [DOI] [PubMed] [Google Scholar]
- 24.Zhang MJ, Qiao B, Xu XB, Zhang JZ. Advances in rapid detection methods for foodborne pathogens. J Microbiol Biotechnol. 2014;24:297–312. doi: 10.4014/jmb.1310.10013. [DOI] [PubMed] [Google Scholar]
- 25.Khoo CH, Cheah YK, Lee LH, Sim JH, Salleh NA, Sidik SM, et al. Virulotyping of Salmonella enterica subsp. enterica isolated from indigenous vegetables and poultry meat in Malaysia using multiplex-PCR. Antonie van Leeuwenhoek. 2009;96:441–457. doi: 10.1007/s10482-009-9358-z. [DOI] [PubMed] [Google Scholar]
- 26.Cheah YK, Salleh NA, Lee L-H, Radu S, Sukardi S, Sim JH. Comparison of PCR fingerprinting techniques for the discrimination of Salmonella enterica subsp. enterica serovar Weltevreden isolated from indigenous vegetables in Malaysia. World J Microbiol Biotechnol. 2008;24:327–335. [Google Scholar]
- 27.Gilbert C, Winters D, O’Leary A, Slavik M. Development of a triplex PCR assay for the specific detection of Campylobacter jejuni Salmonella spp. and Escherichia coli O157:H7. Mol Cell Probes. 2003;17:135–138. doi: 10.1016/s0890-8508(03)00043-4. [DOI] [PubMed] [Google Scholar]
- 28.Germini A, Masola A, Carnevali P, Marchelli R. Simultaneous detection of Escherichia coli O175:H7, Salmonella spp., and Listeria monocytogenes by multiplex PCR. Food Control. 2009;20:733–738. [Google Scholar]
- 29.Kumar TK, Murali H, Batra H. Simultaneous detection of pathogenic B. cereus, S. aureus a nd L. monocytogenes by multiplex PCR. Indian J Microbiol. 2009;49:283–289. doi: 10.1007/s12088-009-0032-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wiwat C, Thiramanas R. Detection of Hemolysin BL Gene of Bacillus cereus Isolates. Mahidol Univ J Pharm Sci. 2014;41:22–30. [Google Scholar]
- 31.Forghani F, Singh P, Seo KH, Oh DH. A novel pentaplex real time (RT)-PCR high resolution melt curve assay for simultaneous detection of emetic and enterotoxin producing Bacillus cereus in food. Food Control. 2016;60:560–568. [Google Scholar]
- 32.Zhang MJ, Qiao B, Xu XB, Zhang JZ. Development and application of a real-time polymerase chain reaction method for Campylobacter jejuni detection. World J Gastroenterol. 2013;19:3090. doi: 10.3748/wjg.v19.i20.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gouws PA, Liedemann L. Evaluation of diagnostic PCR for the detection of Listeria monocytogenes in food products. Food Technol Biotechnol. 2005;43:201–205. [Google Scholar]


