Abstract
Redox regulation is of great importance in chloroplasts. Many chloroplast enzymes, such as those belonging to the Calvin-Benson cycle (CBC), have conserved regulatory cysteines which form inhibitory disulphide bridges when physiological conditions become unfavourable. Amongst these enzymes, cFBP1, the CBC fructose-1,6-bisphosphatase (FBPase) isoform, is well known to be redox activated by thioredoxin f through the reduction of a disulphide bridge involving Cys153 and Cys173. Moreover, data obtained during recent years point to S-nitrosylation as another redox post-translational modification putatively regulating an increasing number of plant enzymes, including cFBP1. In this study we have shown that the Pisum sativum cFBP1 can be efficiently S-nitrosylated by GSNO and SNAP, triggering the formation of the regulatory disulphide. Using in vivo experiments with P. sativum we have established that cFBP1 S-nitrosylation only occurs during the light period and we have elucidated by activity assays with Cys-to-Ser mutants that this enzyme may be inactivated through the S-nitrosylation of Cys153. Finally, in the light of the new data, we have proposed an extended redox-regulation model by integrating the S-nitrosylation and the TRX f-mediated regulation of cFBP1.
Abbreviations: GSNO, S-nitrosoglutathione; SNAP, S-nitroso-N-acetyl-D,L-penicillamine
Keywords: S-nitrosylation; GSNO; Redox regulation; Fructose-1,6-bisphosphatase; Pisum sativum; Calvin-Benson cycle
Highlights
-
•
Only the reduced form of cFBP1 is S-nitrosylated both in vitro and in vivo.
-
•
Cys153 is the target for S-nitrosylation.
-
•
S-nitrosylation triggers the formation of the regulatory disulphide bridge.
1. Introduction
Metabolic plasticity is essential for plant survival. Regarding enzyme activity, post-translational regulation (PTM) is thought to be the fastest regulation level devoted to keeping an optimum environmental adaptation; nevertheless, PTMs can also affect protein sub-cellular localizations, protein-protein interactions, and protein half-life.
Cysteine is frequently observed in functionally important sites of proteins, where it serves catalytic, regulatory, structure-stabilizing, cofactor-binding, and other functions. Due to its redox properties, its sulphur-based functional group is a target for reversible PTMs, including sulfenic acid (Cys-SOH), disulphide bonds (S-S), glutathionylation, and S-nitrosylation (Cys-SNO). Sulfenic acids are frequently the intermediate of thiol-modifications, reacting with neighbouring cysteines to form a disulphide bridge. On the other hand, disulphide bonds can be reduced back to thiols by thioredoxins (TRXs) [1], which are small redox enzymes with a 2-Cys active site and are important players in redox regulation [2].
Redox regulation has an essential role in plant CO2 assimilation. During the day, four Calvin-Benson cycle (CBC) enzymes are reported to be active and hence regulatory cysteines are in the form of thiols, kept oxidized as disulphide bridges at night. However, proteomic analyses have suggested that the whole CBC is redox-regulated through mechanisms which probably involve cysteine residues [3], [4]. In addition, due to their high reactivity and chemical plasticity, cysteines can undergo sequential oxidative PTMs and, during the last decade, gathered evidence suggests that CBC enzymes maybe regulated by multiple redox PTMs, i.e. S-glutathionylation and S-nitrosylation [5]. In recent years, protein S-nitrosylation has been established as an important way via which NO handles its global cellular influence, and as a broad-based mechanism for the post-translational regulation of the main classes of proteins and metabolic pathways [6], [7]. In fact, S-nitrosylated proteins cover nine of the fourteen steps in the CBC in Arabidopsis thaliana [8].
One key CBC enzyme is fructose-1,6-bisphosphatase (FBPase, EC 3.1.3.11), which converts fructose-1,6-bisphosphate into fructose-6-phosphate and Pi. FBPases are homotetrameric enzymes with three different isoforms present in plants, two in chloroplasts (cFBP1 and cFBP2) and one in the cytosol (cyFBP) [9], [10], [11]. Only cFBP1 needs to be redox activated in order to be fully active, whilst cFBP2 is not redox regulated and, despite its activity, resists higher oxidant concentrations than cFBP1, although no physiological function has been described so far [9]. In cFBP1, Cys responsible for redox regulation are Cys153 and Cys173 (pea FBPase numbering) [12]. These cysteines, together with Cys178, take part of a redox domain called the 170's loop, whose tertiary structure has a great influence over the enzyme activity. Once the disulphide bridge has been formed, the protein undergoes structural changes provoking a decrease in the affinity for its cofactor Mg2+, leading to the enzyme inactivation [13], [14]. Although cFBP1 has five more cysteines, no in vivo redox regulation has been attributed so far. Nevertheless, Cys178 has been reported to form a regulatory disulphide bridge in vitro with Cys153 when Cys173 is replaced by a Ser [15]. During the light period, CBC FBPase is activated by TRX f, receiving electrons from ferredoxin (Fd) through ferredoxin-thioredoxin reductase (FTR) [2].
Recently, some authors have proposed chloroplastic FBPase to be S-nitrosylated by S-nitrosoglutathione (GSNO) [16]. In this study we have depicted the GSNO-mediated S-nitrosylation process by (i) identifying the GSNO-targeted cysteine, (ii) analysing how this PTM could affect FBPase activity in vivo, and (iii) propose an extended cFBP1 redox regulation which takes into account the new data.
2. Material and methods
2.1. Plant material
Pisum sativum plants were grown in soil in a light/dark cycle of 16 h /8 h at 22 °C and with a photosynthetically active radiation of 120 μmol photons m−2 s−1.
2.2. Protein purification, directed mutagenesis, and FBPase activity after GSNO treatments
Coding sequences for the mature proteins were amplified by PCR and cloned into the expression vector pET-28b, which adds a His-tag to the N-terminal end of the polypeptide. Recombinant proteins were purified by Ni2+ affinity chromatography (GE Healthcare Life Sciences) according to the manufacturer's instructions. Site-directed mutagenesis was performed by the primer extension method [17]. The primers used for cloning and mutagenesis are listed in Supplementary Table 1.
FBPase activity was determined as previously described by Rojas-González and co-workers [10]. Briefly, 2 μg of FBPase were reduced in a solution containing 100 mM Tris-HCl (pH 8.0) and 50 mM DTT for 30 min. Next, the reducing agent was removed by gel filtration through a Micro Bio-Spin 6 column (BioRad) and the enzyme was then incubated in the dark, for 30 min, in a buffer containing 100 mM Tris–HCl (pH 8.0) and the GSNO concentration specified for each experiment. The assay was done in microtiter plates in a final volume of 200 μl, containing the following components: 2 μg of FBPase, 100 mM Tris-HCl buffer pH (8.0), 1 mM (low Mg2+) or 10 mM (high Mg2+) MgCl2, 0.6 mM fructose-1,6-bisphosphate, 0.3 mM NADP+, 0.7 U phosphoglucose isomerase (ROCHE), and 0.3 U glucose-6-phosphate dehydrogenase (ROCHE). The increase of absorbance at 340 nm (NADPH formation) versus time was read in a microplate reader (Tecan Sunrise™). Incubations and activity assays were done at room temperature (26–27 °C).
2.3. S-nitrosylation assays and biotin switch
Treatment with the reducing agent DTT (50 mM) was carried out for 30 min at room temperature in a solution buffered at pH 8.0 with 100 mM Tris-HCl. After that, the reducing agent was removed by gel filtration through a Micro Bio-Spin 6 column (BioRad). Proteins were incubated with the S-nitrosylating agent GSNO, or S-nitroso-N-acetyl-D,L-penicillamine (SNAP) when indicated, in the dark at room temperature for 30 min. The protein was then incubated with 20 mM methyl-methanethiosulfate (MMTS) and 2.5% SDS at 50 °C for 30 min with frequent vortexing to block free cysteines. Excess MMTS was removed by precipitation with two volumes of cold acetone, and the proteins were solubilised in RB buffer (25 mM HEPES, 1 mM EDTA, and 1% SDS, pH 7.7). After the addition of 1 mM HPDP-biotin (Pierce Protein Biology) and 1 mM ascorbic acid, the mixture was incubated for 1 h at room temperature in the dark with intermittent vortexing. The proteins were then subjected to western blotting analysis using a monoclonal anti-biotin antibody (Sigma-Aldrich).
For the in vivo S-nitrosylation assays, P. sativum leaves were covered with aluminium foil for 2 h, to protect them from light (dark condition), or left uncovered (light condition). Then, leaves were infiltrated with 1 mM GSNO and left for an additional hour under their respective light conditions. Finally, protein of infiltrated leaves was extracted in a cold buffer containing 50 mM HEPES (pH 7.7), 1 mM EDTA, and 0.1% Triton X-100 at a 1:2 ratio (w/v). After centrifugation (11,000g for 20 min at 4 °C), the protein content of the supernatant was determined with the Bradford assay (Bio-Rad), using bovine serum albumin as the standard [18]. Biotin switch was carried out as described before with 1 mg of extracted pea leaves protein.
2.4. Protein extraction methods
Total soluble protein extraction form pea leaves was carried out by cold homogenizing buffer: 50 mM Tris-HCl (pH 7.9), 100 mM potassium acetate, 1 mM EDTA, 1 mM PMSF, and 20% glycerol (1:3 [w/v]). After 30 min of centrifugation at 12,000g, the supernatant protein concentration was determined with the Bradford assay [18].
Before the in vivo GSNO treatments (described above in S-nitrosylation assays and biotin switch”) and in order to establish proper experimental conditions for the GSNO infiltration assay, the redox state of FBPase was analysed in leaves incubated for 2 h under dark (disulphide bonded) or light (reduced cysteines) conditions. For that purpose, leaves were collected, immediately frozen in liquid nitrogen, and cysteine residues in proteins were alkylated in order to avoid unwanted oxidations. The buffer used for the extraction (1:3 [w/v]) contained 100 mM Tris-HCl (pH 8.0), 8 M urea, 1 mM EDTA, and 30 mM iodoacetamide (IAM) and the protein extraction solution was incubated for 1 h at 37 °C. After centrifugation (11,000g for 20 min at 4 °C), protein concentration of the supernatant was determined as indicated before and redox state of FBPase was analysed by western blotting.
2.5. Immunoprecipitation and western blotting
Immunoprecipitation with anti-FBPase antibodies [10] of biotinylated proteins (detailed in "S-nitrosylation assays and biotin switch”) from GSNO-infiltrated leaves was carried out by using the SureBeads™ Magnetic Bead Immunoprecipitation System (BioRad).
Electrophoresis in polyacrylamide gel was performed following Laemmli's protocol [19] (using non-reducing conditions), the proteins were then transferred to a nitrocellulose membrane, and western blotting was performed as in Towbin and co-workers [20]. Bound antibody was visualized with enhanced chemiluminescence (Clarity™ Western ECL Substrate, BioRad).
3. Results
3.1. Reduced FBPase undergoes a rapid oxidation in vitro
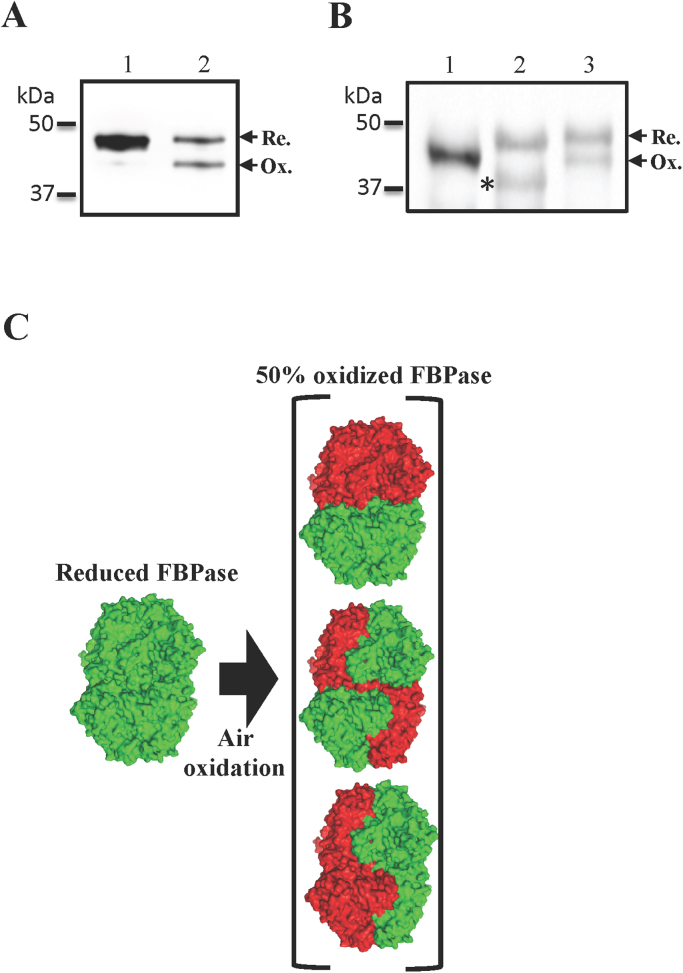
As we expected, cFBP1 needed a high DTT concentration to be fully reduced (higher than 40 mM, Supplementary Fig. 1A), in accordance with the negative values of the mid-point redox potentials (Em) reported in the literature [21]. Taking advantage of the mobility shift between the reduced and the oxidized polypeptides, the redox state of the protein was checked by non-reducing SDS-PAGE [22]. To achieve S-nitrosylation assays, the remaining DTT used for the cFBP1 reduction step had to be removed to avoid interference in the further steps. When we checked the reduction state after gel filtration, we observed that half of the total polypeptide was oxidized (Fig. 1A). A similar oxidation pattern was observed with FBPase in pea protein extracts after reduction and gel filtration (Fig. 1B). Curiously, upon reduction, another electrophoretic band not coincident with the oxidized polypeptide of non-reduced extracts was detected (Fig. 1B, asterisk).
Fig. 1.
Pea chloroplast FBPase oxidation under atmospheric conditions. A, western blot analysis of the in vitro oxidation of the recombinant FBPase. Recombinant cFBP1 was incubated in a solution with 100 mM Tris-HCl (pH 8.0) and 50 mM DTT. After a 30 min incubation at room temperature free cysteines were blocked by adding four volumes of an alkylating solution containing 100 mM Tris-HCl (pH 8.0), 2.7% CHAPS, and 40 mM IAM (lane 1) or after removing excess DTT by gel filtration before adding the alkylating solution (lane 2). 100 ng of the recombinant protein were loaded per lane. B, western blot analysis of the in vitro oxidation of the chloroplast FBPase present in pea extracts. Leaves soluble proteins (detailed extraction protocol in the Section 2) were incubated with DTT and free cysteines alkylated as described in panel A. 10 μg of total soluble protein were loaded per lane. 1, non-reduced protein extract; 2, DTT-incubated protein extract; 3, DTT-incubated protein extract after gel filtration. C, schematic model representing the pea FBPase oxidation exposed to atmospheric air. The green colour represents reduced subunits and the red colour represents oxidized subunits. The structures were prepared using PyMOL Molecular Graphics software (www.pymol.org). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To gain some insight into the redox pattern displayed by cFBP1, we carried out DTT titration experiments (Supplementary Fig. 2A) at pH 8. Curiously, in addition to the expected reduced and oxidized polypeptides, transitory weak signals were evident below the oxidized polypeptide in the titration curves. Disulphide bridge formations involving cysteines of the regulatory redox domain were not the responsible for these signals as they were also observed with the mutants C153S and C153S C173S C178S, lacking one or the three cysteines, respectively, of the redox 170's loop (Supplementary Fig. 2A). As we suspected, after quantifying and plotting the reduced polypeptide fraction we obtained a two-component sigmoid curve (Supplementary Fig. 2B). We were certain of specifically titrating the redox potential of the disulphide Cys153-Cys173 (arrowed as “Oxid.” in Supplementary Fig. 2A) as we did not observe the corresponding polypeptide in C153S nor in C153S C173S C178S. The two calculated Em at pH 8.0 (about −378 mV and −319 mV) were in line with the previously reported estimations for spinach and pea FBPases (−325 mV and −270 mV at pH 6.5 and −350 mV at pH 8.0, respectively) [21]. This result suggests that half of the subunits of the P. sativum FBPase were more prone to oxidation, at least under our experimental conditions. According to our PDB model (Supplementary Fig. 3), Cys153 and Cys173 were closer in subunits A and D (4.6 Å and 7.0 Å, respectively) than in subunits B and C (9.6 Å and 11.9 Å, respectively). This interesting behaviour could be explained on the basis of a structural asymmetry in the CBC FBPase, as has already been reported by some authors [23].
3.2. Chloroplast FBPase is strongly S-nitrosylated in its reduced state
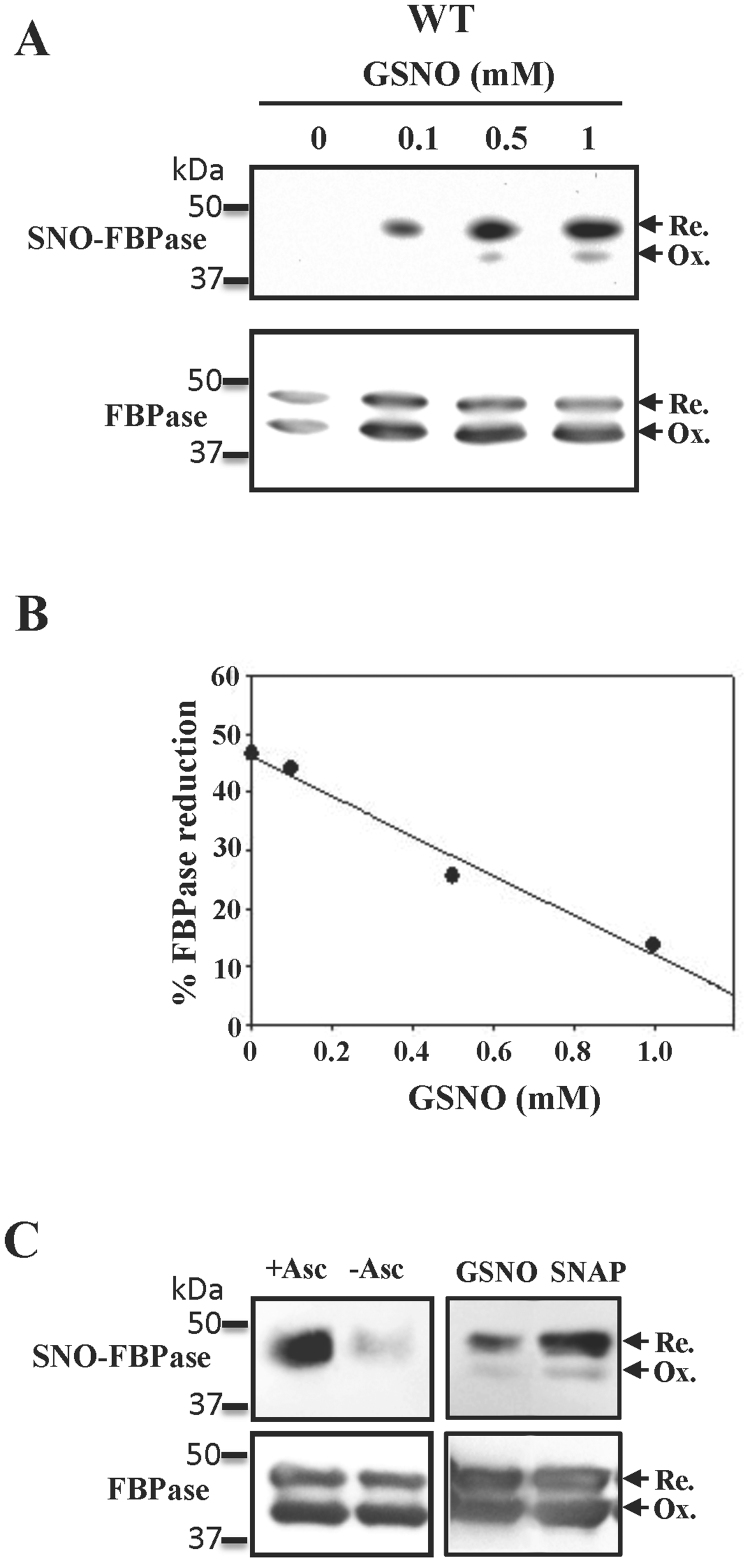
Chloroplast FBPase has been proposed to be S-nitrosylated in P. sativum [16] and A. thaliana [8]. Nevertheless, information regarding cysteine or cysteine residues involved in the S-nitrosylation process was lacking and the nature of the putative regulation still unknown. To answer these questions, we performed S-nitrosylation experiments with increasing GSNO concentrations and by using the biotin-switch method, which introduces a biotin moiety on cysteines that have been S-nitrosylated [24]. Our results confirmed that pea cFBP1 was S-nitrosylated in a GSNO concentration manner (Fig. 2A). Taking advantage of the presence of the two redox states of FBPase resulting after the gel filtration (Fig. 1A), we were able to correlate the strong biotinylation signal which appeared and the reduced state of the polypeptide. Conversely, a comparatively much weaker signal was associated to the oxidized polypeptide (Fig. 2A). This result strongly suggests that Cys153 and/or Cys173 could be efficiently S-nitrosylated by GSNO. However, although much less efficiently, other Cys residues could also be S-nitrosylated in vitro by using these GSNO concentrations as a faint signal corresponding to the oxidized polypeptide was detected. SNAP, another S-nitrosothiol, also turned out to be an effective S-nitrosylating compound (Fig. 2C). As a negative control, the lack of ascorbic acid during the biotin-switch assay provoked a dramatic decrease in the biotinylation signal (Fig. 2C).
Fig. 2.
S-nitrosylation in vitro of the pea chloroplast FBPase. A, western blot analyses of the GSNO-dependent S-nitrosylation of cFBP1. B, Quantification of the FBPase signals shown in panel A (anti-cFBP1 antibodies). Reduced and oxidized polypeptides were quantified with the open-source image analysis software ImageJ. C, western blot analyses of the S-nitrosylation with SNAP and GSNO of FBPase together with the enhancing of the biotin switch reaction by incubating with ascorbic acid incubation (+ Asc) compared to non-incubated samples (−Asc). S-nitrosylation assays are described in the Section 2. Membranes were incubated with antibodies anti-biotin (SNO-FBPase) or anti-cFBP1 (FBPase). 100 ng of recombinant FBPase were loaded per lane.
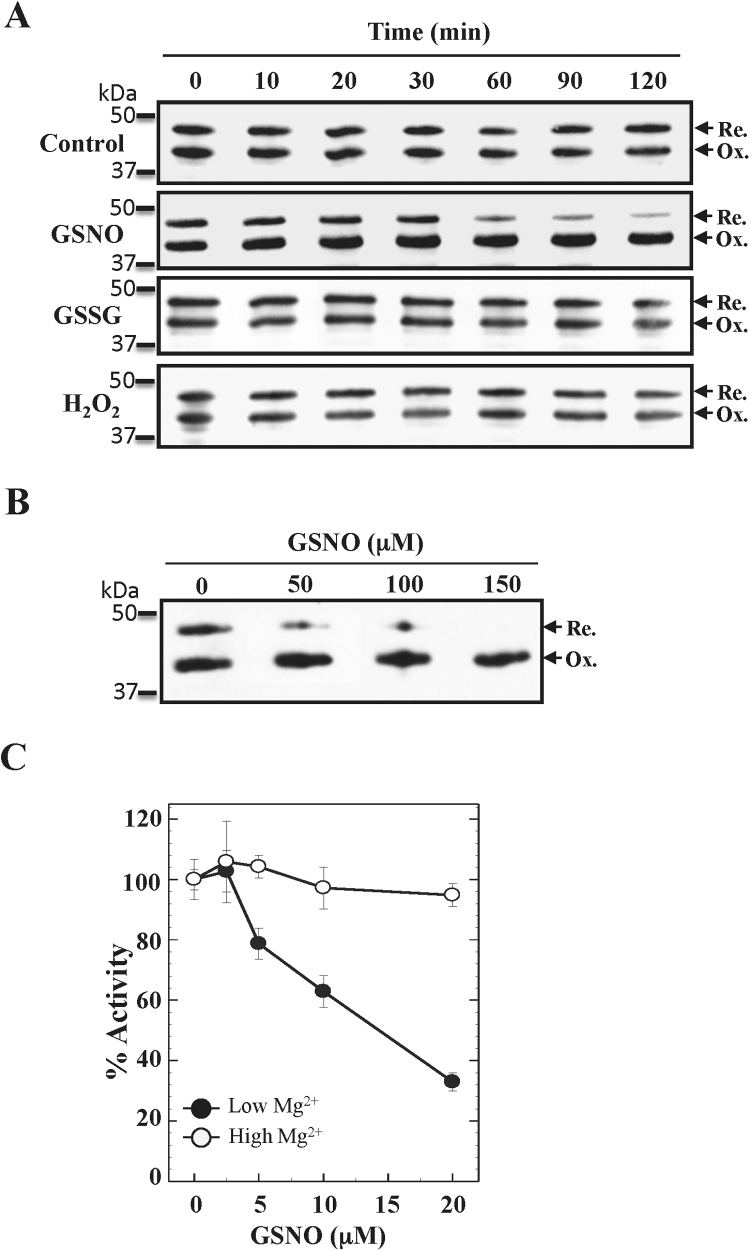
Interestingly, we observed a noticeable decrease in the reduced/oxidized ratio throughout the treatments. The quantification of the signals obtained supported this observation (Fig. 2B), suggesting the existence of a correlation between FBPase oxidation and the presence of GSNO. In order to shed more light on this issue, we carried out FBPase oxidation assays by incubating with equimolar quantities of GSNO and the physiological oxidizing compounds H2O2 and GSSG (Fig. 3A). The result of this assay proved that GSNO was the only compound promoting FBPase oxidation within our incubation times, reaching an almost complete oxidation 2 h after GSNO incubation. In addition, by increasing the GSNO concentration, we obtained similar oxidation results (Fig. 3B). The FBPase activities confirmed that GSNO incubation led to the formation of the regulatory disulphide Cys153-Cys173 (Fig. 3C) as it has been schematized in Fig. 4. It has been well established that this regulation only affects the affinity for the cofactor Mg2+, decreasing the FBPase activity at low concentrations of the cofactor [15], [25].
Fig. 3.
FBPase oxidation by GSNO incubation. A, western blot analyses of the FBPase oxidation after dark incubation with 0.1 mM of the oxidizing molecules GSNO, oxidized glutathione (GSSG), or hydrogen peroxide (H2O2) (in a solution with 100 mM Tris-HCl pH 8.0). No oxidizing compound was added to the control condition. Samples were taken at the indicated times and immediately incubated for 1 h at 37 °C with one volume of alkylating buffer (100 mM Tris-HCl [pH 8.0], 4% CHAPS, and 60 mM IAM). B, effect of GSNO concentration on FBPase oxidation. cFBP1 was incubated as in A, with the indicated GSNO concentrations for 2 h. 100 ng of recombinant FBPase were loaded per lane in A and B. C, inactivation of the FBPase activity following 30 min incubations with GSNO. Activity assays were carried out as described in the Section 2.
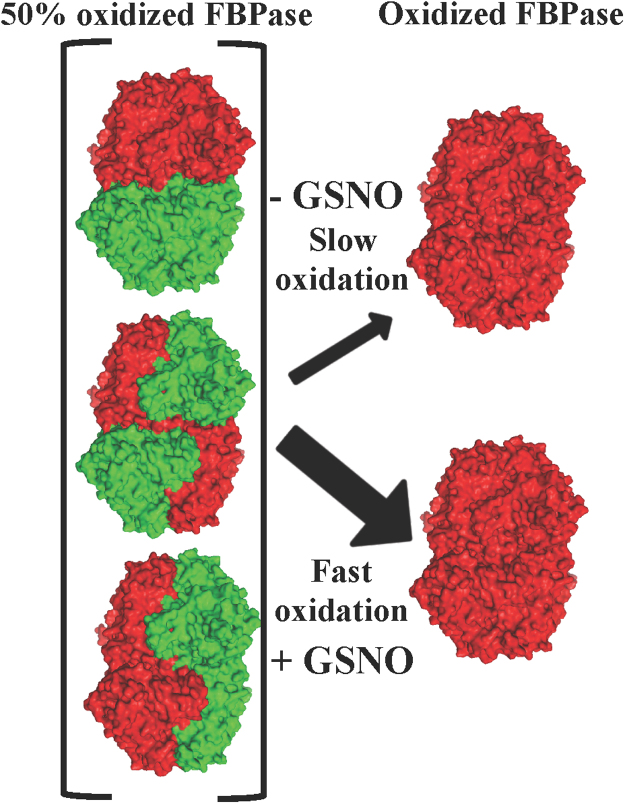
Fig. 4.
Schematic representation of the effect of GSNO on FBPase oxidation based on the in vitro S-nitrosylation assays. The green colour represents reduced subunits and the red colour represents oxidized subunits. The three possible combinations of oxidized and reduced subunits are shown in the 50% oxidized FBPase. The structures were prepared using PyMOL Molecular Graphics software (www.pymol.org). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Cys153 is efficiently S-nitrosylated
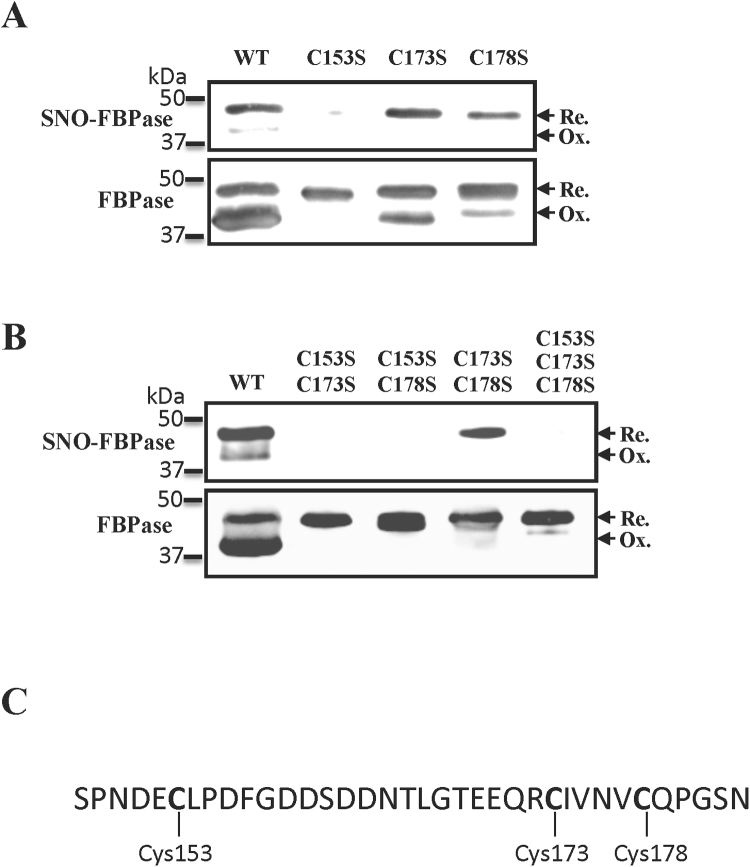
As we found reasonable evidence that regulatory cysteines were putative targets for S-nitrosylation, we obtained a set of FBPase mutants in order to corroborate our hypothesis. In our assays, the S-nitrosylation signal almost disappeared in the C153S mutant (Fig. 5A), with only a very faint signal remaining, evidencing that Cys153, and not Cys173, was the most efficiently S-nitrosylated residue. As has been previously stated by other authors, the existence of an oxidized polypeptide underlines the ability of Cys153 to form a disulphide bridge with Cys178 in the mutant C173S (Fig. 5A) [15]. The use of double and triple mutants reaffirmed the results obtained with the single mutants (Fig. 5B). These results indicate that Cys153 is the best candidate to in vivo undergo GSNO-mediated PTM. The presence of a glutamic residue before the S-nitrosylated Cys153 was in line with the Glu-Cys consensus sequence proposed by some authors [8] (Fig. 5C), the second more represented motif identified in the S-nitrosylated proteins found in this study.
Fig. 5.
S-nitrosylation of FBPase mutants in the Cys of the redox regulatory domain. A, S-nitrosylation assays with single mutants C153S, C173S, and C178S. B, S-nitrosylation assays with double (C153S C173S, C153S C178S, and C173S C178S) and triple (C153S C173S C178S) mutants. Experimental conditions were the same as the shown in the legend of Fig. 1. C, amino acid sequence of the redox regulatory domain indicating the mutated cysteines. S-nitrosylation assays are described in the Section 2. Membranes were incubated with antibodies anti-biotin (SNO-FBPase) or anti-cFBP1 (FBPase). 100 ng of recombinant FBPase were loaded per lane.
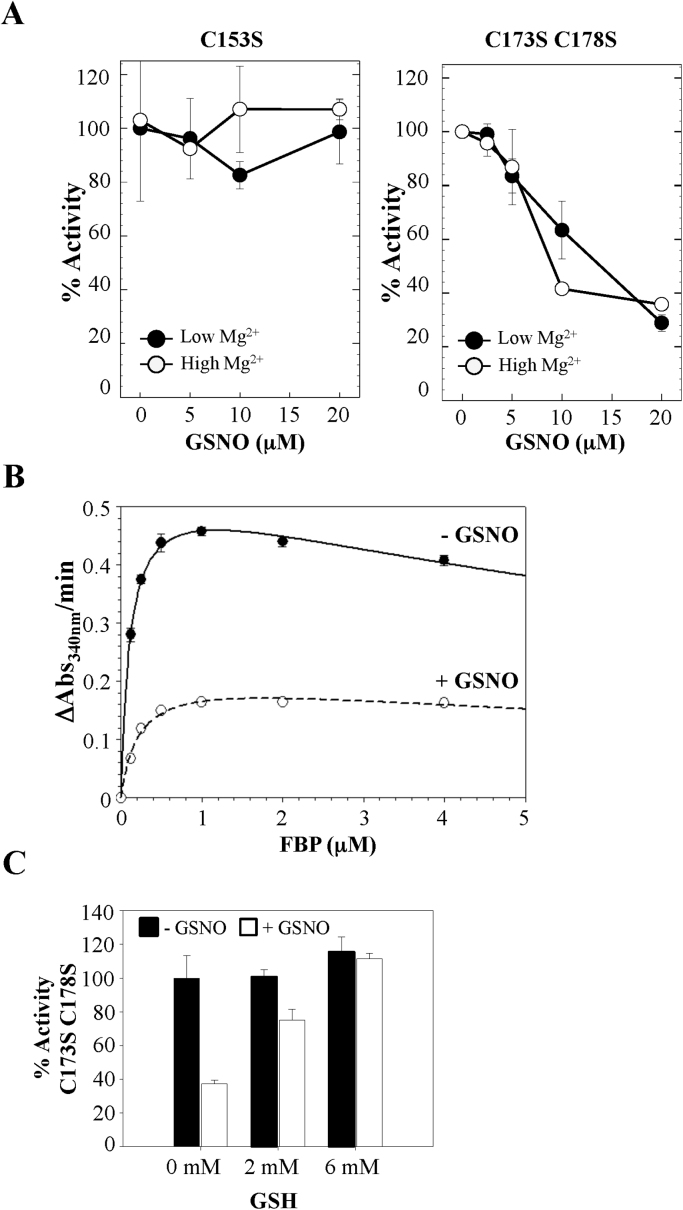
3.4. GSNO inhibits the FBPase activity in the mutant C173S C178S but not in C153S
We have demonstrated that Cys153 S-nitrosylation induces FBPase oxidation in a GSNO concentration manner (Fig. 2, Fig. 3). Besides promoting oxidation, we wondered whether S-nitrosylation affected per se the FBPase activity or was it only an inducing mechanism for the formation of the regulatory disulphide Cys153-Cys173. For this purpose, we carried out S-nitrosylation assays by using the mutants C173S C178S and C153S, unable to form the regulatory disulphides Cys153-Cys173 or Cys153-Cys178. As we expected, incubations with GSNO did not inhibit the activity of the mutant C153S (Fig. 6A). Nonetheless, treatments with 20 μM GSNO produced a 75% activity inhibition in the mutant C173S C178S, compared with the non-treated protein (Fig. 6A). Contrary to WT FBPase, where we only observed an activity decrease at low Mg2+ concentrations, the inhibition of C173S C178S was also observed at the highest Mg2+ concentration, pointing to a different inhibition mechanism. The calculation of the Km for the substrate did not show important differences in the affinity for FBP (0.11 mM ± 0.01 and 0.30 mM ± 0.05 for the non-treated and the GSNO-treated C173S C178S, respectively) but did show a significant loss of the catalytic efficiency in the S-nitrosylated enzyme (Fig. 6B). The results obtained with C153S and C173S C178S suggest that Cys153 S-nitrosylation was sufficient to explain FBPase inhibition. As S-nitrosylation is a reversible process, the incubation of the GSNO-treated C173S C178S with GSH fully recovered the FBPase activity (Fig. 6C) [26].
Fig. 6.
FBPase inhibition by S-nitrosylation of Cys153. A, FBPase activity assays of the FBPase mutants C153S and C173S C178S treated or non-treated with GSNO. B, Effect of GSNO treatment on the affinity of C173S C178S for the fructose-1,6-bisphosphate (FBP). Experimental conditions of panels A and B were the same as shown in the legend of Fig. 3. C, Activity recovery of GSNO-inhibited C173S C178S after incubation (30 min at room temperature) with reduced glutathione (GSH).
3.5. S-nitrosylation in vivo is only detected in the reduced FBPase
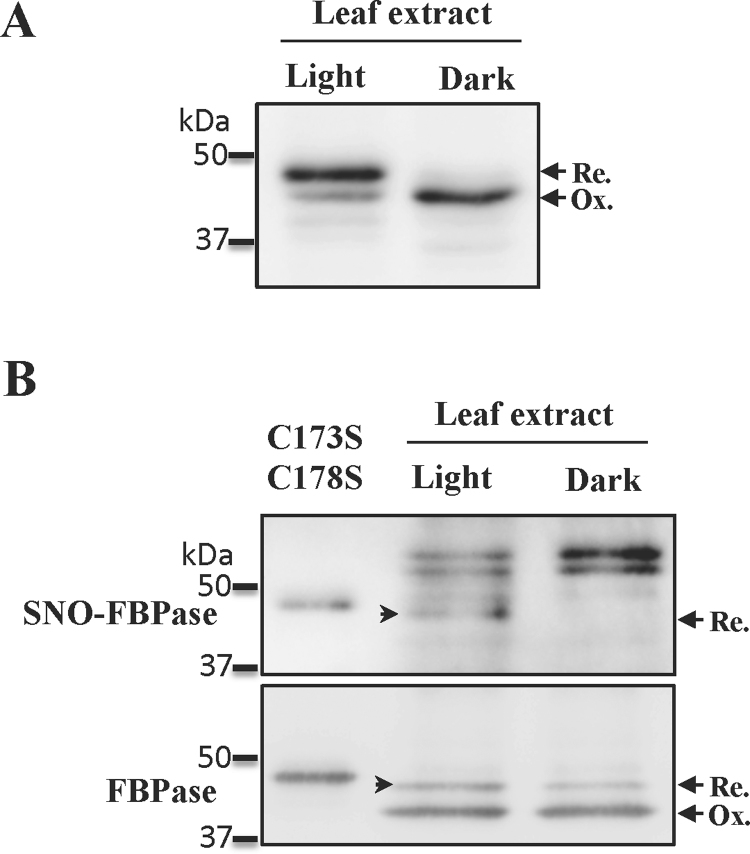
In order to demonstrate that chloroplast FBPase S-nitrosylation was targeting Cys153 in the regulatory 170's loop, according to our in vitro results, we carried out GSNO-infiltration assays in vivo. For that purpose, we took advantage of the different redox states of the CBC FBPase in light- and dark-incubated pea leaves (Fig. 7A). The biotinylated samples were used for pull-down assays with antibodies anti-FBPase and were analysed by western blotting to know whether the FBPase had undergone S-nitrosylation. The results clearly showed (Fig. 7B, arrow) that only the reduced polypeptides were S-nitrosylated in vivo, with no FBPase-associated signal detected in dark-incubated leaves. As we expected, in contrast to the high reduction degree of FBPase observed in light-incubated leaves, after processing the protein extracts the enzyme became highly oxidized, highlighting yet again its oxidation sensitivity. Two more polypeptides with a higher apparent molecular mass were also detected. These signals could originate from the cross-reaction between the mouse anti-IgG antibodies and the heavy chains of the rabbit IgG antibodies used in the pull-down assay. However, we cannot rule out the possibility that these polypeptides may correspond to co-immunoprecipitated S-nitrosylated proteins forming complexes with the CBC FBPase. Further proteomic analyses are necessary to answer this intriguing question.
Fig. 7.
In vivo S-nitrosylation of the chloroplast FBPase from light- and dark-incubated leaves. A, western blot analysis of the redox state of the chloroplast FBPase in light- or dark-incubated leaves (see Section 2). B, western blot analysis of the S-nitrosylation state of the chloroplast FBPase in light- or dark-incubated leaves treated with GSNO (see experimental details in the Section 2).
4. Discussion
cFBP1 is a key enzyme in the Calvin-Benson cycle, where the importance of the redox regulation mediated by the thiol/disulphide switch is well known. In this enzyme, Cys153 and Cys173 are the redox regulatory cysteines; nevertheless, Cys153 is the cysteine which is most exposed to the chloroplast stroma [27]. Additionally, some authors have suggested the formation, under some in vivo conditions, of a second disulphide bridge between Cys49 and Cys190 in the spinach chloroplast FBPase [23]. We have shown that the use of an oxidizing agent provoked the appearance of a double band corresponding to oxidized polypeptides (Supplementary Fig. 1B), underlying the existence of different oxidation states in vitro for pea cFBP1. The question is whether these in vitro observations have an in vivo regulatory/structural role or, if on the contrary, they are just redox artifacts caused by the experimental conditions used. Interestingly, the mutagenesis of Cys153 affects the redox features of C153S and C153S C173S C178S, as an oxidation band was observed close to the reduced polypeptide (Supplementary Fig. 2A). These results highlight the influence over the FBPase structure of the changes provoked in the regulatory redox domain. Due to the high number of cysteine residues present in cFBP1 (seven cysteines), disulphide isomerization would explain the band pattern observed in vitro when using the non-physiological reducer DTT. Knowing that TRX f is responsible for cFBP1 reduction, specifically the reduction of the disulphide bridge Cys153-Cys173, the occurrence of this complex redox pattern in vivo is rather unlikely. Moreover, we have never noticed anything similar when we have analysed the redox state of cFBP1 in plant extracts.
Regarding the rapid oxidation undergone by cFBP1 exposed to the atmospheric air, the two values of Em calculated for the regulatory disulphide suggest an asymmetric quaternary structure, in accordance with that reported for the crystallized FBPase from spinach [23]. The subunits with the higher electronegative Em (approx. −378 mV at pH 8.0) may suffer a rapid oxidation under air-oxidizing conditions upon the removal of DTT (Fig. 1C). When we repeated the assay by using pea protein extracts, we basically observed the same result; ruling out any artifact related to the redox regulation of the recombinant protein (Fig. 1A and B). If we suppose that the regulatory disulphides Cys153-Cys173 of the four FBPase monomers have identical redox potentials, the protein would have continued following a linear oxidation (with a positive slope if we drew a graph representing the oxidation degree versus time). On the contrary, the existence of different Cys153-Cys173 redox potentials within the tetramer would explain our experimental observations, that is, 50% of the polypeptides became oxidized in a matter of less than three minutes (the spin dialysis time) while the remaining polypeptides continued reduced for at least two more hours (Figs. 1A and 3A). This interesting in vitro behaviour leads us to wonder whether there is a physiological meaning for this biochemical feature in pea plants.
Throughout this study we have demonstrated that the chloroplast FBPase isoform cFBP1 is efficiently S-nitrosylated in vitro by using two different NO donors, GSNO and SNAP (Fig. 2C), and we have identified Cys153 as the central NO-targeted cysteine. According to our results, this PTM should only occur during the light period, as we have already shown in vivo (Fig. 7), when chloroplast FBPase is activated by TRX f. Although other cysteine residues could be also S-nitrosylated in vitro, the efficacy of the process was rather low and, in any case, it did not have any influence over the FBPase activity as we can see in Fig. 6A (activity of the protein C153S incubated with GSNO). We were not able to observe any denitrosylation activity by the pea TRX f, at least under the experimental conditions used in this study (Supplementary Fig. 4). Thus, all in all, it seems that TRX- and GSNO-mediated PTMs belong to independent but related signalling pathways converging at cFBP1 regulation. In addition to this enzyme, TRX f regulates other enzymes participating in different processes of the chloroplast. Multiple PTMs acting on TRX-targeted cysteines would modulate redox signalling to reach an optimum metabolic response to external stimuli, which would definitively have positive consequences on plant adaptation [28], [29]. Fine-tuning regulation of the metabolic enzymes is vital for maintaining optimum metabolite values, which constantly fluctuate with the changing environmental conditions. For instance, other CBC enzymes such as ribulose bisphosphate carboxylase (RUBISCO), RUBISCO activase, phosphoglycerate kinase, glyceraldehyde-3-phosphate dehydrogenase, triose phosphate isomerase, sedoheptulose-1,7-bisphosphatase, fructose bisphosphate aldolase, and ribose 5-phosphate isomerase have been reported to be S-nitrosylated [8]. The multiplicity of the incoming stimuli requires orchestrated signalling pathways, some of which could eventually converge at the same targets. For instance, some proteins can be redox regulated at the same time by TRXs (thiol/disulphide switch), glutathionylation, and S-nitrosylation in the photosynthetic organism Chlamydomonas reinhardtii [30]. Currently, we are just beginning to elucidate the complex redox network acting in plant cells.
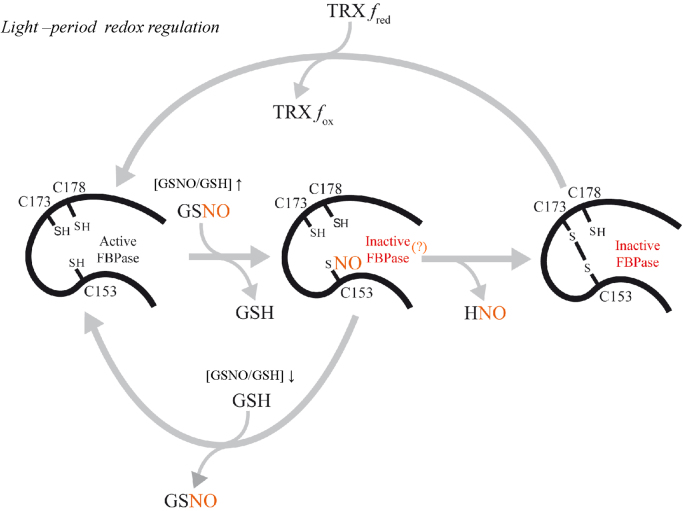
Since Cys153 is S-nitrosylated in the pea chloroplast FBPase, reduction by TRX f would be a sine qua non condition and GSNO-regulation would only affect the activated fraction of cFBP1. GSNO would trigger the formation in vitro of the regulatory disulphide in the wild-type protein (Fig. 3), as we were able to show an FBPase loss of activity at low but not at high Mg2+ concentrations. Apparently, the inhibition mechanism consisted of triggering the formation of the disulphide bridge more efficiently than other physiological oxidants such as H2O2 or GSSG. More than twenty years ago, acceleration of the disulphide bond formation by S-nitrosylation had been proposed as a mechanism to regulate protein function through the intermediacy of nitrosothiols [31]. However, with the aim of dissecting the inhibition mechanism, we performed activity assays with C173S C178S, which only contained Cys153 and were not able to form a regulatory disulphide (Fig. 6A). Our results show that an activity inhibition happened in the GSNO-treated enzyme, independently from the formation of a disulphide bridge. On the contrary, the mutant lacking Cys153 was insensitive to the GSNO treatments (Fig. 6A). Therefore, apart from the induction of the regulatory disulphide, S-nitrosylation provoked a different type of enzyme inactivation, at least in this FBPase mutant, as the activity was not recovered by increasing Mg2+ levels but only with a GSH treatment. The GSH/GSNO ratio in the stroma might be governing the balance between the reduction of the S-nitrosylated protein (high GSH/GSNO ratio) and the formation of the regulatory disulphide (low GSH/GSNO ratio) (Fig. 8). In plants, GSNO can be produced in response to different stress situations [32], for instance high light conditions [33]. Apart from the regulatory role in vivo proposed in this study (Fig. 8), we cannot rule out the possibility that S-nitrosylation may also have a protection function to avoid the irreversible over-oxidation of Cys153 provoked by elevated ROS levels resulting from oxidative stress situations as has been proposed for some human proteins [34], [35].
Fig. 8.
Proposed model showing the redox regulation carried out by TRX f and S-nitrosylation on the chloroplast FBPase. S-nitrosylation of Cys153 mediated by GSNO triggers the formation of the regulatory disulphide Cys153-Cys173 during the light period when FBPase is kept reduced by TRX f.
5. Conclusions
For many years, thiol/disulphide conversion has been the only redox regulatory mechanism known for cFBP1. Nevertheless, in addition to TRX f, S-nitrosylation may also play a complementary regulatory role exclusively limited to the light period. During the day, according to our results, cFBP1 could have three redox states: (i) reduced, (ii) S-nitrosylated and (iii) disulphide bonded. Interestingly, S-nitrosylation of Cys153 spontaneously leads to the formation of the regulatory disulphide bridge, suggesting a transient (and inhibiting) PTM occurring in vivo. S-nitrosylation of FBPase might have evolved as a physiological mechanism to fine-tune plant C metabolism. In this study, we have integrated S-nitrosylation together with the thiol/disulphide switch in the frame of the redox regulation governing cFBP1 activity.
Acknowledgements
The authors thank Trinidad Moreno for the technical support. This work has been funded by research project BIO2012-33292 and BIO2015-65272-C2-1-P, from the Spanish Ministry of Economy and Competitiveness. ALP has been supported by a contract from the CSIC.
Acknowledgments
Conflict of interest
The authors declare that there is no conflict of interest.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2017.10.008.
Contributor Information
Antonio Jesús Serrato, Email: aserrato@eez.csic.es.
María C. Romero-Puertas, Email: maria.romero@eez.csic.es.
Alfonso Lázaro-Payo, Email: alfonso.lazaro@eez.csic.es.
Mariam Sahrawy, Email: mariam.sahrawy@eez.csic.es.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Klomsiri C., Karplus P.A., Poole L.B. Cysteine-based redox switches in enzymes. Antioxid. Redox Signal. 2011;14:1065–1077. doi: 10.1089/ars.2010.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serrato A.J., Fernández-Trijueque J., Barajas-López J.D., Chueca A., Sahrawy M. Plastid thioredoxins: a "one-for-all" redox-signaling system in plants. Front. Plant Sci. 2013;4:463. doi: 10.3389/fpls.2013.00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemaire S.D., Michelet L., Zaffagnini M., Massot V., Issakidis-Bourguet E. Thioredoxins in chloroplasts. Curr. Genet. 2007;51:343–365. doi: 10.1007/s00294-007-0128-z. [DOI] [PubMed] [Google Scholar]
- 4.Lindahl M., Mata-Cabana A., Kieselbach T. The disulfide proteome and other reactive cysteine proteomes: analysis and functional significance. Antioxid. Redox Signal. 2011;14:2581–2642. doi: 10.1089/ars.2010.3551. [DOI] [PubMed] [Google Scholar]
- 5.Michelet L., Zaffagnini M., Morisse S., Sparla F., Pérez-Pérez M.E., Francia F., Danon A., Marchand C.H., Fermani S., Trost P., Lemaire S.D. Redox regulation of the Calvin–Benson cycle: something old, something new. Front. Plant Sci. 2013;4:470. doi: 10.3389/fpls.2013.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindermayr C., Durner J. S-Nitrosylation in plants: pattern and function. J. Proteom. 2009;73:1–9. doi: 10.1016/j.jprot.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Romero-Puertas M.C., Sandalio L.M. Nitric oxide level is self-regulating and also regulates Its ROS partners. Front. Plant Sci. 2016;7:316–320. doi: 10.3389/fpls.2016.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu J., Huang X., Chen L., Sun X., Lu C., Zhang L., Wang Y., Zuo J. Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2015;167:1731–1746. doi: 10.1104/pp.15.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serrato A.J., Yubero-Serrano E.M., Sandalio L.M., Muñoz-Blanco J., Chueca A., Caballero J.L., Sahrawy M. cpFBPaseII, a novel redox-independent chloroplastic isoform of fructose-1,6-bisphosphatase. Plant Cell Environ. 2009;32:811–827. doi: 10.1111/j.1365-3040.2009.01960.x. [DOI] [PubMed] [Google Scholar]
- 10.Rojas-González J.A., Soto-Suárez M., García-Díaz A., Romero-Puertas M.C., Sandalio L.M., Mérida A., Thormahlen I., Geigenberger P., Serrato A.J., Sahrawy M. Disruption of both chloroplastic and cytosolic FBPase genes results in a dwarf phenotype and important starch and metabolite changes in Arabidopsis thaliana. J. Exp. Bot. 2015;66:2673–2689. doi: 10.1093/jxb/erv062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soto-Suárez M., Serrato A.J., Rojas-González J.A., Bautista R., Sahrawy M. Transcriptomic and proteomic approach to identify differentially expressed genes and proteins in Arabidopsis thaliana mutants lacking chloroplastic 1 and cytosolic FBPases reveals several levels of metabolic regulation. BMC Plant Biol. 2016;16:258–274. doi: 10.1186/s12870-016-0945-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacquot J.-P., López-Jaramillo J., Miginiac-Maslow M., Lemaire S., Cherfils J., Chueca A., López-Gorgé J. Cysteine-153 is required for redox regulation of pea chloroplast fructose-1,6-bisphosphatase. FEBS Lett. 1997;401:143–147. doi: 10.1016/s0014-5793(96)01459-7. [DOI] [PubMed] [Google Scholar]
- 13.Chiadmi M., Navaza A., Miginiac-Maslow M., Jacquot J.P., Cherfils J. Redox signalling in the chloroplast: structure of oxidized pea fructose-1,6-bisphosphate phosphatase. EMBO J. 1999;18:6809–6815. doi: 10.1093/emboj/18.23.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai S., Schwendtmayer C., Johansson K., Ramaswamy S., Schürmann P., Eklund H. How does light regulate chloroplast enzymes? Structure-function studies of the ferredoxin/thioredoxin system. Q. Rev. Biophys. 2000;33:67–108. doi: 10.1017/s0033583500003607. [DOI] [PubMed] [Google Scholar]
- 15.Balmer Y., Stritt-Etter A.L., Hirasawa M., Jacquot J.P., Keryer E., Knaff D.B., Schürmann P. Oxidation-reduction and activation properties of chloroplast fructose-1,6-bisphosphatase with mutated regulatory site. Biochemistry. 2001;40:15444–15450. doi: 10.1021/bi011646m. [DOI] [PubMed] [Google Scholar]
- 16.Begara-Morales J.C., López-Jaramillo F.J., Sánchez-Calvo B., Carreras A., Ortega-Muñoz M., Santoyo-González F., Corpas F.J., Barroso J.B. Vinyl sulfone silica: application of an open preactivated support to the study of transnitrosylation of plant proteins by S-nitrosoglutathione. BMC Plant Biol. 2013;13:61–75. doi: 10.1186/1471-2229-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 18.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirasawa M., Schurmann P., Jacquot J.P., Manieri W., Jacquot P., Keryer E., Hartman F.C., Knaff D.B. Oxidation-reduction properties of chloroplast thioredoxins, ferredoxin: thioredoxin reductase, and thioredoxin f-regulated enzymes. Biochemistry. 1999;38:5200–5205. doi: 10.1021/bi982783v. [DOI] [PubMed] [Google Scholar]
- 22.Reichert A., Dennes A., Vetter S., Scheibe R. Chloroplast fructose 1,6-bisphosphatase with changed redox modulation: comparison of the Galdieria enzyme with cysteine mutants from spinach. Biochim. Biophys. Acta. 2003;1645:212–217. doi: 10.1016/s1570-9639(02)00539-3. [DOI] [PubMed] [Google Scholar]
- 23.Villeret V., Huang S., Zhang Y., Xue Y., Lipscomb W.N. Crystal structure of spinach chloroplast fructose-1,6-bisphosphatase at 2.8 A resolution. Biochemistry. 1995;34:4299–4306. doi: 10.1021/bi00013a019. [DOI] [PubMed] [Google Scholar]
- 24.Romero-Puertas M.C., Campostrini N., Mattè A., Righetti P.G., Perazzolli M., Zolla L., Roepstorff P., Delledonne M. Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics. 2008;8:1459–1469. doi: 10.1002/pmic.200700536. [DOI] [PubMed] [Google Scholar]
- 25.Chardot T., Meunier J.C. Properties of oxidized and reduced spinach (Spinacia oleracea) chloroplast fructose-1,6-bisphosphatase activated by various agents. Biochem. J. 1991:787–791. doi: 10.1042/bj2780787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh S.P., Wishnok J.S., Keshive M., Deen W.M., Tannenbaum S.R. The chemistry of the S-nitrosoglutathione/glutathione system. Proc. Natl. Acad. Sci. USA. 1996;93:14428–14433. doi: 10.1073/pnas.93.25.14428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balmer Y., Schurmann P. Heterodimer formation between thioredoxin f and fructose 1,6-bisphosphatase from spinach chloroplasts. FEBS Lett. 2001;492:58–61. doi: 10.1016/s0014-5793(01)02229-3. [DOI] [PubMed] [Google Scholar]
- 28.Silveira N.M., Hancock J.T., Frungillo L., Siasou E., Marcos F.C.C., Salgado I., Machado E.C., Ribeiro R.V. Evidence towards the involvement of nitric oxide in drought tolerance of sugarcane. Plant Physiol. Biochem. 2017;115:354–359. doi: 10.1016/j.plaphy.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 29.Silveira N.M., Marcos F.C.C., Frungillo L., Moura B.B., Seabra A.B., Salgado I., Machado E.C., Hancock J.T., Ribeiro R.V. S-nitrosoglutathione spraying improves stomatal conductance, Rubisco activity and antioxidant defense in both leaves and roots of sugarcane plants under water deficit. Physiol. Plant. 2017;160:383–395. doi: 10.1111/ppl.12575. [DOI] [PubMed] [Google Scholar]
- 30.Zaffagnini M., De Mia M., Morisse S., Di Giacinto N., Marchand C.H., Maes A., Lemaire S.D., Trost P. Protein S-nitrosylation in photosynthetic organisms: a comprehensive overview with future perspectives. Biochim. Biophys. Acta. 2016;1864:952–966. doi: 10.1016/j.bbapap.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 31.Arnelle D.R., Stamler J.S. NO+, NO, and NO- donation by S-nitrosothiols: implications for regulation of physiological functions by S-nitrosylation and acceleration of disulfide formation. Arch. Biochem. Biophys. 1995;318:279–285. doi: 10.1006/abbi.1995.1231. [DOI] [PubMed] [Google Scholar]
- 32.Kovacs I., Lindermayr C. Nitric oxide-based protein modification: formation and site-specificity of protein S-nitrosylation. Front. Plant Sci. 2013;4:137. doi: 10.3389/fpls.2013.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin A., Wang Y., Tang J., Xue P., Li C., Liu L., Hu B., Yang F., Loake G.J., Chu C. Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol. 2012;158:451–464. doi: 10.1104/pp.111.184531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sevilla F., Camejo D., Ortiz-Espín A., Calderón A., Lázaro J.J., Jiménez A. The thioredoxin/peroxiredoxin/sulfiredoxin system: current overview on its redox function in plants and regulation by reactive oxygen and nitrogen species. J. Exp. Bot. 2015;66:2945–2955. doi: 10.1093/jxb/erv146. [DOI] [PubMed] [Google Scholar]
- 35.Sun J., Steenbergen C., Murphy E. S-nitrosylation: no-related redox signaling to protect against oxidative stress. Antioxid. Redox Signal. 2006;8:1693–1705. doi: 10.1089/ars.2006.8.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material