Abstract
Myelin imaging is a form of quantitative magnetic resonance imaging (MRI) that measures myelin content and can potentially allow demyelinating diseases such as multiple sclerosis (MS) to be detected earlier. Although focal lesions are the most visible signs of MS pathology on conventional MRI, it has been shown that even tissues that appear normal may exhibit decreased myelin content as revealed by myelin-specific images (i.e., myelin maps). Current methods for analyzing myelin maps typically use global or regional mean myelin measurements to detect abnormalities, but ignore finer spatial patterns that may be characteristic of MS. In this paper, we present a machine learning method to automatically learn, from multimodal MR images, latent spatial features that can potentially improve the detection of MS pathology at early stage. More specifically, 3D image patches are extracted from myelin maps and the corresponding T1-weighted (T1w) MRIs, and are used to learn a latent joint myelin-T1w feature representation via unsupervised deep learning. Using a data set of images from MS patients and healthy controls, a common set of patches are selected via a voxel-wise t-test performed between the two groups. In each MS image, any patches overlapping with focal lesions are excluded, and a feature imputation method is used to fill in the missing values. A feature selection process (LASSO) is then utilized to construct a sparse representation. The resulting normal-appearing features are used to train a random forest classifier. Using the myelin and T1w images of 55 relapse-remitting MS patients and 44 healthy controls in an 11-fold cross-validation experiment, the proposed method achieved an average classification accuracy of 87.9% (SD = 8.4%), which is higher and more consistent across folds than those attained by regional mean myelin (73.7%, SD = 13.7%) and T1w measurements (66.7%, SD = 10.6%), or deep-learned features in either the myelin (83.8%, SD = 11.0%) or T1w (70.1%, SD = 13.6%) images alone, suggesting that the proposed method has strong potential for identifying image features that are more sensitive and specific to MS pathology in normal-appearing brain tissues.
Keywords: Deep learning, Multiple sclerosis, Myelin water imaging, Machine learning, Magnetic resonance imaging
Graphical Abstract
Highlights
-
•
Deep-learning method to detect MS pathology on normal-appearing brain tissue on MRI
-
•
Unsupervised deep learning and random forest applied to myelin and T1w images
-
•
Deep learning of myelin maps has not been previously attempted.
-
•
Deep-learned features yield more accurate detection (88%) than regional means (74%).
-
•
Joint deep learning improves pathology detection over modality-specific features.
1. Introduction
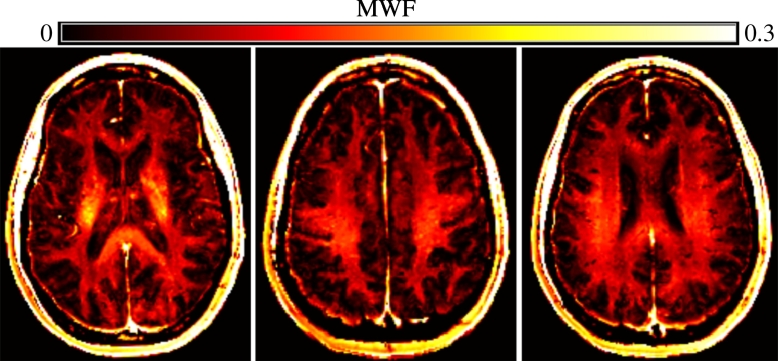
Multiple sclerosis (MS) is an autoimmune disorder characterized by inflammation, demyelination, and degeneration in the central nervous system. Magnetic resonance imaging (MRI) is invaluable for monitoring and understanding the pathology of MS in vivo from the earliest stages of the disease. One promising MR imaging modality is myelin water imaging (MWI) (MacKay et al., 1994), which is a quantitative MRI technique that specifically measures myelin content (Fig. 1) in the form of the myelin water fraction (MWF), which is defined as the ratio of water trapped within myelin over the total amount of water. Although white matter lesions have been traditionally considered the hallmark of MS pathology, histological studies and the MWI technique have shown that MS alterations also occur in tissues that appear normal in conventional MRIs. For example, a study using MWI found that a cohort of MS patients had 16% lower mean global MWF in normal-appearing white matter (NAWM) than healthy controls (Laule et al., 2004). In addition, normal-appearing gray matter (NAGM) has also been shown to have reduced MWF in MS patients (Steenwijk et al., 2014). Although myelin imaging has been indispensable in enhancing our understanding of MS, most analyses to date (e.g., MacKay et al., 1994, Laule et al., 2004, Yoo and Tam, 2013 only use mean myelin measurements, either over the whole brain or in predefined regions, and disregard the fine-scale spatial patterns of myelin content that may potentially be useful for MS diagnosis.
Fig. 1.
An example of a myelin map of a healthy control subject at several different slices in the dataset described in Section 2.1. The intensity reflects the relative amount of myelin present, except for the extraparenchymal areas.
Deep learning (LeCun et al., 2015) is a machine learning approach that uses layered hierarchical, graphical networks to extract features from data at progressively higher levels of abstraction. In recent years, methods based on deep learning have attracted much attention due to their breakthrough performance for classification in many application domains, including image recognition and natural language processing (LeCun et al., 2015). Unsupervised deep learning can be particularly useful in neuroimaging, a domain in which the number of labeled training images is typically limited. For example, unsupervised deep learning of neuroimaging data has been used to perform various tasks such as classification between mild cognitive impairment (MCI) and Alzheimer's disease (AD) (Suk et al., 2014), and to model morphological and lesion variability in MS (Brosch et al., 2014).
In view of this, we employ deep learning to extract latent spatial features in myelin maps, both on their own and combined with structural MRIs, to determine whether the deep-learned features can improve the detection of MS pathology. In doing so, we employ multimodal deep learning (Ngiam et al., 2011) to discover and model correlations between hidden patterns in the normal-appearing brain tissues of coregistered pairs of myelin maps and T1-weighted (T1w) MRIs. Myelin and T1w scans are used to provide complementary information in that the former contain myelin-specific features while the latter contain more general morphological features. Both types of features are known to be impacted by MS, but the benefits of deep learning for extracting myelin or myelin-T1w features are unknown. We hypothesize that deep learning can uncover spatial features in myelin maps that are more sensitive and specific to MS pathology than mean myelin measurements, and that multimodal deep learning can extract more sensitive and specific features than those extracted from either myelin or T1w modality alone.
Our method uses a four-layer deep belief network (DBN) (Hinton et al., 2006) that is applied to 3D image patches of NAWM and NAGM to learn a latent feature representation. The image patches are selected via a voxel-wise t-test that is performed between the MS and healthy control groups. To target only normal-appearing image patches, any patches overlapping with focal MS lesions are excluded, and a feature imputation technique is used to account for missing features originating from regions with focal lesions. We then apply the least absolute shrinkage and selection operator (LASSO) (Tibshirani, 1996) as a feature selection method to construct a sparse feature representation for reducing the risk of overfitting to the training data. The final features are then used to train a random forest (Breiman, 2001) that would discriminate images of MS subjects from those of normal subjects.
2. Material and methods
2.1. Subjects
A cohort of 55 relapsing-remitting MS (RRMS) patients and a cohort of 44 age- and gender-matched normal control (NC) subjects were included in this study. The median age and range for both groups were 45 and 30–60. For the RRMS patients, 63.6% (35/55) were female, and 63.5% (28/44) of the NC subjects were female. The McDonald 2010 criteria (Polman et al., 2011) were used to diagnose the patients for MS. All patients underwent a neurological assessment and were scored on the Expanded Disability Status Scale (EDSS) (Kurtzke, 1983). The median EDSS and range were 4 and 0–5. Informed consent from each participant and ethical approval by the local ethics committee were obtained prior to the study.
2.2. MRI acquisition and pre-processing
The T1w images were acquired with a gradient echo sequence (TR = 28 ms, TE = 4 ms, flip angle = 27°, voxel size = 0.977 × 0.977 × 3.000 mm3 and image dimensions = 256 × 256 × 60). The myelin images were acquired with a 3D GRASE sequence (Prasloski et al., 2012b) (32 echoes, TE = 10, 20, 30,…, 320 ms, TR = 1200 ms, voxel size = 0.958 × 0.958 × 2.500 mm3 and image dimensions = 256 × 256 × 40), and processed with the non-negative least squares fitting algorithm with non-local spatial regularization (Yoo and Tam, 2013) and stimulated echo correction (Prasloski et al., 2012a). All images were acquired on a Philips Achieva 3T scanner with an 8-channel SENSE head coil. Lesion masks were produced for the MS images using a semi-automatic segmentation method (McAusland et al., 2010) applied to T2-weighted and proton density-weighted MRI pairs. The T1w images were preprocessed by applying the N3 inhomogeneity correction method (Sled et al., 1998) iteratively over multiple scales (Jones and Wong, 2002), then followed by denoising and skull-stripping. The multi-scale N3 method works similarly to the N4 algorithm (Tustison et al., 2010), but was optimized to work with the magnetic field of our scanner.
The non-zero T1w intensities were normalized to have a range from 0 to 1, and then standardized to have zero mean and unit standard deviation (SD) to enable the use of Gaussian visible units (explained in Appendix B). In addition, normalization of the T1w intensities made the appearance of high-contrast edge features between the normal-appearing tissue and cerebrospinal fluid more consistent across individuals. In general, this allows the distribution of these edge features to be modeled more accurately during deep learning, which makes training of the networks faster and more stable. The brain masks computed from the T1w images and intensity standardization were also applied to the myelin images. The myelin images were registered to the T1w images using linear registration. Non-linear registration with FSL FNIRT (Jenkinson et al., 2012) was performed on the T1w images to align them to the MNI152 template (Mazziotta et al., 2001), and the computed transforms were also applied to the myelin images.
2.3. Cross-validation procedure
To maximize use of the available data, we performed a cross-validation procedure in which a rotating subset of the subjects acted as the test data, while the rest were used for training. We used an 11-fold cross-validation procedure in which each fold consisted of 9 test subjects (5 MS and 4 NC) and 90 training subjects (50 MS and 40 NC). This partitioning allowed all 99 subjects to be tested once.
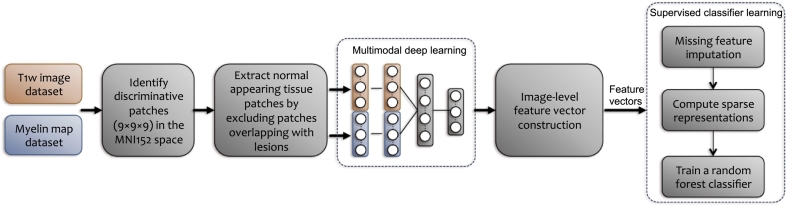
2.4. Overview of the feature learning and classification pipeline
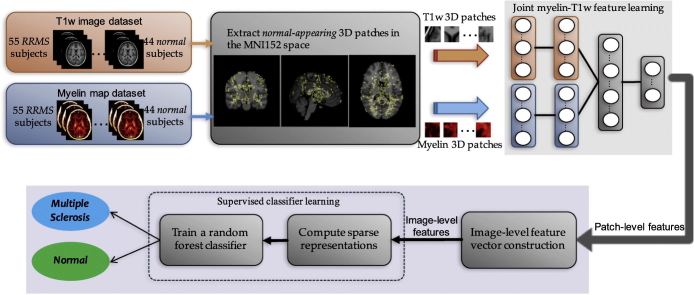
Fig. 2 shows a schematic diagram of our proposed method. The main steps are as follows. First, a common set of class-discriminative patches are extracted from both modalities in the MNI152 template space. Next, for each subject we exclude those patches that overlap with focal lesions. The resulting normal-appearing patches are then used to learn a latent joint myelin-T1w feature representation via unsupervised deep learning. To account for missing features from focal lesions, an imputation method (Marlin, 2008) is performed on the learned feature vectors. To reduce the risk of overfitting by increasing sparsity, we apply LASSO to the feature vectors. We then train a random forest classifier with the joint myelin-T1w features and class labels.
Fig. 2.
A schematic illustration of the proposed algorithm for detecting multiple sclerosis pathology on normal-appearing brain tissues using a latent hierarchical myelin-T1w feature representation.
2.5. Normal-appearing patch extraction
Instead of using all voxels in an image, patch extraction is commonly used for medical image classification to improve discriminative task accuracy and to reduce computational burden (Wu et al., 2015). We extract discriminative candidate patches on normal-appearing brain tissue from the myelin and T1w images in the MNI152 template space using a voxel-wise t-test to determine the statistical significance of the group difference between MS and NC images, as similarly done in previous studies Suk et al., 2014, Tong et al., 2014 for AD/MCI diagnosis. Details on normal-appearing patch extraction are provided in Appendix A.
2.6. Unsupervised deep learning of joint myelin-T1w features
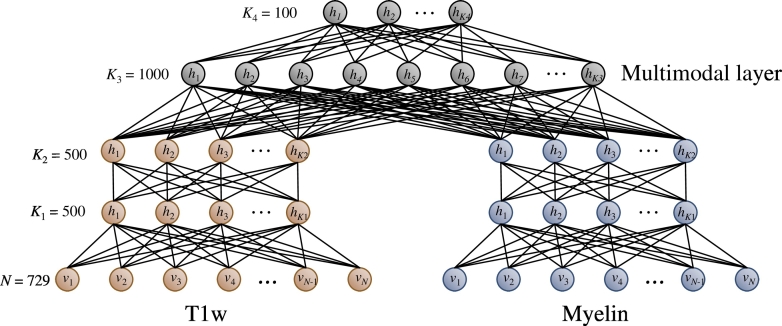
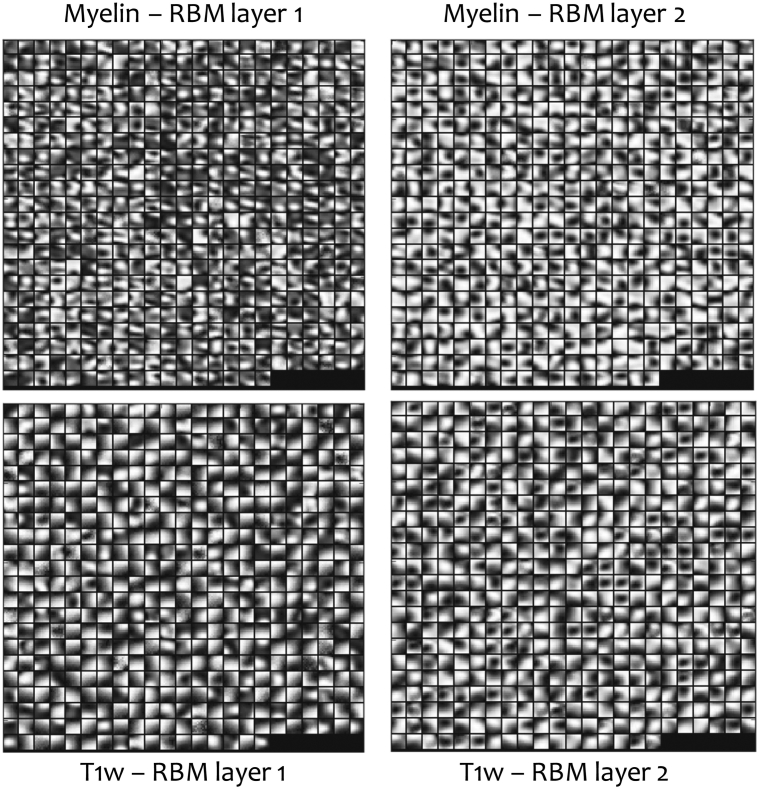
The network architecture (Fig. 3) for unsupervised deep learning consists of two modality-specific DBNs, one for myelin features and the other for T1w features, which are fed into a joint network that learns multimodal features. The number of network layers and number of hidden units were determined from previous literature Suk et al., 2014, Yoo et al., 2014 and a widely used guide (Hinton, 2012) for training restricted Boltzmann machines (RBMs). Fig. 4 shows visualizations of spatial features learned by this network from both myelin and T1w images. Technical details on modeling a joint myelin-T1w feature representation by unsupervised deep learning are described in Appendix B.
Fig. 3.
The multimodal deep learning network architecture used to extract a joint myelin-T1w feature representation.
Fig. 4.
Features at two RBM layers learned from myelin images (top) and T1w images (bottom). The deep network is able to learn a large variety of spatial features from both myelin and T1w images, which supports the hypothesis that myelin maps contain potentially useful structural information for detecting MS pathology.
2.7. Image-level feature vector construction and random forest training
For input into a supervised image-level classifier, single-modality or multi-modality features can be used. Single-modality feature vectors can be constructed by concatenating the second-layer activations from the individual myelin and T1w DBNs for all normal-appearing patches. Joint multimodal feature vectors can be constructed by concatenating the top-level hidden unit activations of the multimodal DBN. For the patches excluded due to lesions, we model each missing feature element using a normal distribution whose parameters μi and σi are estimated as the mean and SD of all feature values from the training dataset, where i = 1,…,m and m = P × K4, and P is the number of patches and K4 is the number of hidden units in the top layer. We then impute each missing feature element with a value sampled from the normal distribution, as previously described (Marlin, 2008).
Since the feature dimension is very high (P × K4, depending on the number of patches P) relative to the number of training samples (90), we construct a sparse representation using a linear regression model to reduce the risk of overfitting during supervised training. In previous work by Kim et al. (2013), it has been shown that applying LASSO (Tibshirani, 1996) as a feature selection method to reduce the dimensionality of the latent features learned by DBNs is beneficial to classification performance. Accordingly, we also explore the impact of using LASSO in our framework. More specifically, LASSO employs the following objective function:
| (1) |
where X ∈ℝT ×m and y ∈ℝT ×1 denote the data matrix and the label vector respectively, and T is the number of subjects used for training. The vector q ∈ℝm ×1 holds the regression coefficients and λ1 is a regularization parameter that controls the sparsity of the model. After LASSO, the non-zero elements in the regression coefficient vector q are used to select the covariates to form a sparse feature representation for each image.
Finally, given the feature vectors and labels, we train a random forest (Breiman, 2001) using the information gain to measure the quality of a split for each node. We also compute the relative importance of each patch for classification between MS and NC by permuting the features for each patch among the training data and computing the generalization error as measured by the out-of-bag error (Breiman, 2001). Then, we relate the feature importance to anatomical regions by using the Harvard-Oxford sub-cortical structural atlas (Desikan et al., 2006), which was derived from the MNI152 template (Mazziotta et al., 2001), and the central voxel of each patch, to enhance the interpretability of the results. The feature importance for each anatomical region is determined by averaging feature importance values from all patches belonging to each anatomical region. The procedure of determining random forest and LASSO parameters is described in Appendix C.
3. Results
3.1. Performance evaluation
Let TP, TN, FP and FN denote True Positive, True Negative, False Positive and False Negative, respectively. The ability of our proposed method to extract discriminative features was evaluated by using the deep-learned features of normal-appearing brain tissues to classify each subject as MS or NC, and measuring several different aspects of classification performance:
-
•
Accuracy = (TP + TN) / (TP + TN + FP + FN)
-
•
Sensitivity = TP / (TP + FN)
-
•
Specificity = TN / (TN + FP)
-
•
Area under the curve (AUC) of the receiver operating characteristic curve
We performed an 11-fold cross-validation procedure as described in Section 2.3. We performed the patch selection, unsupervised deep learning and random forest training using only the training data for each fold.
We used three regional features as baseline comparators: the regional mean T1w intensity, the regional mean myelin content, and the regional mean myelin-T1w features, which were formed by concatenation of the myelin content and T1w intensity feature vectors for each image. All regional means were computed on the same class-discriminative patches as used for unsupervised deep learning. We independently trained the random forest classifier for each mean-based feature. To determine the LASSO regularization parameter λ1, we performed the same nested cross-validation procedure described in Appendix C. The top three rows of Table 1 show the classification performance for each mean-based feature type. To analyze the effect of LASSO, the supervised training was initially done without this regularization. Rows 4 to 6 of Table 1 show the classification performance of the three mean-based features when including the LASSO regularization.
Table 1.
Performance comparison (%) between 6 different feature types with and without LASSO for MS/NC classification on normal-appearing brain tissues. We performed an 11-fold cross-validation on 55 RRMS and 44 NC images and computed the average performance (and standard deviation) for each feature type. The highest value for each measure is in bold. Overall, deep learning improved the classification results over the regional mean-based features across all four measures. In addition, LASSO had a positive effect, but more so for the regional mean-based features than the deep-learned features.
| Feature type | Accuracy | Sensitivity | Specificity | AUC |
|---|---|---|---|---|
| Regional mean without LASSO | ||||
| T1w intensity | 63.6 (16.5) | 74.5 (18.7) | 50.0 (28.2) | 62.3 (16.9) |
| Myelin content | 72.7 (13.7) | 74.6 (18.7) | 68.2 (17.9) | 72.3 (13.8) |
| Myelin-T1w | 67.7 (8.8) | 72.7 (22.5) | 61.4 (14.7) | 67.1 (9.2) |
| Regional mean with LASSO | ||||
| T1w intensity | 66.7 (10.6) | 76.4 (17.2) | 54.5 (24.1) | 65.5 (11.5) |
| Myelin content | 73.7 (13.7) | 76.4 (18.7) | 70.5 (17.9) | 73.4 (12.6) |
| Myelin-T1w | 70.7 (12.8) | 70.9 (21.4) | 70.5 (20.8) | 70.7 (12.0) |
| Deep-learned without LASSO | ||||
| T1w | 70.1 (13.6) | 81.8 (20.9) | 56.8 (22.3) | 69.3 (13.7) |
| Myelin | 83.8 (11.0) | 85.5 (18.0) | 81.8 (14.4) | 83.6 (10.5) |
| Myelin-T1w | 86.9 (9.3) | 85.5 (15.0) | 88.6 (12.5) | 87.0 (9.0) |
| Deep-learned with LASSO | ||||
| T1w | 70.1 (13.6) | 81.8 (20.9) | 56.8 (22.3) | 69.3 (13.7) |
| Myelin | 83.8 (11.0) | 85.5 (18.0) | 81.8 (14.4) | 83.6 (10.5) |
| Myelin-T1w | 87.9 (8.4) | 87.3 (12.9) | 88.6 (12.5) | 88.0 (8.5) |
To determine the effectiveness of feature extraction by deep learning, we compared three deep-learned feature types, which are deep-learned T1w features, deep-learned myelin features, and the output of the multimodal DBN, which combines the deep-learned myelin and T1w features. These features were also tested with and without LASSO regularization, with the results shown in Table 1. Overall, deep learning improved the classification results over the regional mean-based features across all four evaluation metrics. In addition, LASSO had a positive effect, but more so for the mean-based features than the deep-learned features. The accuracy rate attained by the deep-learned myelin-T1w feature with LASSO was statistically better than the ones attained by all of the regional features and the deep-learned T1w feature with LASSO (p < 0.01, two-sided Wilcoxon test), but it was not statistically better than the one attained by the deep-learned myelin features with LASSO.
3.2. Separate analysis in NAWM and NAGM
To determine the relative contributions of white and gray matter to classification performance, we evaluated each deep-learned feature type on predominantly NAWM and NAGM separately. Since LASSO proved to be beneficial for the previous experiments, we applied it to all of the regional NAWM and NAGM analyses. Using the WM and GM masks computed from the T1w MNI152 template, we excluded all patches that did not overlap with WM or GM. For the normal-appearing patches that overlapped with both the WM and GM masks, we labeled each patch as a NAWM patch if the WM voxel count is larger than the GM voxel count, and vice versa.
The separate analysis results computed with each deep-learned feature type are summarized in Table 2. For both NAWM and NAGM, the deep-learned myelin features alone provided the best overall classification performance. Using NAWM patches gave higher performance than that attained by using NAGM patches for the myelin images, while for the T1w images, using NAGM patches gave better performance. This is consistent with the observations that the most discriminative patches in the myelin images came from subcortical WM, while the most discriminative patches in the T1w images came from the cortical and periventricular regions. Overall, the maximum classification performance of using NAWM and NAGM patches separately did not approach that of using all normal-appearing patches together.
Table 2.
Separate analysis results in NAWM and NAGM. The table shows a performance comparison (%) between deep-learned features for MS/NC classification. We performed an 11-fold cross-validation on 55 RRMS and 44 NC images and computed the average performance (and standard deviation).
| Feature type | Accuracy | Sensitivity | Specificity | AUC |
|---|---|---|---|---|
| Deep-learned on NAWM | ||||
| T1w | 66.7 (7.0) | 78.2 (19.2) | 56.8 (24.1) | 67.3 (10.3) |
| Myelin | 82.8 (11.5) | 81.8 (19.9) | 84.1 (11.8) | 83.2 (10.4) |
| Myelin-T1w | 74.7 (13.5) | 74.5 (17.6) | 75.0 (16.3) | 74.8 (13.7) |
| Deep-learned on NAGM | ||||
| T1w | 68.7 (6.7) | 78.2 (19.1) | 59.1 (22.0) | 68.9 (9.9) |
| Myelin | 80.8 (12.6) | 85.5 (15.0) | 75.0 (21.3) | 80.2 (13.0) |
| Myelin-T1w | 73.7 (14.4) | 74.5 (22.5) | 72.7 (14.7) | 73.6 (14.7) |
4. Discussion
The regional mean myelin content features were more discriminative than the regional mean T1w intensity features, which is not surprising given that the T1w sequence is a structural MR sequence designed to show tissue contrast and not for direct quantification. The mean myelin features achieved mean classification performance rates of 73.7% (accuracy, SD 13.7%) and 73.4% (AUC, SD 12.6%) with LASSO, which are approximately 7% (accuracy) and 8% (AUC) higher than those of the regional mean T1w intensity features. However, the regional combined mean myelin-T1w features produced mean classification performance rates of 70.7% for both accuracy and AUC, showing that direct concatenation of the regional mean myelin content and T1w intensity features resulted in reduced classification performance when compared to mean myelin content alone, largely due to reduced specificity, but were still better than the performance achieved using T1w intensity features alone.
Applying LASSO as a feature selection method improved the classification performance for the regional mean features. When including LASSO regularization in the supervised classifier for the regional mean features, the feature dimensionality reduction rate by LASSO was about 60–80%. For the regional mean T1w intensities, LASSO improved classification performance rates by approximately 3% for both mean accuracy and AUC. The impact of LASSO was smaller for the regional mean myelin features, resulting in about a 1% improvement in classification accuracy. LASSO also improved the classification performance of the regional combined mean myelin-T1w features, but did not beyond than that attained by the regional mean myelin features, again suggesting that direct concatenation of heterogeneous modalities is not an effective strategy for improving the classification performance. Overall, the regional mean myelin content feature type was the most accurate, sensitive, and specific regional mean MRI biomarker for distinguishing between MS and NC on normal-appearing brain tissues.
Unsupervised deep learning of the regional myelin contents and T1w intensities yielded superior classification performance over using the regional mean myelin contents and T1w intensities. Without LASSO, the deep-learned T1w features improved the classification performance by about 6% in both mean accuracy and mean AUC over the regional mean T1w intensity features. In addition, the SDs for accuracy and AUC decreased by approximately 3%, showing a more consistent performance across folds. Similarly, the deep-learned myelin features improved the classification performance over the regional mean myelin content features by about 11% in both mean accuracy and mean AUC, demonstrating that spatial feature learning of myelin maps by unsupervised deep learning can produce radiologically useful information associated with MS pathology. Similarly to the deep-learned T1w features, the SDs for accuracy and AUC also decreased by about 3%.
The joint deep-learned regional myelin-T1w features were more discriminative than either of the modality-specific deep-learned feature types, and improved accuracy and AUC by about 4% over the deep-learned myelin features, showing that, in contrast to the case of simple concatenation of regional mean T1w and myelin features, deep-learned joint features improved the classification performance, and decreased the SDs by about 2%. Compared to the regional mean myelin-T1w features, the deep-learned multimodal features improved the classification performance by approximately 17% in mean accuracy and AUC.
We observed a relatively small impact when including the LASSO regularization in the supervised classifier for the deep-learned features. The feature dimensionality reduction rate by LASSO was about 30–50%, which is smaller than the case of regional mean features, suggesting that the deep-learned features had less redundancy. For both the deep-learned T1w features and the deep-learned myelin features, LASSO did not change the classification performance. For the deep-learned joint myelin-T1w features, LASSO improved the classification performance by about 1% in both mean accuracy and AUC. The impact of LASSO was smaller than for the regional mean features. This could be due to the fact that unsupervised deep learning is already capable of extracting less redundant and more independent feature sets which reduced the impact of dimensionality reduction by LASSO.
Overall, the proposed deep-learned joint myelin-T1w features provided the best performance, surpassing all other feature types substantially in accuracy, sensitivity, specificity, and AUC. In addition, they significantly reduced the SDs of both accuracy and AUC compared to the regional mean features, showing a more consistent classification performance across folds. When used independently, the deep-learned myelin features also performed well, surpassing all other regional mean features on all four evaluation measures. All deep-learned features outperformed their regional mean counterparts, with or without LASSO, which indicates that both myelin and T1w modalities contain discriminative latent spatial patterns. Our main conclusion is that the deep-learned myelin features provide valuable pathological information that is more sensitive and specific than the use of regional mean myelin and/or T1w measurements for MS diagnosis on normal-appearing brain tissues, especially when combined jointly with deep-learned T1w features.
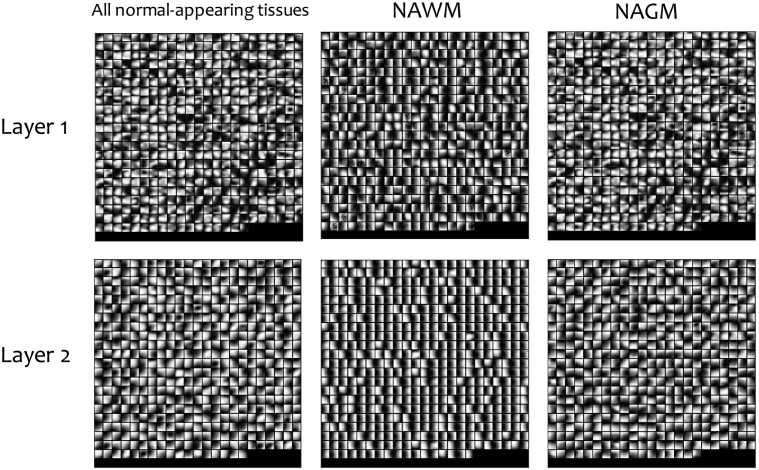
We analyzed the effect of separately using predominantly NAWM and NAGM patches on the various deep learning models we built. The deep-learned myelin features extracted from NAWM patches achieved mean classification performance rates of 82.8% in accuracy (SD 11.5%) and 83.2% in AUC (SD 10.4%), which are approximately 2–3% higher in accuracy and AUC than those of the deep-learned myelin features with NAGM patches, suggesting that the deep-learned myelin features are more pathologically relevant to NAWM than NAGM, which is expected due to the greater myelin content in WM. When using NAWM and NAGM patches separately, the variety of feature patterns learned by the T1w-specific network was reduced compared to those learned by the T1w-specific network with all normal-appearing patches as shown in Fig. 5. The more limited feature set led to classification accuracy rates of 66.7% with NAWM patches and 68.7% with NAGM patches, which are lower than those of the deep-learned T1w features with all normal-appearing patches (70.1%). Since this limited feature set was used as an input to the multimodal myelin-T1w layer, the deep-learned myelin-T1w features did not improve the classification performance in NAWM nor NAGM patches as shown in Table 2.
Fig. 5.
Deep-learned features separately extracted from predominantly NAWM, NAGM and all normal-appearing patches by the T1w modality-specific network. The variety of feature patterns learned by the T1w-specific network with NAWM and NAGM patches is reduced compared to that learned by the T1w-specific network with all normal-appearing patches.
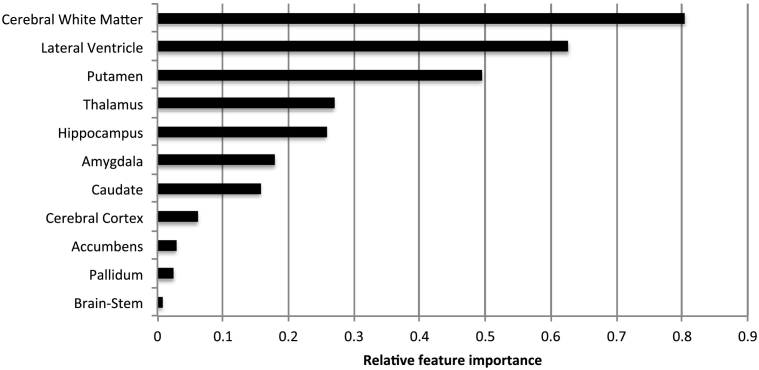
To enhance interpretability of the results and examine their relationship to published MS literature, we determined the relative contribution of the deep-learned joint myelin-T1w features in each patch location, and used the Harvard-Oxford sub-cortical atlas to compute the mean importance values in particular sub-cortical regions and structures. As shown in Fig. 6, the six most discriminative sub-cortical brain regions and structures were found to be the cerebral white matter, lateral ventricles, putamen, thalamus, hippocampus, and amygdala. The most discriminative sub-cortical brain regions were the cerebral white matter and lateral ventricles. The high importance of the cerebral white matter and lateral ventricles are likely due to demyelination in the periventricular region, combined with morphological changes caused by brain atrophy, both of which are strongly associated with MS pathology. The observed importance of the sub-cortical gray matter structures is consistent with previous MS studies (e.g., Hulst and Geurts, 2011), which showed that these specific structures undergo substantial structural and/or chemical changes.
Fig. 6.
The relative importance of the deep-learned joint myelin-T1w features in different sub-cortical brain areas for RRMS vs. NC classification on normal-appearing brain tissues.
It is important to acknowledge limitations of our study. Due to the relatively small training sample size, this study can only provide preliminary results and does not ensure that the proposed model will generalize to produce the exact same results in other cohorts. Secondly, our study only included RRMS patients and did not include progressive MS patients. The proposed model may extract different features for progressive MS cohorts because patients with progressive MS can have different patterns of demyelination and morphological changes throughout the brain. To evaluate this approach for detecting very early MS pathology, our future work should include patients with clinically isolated syndrome, a prodromal stage of MS, with the clinical goal of enabling earlier diagnosis.
As we stated above, the T1w sequence is a structural imaging sequence for displaying tissue contrast and not for direct quantification, and this limitation cannot be corrected by intensity normalization, which is likely a main reason why the regional mean T1w intensity features produced relatively low classification accuracy as shown in Table 1. It could be argued that for complementing the myelin scans, a quantitative T1 relaxometry may be appropriate. However, a primary goal of this study was to determine whether myelin scans contained spatial features finer than regional means that would be useful for distinguishing MS, so a comparison to regional mean intensity features seems appropriate. We believe the primary reason that deep learning on T1w images produced useful features for classification is that the model captured spatial variabilities in the high-contrast boundaries between normal-appearing tissue and cerebrospinal fluid as induced by atrophy and other morphological changes. This is visually verifiable by Fig. 4, Fig. 5, which show that the features extracted by deep learning from the T1w images are mostly high-contrast edges in various shapes and orientations. For deep learning, the intensity normalization procedure is meant to enable the use of Gaussian visible units and to allow the distribution of these edge features to be modeled more accurately which generally makes training of the networks faster and more stable as also stated in Section 2.2. In contrast, deep learning appeared to capture more intensity variations in the myelin images, indicative of changes in myelin content, as shown in Fig. 4, especially in RBM layer 1.
In summary, our experimental results have demonstrated the following for the task of detecting MS pathology on normal-appearing brain tissues:
-
•
The regional mean myelin content features were more discriminative than the regional mean T1w intensity features.
-
•
Direct concatenation of the regional mean myelin content features and the regional mean T1w intensity features did not improve the classification accuracy over using each feature type alone.
-
•
Unsupervised deep learning of the regional myelin contents and T1w intensities yielded superior classification performance over using the regional mean myelin contents and T1w intensities.
-
•
The joint deep-learned regional myelin-T1w features were more discriminative than either of the modality-specific deep-learned feature types.
-
•
Applying LASSO to produce sparser feature representations improved the classification performance for the regional mean features, but the impact of LASSO was marginal for the deep-learned regional features.
-
•
The maximum classification performance of using predominantly NAWM and NAGM patches separately was achieved by the deep-learned regional myelin features. However, it did not approach that of using all normal-appearing patches together.
5. Conclusions
We have demonstrated that unsupervised deep learning of normal-appearing brain tissues on myelin and T1w images can extract information that could be useful for early MS detection and provides superior classification performance to the traditional regional mean MRI measurements when using the same supervised classifier. In addition, we have shown that unsupervised deep learning of joint myelin and T1w features improves the classification performance over deep learning of either modality alone. Using a four-layer multimodal deep learning network to learn latent features, unbiased feature imputation to exclude lesion voxels, a feature selection method (LASSO) to construct sparse feature representations, and a random forest for a supervised classifier, we achieved a mean classification accuracy of 87.9% between RRMS and healthy controls on normal-appearing brain tissues, using an 11-fold cross-validation procedure. The local brain structures that were found to be important for MS classification by our method were consistent with known MS pathology and previous MS literature. In future work, we plan to extend the proposed framework to include other MRI modalities used for studying MS pathology such as multi-echo susceptibility-weighted imaging (Denk and Rauscher, 2010). We also plan to apply our framework to other subgroups of MS patients and longitudinal data for applications in MS prognostication.
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC RGPIN 402202-12); an endMS Doctoral Studentship Award from the Multiple Sclerosis Society of Canada (MS Society Project Number 2739); and the Milan and Maureen Ilich Foundation.
Appendix A. Additional details on normal-appearing patch extraction
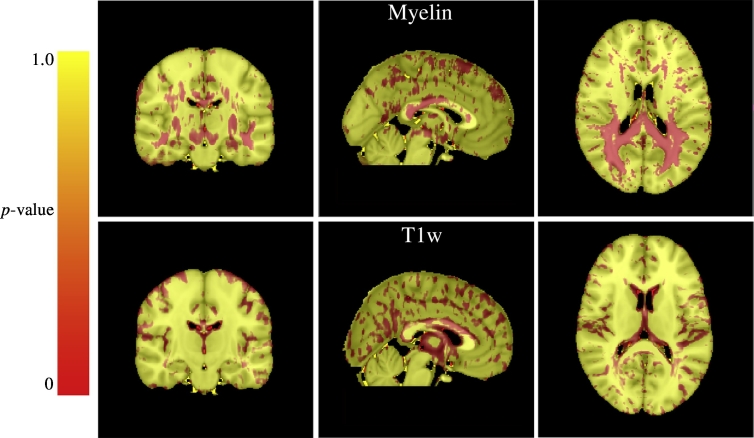
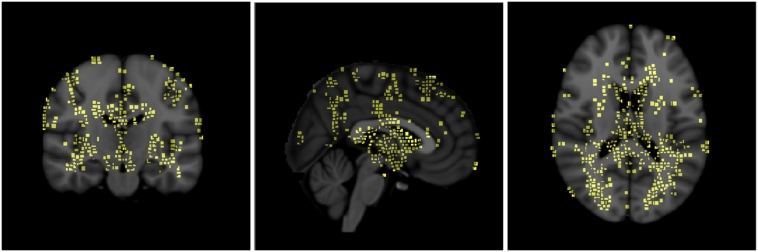
The voxel-wise t-test results for each modality (myelin and T1w) are shown in Fig. A.7. Based on the voxel-wise t-test, the voxels with individual p-values lower than 0.05 are selected as the centers of candidate patches. The mean p-value for each candidate patch is then computed. Starting with the patches with the lowest mean p-values, patches are selected in a greedy manner (Suk et al., 2014) while enforcing an overlap of less than 50% with any previously selected patches. These patches are then further selected by including only those with mean p-values smaller than the average p-value of all candidate patches of both modalities. Finally, the patches overlapping with focal lesions are excluded for each patient in order to retain only the normal-appearing patches. Patch sizes from 7 × 7 × 7 to 11 × 11 × 11 have been suggested to be a good range for capturing local structural information in related work Liu et al., 2014, Suk et al., 2014, Tong et al., 2014. From this perspective, we chose a patch size of 9 × 9 × 9 for our experiments. From the data in this study, the number of selected patches ranged from 8000 to 10000 depending on the images used for training in each cross-validation fold, and on the amount of lesion present. Examples of the selected patches in the MNI152 template are displayed in Fig. A.8.
Fig. A.7.
Voxel-wise t-test results displayed in the MNI152 template showing the most discriminative locations between 55 RRMS patients and 44 normal controls. The red areas indicate statistical significance (p < 0.05). Most of the voxels selected from the myelin maps are located in cerebral white matter regions (the top three images), while most selected from the T1w images are from the cortex and periventricular areas (the bottom three images). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. A.8.
Discriminative patches in the MNI152 template that were extracted from the scans of 50 RRMS patients and 40 normal controls used as training data in one cross-validation fold, using the patch extraction method described in Section 2.5. In this figure, the patches have been rescaled from 9 × 9 × 9 to 3 × 3 × 3 for the purpose of visualization. The patches are used for training the multimodal unsupervised deep learning network with the goal of extracting features that can be used to detect MS pathology on normal-appearing tissues.
Appendix B. Additional details on unsupervised feature learning
We convert the selected patches into one-dimensional vectors v1,… ∈ℝD with D = 729. The number of feature vectors from each image depends on the number of excluded patches due to the presence of lesions. We learn features for the myelin and T1w input vectors independently by using a Gaussian-Bernoulli RBM (Krizhevsky and Hinton, 2009) for each modality. Each RBM has real-valued visible units v of dimension D = 729, binary hidden units h of dimension K1 = 500, and symmetric connections between these two layers as represented by a weight matrix . The energy function of a Gaussian-Bernoulli RBM (Krizhevsky and Hinton, 2009) is defined as
| (B1) |
where bj is the bias for the j-th hidden unit (), σi is the variance term for the i-th visible unit, and ci is the bias for the i-th visible unit (c ∈ℝD). The variance term is set to 1 by standardizing the dataset as described in Section 2.2. The units of the binary hidden layer (conditioned on the visible layer) are independent Bernoulli random variables , where is the sigmoid function. The visible units (conditioned on the hidden layer) are D independent Gaussians with diagonal covariance . In order to capture higher-level correlations between the first-level features, another layer of binary hidden units of dimension K2 = 500 is stacked on top of each RBM to form a DBN for each modality. We follow a standard layer-by-layer approach for training a DBN (Hinton et al., 2006), in which each RBM adopts the previous layer's activations as its input. Fig. 4 shows a large variety of spatial features learned by this network from both myelin and T1w images, which supports the hypothesis that myelin maps contain potentially useful structural information.
We next build a joint model (Fig. 3) that finds multimodal myelin and T1w patterns by modeling the joint distribution between myelin and T1w features. We form a multimodal DBN by adding a layer of K3 = 1000 binary hidden units that are connected to both the myelin and the T1w DBNs, thereby combining their second-layer activations. Finally, we model higher-level multimodal features by stacking another layer of binary hidden units on top of the multimodal RBM. For this multimodal layer, the dimensionality is reduced to K4 = 100 for each patch.
We perform contrastive divergence (Hinton et al., 2006) to approximate gradient descent to update the weights and biases during training. To avoid the difficulty of setting a fixed learning rate and decay schedule, we apply AdaDelta (Zeiler, 2012), which adaptively determines the learning rate for each model parameter and improves the chances of convergence to a global minimum. Given the high dimensionality of the feature vectors and the inherent risk of overfitting to the training data, we use two common regularization approaches during training, consisting of weight decay (Hinton, 2012) with the penalty coefficient 0.0002, which penalizes large weights, and dropout (Srivastava et al., 2014) with a probability of 0.5, which randomly drops hidden units to simulate the effect of using many “thinned” networks to produce an average solution.
Appendix C. Determining random forest and LASSO parameters
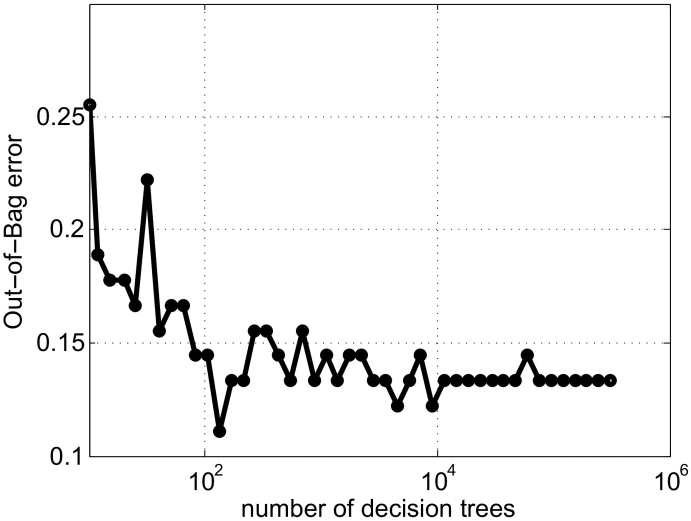
The number of decision trees and their depth determine the generalizability of the random forest. In general, overly shallow trees lead to underfitting while overly deep trees lead to overfitting. We found that tree depths between 20 and 40 produced almost identical out-of-bag errors in our case. From this perspective, the tree depth value was empirically set as 30 to avoid under- and overfitting. To determine a suitable number of trees, we started with 10 and increased it by a step size of 0.2 on a log scale until we observed a stabilization in the out-of-bag error (Fig. C1) using the entire dataset. We determined an appropriate value of 105, which was used for all of our experiments.
Fig. C1.
Influence of the number of decision trees on the generalizability of MS/NC classification, as measured by the out-of-bag error, on normal-appearing brain tissues.
After fixing the random forest parameters, we performed a nested cross-validation procedure to determine the LASSO regularization parameter λ1, which we expected to vary between cross-validation folds. For each of the 11 folds, we performed a nested cross-validation with 10 inner folds. For each inner fold, we varied λ1 between 10 −7 and 1 with a step size of 1 on a log scale, and the λ1 that produced the best mean MS/NC classification accuracy was used for the outer fold.
References
- Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- Brosch T., Yoo Y., Li D.K.B., Traboulsee A., Tam R. Medical Image Computing and Computer-assisted Intervention-MICCAI 2014. Springer; 2014. Modeling the variability in brain morphology and lesion distribution in multiple sclerosis by deep learning; pp. 462–469. [DOI] [PubMed] [Google Scholar]
- Denk C., Rauscher A. Susceptibility weighted imaging with multiple echoes. J. Magn. Reson. Imaging. 2010;31:185–191. doi: 10.1002/jmri.21995. [DOI] [PubMed] [Google Scholar]
- Desikan R.S., Ségonne F., Fischl B., Quinn B.T., Dickerson B.C., Blacker D., Buckner R.L., Dale A.M., Maguire R.P., Hyman B.T., Albert M.S. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Hinton G., Osindero S., Teh Y.-W. A fast learning algorithm for deep belief nets. Neural Comput. 2006;18:1527–1554. doi: 10.1162/neco.2006.18.7.1527. [DOI] [PubMed] [Google Scholar]
- Hinton G.E. Neural Networks: Tricks of the Trade. Springer; 2012. A practical guide to training restricted Boltzmann machines; pp. 599–619. [Google Scholar]
- Hulst H.E., Geurts J.J. Gray matter imaging in multiple sclerosis: what have we learned? BMC Neurol. 2011;11:153. doi: 10.1186/1471-2377-11-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M., Beckmann C.F., Behrens T.E., Woolrich M.W., Smith S.M. FSL. NeuroImage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Jones C., Wong E. Multi-scale application of the N3 method for intensity correction of MR images. Medical Imaging 2002; International Society for Optics and Photonics; 2002. pp. 1123–1129. [Google Scholar]
- Kim Y., Lee H., Provost E.M. Acoustics, Speech and Signal Processing (ICASSP), 2013 IEEE International Conference on. 2013. Deep learning for robust feature generation in audiovisual emotion recognition; pp. 3687–3691. [Google Scholar]
- Krizhevsky A., Hinton G. Tech. Rep. University of Toronto; 2009. Learning multiple layers of features from tiny images. [Google Scholar]
- Kurtzke J.F. Rating neurologic impairment in multiple sclerosis an expanded disability status scale (EDSS) Neurology. 1983;33 doi: 10.1212/wnl.33.11.1444. 1444–1444. [DOI] [PubMed] [Google Scholar]
- Laule C., Vavasour I., Moore G., Oger J., Li D.K.B., Paty D., MacKay A. Water content and myelin water fraction in multiple sclerosis. J. Neurol. 2004;251:284–293. doi: 10.1007/s00415-004-0306-6. [DOI] [PubMed] [Google Scholar]
- LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Liu M., Zhang D., Shen D. Hierarchical fusion of features and classifier decisions for Alzheimer's disease diagnosis. Hum. Brain Mapp. 2014;35:1305–1319. doi: 10.1002/hbm.22254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay A., Whittall K., Adler J., Li D.K.B., Paty D., Graeb D. In vivo visualization of myelin water in brain by magnetic resonance. Magn. Reson. Med. 1994;31:673–677. doi: 10.1002/mrm.1910310614. [DOI] [PubMed] [Google Scholar]
- Marlin B.M. University of Toronto; 2008. Missing data problems in machine learning. Ph.D. thesis. [Google Scholar]
- Mazziotta J., Toga A., Evans A., Fox P., Lancaster J., Zilles K., Woods R., Paus T., Simpson G., Pike B., Holmes C. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos. Trans. R. Soc., B. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAusland J., Tam R., Wong E., Riddehough A., Li D.K.B. Optimizing the use of radiologist seed points for improved multiple sclerosis lesion segmentation. IEEE Trans. Biomed. Eng. 2010;57:2689–2698. doi: 10.1109/TBME.2010.2055865. [DOI] [PubMed] [Google Scholar]
- Ngiam J., Khosla A., Kim M., Nam J., Lee H., Ng A.Y. Proceedings of the 28th International Conference on Machine Learning (ICML-11) 2011. Multimodal deep learning; pp. 689–696. [Google Scholar]
- Polman C.H., Reingold S.C., Banwell B., Clanet M., Cohen J.A., Filippi M., Fujihara K., Havrdova E., Hutchinson M., Kappos L. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann. Neurol. 2011;69:292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasloski T., Mädler B., Xiang Q.-S., MacKay A., Jones C. Applications of stimulated echo correction to multicomponent T2 analysis. Magn. Reson. Med. 2012;67:1803–1814. doi: 10.1002/mrm.23157. [DOI] [PubMed] [Google Scholar]
- Prasloski T., Rauscher A., MacKay A., Hodgson M., Vavasour I.M., Laule C., Mädler B. Rapid whole cerebrum myelin water imaging using a 3D GRASE sequence. NeuroImage. 2012;63:533–539. doi: 10.1016/j.neuroimage.2012.06.064. [DOI] [PubMed] [Google Scholar]
- Sled J.G., Zijdenbos A.P., Evans A.C. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014;15:1929–1958. [Google Scholar]
- Steenwijk M.D., Daams M., Pouwels P.J., Balk L.J., Tewarie P.K., Killestein J., Uitdehaag B.M., Geurts J.J., Barkhof F., Vrenken H. What explains gray matter atrophy in long-standing multiple sclerosis? Radiology. 2014;272:832–842. doi: 10.1148/radiol.14132708. [DOI] [PubMed] [Google Scholar]
- Suk H.-I., Lee S.-W., Shen D., Initiative Alzheimer's Disease Neuroimaging. Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis. NeuroImage. 2014;101:569–582. doi: 10.1016/j.neuroimage.2014.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the LASSO. J. R. Stat. Soc. Ser. B (Methodol.) 1996:267–288. [Google Scholar]
- Tong T., Wolz R., Gao Q., Guerrero R., Hajnal J.V., Rueckert D., Initiative Alzheimer's Disease Neuroimaging. Multiple instance learning for classification of dementia in brain MRI. Med. Image Anal. 2014;18:808–818. doi: 10.1016/j.media.2014.04.006. [DOI] [PubMed] [Google Scholar]
- Tustison N.J., Avants B.B., Cook P.A., Zheng Y., Egan A., Yushkevich P.A., Gee J.C. N4ITK: improved N3 bias correction. IEEE Trans. Med. Imaging. 2010;29:1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Coupé P., Zhan Y., Munsell B., Rueckert D. Springer Verlag. Springer; 2015. Patch-based Techniques in Medical Imaging. [Google Scholar]
- Yoo Y., Brosch T., Traboulsee A., Li D.K., Tam R. International Workshop on Machine Learning in Medical Imaging. Springer; 2014. Deep learning of image features from unlabeled data for multiple sclerosis lesion segmentation; pp. 117–124. [Google Scholar]
- Yoo Y., Tam R. Medical Image Computing and Computer-Assisted Intervention-MICCAI 2013. Springer; 2013. Non-local spatial regularization of MRI T2 relaxation images for myelin water quantification; pp. 614–621. [DOI] [PubMed] [Google Scholar]
- Zeiler, M.D., 2012. ADADELTA: an adaptive learning rate method. arXiv preprint arXiv:1212.5701.