Abstract
Aim
Various bacterial species are differentially prevalent in periodontal health, gingivitis or periodontitis. We tested the independent associations between three bacterial groupings and gingival inflammation in an epidemiological study.
Material and Methods
In 706 Oral Infections and Vascular Disease Epidemiology Study (INVEST) participants ≥55 years, bleeding on probing (BoP), pocket depth (PD) and subgingival plaque samples (n =4866) were assessed in eight sites per mouth. Eleven bacterial species were quantitatively assayed and grouped as follows: (i) aetiologic burden (EB, Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Treponema denticola, Tannerella forsythia); (ii) putative burden (PB, Campylobacter rectus, Eikenella corrodens, Fusobacterium nucleatum, Micromonas micros, Prevotella intermedia); (iii) health-associated burden (HAB, Actinomyces naeslundii, Veillonella parvula).
Results
After mutual adjustment for EB, PB and HAB, the BoP prevalence increased by 45% ( p<0.0001) across increasing quartiles of EB while BoP decreased by 13% ( p<0.0001) across increasing quartiles of HAB. Mean PD increased 0.8 mm and decreased 0.3 mm from the first to fourth quartiles of EB (p<0.0001) and HAB ( p<0.0001), respectively. Among 1214 plaque samples with fourth quartile EB, 60% were collected from sites with PD ≤3 mm.
Conclusion
Bacterial species believed to be aetiologically related to periodontitis were associated with BoP in sites with minimal PD and/or attachment level (AL). Species presumed to be associated with periodontal health demonstrated inverse associations with BoP.
Keywords: bacteria, epidemiology, infection, inflammation, periodontal
The bacterial aetiology of periodontitis has been extensively studied and several putative causal microbes have been identified (Grossi et al. 1994, Haffajee & Socransky 1994, Mombelli et al. 1998, Socransky et al. 1998, Van Winkelhoff et al. 1999, Timmerman et al. 2000, van der Velden et al. 2006). However, much of the population-based research exploring relationships between bacteria and clinical periodontal disease uses pooled bacterial samples and defines bacterial exposure dichotomously by assessing the prevalence (presence/absence) of colonization at the patient level (Grossi et al. 1994, Machtei et al. 1999, Timmerman et al. 2001, Van Winkelhoff et al. 2002, van der Velden et al. 2006).
Epidemiological approaches that measure bacteria quantitatively and use statistical methods to model multiple bacterial exposures simultaneously are necessary for assessing dose-responsiveness and addressing the contributions of various bacterial species to clinical periodontal disease independent of mutual correlations among species. Such approaches may help to clarify the specificity of associations between bacterial species and clinical periodontal variables. This is particularly important in the case of gingival inflammation and bleeding on probing (BoP), which is generally considered to be the result of a non-specific inflammatory response to dental plaque accumulation and its constituent microbiology (Loesche & Grossman 2001).
We previously investigated the association between a priori defined “aetiologic”, “putative” or “health-associated” periodontal bacterial groupings and carotid artery intima-media thickness (c-IMT), in the Oral Infections and Vascular Disease Epidemiology Study (INVEST) (Desvarieux et al. 2005). Our analytical approach allowed for parsimonious statistical models that addressed the specificity of various periodontal microbial colonization patterns to atherosclerosis. We observed the strongest positive association between “aetiologic” bacteria and c-IMT, while an inverse association existed between health-associated bacteria and c-IMT.
In this study, we applied a similar analytical approach to investigate the specificity of the association between selected health- and periodontitis-associated bacterial groupings, and clinical periodontal measures, such as BoP and pocket depth (PD) in the population-based setting of INVEST. We focused on early manifestations of periodontal infections (BoP and PD) to reduce the possibility that these cross-sectional data reflected reversed causality (i.e. that the development of a deep periodontal pocket and/or BoP, preceded and/or potentially contributed to the establishment of a particular subgingival microbial profile).
Material and Methods
INVEST is a randomly sampled prospective population-based cohort study investigating the relationship among oral infections, atherosclerosis and stroke. The selection process has been published (Sacco et al. 1998, Desvarieux et al. 2003). Briefly, 1056 subjects were selected via random digit dialing from Northern Manhattan, an area between 145th Street and 218th Street, bordered westward by the Hudson River and eastward by the Harlem River. Eligibility criteria for INVEST are as follows: (1) White, Black or Hispanic resident (>3 months) of Northern Manhattan (zip-codes 10031, 10032, 10033, 10034 and 10040); (2) contacted by random digit dialing among households with a telephone (all eligible invited); (3) age ≥55; (4) no history of stroke, myocardial infarction or chronic inflammatory conditions; and (5) ability to visit clinic. Baseline full-mouth clinical periodontal exams and subgingival plaque samples were available for 706 subjects.
The Institutional Review Board at Columbia University Medical Center approved the study and all subjects provided informed consent.
Dental examination
Subjects received a full-mouth clinical examination by calibrated examiners. Assessments included number and type of teeth present, presence/absence of BoP, PD in millimetres and location of the gingival margin in relation to the cementoenamel junction at six locations/tooth (mesiobuccal, midbuccal, distobuccal, mesiolingual, midlingual, distolingual) using a UNC-15 manual probe (HuFriedy, Chicago, IL, USA). The latter two assessments were used to compute clinical attachment levels.
Subgingival plaque collection and processing
For each participant, a maximum of eight subgingival plaque samples (median =8) were collected from the two most posterior teeth (mesiolingual in the maxilla; mesiobuccal in the mandible) in each quadrant, yielding a total of 4922 samples. Eleven bacterial species (Agg-regatibacter actinomycetemcomitans, Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola, Fusobacterium nucleatum, Prevotella intermedia, Campylobacter rectus, Micromonas micros, Eikenella corrodens, Veillonella parvula, Actinomyces naeslundii) were assessed using checkerboard DNA–DNA hybridization as described previously (Socransky et al. 1994, Desvarieux et al. 2005).
Statistical analysis
Analyses were performed in PC-SAS for Windows 9.1 and included periodontal sites with concurrent BoP, PD and dental plaque assessments. Unavailable clinical measurements due to restorations or debris led to 4880 and 4866 sites for BoP and PD analyses, respectively.
Bacterial exposure definitions
Laboratory analysis provided a relative quantity of individual bacterial species for each plaque sample in comparison with known standards. Because the distribution of absolute bacterial counts was skewed, values were natural logarithm (ln) transformed. Absolute counts of different species were not comparable across species. Therefore, ln(bacterial count) was averaged within mouth, and then standardized by dividing each respective ln(bacterial count) by the population standard deviation (SD) for the respective species: 1 SD on the ln scale (SDln) was treated as equivalent across microbes; SDln is therefore the unit of each species. For each person, we defined four bacterial groupings in addition to the eleven standardized values for each species. We summed the standardized values for the 11 species to define cumulative burden. Subsets of the cumulative burden were further defined as aetiologic burden (EB), putative burden (PB) and health-associated burden (HAB) (Desvarieux et al. 2005). We utilized (i) the consensus of the 1996 World Workshop in Periodontics identifying three bacterial species as causally related to periodontal disease (P. gingivalis, T. forsythia and A. actinomycetemcomitans) (Consensus Report 1996), and (ii) Socransky’s red complex (Socransky et al. 1998) further identifying T. denticola as a species that closely covaries with P. gingivalis, and T. forsythia in pathological periodontal pockets, to create an EB score, comprising the four species (A. actinomycetem-comitans, P. gingivalis, T. forsythia and T. denticola). The five bacterial species deemed putatively associated with periodontal disease (C. rectus, E. corrodens, F. nucleatum, M. micros and P. intermedia) were grouped as PB (Consensus Report 1996). HAB included two “health-associated” bacterial species, A. naeslundii and V. parvula (Socransky et al. 1998).
All clinical and microbiological data were collected concurrently from periodontal sites. Site was the unit of analysis in all statistical models. Linear and logistic regression analyses tested the linear association between either PD or BoP (dependent variables) and the aforementioned individual bacterial species or bacterial clusters (independent variables). For individual bacterial species we present results from both unadjusted and simultaneously adjusted regression models to show the effect of confounding on the crude parameter estimates. Adjusted results for individual bacterial species stem from regressions that simultaneously modelled all 11 species. This approach provides estimates of the association between each species and PD or BoP while holding constant the levels of other bacterial species in the same periodontal site.
The same approach was used to model bacterial clusters (EB, PB and HAB) as independent variables. In addition, we used ANOVA techniques to model PD and BoP variation across quartiles of all three bacteria burdens. All ANOVA models present the pooled within- and between-mouth estimates. This approach removed the constraints of the linearity assumption imposed by use of continuous variables in regression models and serves as an indicator of goodness-of-fit for linear models. We additionally added attachment loss as a covariate to the models to verify that findings were independent of historical periodontal disease experience.
Because multiple periodontal sites per mouth were included in the analysis and the design was not balanced (not all participants contributed the same number of periodontal sites due to tooth loss), all regressions were performed using either SAS PROC MIXED for continuous PD outcomes or PROC GENMOD for dichotomous BoP outcomes. These procedures allowed us to condition individual participants as random effects to account for the within-mouth correlation (non-independence) of these samples. Therefore, the p-values presented account for the clustered and unbalanced nature of the design. We present both within- and between-mouth estimates of BoP and PD across levels of bacterial burden. To describe within-mouth variability for each bacterial burden, we defined the difference of the bacterial burden at a specific site from the within-mouth mean of that bacterial burden; this difference has mean =0 for each mouth. SDs of these within-mouth differences are presented to characterize within-mouth variability. The within-mouth estimates hold constant all person-level characteristics and are consequently independent of between person variation in measured and unmeasured periodontal disease risk factors (such as age, gender, smoking, race/ethnicity, genetics, etc.) allowing for more precise assessments of associations between clinical and microbiological periodontal parameters. The within-mouth estimates of association between PD or BOP and bacterial species could be biased towards zero because the range of bacterial exposure within-mouth is less than the between-mouth range.
We also used an interaction model to examine whether the association of EB, PB and HAB with BoP was modified by PD.
Results
Participants were 57% Hispanic, 23% Black, 20% White and 60% female with mean ± SD age of 69 ± 9 years and 14 ± 8 missing teeth. Mean PD was 3.0 mm and 35% of sites had BoP. Within- and between-mouth distributional characteristics for all bacterial variables are presented in Table 1, for example, mean ± SD of EB was 29 ± 4 SDln between mouths, with 1.4 SDln within mouth. Within-mouth variation was less for each species and burden score than was between-mouth variation. EB was positively correlated with PB (r =0.81) and HAB (r =0.47). PB and HAB were correlated (r =0.55).
Table 1.
Distributional characteristics of individual bacterial species and bacterial burden scores, between and within mouth
| Bacterial species or burden | Between-mouth | Within-mouth difference from mouth-specific mean | |
|---|---|---|---|
|
|
|
||
| mean | SD | SD | |
| Aetiologic burden | 28.91 | 3.29 | 1.41 |
| A. actinomycetemcomitans | 8.59 | 0.91 | 0.41 |
| P. gingivalis | 5.10 | 0.90 | 0.44 |
| T. denticola | 8.51 | 0.93 | 0.36 |
| T. forsythia | 6.79 | 0.84 | 0.54 |
| Putative burden | 47.03 | 2.87 | 2.33 |
| C. rectus | 9.11 | 0.84 | 0.54 |
| E. corrodens | 8.30 | 0.87 | 0.49 |
| F. nucleatum | 10.15 | 0.79 | 0.62 |
| M. micros | 10.25 | 0.83 | 0.56 |
| P. intermedia | 9.22 | 0.77 | 0.63 |
| Health-associated burden | 20.82 | 1.24 | 1.03 |
| A. naeslundii | 11.52 | 0.85 | 0.53 |
| V. parvula | 9.30 | 0.73 | 0.69 |
Values reported in SDln units.
SD, standard deviation.
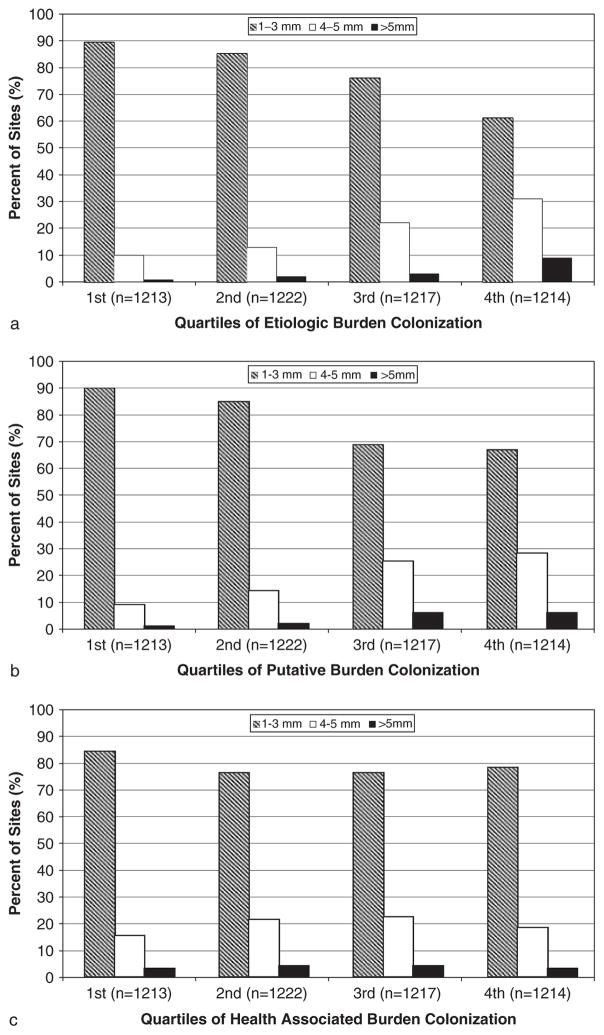
Periodontal plaques with high levels of EB were commonly found in shallow pocketed sites. 60% of plaques with high EB (fourth quartile) were found in 1–3 mm pockets (Fig. 1a). Findings were similar for periodontal plaques with high levels of PB (Fig. 1b) and HAB (Fig. 1c).
Fig. 1.
Distribution of pocket depth according to quartiles of subgingival aetiologic burden, putative burden or health-associated burden (n ~ 1216 periodontal sites per quartile).
The unadjusted within- and between-mouth associations between clinical periodontal disease and each of eleven bacterial species in addition to the three bacteria burden scores are presented in Table 2. Nearly all species demonstrated strong positive associations with PD and BoP. A. naeslundii was inversely related to PD and unrelated to BoP. EB and PB were positively related to both PD and BoP while HAB was only positively associated with between-mouth BoP.
Table 2.
Associations (unadjusted and adjusted)* between selected periodontal bacteria and both pocket depth and bleeding on probing
| Independent variables (bacterial species or burden) | Pocket depth β-coefficients§ | Bleeding on probing odds ratios¶ | ||
|---|---|---|---|---|
|
| ||||
| between mouth | within mouth | between mouth | within mouth | |
| Unadjusted | ||||
| Aetiologic burden | 0.46‡ | 0.16‡ | 3.49‡ | 1.15‡ |
| A. actinomycetemcomitans | 0.28‡ | 0.07‡ | 2.46‡ | 1.09† |
| T. denticola | 0.47‡ | 0.16‡ | 3.18‡ | 1.13‡ |
| T. forsythia | 0.38‡ | 0.13‡ | 2.99‡ | 1.11‡ |
| P. gingivalis | 0.47‡ | 0.18‡ | 3.96‡ | 1.16‡ |
| Putative burden | 0.37‡ | 0.14‡ | 2.65‡ | 1.15‡ |
| C. rectus | 0.34‡ | 0.11‡ | 2.80‡ | 1.13‡ |
| E. corrodens | 0.28‡ | 0.06‡ | 2.52‡ | 1.04 |
| F. nucleatum | 0.37‡ | 0.11‡ | 2.65‡ | 1.12‡ |
| M. micros | −0.12† | 0.10‡ | 0.81† | 1.11‡ |
| P. intermedia | 0.41‡ | 0.17‡ | 1.94‡ | 1.16‡ |
| Health-associated burden | 0.01 | 0.03 | 1.31‡ | 1.00 |
| A. naeslundii | −0.14‡ | −0.003 | 0.92 | 0.97 |
| V. parvula | 0.20‡ | 0.04† | 1.79‡ | 1.02 |
| Adjusted | ||||
| Aetiologic burden | 0.53‡ | 0.16‡ | 5.25‡ | 1.11† |
| A. actinomycetemcomitans | 0.04 | −0.04 | 0.91 | 1.00 |
| T. denticola | 0.13 | 0.07‡ | 1.09 | 1.05 |
| T. forsythia | 0.11 | 0.02 | 1.10 | 0.99 |
| P. gingivalis | 0.58‡ | 0.09‡ | 5.72‡ | 1.05 |
| Putative burden | 0.03 | 0.07† | 0.80 | 1.13† |
| C. rectus | −0.13 | −0.03 | 1.09 | 1.04 |
| E. corrodens | −0.30‡ | 0.03 | 0.55‡ | 0.98 |
| F. nucleatum | 0.02 | 0.001 | 1.21 | 1.05 |
| M. micros | 0.01 | −0.01 | 0.83 | 1.00 |
| P. intermedia | 0.13† | 0.11‡ | 0.76† | 1.07 |
| Health-associated burden | −0.25‡ | −0.09‡ | 0.68‡ | 0.88‡ |
| A. naeslundii | −0.13† | −0.04† | 0.80† | 0.94† |
| V. parvula | −0.05 | −0.02 | 1.04 | 0.96 |
Unadjusted parameter estimates stem from models that only consider each respective bacterial species or burden alone. Adjusted parameter estimates for individual bacterial species are adjusted for each of the other 10 species in this table. Adjusted parameter estimates for bacterial burden scores are adjusted for the other two burden scores. Note that adjusted parameter estimates for individual species are very susceptible to collinearity bias and should interpreted with caution.
p<0.01;
p<0.0001.
β-coefficients represent pocket depth change in millimetre associated with a one SD increase in the standardized bacterial colonization level.
Odds ratios reflect the odds of bleeding on probing versus not bleeding on probing for a one standard deviation increase in the standardized bacterial colonization level.
SDs for all independent variables are presented in Table 1.
SD, standard deviation.
With inclusion of all eleven species in one model, findings for most individual species were attenuated and became non-statistically significant. Within-mouth variations in P. gingivalis, T. denticola and P. intermedia were positively related to PD while A. naeslundii was inversely related to PD (Table 2). A. naeslundii was inversely associated with BoP, but no other individual species was significantly related to BoP within mouth. Results for associations of between-mouth variation in bacterial colonization levels with clinical periodontal disease were similar to those for within-mouth variation yet consistently stronger (Table 2). In between-mouth analyses, T. denticola lost significance and E. corrodens was significantly inversely related to both PD and BOP.
In comparison with unadjusted analyses, mutual adjustments for three bacterial burdens (Table 2), strengthened EB associations, attenuated PB associations and HAB associations, became inverse and statistically significant.
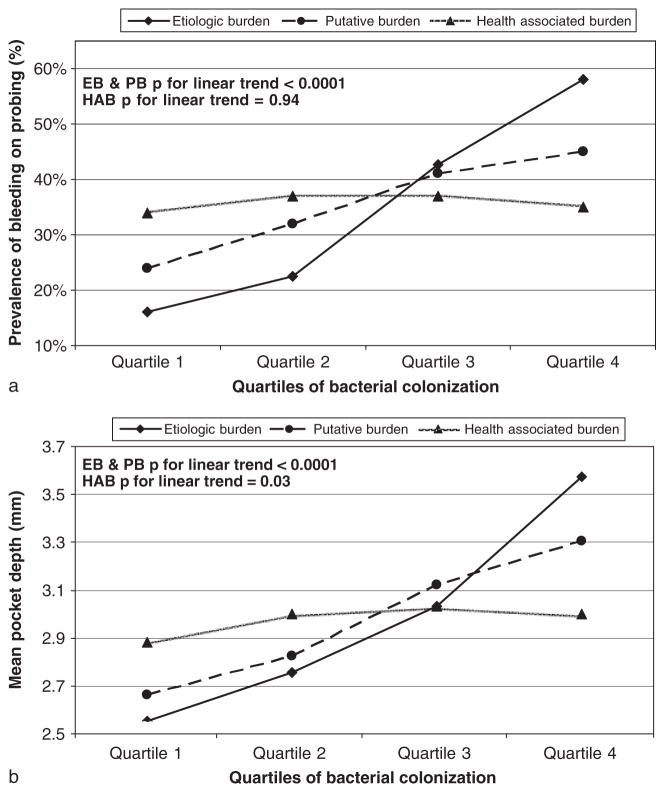
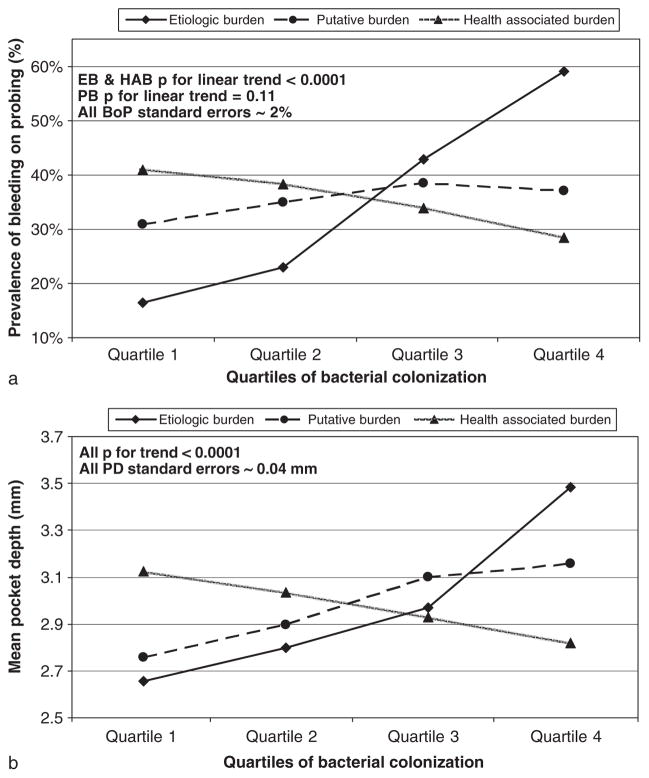
Results for bacterial clusters in Figs 2–4, pool within- and between-mouth effects to provide a sense of the absolute and relative variation in clinical periodontal disease across levels of bacterial clusters. BoP prevalence increased across cumulative burden quartiles from 22% to 48% (first versus fourth quartile; p<0.0001) and mean PD increased from 2.7 to 3.4 mm (first versus fourth quartile; p<0.0001). In unadjusted analyses, both EB and PB demonstrated strong positive associations with BoP and PD (Fig. 2). HAB demonstrated a weak positive association with PD but was not associated with BoP (Fig. 2). After mutual adjustment of each burden for the other burdens, EB maintained a strong positive association with BoP and PD; PB associations were substantially attenuated and HAB became inversely associated with BoP and PD (Fig. 3).
Fig. 2.
Relationship between the level of subgingival colonization by selected bacterial clusters and bleeding on probing or pocket depth; without mutual adjustment for bacterial clusters. *Constituent species of each bacterial grouping: aetiologic burden (EB) =Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola; putative burden (PB) =Fusobacterium nucleatum, Prevotella intermedia, Campylobacter rectus, Micromonas micros, Eikenella corrodens; health-associated burden (HAB) =Veillonella parvula, Actinomyces naeslundii.
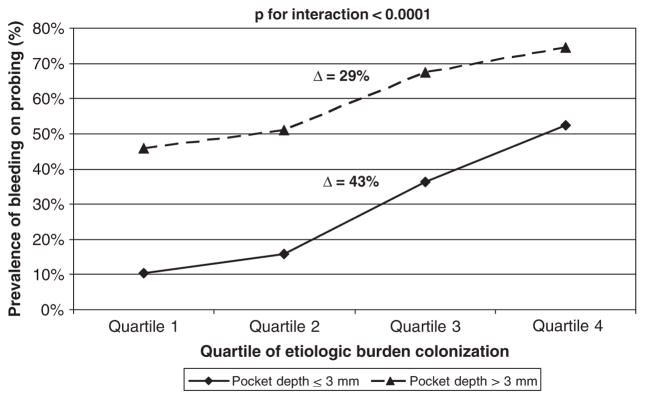
Fig. 4.
Relationship between bleeding on probing and aetiologic burden modified by pocket depth. *Model adjusted for putative burden and health-associated burden; constituent species of each bacterial grouping: aetiologic burden =Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola; putative burden = Fusobacterium nucleatum, Prevotella intermedia, Campylobacter rectus, Micromonas micros, Eikenella corrodens; health-associated burden =Veillonella parvula, Actinomyces naeslundii.
Fig. 3.
Relationship between the level of subgingival colonization by selected bacterial clusters and bleeding on probing or pocket depth; with mutual adjustment for all three bacterial groups. *Constituent species of each bacterial grouping: aetiologic burden (EB) =Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola; putative burden (PB) =Fusobacterium nucleatum, Prevotella intermedia, Campylobacter rectus, Micromonas micros, Eikenella corrodens; health-associated burden (HAB) =Veillonella parvula, Actinomyces naeslundii.
The prevalence of BoP increased across quartiles of EB by 43% (5.3-fold) in pockets ≤3 mm, as compared with 29% (1.6-fold) in pockets ≥4 mm (Fig. 4).
Findings were independent of attachment loss and tooth loss. In a mutually adjusted regression within 1585 shallow pocketed sites with attachment loss 0–2 mm, %BoPs were 3%, 6%, 17% and 25% across quartiles of EB; BoP remained inversely related to HAB (both p<0.002). Findings were unchanged in a subgroup analysis among participants with ≥20 teeth (data not shown).
Moreover, in mutually adjusted regression restricted to sites with PD≤ 3 mm, mean attachment level (AL) values increased across quartiles of EB as follows: 2.1, 2.3, 2.6 and 2.9 mm ( p for linear trend <0.0001). When comparing mean AL between the first and fourth quartiles of either PB or HAB, mean AL increased by 0.5 mm (p for linear trend =0.0003) and decreased by 0.15 mm (p for linear trend =0.10), respectively.
Discussion
Our data demonstrate a strong association between bacterial colonization levels and clinical variables related to gingival inflammation (BoP and PD). The relationship was present when defining subgingival colonization in a cumulative non-specific fashion, which incorporated 11 bacteria. However, formation of three bacterial groupings that distinguished between species previously reported to be related to periodontal health, and putatively or causatively related to periodontitis (Consensus Report 1996, Socransky et al. 1998) demonstrated specificity of the relationship of gingival inflammation to select bacterial groupings. Simultaneous modelling of the levels of EB, PB and HAB as predictors of BoP and PD resulted in a strong positive association for EB; a weak positive association for PB; and an inverse relationship for HAB. These results were only evident if the three bacterial burdens were mutually adjusted. Despite strong positive associations between EB and PD, it was evident that periodontal plaques with high levels of EB are commonly found in shallow periodontal pockets (Fig. 1).
An extensive literature exists on the microbiology of periodontal disease (Haffajee & Socransky 1994). Socransky et al. (1998) explored the interrelationships of 40 species in over 13,000 subgingival plaque samples, providing a comprehensive study of microbial colonization patterns. They reported that colonization patterns of P. gingivalis, T. forsythia and T. denticola were highly correlated and strongly related to PD and BoP. However, the sample was limited to 185 individuals, most of whom had periodontitis. Moreover, bacterial colonization levels tended to be modelled as the dependent variable in relation to clinical periodontal disease and they did not report the independence of these relationships. Importantly, Socransky et al. reported no evident associations between A. naeslundii and either PD or BoP.
In two separate populations lacking exposure to regular dental or periodontal care, Papapanou et al. (1997, 2002) reported strong positive relationships between periodontal bacteria and both PD and attachment loss; however, the relationship between bacterial profiles and BoP was not reported. In one of the largest studies reporting on bacteria-associated periodontal risk, Grossi et al. (1994) found the presence of P. gingivalis and T. forsythia to be risk indicators for attachment loss. However, the analysis was based on pooled plaque samples and bacterial exposure was assessed dichotomously as presence or absence of colonization. Although this approach is common among population-based studies with relatively large sample sizes (Machtei et al. 1999, Timmerman et al. 2001, Van Winkelhoff et al. 2002, van der Velden et al. 2006), it precludes any inference on the site-specific (within-mouth) association between bacterial colonization and clinical phenotype or the assessment of dose-responsiveness. Further, the primary outcome of interest in most studies to date has been AL or PD, as opposed to BoP, which is a clinical sign that may occur earlier in the natural history of periodontal disease and more directly reflects gingival inflammation. Moreover, we are unaware of any study to date that has modelled multiple bacterial groupings simultaneously to assess the independent contribution of each grouping on outcome variables related to periodontal disease.
Our findings confirm the results of previous studies in that many species were positively related to clinical periodontal disease when modelled individually without adjustment for variation in other co-colonizing species. However, the observed associations between individual species and either PD or BoP tended to be markedly changed and frequently non-statistically significant after mutual adjustment. The potential for collinearity to bias the results for 11 mutually adjusted bacterial species was high and emphasizes the potential importance of using bacterial groupings to create more parsimonious statistical models. In doing so, our findings for three bacterial burden scores appear to augment the existing literature regarding bacterial aetiologies of periodontal disease in three ways.
First, by simultaneously modelling three bacterial groupings as predictors of BoP and PD, we minimized the potential for biased results due to high positive correlations among EB, PB and HAB. This statistical control allowed us to examine relative quantities of the various bacterial groupings and clarify the independent contribution of each grouping to gingival inflammation. In unadjusted analyses HAB was positively related to PD and had no relationship with BoP (Fig. 2). Only after adjustment for both EB and PB, did an inverse relationship between HAB and both BoP and PD emerge. In epidemiological terms, the relationships between HAB and both BoP and PD were confounded by levels of EB and PB. These inverse findings for HAB suggest that certain bacterial species appear to assuage the level of gingival inflammation. Also of interest was the finding that PB was unrelated to BoP in adjusted models. Taken together, these findings suggest that bleeding is a consequence of exposure to specific bacterial species and does not support a non-specific nature of gingival inflammation (Loesche & Grossman 2001). The results also suggest that for some species, relative colonization levels might be more important than absolute levels in terms of risk for periodontal inflammation.
Second, while our cross-sectional findings cannot address temporality, our analytical approach minimizes the possibility of spurious findings related to reverse causality. Specifically, the fact that the observed relationship between EB and BoP – an early sign of localized pathology – was stronger in shallow (≤3 mm) than deep pockets reduces the possibility that the formation of a deep pocket and the occurrence of BoP in fact preceded the establishment of the particular microbial colonization pattern. Moreover, the associations between bacterial profiles and BoP were also consistent in two separate subgroup analyses among either (i) periodontal sites with ≤2 mm attachment loss; or (ii) participants with ≥20 teeth present. This further suggests that the findings were not indicative of reverse causality in which gingival tissue with a history of either localized (site level AL) or generalized (advanced tooth loss) disease, fostered environments optimized for bacterial colonization by specific species. Nevertheless, population-based findings such as these will require laboratory-based experiments to precisely elucidate whether (i) a non-inflamed environment becomes inflamed after introduction of selected species; (ii) an inflamed area gets worse after these species grow; and (iii) gingival inflammation is assuaged after the introduction of species believed to be health associated.
Finally, by presenting within-mouth results we substantially minimize the risk of biased results related to between-person characteristics such as age, gender, race, smoking or genetic susceptibility. For example, smokers might be more likely to have oral environments conducive to both bacterial colonization and gingival inflammation regardless of any direct link between colonization and inflammation.
A limitation of the present study is that plaque samples were collected in only eight sites per mouth and thus they may have not adequately represented the microbial profiles of the entire dentition. We minimized this potential bias by always collecting plaque samples from a priori defined teeth, rather than from the most periodontally affected teeth. Further, our microbial assay was limited to the characterization of only 11 bacterial species. Nevertheless, our findings at the site level are consistent with previous reports with more comprehensive bacterial assessments (Socransky et al. 1998).
Our findings support a priori assumptions concerning relationships between selected bacterial species and clinical periodontal disease in the population-based setting of INVEST. The results demonstrate that statistical techniques are required to assess the independent effects of specific subgingival bacterial clusters on clinical periodontal outcomes. Using these techniques, we add to the existing evidence that the bacterial grouping A. actinomycetemcomitans, P. gingivalis, T. forsythia and T. denticola (or correlates of this grouping) are strong predictors of gingival inflammation. We further report that after accounting for the level of colonization by aetiologic bacteria, the levels of putative periodontal pathogens had little bearing on the occurrence of gingival inflammation. Finally, these data show that colonization by health-associated bacteria was inversely related to BoP, suggesting that certain bacteria are potentially protective against manifest gingival inflammation.
Clinical Relevance.
Scientific rationale for the study
We studied the independent associations between levels of selected periodontal bacteria (quantitatively assessed) and gingival inflammation, expressed through BoP.
Principal findings
Bacterial species believed to be aetiologically related to periodontitis were associated with BoP, even in sites with minimal attachment loss. Species presumed to be associated with periodontal health demonstrated inverse associations with BoP.
Practical implications
Species known to be associated with periodontitis are also related to gingival inflammation before the development of destructive disease. Colonization by other species might assuage gingival inflammation, suggesting that the latter is not the mere non-specific response to plaque accumulation.
Acknowledgments
source of funding statement
This research is supported by NIH grant R01DE-13094 (Dr. Desvarieux) and a Chair of Excellence grant from the French Agency for Research and the Institut National de la Santé et de la Recherche Médicale to Dr. Desvarieux (R05115DD). Dr. Demmer was a fellow on T32HL-07779. Patients were seen at the Columbia University General Clinical Research Center, NIH grant RR-00645.
We thank Miriam Herrera-Abreu, Romanita Celenti and Jun Yang for the laboratory analysis of the dental plaque samples; George Loo, Janet DeRosa, Drs. Yira Florez, Mariana Cukier and Shantanu Lal for their devoted patient care; the entire INVEST staff; and importantly the patients. We thank Drs. Aaron Folsom and Mark Herzberg for their intellectual contributions.
Footnotes
Conflict of interest
The authors have no conflicts of interest or financial interests to disclose.
References
- Consensus Report. Periodontal diseases: pathogenesis and microbial factors. Annals of Periodontology. 1996;1:926–932. doi: 10.1902/annals.1996.1.1.926. [DOI] [PubMed] [Google Scholar]
- Desvarieux M, Demmer RT, Rundek T, Boden-Albala B, Jacobs DR, Papapanou PN, Sacco RL. Relationship between periodontal disease, tooth loss, and carotid artery plaque: the Oral Infections and Vascular Disease Epidemiology Study (INVEST) Stroke. 2003;34:2120–2125. doi: 10.1161/01.STR.0000085086.50957.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvarieux M, Demmer RT, Rundek T, Boden-Albala B, Jacobs DR, Sacco RL, Papapanou PN. Periodontal microbiota and carotid intima-media thickness: the Oral Infections and Vascular Disease Epidemiology Study (INVEST) Circulation. 2005;111:576–582. doi: 10.1161/01.CIR.0000154582.37101.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossi SG, Zambon JJ, Ho AW, Koch G, Dunford RG, Machtei EE, Norderyd OM, Genco RJ. Assessment of risk for periodontal disease. I. Risk indicators for attachment loss. Journal of Periodontology. 1994;65:260–267. doi: 10.1902/jop.1994.65.3.260. [DOI] [PubMed] [Google Scholar]
- Haffajee AD, Socransky SS. Microbial etiological agents of destructive periodontal diseases. Periodontology 2000. 1994;5:78–111. doi: 10.1111/j.1600-0757.1994.tb00020.x. [DOI] [PubMed] [Google Scholar]
- Loesche WJ, Grossman NS. Periodontal disease as a specific, albeit chronic, infection: diagnosis and treatment. Clinical Microbiology Reviews. 2001;14:727–752. doi: 10.1128/CMR.14.4.727-752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machtei EE, Hausmann E, Dunford R, Grossi S, Ho A, Davis G, Chandler J, Zambon J, Genco RJ. Longitudinal study of predictive factors for periodontal disease and tooth loss. Journal of Clinical Periodontology. 1999;26:374–380. doi: 10.1034/j.1600-051x.1999.260607.x. [DOI] [PubMed] [Google Scholar]
- Mombelli A, Gmur R, Frey J, Meyer J, Zee KY, Tam JO, Lo EC, Di Rienzo J, Lang NP, Corbet EF. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in young Chinese adults. Oral Microbiology Immunology. 1998;13:231–237. doi: 10.1111/j.1399-302x.1998.tb00701.x. [DOI] [PubMed] [Google Scholar]
- Papapanou PN, Baelum V, Luan WM, Madianos PN, Chen X, Fejerskov O, Dahlen G. Subgingival microbiota in adult Chinese: prevalence and relation to periodontal disease progression. Journal of Periodontology. 1997;68:651–666. doi: 10.1902/jop.1997.68.7.651. [DOI] [PubMed] [Google Scholar]
- Papapanou PN, Teanpaisan R, Obiechina NS, Pithpornchaiyakul W, Pongpaisal S, Pisuithanakan S, Baelum V, Fejerskov O, Dahlen G. Periodontal microbiota and clinical periodontal status in a rural sample in Southern Thailand. European Journal of Oral Sciences. 2002;110:345–352. doi: 10.1034/j.1600-0722.2002.21361.x. [DOI] [PubMed] [Google Scholar]
- Sacco RL, Boden-Albala B, Gan R, Chen X, Kargman DE, Shea S, Paik MC, Hauser WA. Stroke incidence among white, black, and Hispanic residents of an urban community: the Northern Manhattan stroke study. American Journal of Epidemiology. 1998;147:259–268. doi: 10.1093/oxfordjournals.aje.a009445. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL. Microbial complexes in subgingival plaque. Journal of Clinical Periodontology. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Smith C, Martin L, Paster BJ, Dewhirst FE, Levin AE. “Checkerboard” DNA–DNA hybridization. Biotechniques. 1994;17:788–792. [PubMed] [Google Scholar]
- Timmerman MF, Van der Weijden GA, Abbas F, Arief EM, Armand S, Winkel EG, Van Winkelhoff AJ, van der Velden U. Untreated periodontal disease in Indonesian adolescents. Longitudinal clinical data and prospective clinical and microbiological risk assessment. Journal of Clinical Periodontology. 2000;27:932–942. doi: 10.1034/j.1600-051x.2000.027012932.x. [DOI] [PubMed] [Google Scholar]
- Timmerman MF, Van der Weijden GA, Arief EM, Armand S, Abbas F, Winkel EG, Van Winkelhoff AJ, van der Velden U. Untreated periodontal disease in Indonesian adolescents. Subgingival microbiota in relation to experienced progression of periodontitis. Journal of Clinical Periodontology. 2001;28:617–627. doi: 10.1034/j.1600-051x.2001.028007617.x. [DOI] [PubMed] [Google Scholar]
- van der Velden U, Abbas F, Armand S, Loos BG, Timmerman MF, Van der Weijden GA, Van Winkelhoff AJ, Winkel EG. Java project on periodontal diseases. The natural development of periodontitis: risk factors, risk predictors and risk determinants. Journal of Clinical Periodontology. 2006;33:540–548. doi: 10.1111/j.1600-051X.2006.00953.x. [DOI] [PubMed] [Google Scholar]
- Van Winkelhoff AJ, Laine ML, Timmerman MF, Van der Weijden GA, Abbas F, Winkel EG, Arief EM, van der Velden U. Prevalence and serotyping of Porphyromonas gingivalis in an Indonesian population. Journal of Clinical Periodontology. 1999;26:301–305. doi: 10.1034/j.1600-051x.1999.260507.x. [DOI] [PubMed] [Google Scholar]
- Van Winkelhoff AJ, Loos BG, van der Reijden WA, van der Velden U. Porphyromonas gingivalis, Bacteroides forsythus and other putative periodontal pathogens in subjects with and without periodontal destruction. Journal of Clinical Periodontology. 2002;29:1023–1028. doi: 10.1034/j.1600-051x.2002.291107.x. [DOI] [PubMed] [Google Scholar]