ABSTRACT
Respiratory syncytial virus (RSV) is the most important viral agent of severe pediatric respiratory tract disease worldwide, but it lacks a licensed vaccine or suitable antiviral drug. A live attenuated chimeric bovine/human parainfluenza virus type 3 (rB/HPIV3) was developed previously as a vector expressing RSV fusion (F) protein to confer bivalent protection against RSV and HPIV3. In a previous clinical trial in virus-naive children, rB/HPIV3 was well tolerated but the immunogenicity of wild-type RSV F was unsatisfactory. We previously modified RSV F with a designed disulfide bond (DS) to increase stability in the prefusion (pre-F) conformation and to be efficiently packaged in the vector virion. Here, we further stabilized pre-F by adding both disulfide and cavity-filling mutations (DS-Cav1), and we also modified RSV F codon usage to have a lower CpG content and a higher level of expression. This RSV F open reading frame was evaluated in rB/HPIV3 in three forms: (i) pre-F without vector-packaging signal, (ii) pre-F with vector-packaging signal, and (iii) secreted pre-F ectodomain trimer. Despite being efficiently expressed, the secreted pre-F was poorly immunogenic. DS-Cav1 stabilized pre-F, with or without packaging, induced higher titers of pre-F specific antibodies in hamsters, and improved the quality of RSV-neutralizing serum antibodies. Codon-optimized RSV F containing fewer CpG dinucleotides had higher F expression, replicated more efficiently in vivo, and was more immunogenic. The combination of DS-Cav1 pre-F stabilization, optimized codon usage, reduced CpG content, and vector packaging significantly improved vector immunogenicity and protective efficacy against RSV. This provides an improved vectored RSV vaccine candidate suitable for pediatric clinical evaluation.
IMPORTANCE RSV and HPIV3 are the first and second leading viral causes of severe pediatric respiratory disease worldwide. Licensed vaccines or suitable antiviral drugs are not available. We are developing a chimeric rB/HPIV3 vector expressing RSV F as a bivalent RSV/HPIV3 vaccine and have been evaluating means to increase RSV F immunogenicity. In this study, we evaluated the effects of improved stabilization of F in the pre-F conformation and of codon optimization resulting in reduced CpG content and greater pre-F expression. Reduced CpG content dampened the interferon response to infection, promoting higher replication and increased F expression. We demonstrate that improved pre-F stabilization and strategic manipulation of codon usage, together with efficient pre-F packaging into vector virions, significantly increased F immunogenicity in the bivalent RSV/HPIV3 vaccine. The improved immunogenicity included induction of increased titers of high-quality complement-independent antibodies with greater pre-F site Ø binding and greater protection against RSV challenge.
KEYWORDS: bronchiolitis, fusion protein, human parainfluenza virus type 3, intranasal vaccine, live attenuated vaccine, live vaccine, pneumonia, prefusion, respiratory syncytial virus
INTRODUCTION
Human respiratory syncytial virus (RSV) is the leading cause of viral pneumonia and bronchiolitis in infants and young children worldwide (1, 2). It is a major cause of global infant mortality and, as a single agent, is second only to malaria (3). Primary RSV infection occurs early in life, and pediatric infections are responsible for up to 4 million hospitalizations and an estimated 66,000 to 199,000 deaths in children under 5 years of age every year worldwide (4). The global annual death rate at all ages ranges from 200,000 to 500,000 (3). No licensed RSV vaccine or suitable antiviral is available. Formalin-inactivated RSV primed for vaccine-enhanced disease in virus-naive children, and purified RSV subunits appeared to prime for enhanced pulmonary pathology in experimental animals (5–7). However, live attenuated RSV strains and live attenuated parainfluenza virus type 3 (PIV3) vector expressing RSV fusion (F) protein are not associated with this risk (8, 9).
Human PIV3 (HPIV3) is second only to RSV as a major cause of acute viral lower respiratory tract (LRT) infection in young children worldwide (2, 10). An attenuated chimeric bovine/human PIV3 (rB/HPIV3) virus expressing the RSV fusion F glycoprotein, the major conserved RSV neutralization and protective antigen, was developed as a bivalent HPIV3/RSV vaccine (11, 12). The PIV3 genome contains six genes in the order of nucleoprotein (N), phosphoprotein (P), matrix protein (M), fusion glycoprotein (F), hemagglutinin-neuraminidase glycoprotein (HN), and polymerase (L) (Fig. 1). The rB/HPIV3 vector consists of bovine PIV3 (BPIV3), which is attenuated in primates, in which the F and HN genes have been replaced by their HPIV3 counterparts (Fig. 1) (13). rB/HPIV3 expressing unmodified RSV F, called MEDI-534, was previously developed and evaluated by MedImmune in a phase 1 clinical trial in virus-naive children. This construct appeared to be satisfactorily attenuated and well tolerated, which is a major advantage for further development of this vector system, but it was not sufficiently immunogenic for RSV F (8) and thus was not advanced for further clinical evaluation. More recently, we have systematically explored strategies for increasing the immunogenicity of RSV F expressed from this vector, resulting in substantially improved constructs (14, 15).
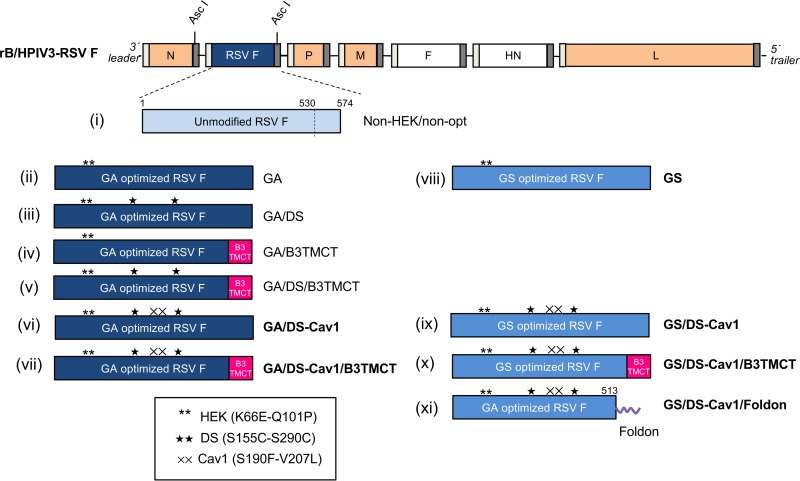
FIG 1.
Design of rB/HPIV3 vectors expressing various forms of RSV F. The genome of rB/HPIV3 is shown at the top. BPIV3 genes are orange, and HPIV3 F and HN genes are unfilled. The gene start and gene end sequences flanking each gene are illustrated as narrow light-gray and dark-gray bars, respectively. RSV F was inserted between the N and P genes at the AscI site. Eleven different forms of RSV F, i to xi, are shown and are described in the Results. Briefly, construct i, non-HEK/non-opt, is unmodified wt RSV F (light blue) and is a close facsimile of MEDI-534, which was previously evaluated in seronegative children (8). All other constructs contain the two HEK assignments (**). Constructs ii to vii were codon optimized by GeneArt (GA; dark blue); constructs viii to xi were codon optimized by GenScript (GS; medium blue). Other modifications (which are all indicated in the names of the constructs) include DS (★) and Cav-1 (××) amino acid substitutions, replacement of the transmembrane and cytoplasmic tail domains of RSV F with their counterparts from BPIV3 F protein (B3TMCT; red), and deletion of the RSV F TMCT and replacement with a phage T4 foldon sequence at the C terminus (horizontal purple squiggle in construct xi). Constructs that are aligned horizontally (ii and viii, vi and ix, and vii and x) are identical except for GA versus GS codon optimization. Names of constructs that were made in the present study (vi to xi) are in boldface; names of previously made constructs are in regular font. The illustration is not drawn to scale.
RSV F mediates fusion of the viral envelope with the host cell plasma membrane during entry through an irreversible conformational change from the prefusion (pre-F) to the postfusion (post-F) form (16–18). The pre- to post-F conformational change is concomitant with the loss of highly sensitive neutralizing epitopes present exclusively in pre-F, including a major neutralization site, called site Ø, as well as other sites identified recently (19–21). Antibodies that bind to pre-F-specific epitopes contribute to most of the neutralizing activity in human sera induced by natural infection (22, 23). The pre-F conformation is intrinsically metastable but can be stabilized with mutations identified by structure-based design (24, 25). One set of pre-F stabilizing mutations (S155C-S290C) introduces a disulfide bond (DS) that enhanced the immunogenicity of RSV F expressed by the rB/HPIV3 vector (14, 15). Additional structure-based mutations that further stabilize the pre-F conformation and/or increase RSV F protein expression may further improve the vector's immunogenicity for RSV. For example, two hydrophobic substitutions (S190F-V207L), called Cav1, fill cavities in the pre-F trimer and further stabilize the pre-F head and site Ø when introduced in addition to DS (25). Recombinant RSV F with DS-Cav1 was more immunogenic than the DS version as a subunit vaccine when injected with an adjuvant in mice and monkeys (25). However, the effect of adding Cav1 to DS in the context of vector-expressed RSV F had not been investigated.
The F glycoproteins of RSV and HPIV3 are packaged into their respective virions through an envelope-anchored transmembrane and a cytoplasmic tail (TMCT) domain (15, 26). Packaging of unmodified RSV F into rB/HPIV3 virions is very inefficient but can be greatly enhanced by replacing its TMCT with that of the vector BPIV3 F protein, resulting in a modification called B3TMCT that has been described previously (15). This modification improved the quantity and quality of the RSV-neutralizing serum antibody response in hamsters and monkeys (15). The RSV F ectodomain, lacking the TMCT domain, is expressed as a secreted post-F form (12, 14, 17). A secreted post-F expressed by rB/HPIV3 vector was much less immunogenic than the membrane-associated or the vector virion-packaged pre-F B3TMCT in hamsters (14). However, the immunogenicity of vector-expressed, secreted stabilized RSV pre-F has been unknown.
Increasing the magnitude of antigen expression by codon optimization can improve the immunogenicity of live viral vaccines and DNA vaccines (27–29). Previously, optimization of RSV F codon usage by GeneArt (Life Technologies) significantly increased its expression in cell culture by the rB/HPIV3 vector (14), but an increase in immunogenicity was not observed in the semipermissive hamster model. Commercial sources of codon optimization typically use different algorithms. For example, some simply avoid the rare codons and use only the most frequent codons, while others resemble the codon usage pattern of the target organism. Their consideration of GC content, secondary mRNA structure, cis-regulatory elements, and other factors might also be different. Altered codon usage may alter the composition of immune-stimulatory CpG elements and potentially immunogenic pathogen-associated molecular pattern (PAMP) enhancer motifs in the viral genome, which can affect the host interferon (IFN) response to infection and consequently the replication and immunogenicity of a live viral vaccine (30–32). Thus, the optimal codon usage for a live vaccine may need to be determined empirically.
In the present study, we systematically evaluated the effects of increased pre-F stabilization, packaging of the improved pre-F into vector virions, expression of secreted soluble pre-F trimers, and alternative algorithms of codon optimization and CpG content for RSV F in the context of the rB/HPIV3 vector. We demonstrated iterative improvements in the immunogenicity of RSV F with some of these modifications and identified a much-improved candidate vaccine suitable for clinical evaluation.
RESULTS
Generation of rB/HPIV3 vectors with modified RSV F constructs.
Figure 1 illustrates the 11 RSV F constructs evaluated in the present study, each expressed from the second gene position of the rB/HPIV3 vector. Constructs i to v are from previous studies (14, 15, 33), and constructs vi to xi were made in the present study. Construct i in Fig. 1, non-HEK/non-opt, encodes unmodified wild-type (wt) RSV F and thus is similar to MEDI-534, which was previously evaluated in seronegative children (see Introduction) (8). The other constructs in Fig. 1, ii to xi, are based on the same wt RSV F amino acid sequence as that for construct i and bear the two HEK amino acid assignments of 66E and 101P that make the encoded F protein identical at the amino acid level to an early passage of strain A2 (34). We previously found that the HEK assignments confer a modestly increased level of expression and substantially reduced fusion activity compared to non-HEK (14); thus, they may make F more stable and less prone to triggering. Other features of constructs ii to xi in Fig. 1 are as follows. Construct ii, GA, and viii, GS, are parallel constructs that encode HEK RSV F that has been codon optimized by, respectively, GeneArt (GA) and GenScript (GS). Thus, these two constructs compare the effects of GA versus GS.
Constructs iii, GA/DS, iv, GA/B3TMCT, and v, GA/DS/B3TMCT, encode GA-optimized HEK RSV F with DS mutations S155C and S290C (iii), the RSV F TMCT replaced with that of BPIV3 F (B3TMCT mutation) (iv), or the combined DS and B3TMCT mutations (v). Thus, these constructs evaluate the effects of DS versus B3TMCT versus DS plus B3TMCT in a GA backbone.
Constructs vi, GA/DS-Cav1, and vii, GA/DS-Cav1/B3TMCT, encode GA-optimized HEK RSV F with the DS mutations plus Cav1 mutations S190F and V207L (DS-Cav1) (vi) or DS-Cav1 combined with B3TMCT (vii). Thus, these constructs compare the effects of DS-Cav1 versus DS-Cav1 plus B3TMCT in a GA background.
Constructs ix, GS/DS-Cav1, and x, GS/DS-Cav1/B3TMCT, encode GS-optimized HEK RSV F, with DS-Cav1 (ix) or DS-Cav1 combined with B3TMCT (x), in a GS background. Thus, these constructs are parallel to constructs vi and vii, differing only in GA (vi and vii) versus GS (ix, x).
Construct xi, GS/DS-Cav1/Foldon, encodes the GA-optimized, 513-residue HEK RSV F ectodomain with DS-Cav1 mutations and the bacteriophage T4 fibritin trimerization domain (called foldon) fused at the C terminus.
These rB/HPIV3-RSV-F vector constructs were designed to be identical apart from the indicated changes to the RSV F open reading frame (ORF) or protein. Each F insert was inserted at the AscI site between the N and P genes in the rB/HPIV3 genome (Fig. 1) and was under the control of the same set of BPIV3 gene start and gene end transcription signals. All constructs were consistent with the “rule of six” (35, 36) for genome length. All of the viruses were readily rescued and grew to titers higher than 7.5 log10 50% tissue culture infectious doses (TCID50)/ml in LLC-MK2 cells (see below). Viral sequences were analyzed by automated sequencing of uncloned reverse transcription-PCR (RT-PCR) products representing the complete genomes, except for the very ends, due to priming there by synthetic oligonucleotides. The constructs were confirmed to be as designed and free of adventitious mutations.
Replication of rB/HPIV3 vectors in cell culture.
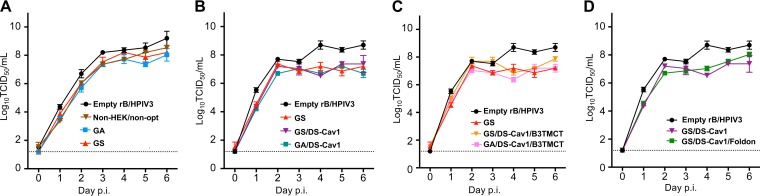
Multicycle replication of rB/HPIV3 vectors was evaluated in Vero cells (Fig. 2). The GA, GS, and non-HEK/non-opt constructs replicated with similar kinetics and were slightly more attenuated than the empty vector (Fig. 2A). Replication in vitro was not significantly affected by the DS-Cav1 mutations (Fig. 2B) or the B3TMCT mutation (Fig. 2C), by GA versus GS optimization (Fig. 2A to C), or by secreted RSV F with the C-terminal foldon (Fig. 2D).
FIG 2.
Multicycle replication of rB/HPIV3 vectors in Vero cells. Vero cells were infected with rB/HPIV3 vectors at an MOI of 0.01 TCID50 per cell. Viral replication over a period of 6 days was examined at 24-h intervals by withdrawing a sample of tissue culture medium for TCID50 titration. Replication kinetics of empty vector rB/HPIV3, non-HEK/non-opt, GA, and GS (A); empty vector, GS, GS/DS-Cav1, and GA/DS-Cav1 (B); empty vector, GS, GS/DS-Cav1/B3TMCT, and GA/DS-Cav1/B3TMCT (C); empty vector, GS/DS-Cav1, and GS/DS-Cav1/Foldon (D).
Expression of modified forms of RSV F.
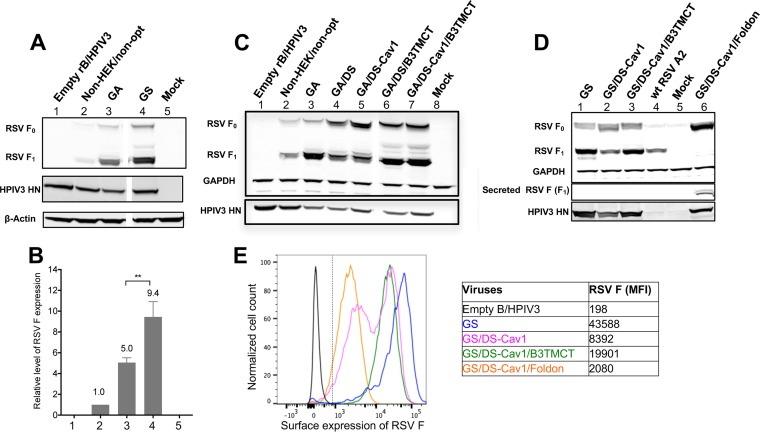
To evaluate the expression of RSV F, Vero cells were infected at a multiplicity of infection (MOI) of 10 TCID50 per cell with various constructs, and the expression of RSV F was evaluated by Western blotting of cell lysates prepared at 48 h postinfection (p.i.) (Fig. 3A to D). RSV F was detected as the 70-kDa F0 uncleaved precursor and the 50-kDa F1 subunit of the cleaved form using a mouse monoclonal antibody (MAb). GA was expressed at a 5-fold higher level than the nonoptimized wt RSV F (non-HEK/non-opt) (Fig. 3A and B, lanes 2 and 3). GS conferred a further 2-fold increase in expression compared to GA (Fig. 3A and B, lanes 3 and 4), yielding a total 9.4-fold increase in protein expression for GS versus non-HEK/non-opt (Fig. 3A and B, lanes 2 and 4). The DS and DS-Cav1 mutations reduced the cleavage efficiency of F0 (Fig. 3C, compare lanes 4 and 5 to lane 3). Inclusion of the B3TMCT mutation significantly increased expression of the GA/DS and GA/DS-Cav1 constructs (Fig. 3C, compare lanes 6 and 7 to lanes 4 and 5, respectively). The effects of DS-Cav1 in reducing F0 cleavage and B3TMCT in increasing RSV F expression were also observed with respective constructs optimized by GS (Fig. 3D, lane 1 to 3). The foldon construct (GS/DS-Cav1/Foldon) was detected predominantly as F0 in cell lysates but only as F1 secreted in the culture medium (Fig. 3D, lane 6). The expression of GS-optimized RSV F by the rB/HPIV3 vector was more efficient than that of unmodified RSV F by wt RSV (Fig. 3D, lanes 1 and 4).
FIG 3.
Expression of various forms of RSV F by rB/HPIV3 vectors. (A to D) Vero cells in 12-well plates were infected with rB/HPIV3 vectors expressing various forms of RSV F at an MOI of 10 TCID50 per cell or with wt RSV at an MOI of 3 PFU per cell as a control. Cultures were harvested 48 h p.i., and cell-associated (cell monolayer) and -secreted (clarified cell culture medium overlay) F protein was evaluated by Western blotting using antibodies conjugated to infrared dyes. (A) Cell-associated expression of RSV F1 and F0 and HPIV3 HN protein in monolayers infected with empty vector (lane 1), non-HEK/non-opt (lane 2), GA (lane 3), GS (lane 4), or mock infected (lane 5). A lane from the original image between the present lanes 3 and 4 was removed to make this figure. The removed lane represented rB/HPIV3 expressing another codon-optimized (DNA2.0) RSV F that was removed because it was not included in the present manuscript. (B) Quantification of RSV F1 + F0 band densities from the experiment shown in panel A, as well as two additional replicate wells per construct from the same experiment, with values normalized to lane 2 (non-HEK/non-opt) set at a value of 1.0. Error bars indicate the standard errors of the means (SEM). (C) Cell-associated expression of RSV F1 and F0 and HPIV3 HN protein in monolayers infected with empty vector (lane 1), non-HEK/non-opt (lane 2), GA (lane 3), GA/DS (lane 4), GA/DS-Cav1 (lane 5), GA/DS/B3TMCT (lane 6), GA/DS-Cav1/B3TMCT (lane 7), or mock infected (lane 8). Two lanes from the original image between the present lanes 5 and 6 were removed to make this figure. The removed lanes represented rB/HPIV3 expressing RSV F GA/DS and GA/DS-Cav1, each containing BPIV3 CT instead of TMCT, and the lanes were removed because these viruses were not included in the present manuscript. (D) Cell-associated (upper and lower) and secreted (middle) expression of RSV F1 and F0 and HPIV3 HN protein in cell cultures infected with GS (lane 1), GS/DS-Cav1 (lane 2), GS/DS-Cav1/B3TMCT (lane 3), wt RSV (lane 4), mock infected (lane 5), and GS/DS-Cav1/Foldon (lane 6). (E) Vero cells in 12-well plates were infected at an MOI of 5 TCID50 per cell with empty rB/HPIV3 vector (black), GS (blue), GS/DS-Cav1 (pink), GS/DS-Cav1/B3TMCT (green), and GS/DS-Cav1/Foldon (orange). Cultures were harvested 48 h p.i., and surface expression of RSV F was determined by flow cytometry analysis of nonpermeabilized cells. The histogram plot (left) shows surface expression of RSV F of live infected single cells, with the y axis indicating the normalized cell count and the x axis the intensity of RSV F staining. The vertical dashed line indicates gating between RSV F negative versus positive cells. The median fluorescence intensities (MFI) are shown in the right panel.
The relative quantity of RSV F on the surface of cells infected with various GS-optimized constructs was determined by flow cytometry analysis (Fig. 3E). Vero cells were infected at an MOI of 5 TCID50 per cell and analyzed at 48 h p.i. As expected, due to the lack of a TM domain, the pre-F foldon construct had the least surface expression (Fig. 3E, orange). The inclusion of the DS-Cav1 mutations substantially reduced the surface expression of RSV F (Fig. 3E, pink), probably due to the reduced cleavage efficiency noted previously. The inclusion of the B3TMCT mutation into DS-Cav1 increased surface expression (Fig. 3E, green), in agreement with the effect on enhancing total expression (Fig. 3C, lanes 6 and 7, and D, lane 3).
Characterization of RSV F packaged in the rB/HPIV3 virions.
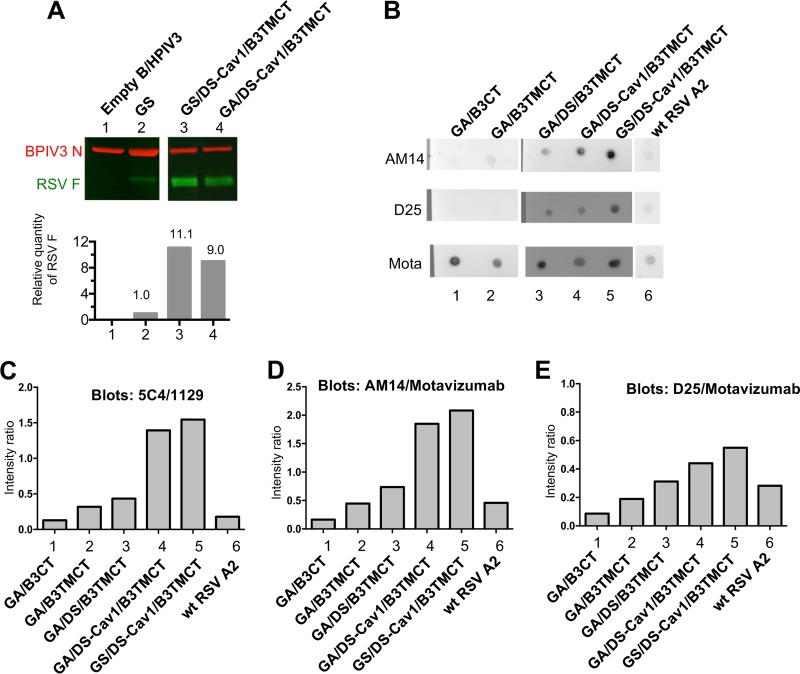
We previously showed that the B3TMCT mutation confers enhanced packaging of RSV F in rB/HPIV3 virions (15). Since the GS modification increased the expression of cell-associated RSV F protein (Fig. 3A), we examined whether it also increased the amount of RSV F packaged into vector virions. The packaging efficiency of GA- and GS-optimized DS-Cav1/B3TMCT was compared by Western blotting of sucrose gradient-purified viruses (Fig. 4A). The GS/DS-Cav1/B3TMCT construct had a 23% increase in packaging compared to GA/DS-Cav1/B3TMCT (Fig. 4A, lane 3 versus lane 4), indicating that increased intracellular expression did result in increased packaging. The packaging of GA/DS-Cav1/B3TMCT and GS/DS-Cav1/B3TMCT was 9.0-fold and 11.1-fold greater, respectively, than that of the GS construct lacking pre-F stabilization or packaging signals (Fig. 4A, lanes 4 and 3 versus lane 2).
FIG 4.
Characterization of RSV F packaged in the rB/HPIV3 virions. (A) Quantification of the relative amount of RSV F packaged in rB/HPIV3 virions. Two micrograms of sucrose gradient-purified virions for r/B/HPIV3 empty vector (lane 1), GS (lane 2), GS/DS-Cav1/B3TMCT (lane 3), and GA/DS-Cav1/B3TMCT (lane 4) was subjected to Western blot analysis as described previously (15). As shown in the upper panel, protein bands were visualized by antibodies conjugated to infrared dyes: RSV F was visualized in green and the BPIV3 N protein in red. The ratio of RSV F to BPIV3 N in each gel lane was calculated and normalized to that of GS set at 1.0 and is shown in the lower panel. Seven lanes from the original image between the present lanes 2 and 3 were removed to make this figure. The removed lanes represented other constructs, controls, and size markers not needed in this figure. (B to E) Determination of the relative content of RSV pre-F protein in virions of GA/B3CT (lane 1), GA/B3TMCT (lane 2), GA/DS/B3TMCT (lane 3), GA/DS-Cav1/B3TMCT (lane 4), GS/DS-Cav1/B3TMCT (lane 5), and wt RSV (lane 6). Viral stocks (clarified cell culture medium supernatants) were blotted as dots on replicate nitrocellulose membrane strips. The dilution factor for each virus was determined as described in Materials and Methods. The dot blots were analyzed with MAbs specific for RSV pre-F protein, namely, D25, AM14, and 5C4, or with MAbs that bind to both pre- and post-F protein, namely, 1129 and motavizumab (Mota). (B) Representative dot blots. (C to E) Relative binding intensities of the MAbs 5C4 (C), AM14 (D), and D25 (E), which quantify pre-F, normalized to the binding intensity of 1129 (C) and motavizumab (D and E), which quantify total F. Each value shown is an average of duplicate blots.
The conformation of virion-packaged RSV F was evaluated with a virion dot blot assay. (Note that all of the constructs in this comparison contained the B3CT or B3TMCT mutations that confer efficient packaging.) Virus stocks were blotted onto a nitrocellulose membrane and then incubated with RSV F epitope-specific MAbs. The MAbs D25 and 5C4 bind to site Ø in pre-F and bind only to pre-F; AM14 spans a quaternary epitope between sites V and IV across two protomers and binds only to trimeric pre-F; 1129 and motavizumab bind to site II, which is present in both pre-F and post-F, and thus quantifies the total amount of F (16, 17, 20, 23, 37). Images of representative dot blots are shown as examples (Fig. 4B). The intensity of binding by D25, 5C4, or AM14 was normalized to that of either 1129 or motavizumab. This quantified the amount of pre-F present in virions relative to the total amount of F (Fig. 4C to E). For comparison, we included a previously described hyperfusogenic RSV F construct, GA/B3CT (15), which consists of GA-optimized RSV F with the CT domain replaced by that of BPIV3 (not shown in Fig. 1).
The hyperfusogenic GA/B3CT had low ratios of 5C4/1129, AM14/motavizumab, and D25/motavizumab, illustrating a low relative proportion of pre-F to total F (Fig. 4C, D, and E, lanes 1). In contrast, modification of RSV F by inclusion of the B3TMCT mutation in place of B3CT increased the proportion of pre-F (Fig. 4C, D, and E, lanes 2). The further inclusion of the DS mutations increased the proportion of pre-F (Fig. 4C, D, and E, lanes 3), and this was further increased by the addition of the Cav1 mutations (lanes 4, GA optimized; lanes 5, GS optimized). Vectors with RSV F stabilized by DS or DS-Cav1, in combination with B3TMCT, had a greater relative amount of packaged pre-F displayed on virions than wt RSV (Fig. 4B to E, lanes 3 to 6).
Replication of rB/HPIV3 vectors in hamsters.
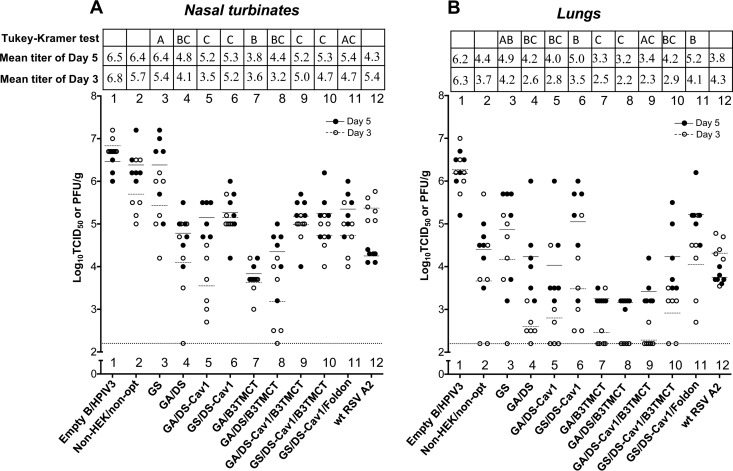
In vivo replication of vectors expressing the various forms of RSV F was evaluated in a hamster model. Hamsters were infected intranasally (i.n.) with 105 TCID50 of the various vectors or 106 PFU of wt RSV. Nasal turbinates (Fig. 5A) and lungs (Fig. 5B) were collected on days 3 and 5, and viral titers were determined by TCID50 titration and plaque titration for vectors and RSV, respectively. In general, vectors containing an RSV F insert replicated to lower titer and more slowly (i.e., day 5 titer was higher than day 3) than the empty vector (Fig. 5A and B, lanes 1 to 11), indicating an attenuating effect of the RSV F insert. The magnitude of the attenuation varied between constructs, although the differences between groups usually were not statistically significant due to the considerable animal-to-animal variation within groups in this outbred animal model. The GS construct replicated with efficiency similar to that of the non-HEK/non-opt construct (Fig. 5A and B, lanes 2 and 3). In comparison, constructs optimized by GA were slightly more attenuated than the respective constructs optimized by GS (Fig. 5A and B, lanes 5 versus 6 and lanes 9 versus 10); this effect was somewhat more evident in the lungs than in the nasal turbinates, although the differences between comparable GA and GS constructs were not statistically significant. The presence of pre-F stabilizing mutations (e.g., Fig. 5A and B, lanes 3 versus 6) or B3TMCT (lanes 4 versus 8) in either GA- or GS-based constructs increased the attenuating effect of the insert. In general, the presence of GA optimization combined with pre-F stabilization and/or the B3TMCT mutation was associated with the greatest attenuation (e.g., Fig. 5A, lanes 7 and 8, and B, lanes 7, 8, and 9). The GS/DS-Cav1/Foldon construct replicated with kinetics similar to those of the other GS-optimized constructs in nasal turbinates (Fig. 5A, lane 11 versus 6 and 10) and was only somewhat less attenuated than the GS-optimized B3TMCT-containing construct in lungs (Fig. 5B, lane 11 versus 10), indicating replacing the TMCT with T4 foldon sequence did not affect vector replication in hamsters.
FIG 5.
Replication of rB/HPIV3 vectors in hamsters. Hamsters were infected i.n. with 105 TCID50 of the indicated rB/HPIV3 construct or 106 PFU of wt RSV. Viral titers in homogenates of the nasal turbinates (A) and lungs (B) from animals sacrificed on days 3 and 5 after inoculation are shown. Titers of the indicated rB/HPIV3 vectors (lanes 1 to 11) and wt RSV (lane 12) were determined by TCID50 and plaque assay, respectively, as described in Materials and Methods. Viral titers on days 3 and 5 for individual animals are shown by open and filled dots, respectively. Mean titers are indicated in the plots by horizontal lines and by numerical values shown in the table at the top. The limit of detection for TCID50 assay was 2.2 log10 TCID50/g, shown as a dashed line. The statistical significance of the difference between groups on day 5 was determined by one-way ANOVA followed by a Tukey-Kramer test. Mean titers of any two groups with the same letter (A, B, or C) are not significantly different.
Immunogenicity and protective efficacy of RSV F expressed by rB/HPIV3 vectors in hamsters.
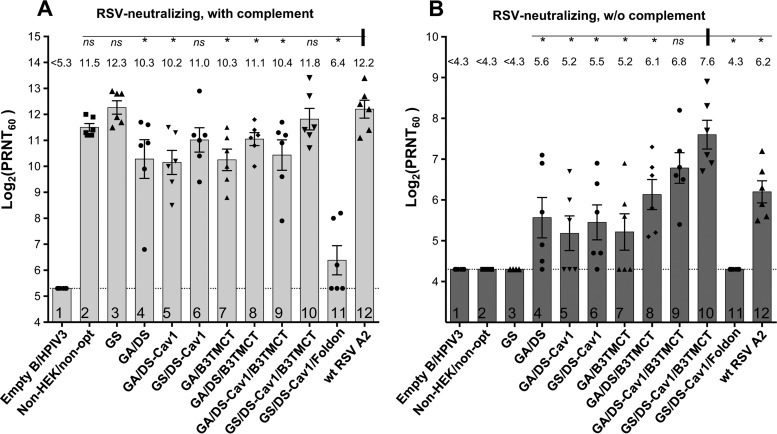
To evaluate the immunogenicity and protective efficacy of the various rB/HPIV3-RSV-F constructs, additional groups of hamsters were inoculated as described above for the replication study, and sera were collected on day 28 p.i. Serum RSV-neutralizing antibody titers were determined by 60% plaque reduction neutralization test (PRNT60) with or without added complement (Fig. 6A and B, respectively). On day 30 p.i., the hamsters were challenged i.n. with 106 PFU of wt RSV. On day 3 postchallenge, wt RSV titers in nasal turbinates and lungs were determined by plaque assay.
FIG 6.
Serum RSV-neutralizing antibody titers induced by rB/HPIV3 vectors and wt RSV in hamsters. Hamster sera collected on day 28 postimmunization were assayed for RSV-neutralizing antibodies by PRNT60. Neutralization titers in the presence (A) or absence (B) of added complement are shown. Mean titers of sera in each group are shown as bars ± standard errors of the means, with mean values also shown on the top. Each dot indicates an individual animal in the group. Statistical significance of the differences between the mean titers of various groups versus that of wt RSV (marked with a vertical bar) (A) and versus GS/DS-Cav1/B3TMCT (marked with a vertical bar) (B) was determined by an unpaired t test. ns, not significant (P > 0.05); *, significant (P < 0.05).
In a PRNT60 performed in the presence of added complement, each of the rB/HPIV3-RSV-F constructs (Fig. 6A, lanes 2 to 10), except for GS/DS-Cav1/Foldon (Fig. 6A, lane 11), induced high titers of RSV-neutralizing antibodies (>10.0 log2 PRNT60). Thus, the secreted soluble form of RSV F was poorly immunogenic despite being stabilized in the highly immunogenic pre-F conformation, as well as in trimers, by the foldon sequence. Three other GS-optimized constructs, including GS, GS/DS-Cav1, and GS/DS-Cav1/B3TMCT (Fig. 6A, lanes 3, 6, and 10), induced RSV-neutralizing antibodies at titers comparable to that induced by wt RSV (Fig. 6A, lane 12). In the case of GS and GS/DS-Cav1, these higher antibody titers correlated with relatively greater replication compared to the other vectors. In contrast, the various GA-optimized constructs (Fig. 6A, lanes 4 and 5 and lanes 7 to 9) induced significantly lower antibody titers than wt RSV (Fig. 6A, lane 12), which correlated with their relatively lower replication compared to those of the other vectors.
RSV-neutralizing serum antibodies also were assayed in the absence of complement (Fig. 6B). Antibodies with neutralizing activity under these conditions were termed “high quality.” As previously reported, the DS and B3TMCT mutations independently enhanced immunogenicity for high-quality neutralizing antibodies (Fig. 6B, lanes 4 and 7) compared to constructs without DS or B3TMCT mutations, which did not induce detectable high-quality RSV-neutralizing antibodies (Fig. 6B, lanes 2 and 3). Furthermore, in combination with the B3TMCT mutation, the further addition of the DS mutation or the DS-Cav1 mutation, or replacement of GA optimization with GS optimization in the presence of DS-Cav1 and B3TMCT, exhibited a trend of additive improvements in the RSV-neutralizing antibody response (Fig. 6B, lanes 7 to 10). It is notable that the GS/DS-Cav1/B3TMCT construct (Fig. 6B, lane 10) induced significantly higher titers of complement-independent RSV-neutralizing antibodies than most of the tested constructs, including a leading candidate developed in a previous study, i.e., GA/DS/B3TMCT (Fig. 6B, lane 8). Furthermore, the GS/DS-Cav1/B3TMCT construct induced a higher titer of complement-independent RSV-neutralizing antibodies than wt RSV, a difference that was statistically significant (Fig. 6B, lane 10 versus 12).
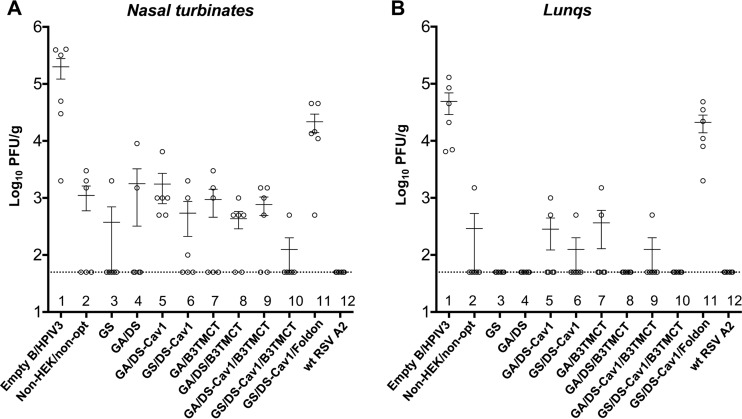
With the exception of the foldon construct (Fig. 7A and B, lanes 11), vectors expressing the various forms of RSV F were protective in hamsters against wt RSV challenge (Fig. 7A and B, lanes 2 to 10). None of the rB/HPIV3-RSV-F constructs conferred complete protection in the nose, although groups immunized with the GS and GS/DS-Cav1/B3TMCT constructs had detectable virus in only one animal each (Fig. 7A, lanes 3 and 10). Several constructs, including GS and GS/DS-Cav1/B3TMCT, conferred complete protection in the lungs. wt RSV provided complete protection in both the nose and lungs (Fig. 7A and B, lanes 12), which was not unexpected, since wt RSV also expresses the additional RSV neutralization antigen G, as well as all of the other RSV proteins, as potential antigens for cellular immunity. The GS-optimized constructs exhibited a trend of improved protection compared to the respective constructs optimized by GA (Fig. 7A and B, lanes 5 versus 6 and lanes 9 versus 10), in agreement with their increased RSV F expression (Fig. 3A and B), enhanced replication in vivo (Fig. 5B), and the stronger induction of RSV-neutralizing antibodies (Fig. 6).
FIG 7.
Protection of hamsters against wt RSV challenge. Hamsters were challenged with 106 PFU of wt RSV i.n. on day 30 postimmunization. Titers of wt RSV on day 3 postchallenge in nasal turbinates (A) and lungs (B) determined by plaque assay are shown. Mean titers with SEMs are shown as horizontal lines. The limit of detection is indicated by the dashed line.
Evaluation of rB/HPIV3 vectors expressing virion-packaged pre-F in nonhuman primates.
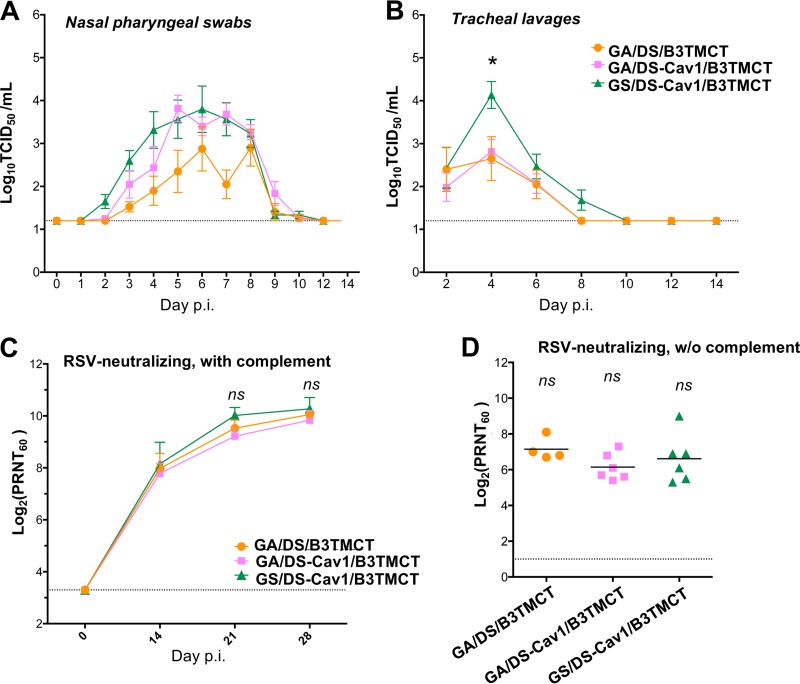
Three selected constructs were evaluated in nonhuman primates, including GA/DS/B3TMCT, i.e., a leading construct identified in a previous study (15), and two constructs made in the present study, GA/DS-Cav1/B3TMCT and GS/DS-Cav1/B3TMCT. Rhesus monkeys were inoculated i.n. and intratracheally (i.t.) with 106 TCID50 of vector per site. Replication of vectors in the upper and lower respiratory tract (URT and LRT, respectively) was monitored for 14 days after immunization by TCID50 titration of nasal pharyngeal swabs and tracheal lavages. Sera were collected on days 0, 14, 21, and 28 and assayed for RSV-neutralizing antibodies in the presence and absence of added complement.
Replication of the three vectors reached peak titers on days 5 to 6 in the URT and day 4 in the LRT (Fig. 8A and B, respectively). The GS/DS-Cav1/B3TMCT construct replicated marginally faster than GA/DS-Cav1/B3TMCT in the URT (Fig. 8A, green and pink, respectively) and reached significantly (P < 0.05) higher titers in the LRT (Fig. 8B), suggesting that GA optimization was more attenuating than GS. This was consistent with the greater attenuation of GA-optimized constructs versus GS-optimized constructs in the lungs of hamsters (Fig. 5B, lanes 9 and 10). Adding the Cav1 mutations to DS/B3TMCT increased replication in the URT but not in the LRT (Fig. 8A and B, pink versus orange), consistent with similar observations in hamsters (Fig. 5A and B, lanes 8 and 9). Despite their differences in replication, the three constructs induce similar titers of serum RSV-neutralizing antibodies (Fig. 8C and D).
FIG 8.
Evaluation of selected rB/HPIV3 vectors in nonhuman primates. Rhesus monkeys were inoculated i.n. and i.t. with 106 TCID50 per site of GA/DS/B3TMCT (4 animals), GA/DS-Cav1/B3TMCT (6 animals), and GS/DS-Cav1/B3TMCT (6 animals). Replication in the upper and lower respiratory tracts was determined by viral titration of nasal pharyngeal (NP) swab and tracheal lavage (TL) samples, respectively, collected over a period of 14 days. (A) Mean titers of vectors in NP swabs. (B) Mean titers of vectors in TLs. An asterisk indicates that the mean TL titer of GS/DS-Cav1/B3TMCT on day 4 is significantly different from that of the other two groups (P < 0.05 by one-way ANOVA followed by a Tukey-Kramer test). (C) Mean titers of serum RSV-neutralizing antibodies in the presence of added complement in monkey sera collected on days 0, 14, 21, and 28 postimmunization. Error bars indicate SEM. (D) Complement-independent serum RSV-neutralizing titers in monkey sera collected on day 28 postimmunization. Each data point is the titer from an individual monkey; horizontal bars indicate the mean titer of each group. ns, the mean titers are not significantly different by t test.
Characterization of conformation-specific and epitope-specific antibody response induced by various forms of RSV F.
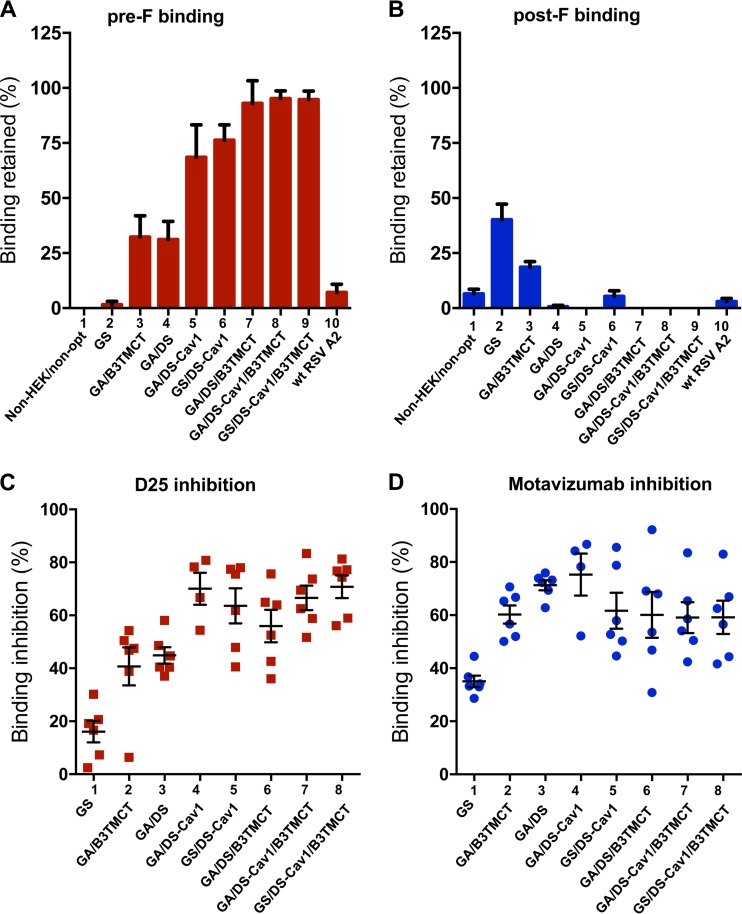
Pre-F-specific antibodies account for most of the RSV-neutralizing activity in the human convalescent serum (22, 23). To evaluate the pre-F-specific response in the present study, the proportions of pre-F-specific antibodies in hamster sera were quantified by competitive pre-F protein binding assays in the presence of excess post-F protein competitor (Materials and Methods) (Fig. 9A). The efficiency of competition by the post-F competitor was validated with a post-F binding assay (Materials and Methods), which confirmed efficient quenching of post-F-binding antibodies by an excess of soluble post-F (Fig. 9B). As expected, vectors expressing RSV F that had neither pre-F stabilization nor the B3TMCT packaging signal failed to induce a robust pre-F-specific antibody response (Fig. 9A, lanes 1 and 2). wt RSV similarly failed to induce a robust pre-F-specific antibody response (Fig. 9A, lane 10). In contrast, the B3TMCT, DS, and DS-Cav1 mutations individually and additively enhanced the pre-F-specific antibody response (Fig. 9A, lanes 3 to 9). In the absence of the B3TMCT mutation, adding Cav1 to DS significantly improved the pre-F-specific response (Fig. 9A, lane 4 versus 5). However, in the presence of the B3TMCT, adding the Cav1 mutation to the DS mutation did not give a further increase in pre-F-specific antibodies (Fig. 9A, lane 7 versus 8), likely because the pre-F-specific response induced by DS/B3TMCT was reaching the maximum detection limit of the assay. GA versus GS codon optimization did not have any detectable effect on the pre-F-specific response, as might have been expected (Fig. 9A, lane 5 versus 6 and lane 8 versus 9).
FIG 9.
Characterization of conformation-specific and epitope-specific hamster serum antibodies induced by various rB/HPIV3 constructs. (A and B) Conformation-specific antibodies. Binding by hamster serum antibodies to immobilized pre-F (A) or post-F (B) protein in the presence of soluble post-F protein competitor, measured by Octet bilayer interferometry, shown as a percentage of binding in the absence of soluble post-F competitor. (C and D) Epitope-specific antibodies. Percentage of pre-F binding by serum antibodies inhibited by the site Ø-specific MAb D25 unique to pre-F (C) or site II-specific motavizumab, which binds pre- and post-F (D).
We then assessed epitope-specific antibody responses using sera from the same hamster experiment. This was done by measuring the ability of MAb D25 (site Ø specific, unique to pre-F) or motavizumab (site II specific, present in pre- and post-F) to compete for the binding of hamster serum antibodies to pre-F protein. Pre-F protein immobilized on the tip of a sensor was preincubated with competing MAb (D25 or motavizumab) or a noncompeting control MAb (310.63) and then assayed for binding by serum antibodies from the individual hamsters in each group. Pre-F binding by sera induced by the GS construct was not efficiently competed by D25 (Fig. 9C, lane 1), indicating a low content of site Ø-specific antibodies in the sera. In contrast, pre-F binding by GA/B3TMCT and GA/DS sera was more efficiently competed by D25 (Fig. 9C, lanes 2 and 3), indicating these sera had an increased content of site Ø-specific antibodies. The competition was even stronger with GA/DS/B3TMCT sera, indicating an even higher proportion of site Ø-specific antibodies in these sera (Fig. 9C, lane 6). These results indicated that the B3TMCT and DS mutations independently and additively stabilized site Ø. Combining the Cav1 and DS mutations also led to greater competition by D25, indicating that Cav1 and DS additively stabilized site Ø (Fig. 9C, lane 3 versus 4). GS versus GA codon optimization did not affect the stability of site Ø, as would be expected (Fig. 9C, lane 4 versus 5 and 7 versus 8). These results are in line with the competitive pre-F binding assay (Fig. 9A). Motavizumab competed with similar efficiency for pre-F binding by serum antibodies induced by the various RSV F constructs, except GS, indicating that the proportions of site II-specific antibodies were similar (Fig. 9D). The lower competition with GS sera by motavizumab was probably due to an overall low level of antibodies binding to pre-F even in the absence of competitor.
We also assessed the epitope-specific antibody responses in the rhesus monkeys immunized with GA/DS/B3TMCT, GA/DS-Cav1/B3TMCT, and GS/DS-Cav1/B3TMCT from the experiment shown in Fig. 8. No significant differences were detected among the three groups for competition by D25 or motavizumab (data not shown), which is consistent with the observations for sera from hamsters infected with the same three constructs (Fig. 9A and B, lanes 7 to 9, and C and D, lanes 6 to 8). Thus, each of the combinations of DS or DS-Cav1 with B3TMCT had a high degree of site Ø maintained on the rB/HPIV3-expressed RSV F able to elicit antibody response.
Reduced replication efficiencies of GA-optimized vectors correlate with increased CpG content and an increased antiviral innate response in vitro.
As described above, the constructs that were GA codon optimized tended to be more restricted for replication than those that were GS codon optimized. In addition, the GS codon-optimized constructs tended to be more immunogenic for serum RSV-neutralizing antibodies, likely as a consequence of the greater replication and antigenic load. We investigated whether any of these effects were associated with differences in the sequence of the RSV F ORF due to codon optimization.
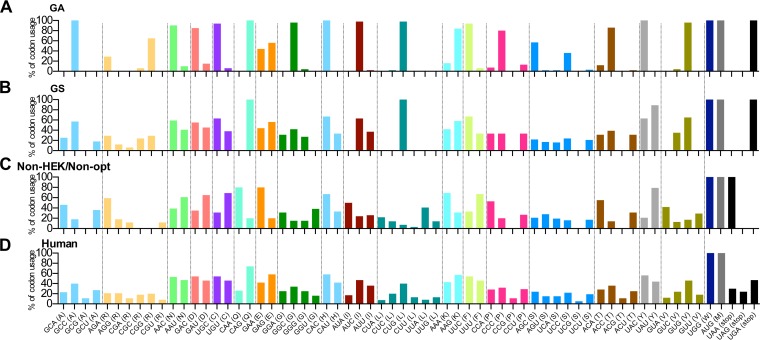
Codon usage in the GA-optimized, GS-optimized, and nonoptimized RSV F ORFs was compared with that of overall human usage (Fig. 10). The codon usage in unmodified wt RSV F (Fig. 10C, non-HEK/non-opt) displayed a different codon preference than human codon usage (Fig. 10D). GS optimization (Fig. 10B) generally mimicked the human codon preferences, except that it has CUG as the sole leucine codon. The reason for the biased preference of CUG is unclear. In contrast, the codon usage of GA optimization (Fig. 10A) was strongly skewed toward the most frequent human codons. More importantly, we compared the GC contents and numbers of CpG dinucleotides in the RSV F ORF versus ORFs encoding structure proteins of rB/HPIV3 (Table 1). This was done because CpG dinucleotides in an RNA virus can be sensed as a pathogen-associated molecular pattern and can trigger the innate IFN-mediated antiviral response. The percentage of GC content of native RSV F was 34.7%, similar to those of the ORFs encoding rB/HPIV3 structural proteins, ranging from 33.3% to 41.3%. GA and GS optimization increased the GC content of RSV F to 57.6% and 49.2%, respectively. The number and frequency of CpG dinucleotides, as potential immunostimulatory CpG elements, were also greatly increased from 12 (0.7%) in the nonoptimized RSV F to 100 (5.8%) in GA-optimized F and 66 (3.83%) in GS-optimized F, suggesting that GA optimization has the potential for a stronger immune-stimulatory effect and more rapid clearance of virus from antigen-expressing cells.
FIG 10.
Comparison of RSV F and human codon usage. The codon usage in the GA-optimized (A), GS-optimized (B), and unmodified wt (C) RSV F ORF was compared with human codon usage (D). The usage of each codon was calculated as the frequency (in percent) of its use among all codons for that amino acid; codons for each particular amino acid were clustered together and are shown as the same color. The codon usage of the different versions of RSV F was analyzed using the software Lasergene; the human codon usage data are from the Codon Usage Database (http://www.kazusa.or.jp/codon/).
TABLE 1.
Analysis of GC content and CpG dinucleotide frequencies in rB/HPIV3-RSV F ORFsa
| ORFb | No. of nucleotides | % GC contentc | No. of CpG dinucleotidesd | CpG dinucleotide frequencye (%) |
|---|---|---|---|---|
| Non-HEK/non-opt | 1,725 | 34.7 | 12 | 0.70 |
| GA | 1,725 | 57.6 | 100 | 5.80 |
| GS | 1,725 | 49.2 | 66 | 3.83 |
| N | 1,548 | 41.3 | 28 | 1.81 |
| P | 1,791 | 41.1 | 31 | 1.73 |
| M | 1,056 | 40.0 | 13 | 1.23 |
| F | 1,620 | 35.6 | 17 | 1.05 |
| HN | 1,719 | 36.1 | 16 | 0.93 |
| L | 6,702 | 33.4 | 68 | 1.01 |
Sequences were analyzed with the GeneQuest module in the Lasergene 13 software package (DNASTAR).
Coding sequences of ORFs of RSV F (non-HEK/non-opt, GA, and GS) and rB/HPIV3 structural proteins (N, P, M, F, HN, and L) as illustrated in Fig. 1 were analyzed.
Percentage of G and C nucleotides in the ORF.
Number of CpG dinucleotides in the ORF.
Frequency of CpG dinucleotides in the ORF.
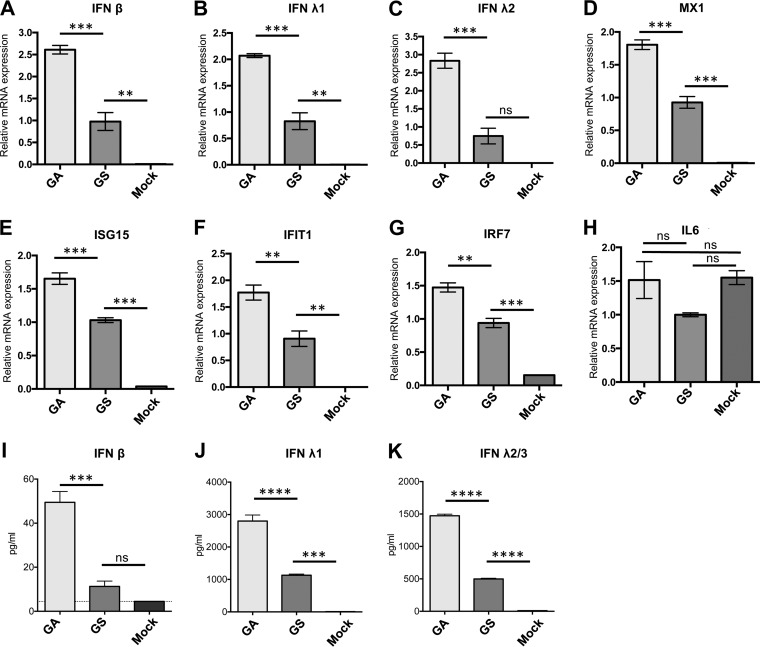
To determine whether this difference in CpG content indeed affected immune stimulation, the innate immune response induced by infection with GA- and GS-optimized DS-Cav1/B3TMCT was evaluated in an IFN-competent human lung epithelial A549 cell line. Cells were infected with sucrose-purified viruses at an MOI of 10 PFU per cell. At 18 h p.i., the expression of IFN-β (type I), IFN-λ1, IFN-λ2 (type III), and selected IFN-stimulated genes (ISGs) was analyzed by quantitative RT-PCR (qRT-PCR). In addition, at 24 h p.i., the amount of IFNs secreted into the cell culture medium was measured by enzyme-linked immunosorbent assay (ELISA). The level of transcripts in GS was used for normalization (assigned a value of 1.0), and transcript levels for GA are shown as fold difference relative to GS. The GA-optimized construct induced 2- to 3-fold higher levels of IFN-β and IFN-λ1-2 mRNA expression than the GS-optimized construct (Fig. 11A to C). A similar trend was also observed for the transcripts of ISGs, such as MX1 (Fig. 11D), ISG15 (Fig. 11E), IFIT1 (Fig. 11F), and IRF7 (Fig. 11G). Expression of the proinflammatory cytokine interleukin-6 (IL-6) was similar for GA, GS, and mock infection and was not induced by infection at 18 h p.i. (Fig. 11H). Consistent with mRNA expression, GA-optimized DS-Cav1/B3TMCT induced 3- to 4-fold higher levels of secreted IFN-β and IFN-λ1-3 than the construct optimized by GS (Fig. 11I to K).
FIG 11.
Cytokine responses in A549 cells infected with rB/HPIV3 vector expressing GA- or GS-optimized RSV F. Human lung alveolar epithelial A549 cells were infected with sucrose-purified GA- versus GS-optimized DS-Cav1/B3TMCT viruses at an MOI of 10 TCID50 per cell. (A to H) Three replicate wells of cell lysates were harvested 18 h p.i. Total cell-associated RNA was extracted and reverse transcribed to cDNA, followed by qRT-PCR to quantify the relative expression of mRNAs for IFN-β (A), IFN-λ1 (B), IFN-λ2 (C), and ISGs, including MX1 (D), ISG15 (E), IFIT1 (F), IRF7 (G), and IL-6 (H). Each column represents the mean value of relative mRNA expression from three replicate samples. Error bars indicate SEM. (I to K) From another set of three replicate wells, cell culture medium supernatants were harvested at 24 h p.i. and quantified by ELISA for the following proteins: IFN-β (I), IFN-λ1 (J), and IFN-λ2/3 (K). Each column represents the average concentration (in picograms per milliliter) of secreted IFN in the medium supernatants of three replicate wells. Error bars indicate SEM. The statistical significance of the difference between groups was determined by one-way ANOVA followed by Tukey-Kramer test. The P values for the statistical significance of differences between two groups are indicated by asterisks: **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
DISCUSSION
A major challenge for the development of a pediatric RSV vaccine is to induce a robust and protective response in infants and young children with a virus that is sufficiently attenuated to be well tolerated (38). Live attenuated RSV vaccine candidates for pediatric use are being actively developed. RSV bearing a deletion of M2-2 ORF was found to express increased amounts of viral antigens and was highly attenuated and quite immunogenic in a clinical trial in seronegative children (39). Several versions of M2-2 deletion viruses are presently being evaluated in clinical trials (e.g., Clinicaltrials.gov registration no. NCT02237209). RSV vaccine candidates in preclinical development include thermostable RSV with deletions and modifications of multiple proteins (40), RSV modified by codon pair deoptimization (41), RSV bearing deletions of the NS2 gene, and the codon for amino acid S1313 of the L protein (42), or RSV that expresses a less fusogenic F protein (43).
Compared to a live attenuated RSV strain, an HPIV vector expressing RSV F protein engineered to be stabilized in the pre-F conformation offers several unique advantages. (i) While RSV does not replicate efficiently in cell culture and can readily lose infectivity unless handled carefully, HPIVs grow much more efficiently and are physically more stable. This should greatly facilitate manufacture, distribution, and use, and in particular it would facilitate their use in resource-limited settings where 99% of RSV-related childhood mortality occurs and a vaccine is most needed. (ii) Pre-F protein appears to be the optimal antigen for inducing highly neutralizing antibodies, and the use of a stabilized form provides a “better-than-nature” antigen that induces an antibody response that is strongly skewed in favor of these desirable antibodies. When used in the first exposure in life to RSV F, this might prime for superior lifelong antibody responses. The F protein of an attenuated RSV strain cannot be substantially stabilized in the pre-F conformation because it would be inactive in fusion. (iii) Whereas immunity to RSV, such as that from a prior vaccination or a passive antibody strategy, would strongly restrict the replication of attenuated RSV strains, it would provide little or no restriction of an HPIV vector. Thus, an HPIV-vectored RSV vaccine should be particularly well suited for use in combination with passive antibody strategies, such as parenterally administered MAbs or maternal immunization, and also should be effective for booster immunizations following the use of a live attenuated RSV vaccine. (iv) The rB/HPIV3 vector has already been shown to be attenuated and well tolerated in seronegative infants and children, both as the empty vector and expressing wt (unmodified) RSV F (8, 44), so this vector expressing an improved RSV F protein can be expected to have a desirable safety profile that would expedite development.
Because rB/HPIV3 expressing unmodified RSV F, called MEDI-534, was attenuated and well tolerated in seronegative children and was immunogenic against HPIV3 but unsatisfactorily immunogenic against RSV (8), we have been evaluating a number of parameters to improve the immunogenicity of RSV F expressed from this vector. These include altering the insert position for RSV F in the vector genome, optimizing codon usage, using the RSV F sequence from an early-passage virus, comparing membrane-anchored versus secreted F, increasing RSV F packaging in the vector virion, and stabilizing RSV F in the pre-F conformation (14, 15, 33). We recently described an optimized candidate, GA/DS/B3TMCT, that induced higher quantity and quality of RSV-neutralizing antibodies in monkeys than a construct resembling MEDI-534 (15). In the present study, we sought to further enhance RSV F immunogenicity by evaluating the effect of (i) increased pre-F stabilization, (ii) different codon-optimized sequences of RSV F, and (iii) secreted soluble pre-F trimers.
Our previous candidate GA/DS/B3TMCT, noted above, which expresses an F protein that was partially stabilized in the pre-F conformation by DS mutations, was more immunogenic than unmodified F or post-F (14, 15), indicating that pre-F stabilization enhanced the immunogenicity of vector-expressed RSV F. In the present study, further stabilization of pre-F by adding the Cav1 mutations to DS further enhanced RSV F immunogenicity. The pre-F-stabilizing effect of Cav1 was confirmed in three ways. First, a virion dot blot assay was used to directly measure the proportion of pre-F packaged in virions based on reactivity with MAbs that bind only pre-F protein versus MAbs that bind both pre- and post-F. Second, sera from hamsters immunized with the various constructs were assayed for antibodies specific to pre-F; this was done by measuring binding to immobilized pre-F or post-F in the presence or absence of soluble post-F competitor. Third, sera from hamsters and rhesus monkeys immunized with the various constructs were further assayed for antibodies specific to site Ø in pre-F; this was done by binding assays that measured the ability of hamster serum antibodies to compete with MAb D25 (which binds site Ø, which is unique to pre-F) versus motavizumab (which binds site II, present in both pre- and post-F) for binding to immobilized pre-F protein. All three assays demonstrated that the addition of the Cav1 mutations to the DS mutations provided substantial improvement in stabilizing the pre-F conformation. Consistent with its increased pre-F stabilization, RSV F with DS-Cav1 exhibited a trend of increased induction of high-quality RSV-neutralizing antibodies in hamsters compared to DS alone, defined as the ability to neutralize RSV in vitro without added complement. A similar trend toward higher antibody titers was not observed in a less stringent complement-dependent RSV neutralization assay, suggesting that there was an increase in antibody quality more than quantity. Recently, other mutations that further stabilize pre-F in combination with DS-Cav1 have been identified by structure-based design (45). Some of these mutations also increase the efficiency of F protein expression. We are presently investigating whether introducing these next-generation pre-F stabilizing mutations will further improve the immunogenicity of RSV F expressed from the rB/HPIV3 vector.
Another major variable examined in the present study was the effect of alternative codon usage for RSV F. Previously, GA optimization of codon usage increased RSV F expression by 2-fold (14). GS optimization instead of GA optimization conferred 2-fold greater expression than GA. Combined with the further effect of the HEK assignments, this resulted in a total of an ∼10-fold increase compared to the original nonoptimized RSV F. In hamsters, GS-optimized constructs replicated more efficiently than the respective GA-optimized constructs, with a trend particularly apparent in the lower respiratory tract. This also was associated with a trend of increased induction of RSV-neutralizing serum antibodies compared to the respective GA constructs (both in the presence and absence of added complement). This increased immunogenicity likely reflected increased antigenic load resulting from increased expression from the GS- versus GA-optimized ORF as well as from increased vector replication. However, these effects were not always evident. For example, in monkeys, the GS construct replicated more efficiently than the respective GA construct, but no substantial increase of neutralization antibodies was detected. These inconsistencies likely reflect the limitations and variability of these semipermissive, outbred animal models, as well as the modest nature of the differences.
Another consequence of changing the codon usage of the RSV F ORF in the present study is that this changed its content of CpG dinucleotides. Specifically, while the native RSV F ORF contained only 12 CpG dinucleotides, the GS- and GA-optimized RSV F ORFs contained 66 and 100 CpG dinucleotides, respectively. It has long been recognized that CpG dinucleotides are underrepresented in RNA viruses (46). Various mechanisms may account for this, depending on the virus (47). In particular, an increase of CpG dinucleotides even in a limited region of a viral RNA genome can stimulate an increased host innate immune response, resulting in increased restriction of viral replication (30, 48), which thus presents a selective pressure favoring underrepresentation. Indeed, increased CpG content has been suggested to be a contributing mechanism for attenuation by codon or codon pair deoptimization (32, 49). These observations suggested that the greater restriction of replication associated with GA optimization compared to GS optimization may be due to its higher CpG dinucleotide content and induction of a stronger innate antiviral response. Consistent with this, infection of human lung epithelial cells with a GA construct induced significantly higher levels of IFNs and ISGs than a parallel GS construct. Thus, we hypothesize that the induction of a stronger innate antiviral response by GA optimization compared to GS optimization indeed was a consequence of an increased number of CpG dinucleotides, and that this contributed to increased attenuation of the GA construct. While the increase in innate immune stimulation could be expected to increase the adaptive immune responses, there was no evidence for this. These results suggest that while codon optimization yielded an increase in the expression of RSV F protein due to increased efficiency of translation, it did so at the expense of reducing viral replication because of increased CpG content and increased innate immune responses. In the future, we will further investigate these effects by making and evaluating a vector construct in which the RSV F insert bears the HEK, DS-Cav1, and B3TMCT mutations but is not codon optimized. This construct might exhibit increased replication and immunogenicity while retaining the satisfactory level of attenuation of MEDI-534, which also bore an RSV F insert that was not codon optimized.
In addition, we also evaluated the immunogenicity of a secreted soluble pre-F trimer. Antigens presented in the form of virus-like particles (VLPs) or in oligomerized or aggregated forms typically are more immunogenic than monomeric and nonaggregated forms (50–53). In a previous study (14), we compared two versions of RSV F that had been designed to lack the TM and CT domains, which would favor secretion. One version (called postfusion) was engineered to have the postfusion conformation, and the other (called Ecto) contained the complete RSV F ectodomain but also had a postfusion conformation based on antibody binding studies. Postfusion F was efficiently secreted and poorly immunogenic, while Ecto F remained partly cell associated and had stronger immunogenicity. We suspected that Ecto F retained an ability to aggregate with the cell and with itself due to the presence of the hydrophobic fusion peptide that was lacking in the postfusion construct. These results suggested that aggregates of RSV F protein are more immunogenic than the soluble trimers. In addition, as already discussed (see Introduction), we previously showed that packaging the RSV F protein into the vector particle via the B3TMCT mutation greatly increased its immunogenicity (15). In the present study, we investigated the effect of trimerization of the F protomers. Specifically, we made a construct in which the ectodomain of RSV F was GS optimized and contained the DS-Cav1 mutations and, in addition, contained a phage T4 foldon domain added at the C terminus. This foldon was previously shown to direct the assembly of RSV F protomers into native trimers when expressed in a mammalian system, and this antigen was immunogenic in mice when evaluated as a subunit vaccine (25). However, in the present study, this GS/DS-Cav1/Foldon construct expressed from the rB/HPIV3 vector was poorly immunogenic compared to constructs in which RSV F retained its native TMCT domains and was expressed at the cell surface, or it contained the B3TMCT domains and was expressed at the cell surface and also efficiently packaged in the vector particle. The reason for the lower immunogenicity of the vector-expressed trimers is unclear but suggests that the incorporation of RSV-F into larger antigenic structures, such as association with cells or assembly into vector particles, is much more immunogenic than secreted monomers or stabilized trimers.
The new candidate, GS/DS-Cav1/B3TMCT, was found to be the most immunogenic vector to date for serum RSV-neutralizing antibodies and the most protective against RSV challenge. It was substantially more attenuated than a construct similar to MEDI-534 and induced titers of high-quality RSV-neutralizing antibodies that were significantly higher (∼3-fold) than those of wt RSV and also higher than those of the previous leading candidate, GA/DS/B3TMCT. It was essentially equivalent in protection to wt RSV. That the vector compared favorably to wt RSV, used as a positive control, is noteworthy, since RSV was given at a 10-fold higher dose: we used a lower dose for the vectors in order to increase our ability to detect differences in their relative immunogenicity and protective efficacy. In addition, the RSV control was not attenuated, expresses the neutralization and protective antigen G in addition to F, and expresses all of the RSV proteins; thus, it should induce a broader T cell response, which is known to restrict RSV challenge but wanes within several months (and thus might augment the relative effectiveness of RSV versus vector in a short-term challenge experiment). In any event, the relative performance of vector versus RSV in experimental animals is not necessarily directly predictive of relative performance in humans. For example, immunogenicity and protective efficacy can be influenced by replication efficiency, and the relative replication and immunogenicity of an rB/HPIV3-RSV-F vector versus an attenuated RSV strain may be different in humans than the present comparison in experimental animals.
In conclusion, the rB/HPIV3-RSV-F vector system has the major advantage of being safe and well tolerated in seronegative infants and young children and thus remains an important vaccine candidate. We have been systematically working to increase the immunogenicity for the RSV F insert. Since immune protection against RSV in infancy frequently is incomplete even following infection with wt RSV, any increase in the immunogenicity of an RSV vaccine is desirable. This study demonstrated that immunogenicity of an RSV F-based vectored vaccine can be enhanced by improving pre-F stabilization, increasing pre-F expression, and using RSV F coding sequence containing fewer CpG elements to achieve optimal levels of vector replication in vivo. This identified a further improved RSV vaccine candidate suitable for pediatric clinical trials.
MATERIALS AND METHODS
Cells and viruses.
African green monkey Vero cells (ATCC CCL-81), rhesus monkey LLC-MK2 cells (ATCC CCL-7), and baby hamster kidney BHK BSR-T7/5 cells were maintained as previously described (33). The human lung epithelial A549 cells (ATCC CCL-185) were grown in F-12K medium (ATCC) with 10% fetal bovine serum at 37°C. Wild-type (wt) RSV used in the study was strain A2 (GenBank accession number KT992094). The rB/HPIV3 vector and methods of propagation were described previously (11, 33).
Eleven versions of rB/HPIV3-expressing RSV F were evaluated in the present study; these are summarized in Fig. 1 and described in Results. The RSV F sequence is based on that of wt recombinant RSV strain A2 in GenBank accession number KT992094, and the rB/HPIV3 vector is that of GenBank accession number BD291436.1 (13), as previously described (14, 15, 33). In all of the constructs except i, the RSV F sequence was modified by the addition of the two HEK assignments of amino acids 66E and 101P (34). The DS (disulfide bond) stabilization of RSV pre-F involved the mutations S155C and S290C (25). The DS-Cav1 stabilization of RSV pre-F involved the DS mutations combined with the Cav1 mutations S190F and V207L (25). Optimization of codon usage for efficient translation in humans was performed by GeneArt (GA) or GenScript (GS). The foldon trimerization domain in the construct GS/DS-Cav1/Foldon (Fig. 1, construct xi) comprised the bacteriophage T4 fibritin trimerization domain (GYIPEAPRDGQAYVRKDGEWVLLSTFL) (PDB entry 1RFO), which was fused to the C-terminal end of the 513-residue secreted RSV pre-F ectodomain via a linker (SAIG) (54).
Constructs i to v were made in previous studies (14, 15, 33), and the other six constructs, vii to ix, were made in the present study in a directly compatible fashion. The F inserts were made synthetically by GeneArt or GenScript. The inserts were designed to be inserted into the AscI site in the downstream noncoding region of the rB/HPIV3 N gene and designed to be flanked by BPIV3 gene start and gene end signals. Details of the inserts were the same as those in a construct named rB/HPIV3-F2, illustrated in Fig. 1 of a previous publication (33).
All rB/HPIV3 vectors with RSV F were rescued in BHK BSR-T7/5 cells constitutively expressing T7 RNA polymerase as described previously (33, 55). Rescued viruses were passaged twice on LLC-MK2 cells at 32°C. Titers were determined by 50% tissue culture infective dose (TCID50) assay in LLC-MK2 cells using hemadsorption in 96-well plates. The percentage of PFU in each viral stock that expressed RSV F protein was determined by a fluorescence double-staining plaque assay in Vero cells as described previously (14). All stocks used for the study had 99 to 100% of PFU expressing RSV F. Viral sequences were confirmed in their entirety (except for the 30 and 120 nucleotides at the 3′- and 5′-terminal ends, respectively, due to the positioning of the sequencing primers) with automated Sanger sequencing analysis of uncloned reverse transcription-PCR (RT-PCR) products amplified from viral RNA genome.
Multicycle replication of rB/HPIV3 vectors in cell culture.
Vero cells in 6-well plates were infected in triplicates with rB/HPIV3 vectors at a multiplicity of infection (MOI) of 0.01 TCID50 per cell. After infection, cell monolayers were washed twice with culture medium. A total volume of 2 ml medium was added into each well. Infected cells were incubated at 32°C for 6 days; 0.5 ml of culture medium was collected every 24 h and replaced with 0.5 ml of fresh medium after each collection. All harvested samples were titrated together by TCID50 assay in LLC-MK2 cells using hemadsorption.
Western blot analysis for RSV F expression.
Vero cells in 12-well plates (∼2 × 105 cells per well) were infected with rB/HPIV3 vectors at an MOI of 10 TCID50 per cell or wt RSV at an MOI of 3 PFU per cell. Infected cells were incubated at 32°C. At 48 h p.i., culture medium supernatant (0.5 ml per well) was collected and clarified by centrifugation at 10,000 × g for 1 h at 4°C. Clarified medium supernatants were then adjusted (using a 4× stock) to contain 1× LDS buffer (Thermo Fisher Scientific) with reducing reagent (Thermo Fisher Scientific) and were heated for 5 min at 95°C. Cell monolayers were washed once with ice-cold phosphate-buffered saline (PBS), lysed in wells with 200 μl 1× LDS buffer, and heated with added reducing reagent for 5 min at 95°C. Heated lysates or medium supernatants (20 μl) were loaded on the 4 to 12% Bis Tris NuPAGE gels (Thermo Fisher Scientific) with added antioxidant (Thermo Fisher Scientific) in the cathode chamber. After transferring to polyvinylidene difluoride (PVDF) membranes with the iBlot system (Thermo Fisher Scientific), membranes were incubated with blocking buffer (LiCor) containing primary antibodies at 1:1,000 dilution, followed by incubation with blocking buffer containing infrared dye-labeled secondary antibodies (LiCor) at a 1:10,000 dilution. Images of blots were captured and analyzed by an Odyssey Imaging system (LiCor). Primary antibodies included murine anti-RSV F (ab43812; Abcam), rabbit anti-HPIV3 antiserum (15), rabbit anti-HPIV3 HN (raised against a synthetic peptide) (33), and murine anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; G8795; Sigma-Aldrich); secondary antibodies included donkey anti-mouse IRDye 800CW and donkey anti-rabbit IRDye680 (LiCor).
Flow cytometry analysis of RSV F expression on cell surface.
Vero cells in 12-well plates (∼2× 105 cells/well) were infected at an MOI of 5 TCID50 per cell. After 48 h of incubation at 32°C, cells were made into a suspension with 50 mM EDTA in PBS. After washing twice with fluorescence-activated cell sorter (FACS) buffer (PBS with 2% fetal bovine serum [FBS]), cells were resuspended and incubated in FACS buffer containing RSV F murine monoclonal antibody (MAb) 1129 conjugated with Alexa Fluor 488 as described previously (14). After washing twice in ice-cold FACS buffer, cells were incubated in near-infrared LIVE/DEAD dye (Thermo Fisher Scientific) diluted 1:1,000 in PBS to discriminate dead cells. Stained cells were washed twice and analyzed with a BD FACSCanto II flow cytometer.
Virion dot blot antibody binding assay.
The dot blot antibody binding analysis was performed to assess the stability of pre-F-specific antigenic sites by quantifying binding of site-specific MAbs to virions of various vector constructs. To identify appropriate viral dilutions, virus stocks were 4-fold serially diluted in 1% bovine serum albumin (BSA) in PBS, blotted onto a nitrocellulose membrane, and dried. The membranes were blocked in 10% nonfat dry milk for 2 h at room temperature. Subsequently, membranes were washed twice in Tris-buffered saline (TBS), pH 7.6, containing 0.2% Tween (TBS-T) and once in PBS for 10 min. Washed membranes were incubated on a shaker for 1 h with 10 μg/ml motavizumab in 5% nonfat dry milk. Following the washing steps described above, the membranes were incubated with 1:3,000 diluted anti-human horseradish peroxidase (HRP)-conjugated antibodies in 5% nonfat dry milk for 1 h. After washing as described above, membranes were treated with 1.2 ml of ECL Prime Western blotting detection reagent (GE Healthcare) for 5 min. Images were captured with a GeneSys System. A dilution was chosen for each virus that produced signal intensity similar to dilutions selected for the other viruses on the same blot. Three microliters of diluted viruses, and purified pre-F protein (0.94 μg/ml) as a control, then were blotted onto nitrocellulose membranes in duplicate and assessed for binding by MAb D25, AM14, and 5C4 (specific to pre-F) and motavizumab and 1129 (which bind both pre- and post-F) using the conditions described above. Secondary HRP-conjugated antibodies of matching species (1:3,000) were used, followed by ECL detection as described above. Densitometry readings for each virus were normalized to pre-F protein control signal for that blot. All reported values per virus were an average of duplicates.
Hamster studies.
Hamster studies were carried out by following protocols approved by the National Institutes of Health (NIH) Institutional Animal Care and Use Committee (IACUC). Groups (n = 6) of 6-week-old Golden Syrian hamsters (Envigo Laboratories), confirmed to be seronegative for RSV and HPIV3 by plaque reduction neutralization assay using RSV expressing green fluorescent protein (GFP) and hemagglutination inhibition (HAI) assay, respectively, were anesthetized and intranasally (i.n.) inoculated with 105 TCID50 of rB/HPIV3 vectors or 106 PFU of wt RSV in 0.1 ml of Leibovitz's L15 medium (Thermo Fisher Scientific). After euthanasia, nasal turbinates and lungs were collected on days 3 or 5 after inoculation in two separate studies. Virus replication was assessed by determining virus titers in tissue homogenates by TCID50 assay in 96-well plates of LLC-MK2 cells for rB/HPIV3 vectors, detected by hemadsorption, and plaque assay in 24-well plates of Vero cells for wt RSV, detected by immunostaining (33). In a separate study, hamsters in equivalent groups were inoculated as described above, and sera were collected on day 28 to evaluate the RSV- and HPIV3-specific neutralizing serum antibody titers by 60% plaque reduction neutralization test (PRNT60) in 24-well plates of Vero cells in the presence or absence of added guinea pig complement (Lonza), using RSV and HPIV3 recombinants expressing GFP. To evaluate the protective efficacy against RSV, hamsters were challenged i.n. on day 30 with 106 PFU of wt RSV A2 strain. Nasal turbinates and lungs were harvested on day 3 postchallenge, homogenates were prepared, and titers of challenge RSV were determined by plaque assay in 24-well plates of Vero cells using immunostaining.
Nonhuman primate study.
The nonhuman primate study was approved by the NIH Institutional Animal Care and Use Committee (IACUC). The NIH's Public Health Service Policy on the Humane Care and Use of Laboratory Animals served as the guidelines for the care and use of animals in this study (56). A total of 14 rhesus macaques seronegative for HPIV3 and RSV (confirmed by assays described above) each were inoculated i.n. and intratracheally (i.t.) with 106 TCID50 of rB/HPIV3 vectors. Three groups consisting of four, six, and six monkeys were inoculated with GA/DS/B3TMCT, GA/DS-Cav1/B3TMCT, and GS/DS-Cav1/B3TMCT, respectively. Nasopharyngeal (NP) swab samples were collected daily from days 0 to 10 and days 12 and 14; tracheal lavage (TL) samples were collected every other day from day 2 to day 14. Sera were collected on days 0, 14, 21, and 28. Titers of replicated vectors in NP and TL samples were determined by TCID50 assay in 96-well plates of LLC-MK2 cells detected using hemadsorption. Serum RSV-neutralizing antibody titers were determined by PRNT60 with RSV-GFP as described above.
Octet assay for pre-F-specific antibodies.
Hamster sera were assessed for the presence of antibodies binding to pre-F or post-F in the presence or absence of competing soluble post-F by bilayer interferometry (BLI) using an Octet HTX instrument (ForteBio). Briefly, RSV pre-F or post-F protein was loaded onto HIS1K biosensors (ForteBio) through polyhistidine tags by incubation for 300 s in 30 μg/ml of purified pre-F or post-F in PBS with 1% BSA. Typical capture levels were between 2.0 and 2.3 nm, and variability within a row of 8 tips did not exceed 0.2 nm. The loaded biosensor tips were then equilibrated for 60 s in 1% BSA in PBS, and the biosensors were then incubated for 300 s with diluted serum samples (diluted 1:20 in 1% BSA in PBS) in the presence or absence of 4 μg untagged post-F. The percentage of pre-F or post-F binding retained in the presence of competing soluble post-F was calculated relative to the absence of competition. Octet assays were performed at 30°C.
Octet assay for epitope-specific RSV F antibodies.
The epitope specificity of antibodies in hamster sera was evaluated by a competitive binding assay with D25 (specific for pre-F) and motavizumab (which binds both pre- and post-F) Fabs, performed by BLI using an Octet HTX instrument. Briefly, RSV pre-F protein trimers were loaded onto HIS1K biosensors and equilibrated as described above. They were then incubated for 300 s with D25 or motavizumab Fab or an unrelated control, Fab 310.63 (30 μg/ml in 1% BSA in PBS), followed by a baseline equilibration of 60 s. This was followed by incubation for 300 s with individual test hamster serum specimens (diluted 1:20 in 1% BSA in PBS). The percent inhibition of hamster serum binding to pre-F was determined with the equation [100 − (serum binding in the presence of RSV Fab/serum binding in the presence of Fab 310.63 × 100)].
Quantification of the IFN response in infected A549 cells.
Human lung alveolar epithelial A549 cells were seeded in 24-well plates and infected with viruses at an MOI of 10 PFU per cell. Six wells of A549 cells were infected by each virus or mock infected with cell culture medium. At 18 h p.i., cells in triplicate wells were lysed in RLT buffer and total RNA was extracted using the RNeasy minikit (Qiagen), followed by treatment with RNase-Free DNase I (Qiagen) and reverse transcription using the high-capacity cDNA reverse transcription kit (Applied Biosystems). Transcripts encoding various IFN-stimulated genes (ISGs) were quantified by real-time PCR (relative quantification) using the TaqMan universal PCR master mix (Applied Biosystems) on a 7900HT Fast real-time PCR system. Each sample of cDNA was analyzed in duplicate by real-time PCR. The threshold cycle (CT) values for GS were used as a calibrator (given a value of 1.0), and those for GA were calculated as fold change relative to GS with the 2−ΔΔCT method using RQ Manager software (Applied Biosystems). TaqMan assays of the following ISG (Thermo Fisher Scientific), with their identities in parentheses, were used: IFN-α1 (Hs00256882_s1), IFN-α2 (Hs00265051_s1), IFN-β (Hs01077958_s1), IFN-λ1 (Hs00601677_g1), IFN-λ2 (Hs00820125_g1), MX1 (Hs00895608_m1), ISG15 (Hs09121425_s1), IFIT1 (Rh00929909_m1), IRF7 (Hs00185375_m1), IL-6 (Hs00985641_m1), and GAPDH (Hs99999905_m1). From another set of triplicate wells, the cell culture medium supernatants were collected at 24 h p.i. and clarified, and secreted type 1 (IFN-β) and type 3 (IFN-λ1, λ2/3) IFNs were quantified by ELISA (BioLegend).
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of the Division of Intramural Research and the Vaccine Research Center, NIAID, NIH.
We thank Ashley D. Hackenberg from the Experimental Primate Virology Section and other technical personnel of the Comparative Medicine Branch, NIAID, NIH, for assistance during animal experiments and for the care and management of animals. We also thank Ursula J. Buchholz for kindly providing BHK BSR-T7/5 cells, wt RSV, and RSV expressing GFP.
REFERENCES
- 1.Weber MW, Mulholland EK, Greenwood BM. 1998. Respiratory syncytial virus infection in tropical and developing countries. Trop Med Int Health 3:268–280. doi: 10.1046/j.1365-3156.1998.00213.x. [DOI] [PubMed] [Google Scholar]
- 2.Murphy BR, Prince GA, Collins PL, Van Wyke Coelingh K, Olmsted RA, Spriggs MK, Parrott RH, Kim HW, Brandt CD, Chanock RM. 1988. Current approaches to the development of vaccines effective against parainfluenza and respiratory syncytial viruses. Virus Res 11:1–15. doi: 10.1016/0168-1702(88)90063-9. [DOI] [PubMed] [Google Scholar]
- 3.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahodwala N, De Leo D, Degenhardt L, Delossantos A, Denenberg J, Des Jarlais DC, Dharmaratne SD, Dorsey ER, Driscoll T, Duber H, Ebel B, Erwin PJ, Espindola P, Ezzati M, Feigin V, Flaxman AD, Forouzanfar MH, Fowkes FG, Franklin R, Fransen M, Freeman MK, Gabriel SE, Gakidou E, Gaspari F, Gillum RF, Gonzalez-Medina D, Halasa YA, Haring D, Harrison JE, Havmoeller R, Hay RJ, Hoen B, Hotez PJ, Hoy D, Jacobsen KH, James SL, Jasrasaria R, Jayaraman S, Johns N, Karthikeyan G, Kassebaum N, Keren A, Khoo JP, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Lipnick M, Lipshultz SE, Ohno SL, Mabweijano J, MacIntyre MF, Mallinger L, March L, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, McAnulty JH, McDermott MM, McGrath J, Mensah GA, Merriman TR, Michaud C, Miller M, Miller TR, Mock C, Mocumbi AO, Mokdad AA, Moran A, Mulholland K, Nair MN, Naldi L, Narayan KM, Nasseri K, Norman P, O'Donnell M, Omer SB, Ortblad K, Osborne R, Ozgediz D, Pahari B, Pandian JD, Rivero AP, Padilla RP, Perez-Ruiz F, Perico N, Phillips D, Pierce K, Pope CA III, Porrini E, Pourmalek F, Raju M, Ranganathan D, Rehm JT, Rein DB, Remuzzi G, Rivara FP, Roberts T, De Leon FR, Rosenfeld LC, Rushton L, Sacco RL, Salomon JA, Sampson U, Sanman E, Schwebel DC, Segui-Gomez M, Shepard DS, Singh D, Singleton J, Sliwa K, Smith E, Steer A, Taylor JA, Thomas B, Tleyjeh IM, Towbin JA, Truelsen T, Undurraga EA, Venketasubramanian N, Vijayakumar L, Vos T, Wagner GR, Wang M, Wang W, Watt K, Weinstock MA, Weintraub R, Wilkinson JD, Woolf AD, Wulf S, Yeh PH, Yip P, Zabetian A, Zheng ZJ, Lopez AD, Murray CJ, Al Mazroa MA, Memish ZA. 2012. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connors M, Collins PL, Firestone CY, Sotnikov AV, Waitze A, Davis AR, Hung PP, Chanock RM, Murphy BR. 1992. Cotton rats previously immunized with a chimeric RSV FG glycoprotein develop enhanced pulmonary pathology when infected with RSV, a phenomenon not encountered following immunization with vaccinia–RSV recombinants or RSV. Vaccine 10:475–484. doi: 10.1016/0264-410X(92)90397-3. [DOI] [PubMed] [Google Scholar]
- 6.Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, Jensen K, Parrott RH. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol 89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 7.Schneider-Ohrum K, Cayatte C, Snell Bennett A, Manohar Rajani G, McTamney P, Nacel K, Hostetler L, Cheng L, Ren K, O'Day T, Prince GA, McCarthy MP. 1 February 2017. Immunization with low doses of recombinant post-fusion or pre-fusion RSV F primes for vaccine-enhanced disease in the cotton rat model independent of the presence of a Th1-biasing (GLA-SE) or Th2-biasing (Alum) adjuvant. J Virol doi: 10.1128/JVI.02180-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein D, Malkin E, Abughali N, Falloon J, Yi T, Dubovsky F. 2012. Phase 1 study of the safety and immunogenicity of a live, attenuated respiratory syncytial virus and parainfluenza virus type 3 vaccine in seronegative children. Pediatr Infect Dis J 31:109–114. doi: 10.1097/INF.0b013e31823386f1. [DOI] [PubMed] [Google Scholar]
- 9.Wright P, Karron R, Belshe R, Shi J, Randolph V, Collins P, O'Shea A, Gruber W, Murphy B. 2007. The absence of enhanced disease with wild type respiratory syncytial virus infection occurring after receipt of live, attenuated, respiratory syncytial virus vaccines. Vaccine 25:7372–7378. doi: 10.1016/j.vaccine.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein MI, Coviello S, Bauer G, Benitez A, Serra ME, Schiatti MP, Delgado MF, Melendi GA, Novalli L, Pena HG, Karron RA, Kleeberger SR, Polack FP. 2006. The impact of infection with human metapneumovirus and other respiratory viruses in young infants and children at high risk for severe pulmonary disease. J Infect Dis 193:1544–1551. doi: 10.1086/503806. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt AC, McAuliffe JM, Murphy BR, Collins PL. 2001. Recombinant bovine/human parainfluenza virus type 3 (B/HPIV3) expressing the respiratory syncytial virus (RSV) G and F proteins can be used to achieve simultaneous mucosal immunization against RSV and HPIV3. J Virol 75:4594–4603. doi: 10.1128/JVI.75.10.4594-4603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang RS, MacPhail M, Schickli JH, Kaur J, Robinson CL, Lawlor HA, Guzzetta JM, Spaete RR, Haller AA. 2004. Parainfluenza virus type 3 expressing the native or soluble fusion (F) protein of respiratory syncytial virus (RSV) confers protection from RSV infection in African green monkeys. J Virol 78:11198–11207. doi: 10.1128/JVI.78.20.11198-11207.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt AC, McAuliffe JM, Huang A, Surman SR, Bailly JE, Elkins WR, Collins PL, Murphy BR, Skiadopoulos MH. 2000. Bovine parainfluenza virus type 3 (BPIV3) fusion and hemagglutinin-neuraminidase glycoproteins make an important contribution to the restricted replication of BPIV3 in primates. J Virol 74:8922–8929. doi: 10.1128/JVI.74.19.8922-8929.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang B, Surman S, Amaro-Carambot E, Kabatova B, Mackow N, Lingemann M, Yang L, McLellan JS, Graham BS, Kwong PD, Schaap-Nutt A, Collins PL, Munir S. 2015. Enhanced neutralizing antibody response induced by respiratory syncytial virus prefusion F protein expressed by a vaccine candidate. J Virol 89:9499–9510. doi: 10.1128/JVI.01373-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang B, Ngwuta JO, Herbert R, Swerczek J, Dorward DW, Amaro-Carambot E, Mackow N, Kabatova B, Lingemann M, Surman S, Yang L, Chen M, Moin SM, Kumar A, McLellan JS, Kwong PD, Graham BS, Schaap-Nutt A, Collins PL, Munir S. 2016. Packaging and prefusion stabilization separately and additively increase the quantity and quality of respiratory syncytial virus (RSV)-neutralizing antibodies induced by an RSV fusion protein expressed by a parainfluenza virus vector. J Virol 90:10022–10038. doi: 10.1128/JVI.01196-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, Kumar A, Modjarrad K, Zheng Z, Zhao M, Xia N, Kwong PD, Graham BS. 2013. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLellan JS, Yang Y, Graham BS, Kwong PD. 2011. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol 85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swanson KA, Settembre EC, Shaw CA, Dey AK, Rappuoli R, Mandl CW, Dormitzer PR, Carfi A. 2011. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc Natl Acad Sci U S A 108:9619–9624. doi: 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corti D, Bianchi S, Vanzetta F, Minola A, Perez L, Agatic G, Guarino B, Silacci C, Marcandalli J, Marsland BJ, Piralla A, Percivalle E, Sallusto F, Baldanti F, Lanzavecchia A. 2013. Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature 501:439–443. doi: 10.1038/nature12442. [DOI] [PubMed] [Google Scholar]
- 20.Gilman MS, Moin SM, Mas V, Chen M, Patel NK, Kramer K, Zhu Q, Kabeche SC, Kumar A, Palomo C, Beaumont T, Baxa U, Ulbrandt ND, Melero JA, Graham BS, McLellan JS. 2015. Characterization of a prefusion-specific antibody that recognizes a quaternary, cleavage-dependent epitope on the RSV fusion glycoprotein. PLoS Pathog 11:e1005035. doi: 10.1371/journal.ppat.1005035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilman MS, Castellanos CA, Chen M, Ngwuta JO, Goodwin E, Moin SM, Mas V, Melero JA, Wright PF, Graham BS, McLellan JS, Walker LM. 2016. Rapid profiling of RSV antibody repertoires from the memory B cells of naturally infected adult donors. Sci Immunol 1:eaaj1879. doi: 10.1126/sciimmunol.aaj1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magro M, Mas V, Chappell K, Vazquez M, Cano O, Luque D, Terron MC, Melero JA, Palomo C. 2012. Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc Natl Acad Sci U S A 109:3089–3094. doi: 10.1073/pnas.1115941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ngwuta JO, Chen M, Modjarrad K, Joyce MG, Kanekiyo M, Kumar A, Yassine HM, Moin SM, Killikelly AM, Chuang GY, Druz A, Georgiev IS, Rundlet EJ, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Nason MC, Capella C, Peeples ME, Ledgerwood JE, McLellan JS, Kwong PD, Graham BS. 2015. Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci Transl Med 7:309ra162. doi: 10.1126/scitranslmed.aac4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krarup A, Truan D, Furmanova-Hollenstein P, Bogaert L, Bouchier P, Bisschop IJ, Widjojoatmodjo MN, Zahn R, Schuitemaker H, McLellan JS, Langedijk JP. 2015. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat Commun 6:8143. doi: 10.1038/ncomms9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Chen L, Srivatsan S, Zheng A, Zhou T, Graepel KW, Kumar A, Moin S, Boyington JC, Chuang GY, Soto C, Baxa U, Bakker AQ, Spits H, Beaumont T, Zheng Z, Xia N, Ko SY, Todd JP, Rao S, Graham BS, Kwong PD. 2013. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zimmer G, Bossow S, Kolesnikova L, Hinz M, Neubert WJ, Herrler G. 2005. A chimeric respiratory syncytial virus fusion protein functionally replaces the F and HN glycoproteins in recombinant Sendai virus. J Virol 79:10467–10477. doi: 10.1128/JVI.79.16.10467-10477.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carnero E, Li W, Borderia AV, Moltedo B, Moran T, Garcia-Sastre A. 2009. Optimization of human immunodeficiency virus gag expression by Newcastle disease virus vectors for the induction of potent immune responses. J Virol 83:584–597. doi: 10.1128/JVI.01443-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao F, Li Y, Decker J, Peyerl F, Bibollet Ruche F, Rodenburg C, Chen Y, Shaw D, Allen S, Musonda R, Shaw G, Zajac A, Letvin N, Hahn B. 2003. Codon usage optimization of HIV type 1 subtype C gag, pol, env, and nef genes: in vitro expression and immune responses in DNA-vaccinated mice. AIDS Res Hum Retrovir 19:817–823. doi: 10.1089/088922203769232610. [DOI] [PubMed] [Google Scholar]
- 29.Kim S, Jang JE, Yu JR, Chang J. 2010. Single mucosal immunization of recombinant adenovirus-based vaccine expressing F1 protein fragment induces protective mucosal immunity against respiratory syncytial virus infection. Vaccine 28:3801–3808. doi: 10.1016/j.vaccine.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 30.Jimenez-Baranda S, Greenbaum B, Manches O, Handler J, Rabadan R, Levine A, Bhardwaj N. 2011. Oligonucleotide motifs that disappear during the evolution of influenza virus in humans increase alpha interferon secretion by plasmacytoid dendritic cells. J Virol 85:3893–3904. doi: 10.1128/JVI.01908-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J, Mercado-Lopez X, Grier JT, Kim WK, Chun LF, Irvine EB, Del Toro Duany Y, Kell A, Hur S, Gale M Jr, Raj A, Lopez CB. 2015. Identification of a natural viral RNA motif that optimizes sensing of viral RNA by RIG-I. mBio 6:e01265-15. doi: 10.1128/mBio.01265-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burns CC, Campagnoli R, Shaw J, Vincent A, Jorba J, Kew O. 2009. Genetic inactivation of poliovirus infectivity by increasing the frequencies of CpG and UpA dinucleotides within and across synonymous capsid region codons. J Virol 83:9957–9969. doi: 10.1128/JVI.00508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang B, Munir S, Amaro-Carambot E, Surman S, Mackow N, Yang L, Buchholz UJ, Collins PL, Schaap-Nutt A. 2014. Chimeric bovine/human parainfluenza virus type 3 expressing respiratory syncytial virus (RSV) F glycoprotein: effect of insert position on expression, replication, immunogenicity, stability, and protection against RSV infection. J Virol 88:4237–4250. doi: 10.1128/JVI.03481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitehead SS, Juhasz K, Firestone CY, Collins PL, Murphy BR. 1998. Recombinant respiratory syncytial virus (RSV) bearing a set of mutations from cold-passaged RSV is attenuated in chimpanzees. J Virol 72:4467–4471. http://jvi.asm.org/content/72/5/4467.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calain P, Roux L. 1993. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol 67:4822–4830. http://jvi.asm.org/content/67/8/4822.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L. 1998. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol 72:891–899. http://jvi.asm.org/content/72/2/891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwakkenbos MJ, Diehl SA, Yasuda E, Bakker AQ, van Geelen CM, Lukens MV, van Bleek GM, Widjojoatmodjo MN, Bogers WM, Mei H, Radbruch A, Scheeren FA, Spits H, Beaumont T. 2010. Generation of stable monoclonal antibody-producing B cell receptor-positive human memory B cells by genetic programming. Nat Med 16:123–128. doi: 10.1038/nm.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collins PL, Melero JA. 2011. Progress in understanding and controlling respiratory syncytial virus: still crazy after all these years. Virus Res 162:80–99. doi: 10.1016/j.virusres.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karron RA, Luongo C, Thumar B, Loehr KM, Englund JA, Collins PL, Buchholz UJ. 2015. A gene deletion that up-regulates viral gene expression yields an attenuated RSV vaccine with improved antibody responses in children. Sci Transl Med 7:312ra175. doi: 10.1126/scitranslmed.aac8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stobart CC, Rostad CA, Ke Z, Dillard RS, Hampton CM, Strauss JD, Yi H, Hotard AL, Meng J, Pickles RJ, Sakamoto K, Lee S, Currier MG, Moin SM, Graham BS, Boukhvalova MS, Gilbert BE, Blanco JC, Piedra PA, Wright ER, Moore ML. 2016. A live RSV vaccine with engineered thermostability is immunogenic in cotton rats despite high attenuation. Nat Commun 7:13916. doi: 10.1038/ncomms13916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le Nouen C, McCarty T, Brown M, Smith ML, Lleras R, Dolan MA, Mehedi M, Yang L, Luongo C, Liang B, Munir S, DiNapoli JM, Mueller S, Wimmer E, Collins PL, Buchholz UJ. 2017. Genetic stability of genome-scale deoptimized RNA virus vaccine candidates under selective pressure. Proc Natl Acad Sci U S A 114:E386–E395. doi: 10.1073/pnas.1619242114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luongo C, Winter CC, Collins PL, Buchholz UJ. 2013. Respiratory syncytial virus modified by deletions of the NS2 gene and amino acid S1313 of the L polymerase protein is a temperature-sensitive, live-attenuated vaccine candidate that is phenotypically stable at physiological temperature. J Virol 87:1985–1996. doi: 10.1128/JVI.02769-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rostad CA, Stobart CC, Gilbert BE, Pickles RJ, Hotard AL, Meng J, Blanco JC, Moin SM, Graham BS, Piedra PA, Moore ML. 2016. A recombinant respiratory syncytial virus vaccine candidate attenuated by a low-fusion F protein is immunogenic and protective against challenge in cotton rats. J Virol 90:7508–7518. doi: 10.1128/JVI.00012-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karron RA, Thumar B, Schappell E, Surman S, Murphy BR, Collins PL, Schmidt AC. 2012. Evaluation of two chimeric bovine-human parainfluenza virus type 3 vaccines in infants and young children. Vaccine 30:3975–3981. doi: 10.1016/j.vaccine.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joyce MG, Zhang B, Ou L, Chen M, Chuang GY, Druz A, Kong WP, Lai YT, Rundlet EJ, Tsybovsky Y, Yang Y, Georgiev IS, Guttman M, Lees CR, Pancera M, Sastry M, Soto C, Stewart-Jones GB, Thomas PV, Van Galen JG, Baxa U, Lee KK, Mascola JR, Graham BS, Kwong PD. 2016. Iterative structure-based improvement of a fusion-glycoprotein vaccine against RSV. Nat Struct Mol Biol 23:811–820. doi: 10.1038/nsmb.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greenbaum BD, Rabadan R, Levine AJ. 2009. Patterns of oligonucleotide sequences in viral and host cell RNA identify mediators of the host innate immune system. PLoS One 4:e5969. doi: 10.1371/journal.pone.0005969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng X, Virk N, Chen W, Ji S, Ji S, Sun Y, Wu X. 2013. CpG usage in RNA viruses: data and hypotheses. PLoS One 8:e74109. doi: 10.1371/journal.pone.0074109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gaunt E, Wise HM, Zhang H, Lee LN, Atkinson NJ, Nicol MQ, Highton AJ, Klenerman P, Beard PM, Dutia BM, Digard P, Simmonds P. 2016. Elevation of CpG frequencies in influenza A genome attenuates pathogenicity but enhances host response to infection. eLife 5:e12735. doi: 10.7554/eLife.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tulloch F, Atkinson NJ, Evans DJ, Ryan MD, Simmonds P. 2014. RNA virus attenuation by codon pair deoptimisation is an artefact of increases in CpG/UpA dinucleotide frequencies. eLife 3:e04531. doi: 10.7554/eLife.04531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Julik E, Reyes-Del Valle J. 2016. Generation of a more immunogenic measles vaccine by increasing its hemagglutinin expression. J Virol 90:5270–5279. doi: 10.1128/JVI.00348-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rybniker J, Nowag A, Janicki H, Demant K, Hartmann P, Buning H. 2012. Incorporation of antigens into viral capsids augments immunogenicity of adeno-associated virus vector-based vaccines. J Virol 86:13800–13804. doi: 10.1128/JVI.01708-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomusange K, Wijesundara D, Gummow J, Garrod T, Li Y, Gray L, Churchill M, Grubor-Bauk B, Gowans EJ. 2016. A HIV-Tat/C4-binding protein chimera encoded by a DNA vaccine is highly immunogenic and contains acute EcoHIV infection in mice. Sci Rep 6:29131. doi: 10.1038/srep29131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Denis J, Majeau N, Acosta-Ramirez E, Savard C, Bedard MC, Simard S, Lecours K, Bolduc M, Pare C, Willems B, Shoukry N, Tessier P, Lacasse P, Lamarre A, Lapointe R, Lopez Macias C, Leclerc D. 2007. Immunogenicity of papaya mosaic virus-like particles fused to a hepatitis C virus epitope: evidence for the critical function of multimerization. Virology 363:59–68. doi: 10.1016/j.virol.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 54.Frank S, Kammerer RA, Mechling D, Schulthess T, Landwehr R, Bann J, Guo Y, Lustig A, Bachinger HP, Engel J. 2001. Stabilization of short collagen-like triple helices by protein engineering. J Mol Biol 308:1081–1089. doi: 10.1006/jmbi.2001.4644. [DOI] [PubMed] [Google Scholar]
- 55.Buchholz UJ, Finke S, Conzelmann K-K. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol 73:251–259. http://jvi.asm.org/content/73/1/251.full.pdf+html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.National Institutes of Health. 2015. Public Health Service policy on humane care and use of laboratory animals. Office of Laboratory Animal Welfare, National Institutes of Health, Bethesda, MD: https://grants.nih.gov/grants/olaw/references/phspolicylabanimals.pdf. [Google Scholar]