Significance
Viruses with large DNA genomes, such as herpesviruses, are difficult to manipulate with existing genetic tools. We describe an application of synthetic genomics assembly tools that enables rapid and efficient generation of combinatorial mutations in herpesvirus genomes. The method provides the capacity to design, generate, and test numerous multiloci mutants in parallel, which can help us understand basic virus biology, facilitate vaccine development, and aid development of next-generation virus-based delivery systems. This class of viruses is being used as vectors for therapeutics and vaccines, with an oncolytic herpesvirus approved for the treatment of melanoma. Although such improvements in genome assembly and manipulation raise dual-use concerns, we believe the potential benefits substantially outweigh the risks.
Keywords: herpesvirus, herpes simplex virus type 1, synthetic genomics, genome-wide engineering, tegument genes
Abstract
Here, we present a transformational approach to genome engineering of herpes simplex virus type 1 (HSV-1), which has a large DNA genome, using synthetic genomics tools. We believe this method will enable more rapid and complex modifications of HSV-1 and other large DNA viruses than previous technologies, facilitating many useful applications. Yeast transformation-associated recombination was used to clone 11 fragments comprising the HSV-1 strain KOS 152 kb genome. Using overlapping sequences between the adjacent pieces, we assembled the fragments into a complete virus genome in yeast, transferred it into an Escherichia coli host, and reconstituted infectious virus following transfection into mammalian cells. The virus derived from this yeast-assembled genome, KOSYA, replicated with kinetics similar to wild-type virus. We demonstrated the utility of this modular assembly technology by making numerous modifications to a single gene, making changes to two genes at the same time and, finally, generating individual and combinatorial deletions to a set of five conserved genes that encode virion structural proteins. While the ability to perform genome-wide editing through assembly methods in large DNA virus genomes raises dual-use concerns, we believe the incremental risks are outweighed by potential benefits. These include enhanced functional studies, generation of oncolytic virus vectors, development of delivery platforms of genes for vaccines or therapy, as well as more rapid development of countermeasures against potential biothreats.
Herpesviruses are major human pathogens that cause lifelong persistent infections, with clinical manifestations that range from a mild cold sore to cancer. Herpes simplex virus (HSV) is a frequently encountered pathogen. Infection with HSV-1 results in orolabial disease but can also cause ocular disease (keratitis) that can lead to blindness and encephalitis (1). HSV-2 infections are characterized by genital ulcerative disease and can result in serious infections in newborns (2).
The use of bacterial artificial chromosome (BAC) cloning technology significantly advanced the capacity of herpesvirus researchers to manipulate virus genomes, following the work of Messerle et al. (3) to clone the mouse cytomegalovirus genome using this system. Many herpesvirus genomes have now been cloned as BAC plasmids (4) and, once in Escherichia coli, can be modified by recombineering methods (5). This method enabled the introduction of reverse genetics—the ability to engineer desired changes in the genome and assess the phenotypic consequences—to many herpesviruses that previously could not be easily manipulated (4, 6). However, several issues, including genome instability, regarding herpesvirus-BAC clones have been reported (7, 8). In addition, the allelic exchange methods used to make changes in the herpesvirus BAC expose the whole genome to very active recombinases, which could potentially generate unwanted changes. Furthermore, each single modification with the BAC-based system takes weeks to complete. If multiple changes in the genome are desired, each must be made sequentially, greatly increasing the timeframe of making mutant viruses.
The synthetic genomics assembly method described herein has many potential advantages over the BAC-based system (9). It is an application of existing tools to engineer large virus genomes and uses yeast genetics to separate the genome into multiple parts. Like an automobile assembly line, these discrete parts can be modified as independent modules with a variety of genetic tools in vitro, in E. coli, and in yeast and then returned to the yeast-based assembly line to produce the complete genome. This type of assembly method significantly reduces the time needed to make multiple changes in the genome because the parts can be manipulated in parallel and combined with other wild-type or mutant parts to obtain a modified complete genome to rapidly generate genome-wide alterations. It also provides a more stable system for engineering these genomes by isolating potentially unstable sequences, such as the complex DNA repeat sequences as well as origins of DNA replication, into separate cloned fragments, which are only present together at the final step when the complete genome is assembled in yeast.
This technique is based on technology developed to clone large regions of the human genome using homologous recombination in Saccharomyces cerevisiae (10) and was applied to complete the synthesis of the Mycoplasma mycoides subspecies capri genome (11). Yeast-based assembly is very robust; Gibson et al. (12, 13) have demonstrated that this method can be used to assemble a 500-kb bacterial genome from a few large DNA fragments (>140 kb) or from many (up to 25) smaller fragments. Small virus genomes have been synthesized using various in vitro methods (14–16); however, we do not believe these in vitro methods could efficiently assemble the genomes of herpesviruses, due to their large size, high GC content, and complex repeat regions. The boundaries of synthetic genomics are also being pushed by ongoing projects to synthesize the chromosomes of eukaryotes (17–25). Approaches to assemble large DNA virus genomes, along with the methods used to create the first synthetic bacterial cell (11) at the J. Craig Venter Institute enable scientists to program or to produce organisms for a wide variety of beneficial uses previously difficult or impossible to achieve. We note that the J. Craig Venter Institute has also been at the forefront of developing options for responsible governance of this powerful technology (26).
Here, we demonstrate an application of this modular yeast-based assembly approach, as a proof of concept, to make rapid genome-wide modifications of virion structural genes in HSV-1. This method has applicability in efficiently generating and testing engineered candidates to improve large DNA virus-based therapeutics, such as oncolytic herpesviruses (27), and for the delivery of genes expressing antigens for potential vaccines.
Results
Design of the Assembly of the HSV-1 Genome.
The design for the distinct HSV-1 fragments and the assembled complete genome was based on the HSV-1 strain KOS sequence and KOS-37 BAC (KOS genome cloned as a BAC plasmid) as models; strain KOS is a well-characterized and widely used laboratory strain (28, 29). The HSV-1 genome (152 kb) consists of two covalently linked components, the unique long (UL) and the unique short (US), that are each flanked by inverted repeat sequences. Terminal repeat long (TRL) and internal repeat long (IRL) border UL, and terminal repeat short (TRS) and internal repeat short (IRS) border US. The genome was divided into 11 fragments, which are 14 kb on average, cover the entire HSV-1 KOS genome, and encompass each inverted repeat region within a single fragment (Fig. 1A). Each fragment overlaps the neighboring fragment by at least 80 unique base pairs on each side. The BAC vector sequence in fragment 7 was modified to contain both a BAC and yeast centromeric plasmid (YCp) sequence at the same site for growth in E. coli and S. cerevisiae. The complete assembled HSV-1 genome was designated KOSYA (KOS yeast assembled).
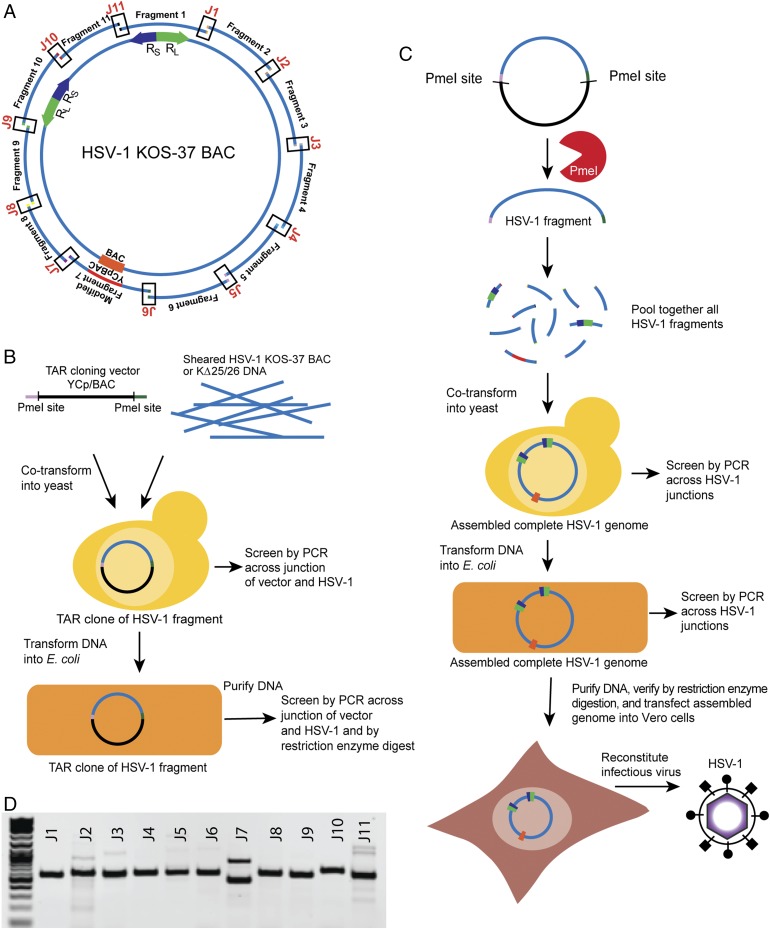
Fig. 1.
Cloning of overlapping fragments of HSV-1 strain KOS and assembly of the complete virus genome using TAR in yeast. (A) Schematic of HSV-1 KOS-37 BAC showing the 11 overlapping fragments of the genome. J1 through J11 denote the junctions between fragments. The locations of the long repeat (RL, green) and short repeat (RS, purple) are shown. The YCp/BAC, which allows for growth in E. coli and yeast is located in fragment 7 (orange). (B) Diagram of TAR cloning of HSV-1 fragments from KOS-37 BAC or KΔ25/26 virus DNA. A linear TAR cloning vector containing BAC and YCp sequences with 40 bp of homology to the desired fragment of HSV-1 and PmeI restriction sites was cotransformed with HSV-1 template genomic DNA into yeast cells. Yeast transformants were screened by PCR and positive candidate DNA was transferred into E. coli. E. coli transformants were also screened by PCR and restriction enzyme digestion analysis of the cloned DNA. (C) Assembly of a complete HSV-1 genome from the 11 overlapping fragments starts with PmeI digestion of the TAR clones to release the HSV-1 sequences followed by cotransformation of all fragments into yeast. Transformants were screened by PCR for the junctions between fragments. DNA was isolated from positive yeast clones and transformed into E. coli, which were similarly screened by PCR. The presence of complete HSV-1 genomic DNA in E. coli was further confirmed by sequencing or restriction enzyme digestion analysis. Yeast assembled genomes were transfected into Vero cells to reconstitute infectious HSV-1 virus. (D) A representative agarose gel following PCR amplification showing the presence of all junctions (J1–J11), indicative of a complete circular genome. The J7 PCR product corresponds to the lower band.
Transformation-Associated Recombination Cloning of HSV-1 Fragments.
The 11 overlapping HSV-1 fragments were isolated by transformation-associated recombination (TAR) cloning, which uses the natural propensity of S. cerevisiae to undergo homologous recombination for the repair of double-strand DNA (dsDNA) breaks to clone large regions of DNA (10). A TAR cloning vector with a BAC and YCp sequences was generated by PCR amplification (SI Appendix, Table S1). The 3′ and 5′ ends of each vector contain 40 bp of homology to a targeted, unique HSV-1 region, as well as sites for the PmeI restriction enzyme, which does not cleave in the HSV-1 genome, to allow for release of the cloned HSV-1 fragments. Each of the 11 fragments was cloned in independent reactions by cotransforming the TAR cloning vector into yeast cells with sheared HSV-1 genome template DNA, either from KOS-37 BAC DNA purified from E. coli or DNA from KΔ25/26- (a mutant KOS virus that contains deletions in both the UL25 and UL26 genes) infected cells (Fig. 1B). DNA from positive TAR clones, identified by PCR with detection primers (SI Appendix, Table S1) across the junction of the vector and HSV-1 fragment, were transformed into E. coli and were further confirmed by restriction enzyme digestion analysis (SI Appendix, Fig. S1). Positive clones were designated with the strain and clone number (e.g., KOS TAR-4 for fragment 4). The KOS TAR clones are stable through several passages of growth in E. coli (SI Appendix, Fig. S2). KOS TAR-1, which contains the terminal repeats TRS and TRL, was difficult to isolate from KOS-37 BAC DNA but easily isolated from KΔ25/26-infected cell DNA. Independent TAR-1 isolates contained variable sequences, which are known to occur in the repeat regions of replicating HSV-1 DNA (30). KOS TAR-7, which contained the BAC sequence, was reengineered to include both BAC and YCp sequences at the same UL37/UL38 intergenic site used in KOS-37 BAC (29). At the junction of HSV-1 and vector sequences, loxP sites were included in direct orientation to facilitate Cre-mediated excision of the vector sequence (SI Appendix, Fig. S3).
Assembly of a Complete HSV-1 Genome in Yeast from 11 DNA Fragments.
All 11 HSV-1 fragments were released from the vector sequence by PmeI digestion, cotransformed, and assembled by TAR in S. cerevisiae, which, through homologous recombination, produces a scarless junction without any nonnative HSV-1 sequence or PmeI restriction sites (Fig. 1C). The resulting transformants were screened by PCR for the presence of each junction between adjacent fragments, indicating the complete assembly of the HSV-1 genome (Fig. 1D and SI Appendix, Table S1). Sanger sequencing of PCR products across the 11 assembly junctions of four independent clones demonstrated that assembly in yeast consistently produced genomes without extraneous sequence at the junctions. We have assembled 24 independent HSV-1 genomes using both yeast spheroplast and lithium acetate transformation methods with an average, for both methods, of 7% of transformants containing DNA that was positive for all 11 junctions by PCR screening. When the concentrations of the fragment DNAs were optimized (20 fmol of fragment 7, which contains the BAC/YCp vector, and 50–250 fmol of the other 10 fragments), the efficiency of assembly in yeast rose to 36%. DNA isolated from yeast clones positive for all junctions was transformed into E. coli and the resulting transformants were similarly screened for all 11 HSV-1 fragment junctions. Assembled HSV-1 genome DNA was purified from positive E. coli clones. Genomes were verified by several different restriction enzyme digestion analyses and there were no discernable rearrangements of the genome (SI Appendix, Figs. S4–S6). However, we observed a variation in the EcoRI L fragment, which resides in TAR-6 (SI Appendix, Fig. S4B). This difference was likely due to variations in the PQ repeat region of the UL36 gene product (VP1-2) in this fragment, which has been observed in the laboratory and also documented by recent high-throughput sequencing of HSV-1 genomes (31, 32). While a larger variant is predominant in K∆25/26 genomic DNA, we may have isolated a smaller variant during TAR-6 cloning, which was maintained during assembly (SI Appendix, Fig. S4B). Sequence analysis (see below) of this region of KOSYA showed a 467-bp deletion in the PQ repeat region of VP1-2. While this deletion suggests a frameshift at the C-terminal end of the UL36 gene, a functional VP1-2 is essential and must be produced in KOSYA (33, 34).
Reconstitution of Infectious Virus and Growth Properties of KOSYA.
The complete assembled HSV-1 genome, KOSYA, was transferred into mammalian cells to verify virus infectivity. Purified DNA was transfected into Vero cells and, after incubation, plaque formation was observed (Fig. 2A). KOSYA produced viable HSV-1 virus, which was amplified.
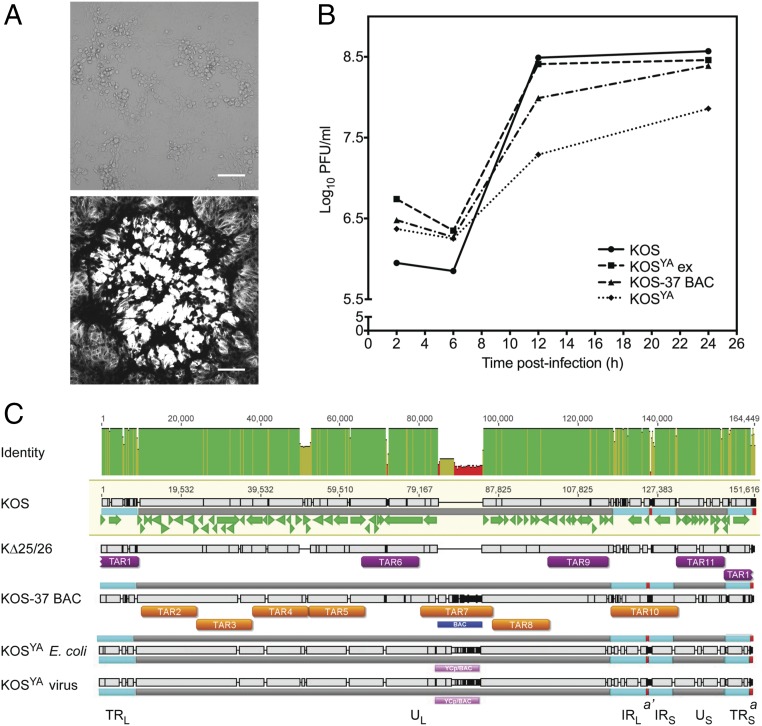
Fig. 2.
Reconstitution and replication properties of an infectious clone of KOSYA. (A) The KOSYA genome was transfected into Vero cells and gave rise to typical cytopathic effects of productive HSV-1 infection in cell culture (Upper, brightfield image). KOSYA was used to infect Vero cell monolayers, stained with Crystal violet, and a single plaque is imaged in grayscale (Lower image). (Scale bars, 100 μm.) (B) KOS, KOSYA, KOS-37 BAC, and KOSYA ex were analyzed by single step growth curves in Vero cells. Virus yields that were produced from 5 × 105 Vero cells infected at a multiplicity of infection (MOI) of 10 plaque forming units (PFU) per cell were enumerated by plaque assays and data plotted on the graph. Infected cells were harvested at 2, 6, 12, and 24 hpi. Data are an average of three independent infections. (C) PacBio sequencing methods were used to determine the sequence of template and final, assembled genomes. For KOS-37 BAC, KΔ25/26, and KOSYA DNA isolated from E. coli, de novo assemblies were generated. Long sequencing reads of infected cell DNA from KOSYA virus were mapped to the KOSYA from E. coli sequence. The presence of insertions and deletions are indicated by the black and gray bars. The green arrows indicate the HSV-1 ORFs from the GenBank annotation of KOS (accession no. JQ673480). The identity across the coordinates of the alignment between the sequences is displayed at the Top (green is 100% identity, yellow is less than complete identity, and red is very low identity). Regions of lower identity can be attributed to variations in variable number tandem repeat regions (such as the PQ repeat variations in the UL36 gene), an engineered deletion of the UL25/UL26 locus in KΔ25/26, and engineered changes to the YCp/BAC sequences (blue and pink blocks) in different viral genomes. KOS TAR clone regions are displayed with the corresponding DNA cloning template (purple blocks for KΔ25/26, orange blocks for KOS-37 BAC). The following features are labeled: IRL, internal repeat long; IRS, internal repeat short; TRL, terminal repeat long; TRS, the terminal repeat short; UL, unique long; US, unique short. Small red squares indicate the a and a′ sequences.
We observed that KOSYA had a reduced growth rate compared with wild-type KOS and KOS-37 BAC (Fig. 2B). We hypothesized that the presence of the YCp/BAC sequence was causing the decreased growth rate (29, 35). To confirm this, we excised the YCp/BAC sequence from KOSYA DNA by in vitro loxP Cre-mediated excision. Reactions were directly transfected into Vero cells and virus was purified by limiting dilution. We obtained a 55% efficiency of YCp/BAC excision, as judged by PCR screening of infected cell DNA (SI Appendix, Table S1). Sequencing of the PCR product showed that the YCp/BAC excision was scarless other than the presence of a single loxP site. A positive purified virus was designated KOSYA ex. Restriction enzyme digestion analyses of viral DNA from cells infected with KOSYA ex showed that the HSV-1 sequence was unaffected by excision (SI Appendix, Fig. S4). All of the viruses—KOS, KOS-37 BAC, KOSYA, and KOSYA ex—were examined using a single-step growth assay. In contrast to KOSYA, KOSYA ex had titers greater than that of KOS-37 BAC and similar to those of wild-type KOS (Fig. 2B). This demonstrated that our engineered virus has the same growth properties as KOS in cell culture.
Sequence Analysis of KOSYA.
Following confirmation that we had generated an infectious clone of KOSYA, the template genomes used for TAR cloning (KOS-37 BAC and KΔ25/26) and the assembled KOSYA genomes isolated from E. coli and the reconstituted virus were sequenced on the PacBio platform. The template and assembled sequences are each >92% identical to the reported HSV-1 strain KOS reference sequence (28) (GenBank accession no. JQ673480) (SI Appendix, Table S2). Some of this variation is probably due to passaging of the virus or BAC plasmid. However, there are also engineered differences (BAC insertion, UL25/UL26 deletion) that contribute to this variation. The junctions between fragments of the KOSYA genomes did not contain any intervening sequence, including the PmeI restriction sites.
When we compare the KOSYA sequence of each region corresponding to a particular TAR cloned fragment to the template (either KOS-37 BAC or KΔ25/26) from which it was cloned, the identity is 100% (0 or 1 nucleotide differences) for 7 of the 11 TAR regions (SI Appendix, Table S3). Four of the TAR regions—corresponding to TAR fragments 1, 6, 10, and 11—contain changes between sequence of the assembled genome and the appropriate template in variable number tandem repeats (VNTRs), which are concentrated in the inverted repeat regions, are known regions of variability, and are very difficult to accurately sequence (31, 32, 36–38) (Fig. 2C and SI Appendix, Table S3). TAR-1 contains four VNTR regions that diverge in sequence from KΔ25/26 in the TRL and TRS. Similarly, TAR-10 contains four VNTR regions within the internal repeat long (IRL) and internal repeat short (IRS) that change between the template, KOS-37 BAC, and KOSYA genomes. According to the sequencing analysis, the TAR-10 region also has a 297-bp deletion in the a′ sequence between IRL and IRS. However, this disagrees with restriction enzyme digestion analysis, which indicates that the a′ sequence in the internal inverted repeats is not shorter than expected from the reference sequence (SI Appendix, Fig. S5A). It is important to note that this region is also difficult to sequence (32). There is also a 6-bp deletion in the intron of the RL2 gene in the TAR-10 region. TAR-6 and TAR-11 each have a deletion in a single VNTR region, in the PQ repeat of VP1-2, and in the intergenic region between US9 and US10, respectively.
The template DNA (KOS-37 BAC or KΔ25/26) and KOSYA genome isolated from E. coli only differ by seven SNPs that cannot be attributed to potential changes in copy number or pattern of VNTRs or sequencing artifacts in homopolymer stretches. All of the changes we identified, except for the deletion in UL36, between the template and KOSYA genomes are in noncoding regions. The KOSYA genomes that were isolated from E. coli or reconstituted virus were sequenced independently and contain 12 noncoding nucleotide differences across the whole genome, demonstrating that the genome is stable through low passage in cell culture (SI Appendix, Table S2). Overall, this confirms that, although there is variation in some noncoding regions, the KOSYA genome was assembled correctly.
Fusion of a Spectrum of Fluorescent Proteins to Three Structural Proteins of HSV-1.
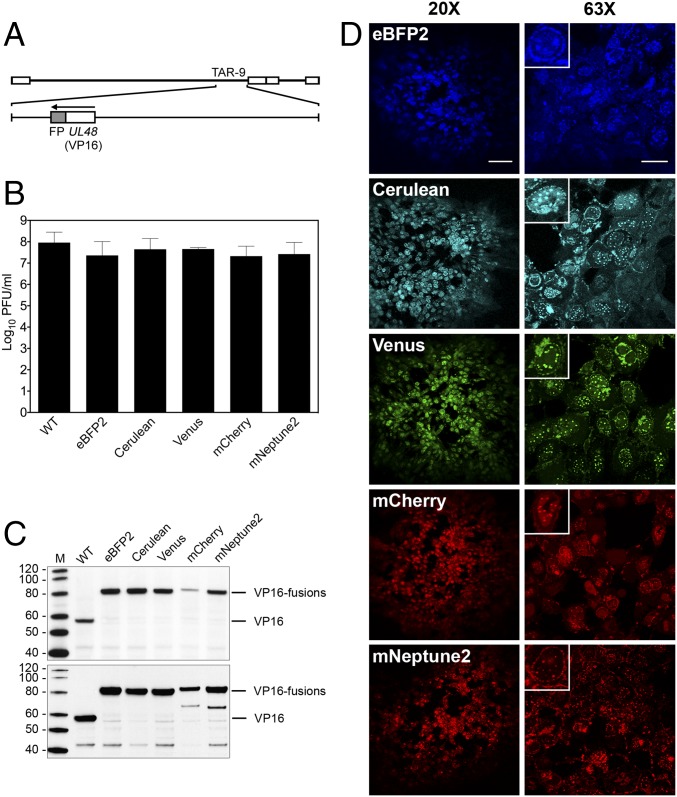
We chose to engineer fluorescent protein (FP) fusions to three HSV-1 virion proteins, as these types of viruses are useful for live cell imaging (39–41) and to demonstrate the utility and efficiency of our genomic assembly method. We generated five independent fusions of genes encoding FPs (eBFP2, Cerulean, Venus, mCherry, and mNeptune2) to the C terminus of VP16, which is encoded by the UL48 gene (Fig. 3A). Previous studies demonstrated that VP16GFP fusions could be made at either the N or C terminus without affecting virus replication (42). The fusions were generated in the HSV-1 KOS TAR-9 fragment using a variety of TAR assembly methods in yeast (SI Appendix, Fig. S7). An in vitro CRISPR-Cas9 method was the most efficient (up to 75% of transformants were positive by PCR screening) and used a single site-specific DNA cleavage of TAR-9 and TAR assembly in yeast to insert FP ORF sequences. Complete HSV-1 genomes containing each of the VP16FP fusions were assembled using the modified TAR-9 fragments along with the remaining wild-type parts.
Fig. 3.
Analysis of recombinant KOSYA genomes expressing five different VP16 FP fusions. (A) A linear map of the KOS genome shows TAR-9, which was modified to contain five different FP fusions (shaded gray) to the C terminus of VP16, which is encoded by UL48. (B) Single-step growth curve analysis was performed for each VP16FP fusion virus and compared with KOSYA (WT). Vero cells were infected at a MOI of 10 PFU per cell and harvested 24 hpi. The titer of virus produced from infected cells after a single cycle of growth was enumerated by plaque assay and the results plotted on the graph. Data are an average of two independent infections. Error bars represent 1 SD. (C) Western blot analysis of proteins from infected cell lysates. Cells were harvested at 8 hpi (Upper) and 24 hpi (Lower) and the infected cell proteins were probed with anti-VP16 antibodies (LP1) following transfer to membranes. Protein standards are in lane M and are labeled in kilodaltons. (D) Confocal analysis was used to analyze KOSYA viruses expressing a range of FP fusions to VP16. Vero cell monolayers were infected at a low MOI and at 72-hpi images were taken with 20× and 63× objective lenses. [Scale bars, 100 µm (Left) and 25 µm (Right).] Insets show a single infected cell. Inset images were cropped from the 63× images to highlight a single infected cell and were each equally digitally enlarged for visibility.
After confirmation by PCR and restriction enzyme digestion analyses (SI Appendix, Figs. S5 and S6), all of the VP16-tagged KOSYA DNAs were transfected into Vero cells to reconstitute the virus. All five genomes generated plaques with the correct fluorescent signal and were amplified. Using single-step growth curve analysis in Vero cells, all of the FP recombinant viruses replicated to titers comparable to KOSYA (Fig. 3B). Immunoblot analysis, using an anti-VP16 antibody, of infected cell proteins harvested at 8 and 24 h postinfection (hpi) showed stable accumulation of all of the VP16FP polypeptides, except for the VP16mCherry protein, which showed impaired expression/accumulation at both time points (Fig. 3C). Confocal microscopy images of virus plaques on Vero cells confirmed the cellular localization previously reported for a VP16GFP recombinant virus (42). Nuclear foci and juxtanuclear localization patterns were observed for each virus, indicative of the role of VP16 as a nuclear transactivator (43) and incorporation into the virion at a cytoplasmic site (42) (Fig. 3D).
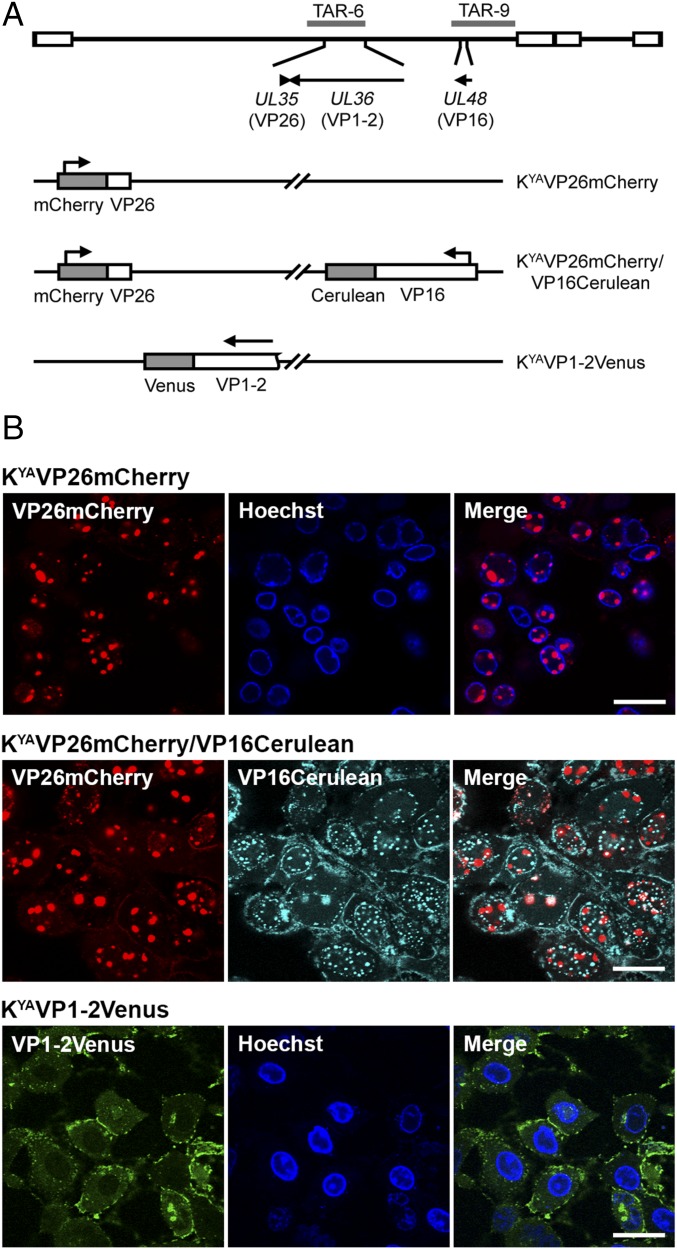
We generated additional FP fusions to the small capsid protein (VP26) and large tegument protein (VP1-2), which are both contained in KOS TAR-6 (Fig. 4A). VP26 (encoded by UL35) decorates capsid structures (44, 45) and is commonly used as a marker for capsid trafficking (39). VP1-2, which is encoded by the UL36 gene, is an essential multifunctional component of the virion tegument and is important in capsid trafficking and virus maturation (33, 46, 47). Based on previous studies (39, 48), we inserted the mCherry ORF between the fourth and fifth amino acid residues of VP26 and the Venus ORF at the C terminus of VP1-2. Using in vitro CRISPR-Cas9 editing, the modifications were generated within TAR-6. These modified TAR clones were used independently with the remaining complement of TAR fragments to assemble the KOSYA genomes, KYAVP26mCherry and KYAVP1-2Venus. To demonstrate the ability to mix and match TAR clones containing HSV-1 gene modifications, we used the VP16Cerulean fusion (contained in TAR-9) and VP26mCherry (contained in TAR-6) to create KYAVP26mCherry/VP16Cerulean (Fig. 4A). Following reconstitution and amplification of these viruses, plaques were analyzed by confocal microscopy in Vero cells (Fig. 4B). VP26mCherry was observed to localize to nuclear puncta in cells infected with both the single- and dual-tagged viruses, similar to previous studies (49). Localization of VP16Cerulean was also similar to that seen in Fig. 3D. Cells infected with KYAVP1-2Venus were observed with predominantly cytoplasmic localization of VP1-2Venus with aggregates at juxtanuclear and plasma membrane sites (50).
Fig. 4.
FP fusions to the largest viral structural protein and the small capsid protein. (A) A map of the KOS genome depicts the gene locations of VP26 (encoded by UL35), VP1-2 (encoded by UL36), VP16 (encoded by UL48), and the corresponding KOS TAR clone. Viruses assembled are listed and FP fusions to each gene are shown by shaded blocks. (B) Vero cell monolayers were infected with KOSYA recombinant viruses at low MOI and incubated for 72 h before imaging on a confocal microscope using a 63× objective lens. (Scale bars, 25 µm.) Hoechst dye was used as a nuclear marker for some infections.
Combinatorial Deletion of Tegument Genes.
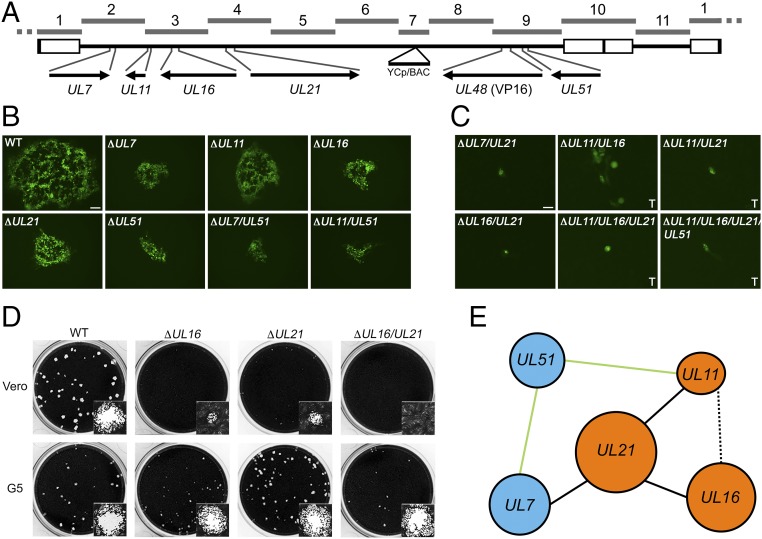
The herpesvirus virion is comprised of three structural components: an icosahedral capsid; the tegument, which immediately surrounds the capsid; and an outer envelope (51, 52). Most virion proteins reside in the tegument layer and this structure, because of the different constituents, plays an important role in all stages of the virus life cycle (53, 54). To further demonstrate our ability to quickly generate mutants across the HSV-1 genome with this method, we constructed a series of single, double, triple, and quadruple deletions in five genes, conserved in all herpesviruses, encoding virion structural proteins: UL7, UL11, UL16, UL21, and UL51 (Fig. 5A and SI Appendix, Fig. S8). The proteins of UL7–UL51 and UL11–UL16–UL21, respectively, are known to physically interact (55–57) and are resident or associated with the tegument structure of the mature virion (58). All five of these genes had been individually deleted previously and shown to be nonessential for growth in cell culture (59–64). However, combinatorial deletions of these genes have not been reported in the literature except recently for UL7 and UL51 (65). We produced a total of 13 HSV-1 genomes with single or combinatorial deletions of these five tegument genes. The KOS TAR clones were edited by a variety of TAR assembly methods (SI Appendix, Fig. S8). Complete ORFs of UL16 and UL21 were deleted using the in vitro CRISPR-Cas9 protocol. The other genes were truncated to avoid disrupting overlapping genes or adjacent promoter sequences. Genes UL16 and UL11 are located in the same KOS TAR clone; therefore, single and double deletions were generated in TAR-3. The UL51 deletion was generated in a TAR-9 clone with the VP16Venus fusion. All KOSYA genomes with deletions were generated with a VP16Venus tag, which served as a marker for delivery of the DNA to cells and virus gene expression. Each of these modified TAR clones were mixed and matched to assemble complete KOSYA genomes with the desired combination of mutations.
Fig. 5.
Replication of viruses containing combinatorial deletions of five tegument genes. (A) A linear map of the HSV-1 genome shows the locus of each gene targeted for modification and its location in the respective TAR fragments. The UL48 gene encodes the VP16 protein. (B) Recombinant viruses, each containing a different tegument gene deletion or combination of deletions and the VP16Venus fusion, which replicate in Vero cells are shown. Plaques were visualized using wide-field fluorescence light microscopy 3 d postinfection. (Scale bar, 100 μm.) Wild-type is KYAVP16Venus and contains no other gene modifications. (C) KOSYA mutant viruses that did not exhibit productive infection in Vero cells were infected and imaged as indicated in B or recombinant genomes were transfected (marked “T”) into Vero cells and imaged 6 d posttransfection. (Scale bar, 25 μm.) (D) KOSYA viruses containing a deletion of UL16, UL21, or combination thereof were infected at low MOI on Vero and G5 cell monolayers in 12-well trays. Cells were stained with Crystal violet 3 d postinfection to visualize plaque formation. Insets show a single plaque. Stained plaques were imaged using the CCD (charge-coupled device) camera of a Gel Doc XR+ (Bio Rad) and insets show a representative single plaque imaged with the brightfield settings of a Zoe Fluorescent Cell Imager (Bio Rad) with a 20× objective lens. (E) Each of the genes analyzed in this study are shown and blue or orange is used to highlight previously discovered physically interacting complexes (55). Solid black lines indicate essential genetic pairs (i.e., UL21 and UL7), whereas solid green lines indicate nonessential pairs (i.e., UL51 and UL11). A dashed black line represents a pair, UL11 and UL16, resulting in severely diminished, but partial, biological activity.
The HSV-1 genomes with tegument gene deletions, which were confirmed by PCR and restriction digestion analysis (SI Appendix, Figs. S5 and S6), were transfected into Vero cells. However, only some viruses were amplified due to replication deficiencies of the mutants. Genomes with UL16 and UL21 deletions were also transfected into the transformed Vero cell line G5, which complements UL16 and UL21 in trans (66, 67). Viruses that had evident cell-to-cell spread on either Vero or G5 cells were amplified. Finally, viruses were used to infect Vero cells, or transfected, and plaque formation was observed using the VP16Venus marker and wide-field microscopy (Fig. 5 B and C). As was expected, all viruses carrying a single deletion were replication competent, albeit cell-to-cell spread was limited compared with the wild-type (KYA-VP16Venus), as well as two viruses carrying double deletions: KYAΔUL7/UL51 and KYAΔUL11/UL51. All other viruses generated in this study were not observed to replicate in Vero cells, aside from KYAΔUL11/UL16, which had a severe deficiency in cell-to-cell spread. We showed that KYAΔUL16, KYAΔUL21, and KYAΔUL16/UL21 replication can be recovered by plaque assay on G5 cells, indicating that reduced titers are due to the designated deletion and not from any unintended mutation in the genome (Fig. 5D). Our data are summarized in a genetic interaction map, which depicts essential genetic interaction pairs that were discovered (Fig. 5E).
Discussion
Advantages of Our Method.
We have described an application of a yeast-based modular assembly technique to make rapid and efficient genome-wide changes in HSV-1. The genome was separated into 11 overlapping fragments that were cloned as plasmids in yeast by TAR cloning. Discrete regions of the genome were isolated from each other so that mutagenesis could be carried out on individual fragments using synthetic biology methods in yeast, E. coli, or in vitro. By developing a library of overlapping HSV-1 fragments that can be stored as sequence-validated parts, many HSV-1 mutants can be rapidly generated through combinatorial and parallel assembly. This method also isolates potentially problematic or “difficult sequences,” such as the inverted repeat sequences, from the other fragments so that during the engineering of a gene, these sequences are not involved. As proof of principle, wild-type and mutant HSV-1 genomes were assembled from the overlapping fragments in yeast and reconstituted in mammalian cells. The assembled HSV-1 viruses recapitulated the expected growth phenotypes for wild-type KOS, which indicates identical biological function between the parental KOS and KOSYA in a cell culture system. We used a total of 23 unique wild-type and mutant parts for this study, which include fluorescent protein fusions, deletions, and modifications to the plasmid backbone to facilitate replication in yeast and E. coli. The ability to mix and match HSV-1 fragments is one of the biggest advances of this method; once recombinant or genomic fragments of interest have been generated by mutagenesis or TAR cloning, new HSV-1 genomes can be assembled quickly (within about 2 wk) by combining previously generated fragments with modified fragments. If desired, mutagenesis of the complete genome with the YCp sequence could also be carried out in yeast using in vivo CRISPR-Cas9 tools. However, this negates the benefits of maintaining the repeat regions separately and the flexibility of manipulating fragments in parallel for combinatorial use in “mix and match” assemblies. Our approach is complementary to current BAC technologies and the synthetic genomics methods can be easily translated by other laboratories without significant investment in equipment or reagents since the genome assembly method uses a yeast genetics system. We have a commitment to share and distribute our reagents to the community as evident from our past gifts of viruses and cell lines.
Remaining Challenges.
There were some issues with our protocol that we are working to resolve and understand in more detail. KOS TAR-1 and TAR-10, which contain the terminal and internal inverted repeats and extensive VNTRs, were the most difficult to clone. We isolated several clones of TAR-1, which show sequence variation when examined by restriction enzyme digestion analyses. Variations in the inverted repeat sequence have been observed previously (30). We were only able to isolate a single clone of the TAR-10 fragment; the reason for the difficulty is unknown. PacBio sequencing analysis of the TAR-1 and TAR-10 regions from the assembled KOSYA and the template genomes show variation in the copy number of the VNTRs, which are abundant in the inverted repeat regions. However, the variation described by the sequencing analyses in the TRs and IRs is not consistently supported by the restriction enzyme digestion analyses of the same regions. For example, restriction enzyme fragments observed indicate the junction of the long and short inverted repeats and the a sequence in TAR-1 is shorter than same region in TAR-10 (SI Appendix, Fig. S5A); however, the sequencing shows a deletion in the a′ sequence of TAR-10 and not in the a sequence of TAR-1. Obtaining high-quality sequencing of the inverted repeats and VNTRs in HSV-1 genomes has been fraught with difficulties. Genomes sequenced with the Illumina or Roche 454 platforms, were not able to resolve the TRs and IRs independently (68) or the copy number of some of the VNTRs (32, 37, 69) or both (28, 31, 38, 70). However, often the sequences of these regions are still reported. For the inverted repeats, the sequence reads are often all aligned to a single copy of the inverted repeats and then duplicated. For VNTR regions, the reads are mapped to a reference genome, HSV-1 strain 17 (GenBank accession no. NC_001806), to check for consensus and the copy number is adjusted to reflect the copy number of the VNTRs in the strain 17 reference or to reflect the depth of the sequencing coverage. TAR-6 and TAR-11 also contain VNTRs that differ between the template and the final assembled KOSYA genomes. There is some doubt about the length of the deletion in TAR-6; as reported, it would cause a frameshift in the PQ region of VP1-2. However, this gene is essential and, therefore, a functional gene product has to be made in KOSYA. The majority of the sequence variation in all four of these regions, and therefore the genome, is clustered at the VNTRs with rare single-nucleotide polymorphisms identified throughout the genome.
Based on analysis of the sequences, we believe there are three plausible explanations for the variable copy number of the VNTRs observed. First, we may have isolated a single variant containing a set number of VNTRs of several variants present in the template virus genomic DNA. This does not explain discrepancies we see between the consensus sequences generated and the restriction enzyme digestion analyses. Second, the sequence of these regions could have been altered during growth of the HSV-1 fragments or complete genomes as plasmids in the S. cerevisiae or E. coli hosts. Finally, due to the complexity of sequencing genomes with high GC contents and repeated sequences, the consensus sequences generated from the PacBio platform could be incorrect. We did encounter sequencing artifacts of incorrect stretches of G or C homopolymer base repeats in many locations throughout the genome (Materials and Methods). While additional work is needed to address these issues, the wild-type behavior of KOSYA in cell culture indicates this method is useful for the construction of HSV-1 mutants, despite these potential changes.
Multiple isomers of the HSV-1 genome, containing inversions of the UL or US regions, have been observed in virus populations (71). Using our assembly protocol, we find that only one genomic isomer can be assembled and believe this should remain “fixed” in yeast and E. coli. Restriction enzyme digestion analyses of all full-length assembled HSV-1 genomes indicate that only one isomer is present, with UL and US oriented in the same direction (SI Appendix, Fig. S6). This contradicts our de novo sequence assembly of KOSYA, which shows UL and US oriented in opposite directions. We reversed the orientation of UL in all of the de novo sequence assemblies to match the reference sequence and the restriction enzyme digestion analyses. Other researchers have mapped reads or contigs onto HSV-1 reference genomes and therefore not determined the orientation of the unique regions (28, 31, 38). The reason for this inconsistency is unknown.
Another issue that we have encountered has been the low efficiency of transfer of intact HSV-1 genomes into E. coli after assembly in yeast, likely due to shearing of the genomes during DNA isolation. Growth in E. coli was used to produce high-concentration DNA stocks that can be used for transfection into mammalian cells. In the future, this step may be skipped altogether by fusing yeast and mammalian cells directly using an improved fusion method (72).
Genetic Interaction Between Tegument Genes UL7 and UL21.
The herpesvirus tegument is a complex structure, which contains proteins that specify diverse functions during all stages of the viral life cycle. While deletions of each of the tegument proteins encoded by UL7, UL11, UL16, UL21, and UL51 have been analyzed (57, 59–64, 67, 73, 74), the redundant nature of these proteins makes it harder to define their functions in the cell (55, 75). Our assembly method allowed us to study interactions of each by generating many combinatorial deletions of these genes in an efficient manner. Our data showed that the plaque size of KYAΔUL7, KYAΔUL51, and KYAΔUL7/UL51 were all similar, demonstrating that while the UL7–UL51 complex is required for optimal growth of the virus but is not essential for virus replication in tissue culture. This result is similar to data reported recently (65). Combinatorial deletions of any group of genes within the UL11–UL16–UL21 complex have detrimental effects on virus replication. A novel finding was that a virus carrying both UL7 and UL21 deletions was not infectious, suggesting an interaction of the UL7–UL51 and UL11–UL16–UL21 pathways during infection. Our modular assembly system enabled us to engineer a series of modified viruses that would have been challenging to construct by traditional methods.
Potential Medical Applications.
The ability to rapidly manipulate viruses with synthetic genomics tools can accelerate vaccine development for herpesviruses and large dsDNA viruses. Of the eight known human herpesviruses, an effective, approved vaccine exists only for varicella-zoster virus, the causative agent of chickenpox and shingles (76). The development of additional herpesvirus vaccines would improve global public health and lower health-care costs. For example, an estimated 11.3% of adults world-wide aged 19–54 y are living with HSV-2 infection (77) and, in the United States, the total lifetime direct medical cost for HSV-2 infections is $540 million (in 2010 US dollars) (78). The ability to rapidly engineer alphaherpesviruses, such as the closely related HSV-1 used here, may lead to an effective HSV-2 vaccine. Additionally, these tools can be extended to other herpesviruses and aid in the development of vaccines to prevent cancers associated with Epstein–Barr virus and Kaposi’s sarcoma-associated herpesvirus, which together account for >250,000 cases of cancer globally every year (79, 80). However, another key application of this platform is the capacity to reconstitute a clinical strain (if the sequence is known) that is impossible to isolate through tissue culture, as we have accomplished for a primary isolate of human cytomegalovirus (81).
The assembly of fragments of HSV-1 can be used to quickly engineer HSV-1 platforms for vaccines, gene therapy, or oncolytic vectors. An engineered strain of HSV-1 is currently approved as an oncolytic virus for cancer treatment (27). The ability to rapidly generate new recombinant viruses with novel combinations of therapeutic modules for testing would enhance the development of improved HSV-1 oncolytic therapeutics for personalized treatments. Our HSV-1 assembly platform can also be used to develop virus platforms to deliver genes either for DNA vaccines or for gene therapy.
Dual-Use Concerns.
Benefits notwithstanding, advances in synthetic genomics methods, including the method described herein, raise several dual-use concerns. For viruses, the most obvious concern is the application of these techniques to recreate genomes of viruses categorized as select agents, such as variola virus, which causes smallpox, from synthetic DNA. This capability, however, existed before our current research, a conclusion reached by an Independent Advisory Group commissioned by the WHO in 2015 (82) and speculated by others (83, 84) a decade earlier. Although not yet published in a peer-reviewed journal, researchers in Canada have reconstituted horsepox virus, a close relative of variola, from synthetic DNA (85). As reviewed in the introduction, the synthetic genomics techniques, applied here to large dsDNA viruses, were developed for synthesis of the M. mycoides genome in 2010 (11) and have been used by other groups since that time (86, 87). Moreover, recreating a virus genome requires starting material (i.e., genomic DNA or synthetic DNAs), which is not readily available for variola virus and many other regulated pathogens. Most companies that commercially synthesize DNA follow a “Screening Guidance” issued in 2010 by the US Department of Health and Human Services (88), which specify procedures for screening synthetic DNA orders for potentially dangerous sequences, including pathogens and toxin genes from the US Select Agents and Toxins List, and for customers who might have nefarious intent. By 2015, ≈80% of synthetic DNA providers followed such Screening Guidance; to keep compliance high as synthetic DNA prices continue to fall, it will be important for the US Department of Health and Human Services to modify the Screening Guidance to keep pace with changes in technology (89). While concerns about the ability to recreate viral genomes are real, we believe that the methods described in our study do not significantly elevate this particular dual-use concern.
A second category of concerns relates to the use of our approach to increase the virulence, transmissibility, or resistance to therapeutic interventions of HSV-1 or other pathogenic DNA viruses. For example, this approach could be used to incorporate toxins, oncogenes, or other virulence factors into HSV-1 with the intent of using HSV-1 as a delivery mechanism for causing harm. For simple modifications, such nefarious aims can already be accomplished with current BAC technology, although at a longer timeline than our method. For more complex modifications—for example, the multiple simultaneous changes for which our method was developed and excels—the controlling factor is the knowledge about which genes or pathways to modify to make viruses more dangerous, rather than the technical capability to modify their genomes. The latter is a legitimate concern, but considering the difficulty of discovering new pathways to harm, perhaps less enticing or useful to a potential “bad actor” than it might at first appear. Quite a few highly pathogenic DNA viruses are already listed as Federal Select Agents. It is probably easier for bad actors to isolate these viruses from nature or laboratory stocks than to rationally engineer less-dangerous ones for malicious intent. In fact, we expect that our work will be of greater help to researchers developing countermeasures for pathogenic virus isolates from nature or from synthetic genomics technologies used for ill.
While we have outlined several dual-use concerns, we emphasize that, as discussed above in more detail, these are outweighed by the beneficial uses of synthetic genomics technology for safeguarding and improving public health. The WHO Advisory Committee on Variola Virus Research concurs with this point in their 2017 report and notes that “on balance, the historical record has clearly demonstrated that society gains far more that it loses by harnessing and building on these scientific technologies” (90). We firmly believe that the method we are reporting on falls within this realm.
Future of Synthetic Viral Genomics.
The system we have developed is a proof-of-concept. We used HSV-1 as a model viral system for herpesviruses and other large dsDNA viruses. We envision the overall cloning and assembly protocol will be used to introduce reverse genetics into more difficult systems and facilitate basic research. Genetic engineering of other herpesviruses, such as Epstein–Barr virus and Kaposi’s sarcoma-associated herpesvirus, is more difficult compared with HSV-1. Beyond the herpesviruses, we believe this modular assembly approach can be a general platform for other large dsDNA viruses, including African swine fever virus and Vaccinia virus, as well as single-stranded DNA and RNA viruses.
Materials and Methods
Cells, Viruses, and Strains.
Vero cells and the G5 cell line (66) were grown in minimum essential medium-α medium supplemented with 10% FBS (Gemini) and passaged, as described by Desai et al. (91).
All HSV-1 KOS and yeast assembled virus stocks were prepared as previously described (91). The KOS mutant virus KΔ25/26, which contains deletions in both UL25 and UL26 genes, was passaged in the F3 complementing Vero cell line (92).
The yeast strain VL6-48N (93) (MATα, his3-Δ200, trp1-Δ1, ura3-Δ1, lys2, ade2–101, met14, cir°) was used for all yeast transformations and grown in YPD media supplemented with adenine. Yeast transformed with a YCp was grown in synthetic dropout media without histidine (−HIS) or without uracil (−URA) and supplemented with adenine.
E. coli strains Epi300 (Epicentre), DH10B (Thermo Fisher), and TOP10 (Thermo Fisher) were used for production of DNA stocks of HSV-1 TAR clones and assembled complete genomes. Cultures were grown overnight at 37 °C in Luria-Bertani (LB) or 2-YT media with chloramphenicol.
Plasmids.
KOS-37 BAC was a kind gift from David Leib, Dartmouth Geisel School of Medicine, Hanover, NH (29). pCC1BAC-ura3 and pCC1BAC-his3 (11) and pRS313 (94) have been previously reported. FP ORFs were amplified from pEBFP2-N1 (eBFP2), pCerulean-VSVG51 (Cerulean), pVenus-VSVG51 (Venus), pFB-ORF67mCherry52 (mCherry), and pmNeptune2-N1 (mNeptune2). pEBFP2-N1 (Plasmid #54595; Addgene) and pmNeptune2-N1 (Plasmid #54837; Addgene) were gifts from Michael Davidson, Florida State University, Tallahassee, FL. pCerulean-VSVG (Plasmid #11913; Addgene) and pVenus-VSVG (Plasmid #11914; Addgene) were gifts from Jennifer Lippincott-Schwartz, National Institutes of Health, Bethesda.
Additional methods can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank the following colleagues for their assistance and support: David Leib (Dartmouth Geisel School of Medicine) for the gift of KOS-37 BAC; Robert Sebra (Mount Sinai) for performing PacBio sequencing; Klaus Früh and his group at Oregon Health and Science University for stimulating discussions and their collaboration on human cytomegalovirus; Reed Shabman, Granger Sutton, and Derrick Fouts (the J. Craig Venter Institute, JCVI) for their assistance with sequencing analysis; Chuck Merryman (JCVI) for assistance with in vitro CRISPR-Cas9 editing; Erin Pryce (The Johns Hopkins University Integrated Imaging Center) for assistance with confocal analysis; Leslie Mitchel and Jef Boeke (New York University, Langone Medical Center) for help with yeast DNA protocols; Prof. Tony Minson (University of Cambridge) for providing the LP1 antibodies; and Suchismita Chandran and Nacrya Assad-Garcia (JCVI) for helpful discussions. These studies were supported by NIH Grants R21AI109418 (to S.V.), R21AI109338 (to P.J.D. and S.V.), R21AI107537 (to P.J.D.), and R33CA186790 (to P.J.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the NCBI BioProject database, https://www.ncbi.nlm.nih.gov/bioproject (accession no. PRJNA386147).
See Commentary on page 11006.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700534114/-/DCSupplemental.
References
- 1.Whitley RJ, Roizman B. Herpes simplex virus infections. Lancet. 2001;357:1513–1518. doi: 10.1016/S0140-6736(00)04638-9. [DOI] [PubMed] [Google Scholar]
- 2.Sacks SL, et al. HSV-2 transmission. Antiviral Res. 2004;63(Suppl 1):S27–S35. doi: 10.1016/j.antiviral.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Messerle M, Crnkovic I, Hammerschmidt W, Ziegler H, Koszinowski UH. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA. 1997;94:14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall RN, Meers J, Fowler E, Mahony T. Back to BAC: The use of infectious clone technologies for viral mutagenesis. Viruses. 2012;4:211–235. doi: 10.3390/v4020211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tischer BK, Smith GA, Osterrieder N. En passant mutagenesis: A two step markerless red recombination system. Methods Mol Biol. 2010;634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 6.Warden C, Tang Q, Zhu H. Herpesvirus BACs: Past, present, and future. J Biomed Biotechnol. 2011;2011:124595. doi: 10.1155/2011/124595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanda T, et al. Unexpected instability of family of repeats (FR), the critical cis-acting sequence required for EBV latent infection, in EBV-BAC systems. PLoS One. 2011;6:e27758. doi: 10.1371/journal.pone.0027758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yakushko Y, et al. Kaposi’s sarcoma-associated herpesvirus bacterial artificial chromosome contains a duplication of a long unique-region fragment within the terminal repeat region. J Virol. 2011;85:4612–4617. doi: 10.1128/JVI.02068-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montague MG, Lartigue C, Vashee S. Synthetic genomics: Potential and limitations. Curr Opin Biotechnol. 2012;23:659–665. doi: 10.1016/j.copbio.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 10.Kouprina N, Larionov V. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat Protoc. 2008;3:371–377. doi: 10.1038/nprot.2008.5. [DOI] [PubMed] [Google Scholar]
- 11.Gibson DG, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 12.Gibson DG, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 13.Gibson DG, et al. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Natl Acad Sci USA. 2008;105:20404–20409. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith HO, Hutchison CA, 3rd, Pfannkoch C, Venter JC. Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. Proc Natl Acad Sci USA. 2003;100:15440–15445. doi: 10.1073/pnas.2237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 16.Cello J, Paul AV, Wimmer E. Chemical synthesis of poliovirus cDNA: Generation of infectious virus in the absence of natural template. Science. 2002;297:1016–1018. doi: 10.1126/science.1072266. [DOI] [PubMed] [Google Scholar]
- 17.Annaluru N, et al. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344:55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mercy G, et al. 3D organization of synthetic and scrambled chromosomes. Science. 2017;355:eaaf4597. doi: 10.1126/science.aaf4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell LA, et al. Synthesis, debugging, and effects of synthetic chromosome consolidation: synVI and beyond. Science. 2017;355:eaaf4831. doi: 10.1126/science.aaf4831. [DOI] [PubMed] [Google Scholar]
- 20.Richardson SM, et al. Design of a synthetic yeast genome. Science. 2017;355:1040–1044. doi: 10.1126/science.aaf4557. [DOI] [PubMed] [Google Scholar]
- 21.Shen Y, et al. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome. Science. 2017;355:eaaf4791. doi: 10.1126/science.aaf4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Y, et al. Bug mapping and fitness testing of chemically synthesized chromosome X. Science. 2017;355:eaaf4706. doi: 10.1126/science.aaf4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie ZX, et al. “Perfect” designer chromosome V and behavior of a ring derivative. Science. 2017;355:eaaf4704. doi: 10.1126/science.aaf4704. [DOI] [PubMed] [Google Scholar]
- 24.Zhang W, et al. Engineering the ribosomal DNA in a megabase synthetic chromosome. Science. 2017;355:eaaf3981. doi: 10.1126/science.aaf3981. [DOI] [PubMed] [Google Scholar]
- 25.Boeke JD, et al. GENOME ENGINEERING. The genome project-write. Science. 2016;353:126–127. doi: 10.1126/science.aaf6850. [DOI] [PubMed] [Google Scholar]
- 26.Garfinkel MS, Endy D, Epstein GL, Friedman RM. Synthetic genomics | Options for governance. Biosecur Bioterror. 2007;5:359–362. doi: 10.1089/bsp.2007.0923. [DOI] [PubMed] [Google Scholar]
- 27.Andtbacka RH, et al. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol. 2015;33:2780–2788. doi: 10.1200/JCO.2014.58.3377. [DOI] [PubMed] [Google Scholar]
- 28.Macdonald SJ, Mostafa HH, Morrison LA, Davido DJ. Genome sequence of herpes simplex virus 1 strain KOS. J Virol. 2012;86:6371–6372. doi: 10.1128/JVI.00646-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gierasch WW, et al. Construction and characterization of bacterial artificial chromosomes containing HSV-1 strains 17 and KOS. J Virol Methods. 2006;135:197–206. doi: 10.1016/j.jviromet.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 30.Post LE, Conley AJ, Mocarski ES, Roizman B. Cloning of reiterated and nonreiterated herpes simplex virus 1 sequences as BamHI fragments. Proc Natl Acad Sci USA. 1980;77:4201–4205. doi: 10.1073/pnas.77.7.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szpara ML, et al. Evolution and diversity in human herpes simplex virus genomes. J Virol. 2014;88:1209–1227. doi: 10.1128/JVI.01987-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colgrove RC, et al. History and genomic sequence analysis of the herpes simplex virus 1 KOS and KOS1.1 sub-strains. Virology. 2016;487:215–221. doi: 10.1016/j.virol.2015.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desai PJ. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J Virol. 2000;74:11608–11618. doi: 10.1128/jvi.74.24.11608-11618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Batterson W, Furlong D, Roizman B. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J Virol. 1983;45:397–407. doi: 10.1128/jvi.45.1.397-407.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner M, Jonjic S, Koszinowski UH, Messerle M. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J Virol. 1999;73:7056–7060. doi: 10.1128/jvi.73.8.7056-7060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norberg P, Bergström T, Rekabdar E, Lindh M, Liljeqvist JA. Phylogenetic analysis of clinical herpes simplex virus type 1 isolates identified three genetic groups and recombinant viruses. J Virol. 2004;78:10755–10764. doi: 10.1128/JVI.78.19.10755-10764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szpara ML, Parsons L, Enquist LW. Sequence variability in clinical and laboratory isolates of herpes simplex virus 1 reveals new mutations. J Virol. 2010;84:5303–5313. doi: 10.1128/JVI.00312-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macdonald SJ, Mostafa HH, Morrison LA, Davido DJ. Genome sequence of herpes simplex virus 1 strain McKrae. J Virol. 2012;86:9540–9541. doi: 10.1128/JVI.01469-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Desai P, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol. 1998;72:7563–7568. doi: 10.1128/jvi.72.9.7563-7568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elliott G, O’Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 41.Hogue IB, et al. Fluorescent protein approaches in alpha herpesvirus research. Viruses. 2015;7:5933–5961. doi: 10.3390/v7112915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.La Boissière S, Izeta A, Malcomber S, O’Hare P. Compartmentalization of VP16 in cells infected with recombinant herpes simplex virus expressing VP16-green fluorescent protein fusion proteins. J Virol. 2004;78:8002–8014. doi: 10.1128/JVI.78.15.8002-8014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Triezenberg SJ, LaMarco KL, McKnight SL. Evidence of DNA: Protein interactions that mediate HSV-1 immediate early gene activation by VP16. Genes Dev. 1988;2:730–742. doi: 10.1101/gad.2.6.730. [DOI] [PubMed] [Google Scholar]
- 44.Zhou ZH, et al. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2:1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]
- 45.Trus BL, et al. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: Structural authenticity and localization of VP26. J Virol. 1995;69:7362–7366. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luxton GW, Lee JI, Haverlock-Moyns S, Schober JM, Smith GA. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J Virol. 2006;80:201–209. doi: 10.1128/JVI.80.1.201-209.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shanda SK, Wilson DW. UL36p is required for efficient transport of membrane-associated herpes simplex virus type 1 along microtubules. J Virol. 2008;82:7388–7394. doi: 10.1128/JVI.00225-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luxton GW, et al. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc Natl Acad Sci USA. 2005;102:5832–5837. doi: 10.1073/pnas.0500803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Desai P, Akpa JC, Person S. Residues of VP26 of herpes simplex virus type 1 that are required for its interaction with capsids. J Virol. 2003;77:391–404. doi: 10.1128/JVI.77.1.391-404.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abaitua F, O’Hare P. Identification of a highly conserved, functional nuclear localization signal within the N-terminal region of herpes simplex virus type 1 VP1-2 tegument protein. J Virol. 2008;82:5234–5244. doi: 10.1128/JVI.02497-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wildy P, Russell WC, Horne RW. The morphology of herpes virus. Virology. 1960;12:204–222. doi: 10.1016/0042-6822(60)90195-1. [DOI] [PubMed] [Google Scholar]
- 52.Grünewald K, et al. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302:1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 53.Mettenleiter TC, Klupp BG, Granzow H. Herpesvirus assembly: An update. Virus Res. 2009;143:222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 54.Johnson DC, Baines JD. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol. 2011;9:382–394. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 55.Owen DJ, Crump CM, Graham SC. Tegument assembly and secondary envelopment of alphaherpesviruses. Viruses. 2015;7:5084–5114. doi: 10.3390/v7092861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han J, Chadha P, Starkey JL, Wills JW. Function of glycoprotein E of herpes simplex virus requires coordinated assembly of three tegument proteins on its cytoplasmic tail. Proc Natl Acad Sci USA. 2012;109:19798–19803. doi: 10.1073/pnas.1212900109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roller RJ, Fetters R. The herpes simplex virus 1 UL51 protein interacts with the UL7 protein and plays a role in its recruitment into the virion. J Virol. 2015;89:3112–3122. doi: 10.1128/JVI.02799-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loret S, Guay G, Lippé R. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J Virol. 2008;82:8605–8618. doi: 10.1128/JVI.00904-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baines JD, Roizman B. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J Virol. 1991;65:938–944. doi: 10.1128/jvi.65.2.938-944.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baines JD, Roizman B. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J Virol. 1992;66:5168–5174. doi: 10.1128/jvi.66.8.5168-5174.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baird NL, Starkey JL, Hughes DJ, Wills JW. Myristylation and palmitylation of HSV-1 UL11 are not essential for its function. Virology. 2010;397:80–88. doi: 10.1016/j.virol.2009.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanaka M, Sata T, Kawaguchi Y. The product of the Herpes simplex virus 1 UL7 gene interacts with a mitochondrial protein, adenine nucleotide translocator 2. Virol J. 2008;5:125. doi: 10.1186/1743-422X-5-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mbong EF, Woodley L, Frost E, Baines JD, Duffy C. Deletion of UL21 causes a delay in the early stages of the herpes simplex virus 1 replication cycle. J Virol. 2012;86:7003–7007. doi: 10.1128/JVI.00411-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roller RJ, Haugo AC, Yang K, Baines JD. The herpes simplex virus 1 UL51 gene product has cell type-specific functions in cell-to-cell spread. J Virol. 2014;88:4058–4068. doi: 10.1128/JVI.03707-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Albecka A, et al. Dual function of the pUL7-pUL51 tegument protein complex in herpes simplex virus 1 infection. J Virol. 2017;91:e02196-16. doi: 10.1128/JVI.02196-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Desai P, DeLuca NA, Glorioso JC, Person S. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J Virol. 1993;67:1357–1364. doi: 10.1128/jvi.67.3.1357-1364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Starkey JL, Han J, Chadha P, Marsh JA, Wills JW. Elucidation of the block to herpes simplex virus egress in the absence of tegument protein UL16 reveals a novel interaction with VP22. J Virol. 2014;88:110–119. doi: 10.1128/JVI.02555-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watson G, et al. Sequence and comparative analysis of the genome of HSV-1 strain McKrae. Virology. 2012;433:528–537. doi: 10.1016/j.virol.2012.08.043. [DOI] [PubMed] [Google Scholar]
- 69.Rastrojo A, López-Muñoz AD, Alcamí A. Genome sequence of herpes simplex virus 1 strain SC16. Genome Announc. 2017;5:e01392-16. doi: 10.1128/genomeA.01392-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Danaher RJ, et al. HSV-1 clinical isolates with unique in vivo and in vitro phenotypes and insight into genomic differences. J Neurovirol. 2017;23:171–185. doi: 10.1007/s13365-016-0485-9. [DOI] [PubMed] [Google Scholar]
- 71.Hayward GS, Jacob RJ, Wadsworth SC, Roizman B. Anatomy of herpes simplex virus DNA: Evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci USA. 1975;72:4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brown DM, et al. Efficient size-independent chromosome delivery from yeast to cultured cell lines. Nucleic Acids Res. 2017;45:e50. doi: 10.1093/nar/gkw1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Muto Y, Goshima F, Ushijima Y, Kimura H, Nishiyama Y. Generation and characterization of UL21-null herpes simplex virus type 1. Front Microbiol. 2012;3:394. doi: 10.3389/fmicb.2012.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baines JD, Koyama AH, Huang T, Roizman B. The UL21 gene products of herpes simplex virus 1 are dispensable for growth in cultured cells. J Virol. 1994;68:2929–2936. doi: 10.1128/jvi.68.5.2929-2936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nishiyama Y. Herpes simplex virus gene products: The accessories reflect her lifestyle well. Rev Med Virol. 2004;14:33–46. doi: 10.1002/rmv.409. [DOI] [PubMed] [Google Scholar]
- 76.Oxman MN, et al. Shingles Prevention Study Group A vaccine to prevent herpes zoster and postherpetic neuralgia in older adults. N Engl J Med. 2005;352:2271–2284. doi: 10.1056/NEJMoa051016. [DOI] [PubMed] [Google Scholar]
- 77.Looker KJ, et al. Global and regional estimates of prevalent and incident herpes simplex virus type 1 infections in 2012. PLoS One. 2015;10:e0140765. doi: 10.1371/journal.pone.0140765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Owusu-Edusei K, Jr, et al. The estimated direct medical cost of selected sexually transmitted infections in the United States, 2008. Sex Transm Dis. 2013;40:197–201. doi: 10.1097/OLQ.0b013e318285c6d2. [DOI] [PubMed] [Google Scholar]
- 79.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 80.Cohen JI, Fauci AS, Varmus H, Nabel GJ. Epstein-Barr virus: An important vaccine target for cancer prevention. Sci Transl Med. 2011;3:107fs7. doi: 10.1126/scitranslmed.3002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vashee S, et al. Cloning, assembly and modification of the primary human cytomegalovirus isolate Toledo by yeast-based transformation-associated recombination. mSphere. 2017 doi: 10.1128/mSphereDirect.00331-17. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.World Health Organization 2015 The independent advisory group on public health implications of synthetic biology technology related to smallpox. WHO/HSE/PED/2015 (World Health Organization, Geneva). Available at apps.who.int/iris/bitstream/10665/198357/1/WHO_HSE_PED_2015.1_eng.pdf. Accessed July 12, 2017.
- 83.Baric RS. Synthetic viral genomics: Risks and benefits for science and society. In: Garfinkel MS, Endy D, Epstein GL, Friedman RM, editors. Working Papers for Synthetic Genomics: Risks and Benefits for Science and Society. J. Craig Venter Institute; Rockville, MD: 2007. pp. 35–81. [Google Scholar]
- 84.Collett MS. Impact of synthetic genomics on the threat of bioterrorism with viral agents. In: Garfinkel MS, Endy D, Epstein GL, Friedman RM, editors. Working Papers for Synthetic Genomics: Risks and Benefits for Science and Society. J. Craig Venter Institute; Rockville, MD: 2007. pp. 83–103. [Google Scholar]
- 85.Kupferschmidt K. Labmade smallpox is possible, study shows. Science. 2017;357:115–116. doi: 10.1126/science.357.6347.115. [DOI] [PubMed] [Google Scholar]
- 86.Jaschke PR, Lieberman EK, Rodriguez J, Sierra A, Endy D. A fully decompressed synthetic bacteriophage øX174 genome assembled and archived in yeast. Virology. 2012;434:278–284. doi: 10.1016/j.virol.2012.09.020. [DOI] [PubMed] [Google Scholar]
- 87.Ando H, Lemire S, Pires DP, Lu TK. Engineering modular viral scaffolds for targeted bacterial population editing. Cell Syst. 2015;1:187–196. doi: 10.1016/j.cels.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Health UDo & Services H Screening framework guidance for providers of synthetic double-stranded DNA. Fed Regist. 2010;75:62820–62832. [Google Scholar]
- 89.Carter SR, Friedman RM. 2015 DNA synthesis and biosecurity: Lessons learned and options for the future. Available at www.jcvi.org/cms/fileadmin/site/research/projects/dna-synthesis-biosecurity-report/report-complete.pdf. Accessed July 27, 2017.
- 90.World Health Organization 2017 WHO Advisory committee on variola virus research: Report of the 18th meeting, 2–3 November 2016 (World Health Organization, Geneva). Available at www.who.int/csr/resources/publications/smallpox/18-ACVVR-Final.pdf. Accessed July 12, 2017.
- 91.Desai P, DeLuca NA, Person S. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology. 1998;247:115–124. doi: 10.1006/viro.1998.9230. [DOI] [PubMed] [Google Scholar]
- 92.Desai P, Watkins SC, Person S. The size and symmetry of B capsids of herpes simplex virus type 1 are determined by the gene products of the UL26 open reading frame. J Virol. 1994;68:5365–5374. doi: 10.1128/jvi.68.9.5365-5374.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Noskov V, et al. A genetic system for direct selection of gene-positive clones during recombinational cloning in yeast. Nucleic Acids Res. 2002;30:E8. doi: 10.1093/nar/30.2.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.