Abstract
Biology is marked by a hierarchical organization: all life consists of cells; in some cases, these cells assemble into groups, such as endosymbionts or multicellular organisms; in turn, multicellular organisms sometimes assemble into yet other groups, such as primate societies or ant colonies. The construction of new organizational layers results from hierarchical evolutionary transitions, in which biological units (e.g., cells) form groups that evolve into new units of biological organization (e.g., multicellular organisms). Despite considerable advances, there is no bottom-up, dynamical account of how, starting from the solitary ancestor, the first groups originate and subsequently evolve the organizing principles that qualify them as new units. Guided by six central questions, we propose an integrative bottom-up approach for studying the dynamics underlying hierarchical evolutionary transitions, which builds on and synthesizes existing knowledge. This approach highlights the crucial role of the ecology and development of the solitary ancestor in the emergence and subsequent evolution of groups, and it stresses the paramount importance of the life cycle: only by evaluating groups in the context of their life cycle can we unravel the evolutionary trajectory of hierarchical transitions. These insights also provide a starting point for understanding the types of subsequent organizational complexity. The central research questions outlined here naturally link existing research programs on biological construction (e.g., on cooperation, multilevel selection, self-organization, and development) and thereby help integrate knowledge stemming from diverse fields of biology.
Keywords: major evolutionary transitions, hierarchical evolutionary transitions, bottom-up approach, life cycle, animal sociality
From a primordial soup of elements to the emergence of protocells, from single cells to multicellular organisms, and from multicellular organisms to animal groups, evolution has been punctuated by hierarchical evolutionary transitions (HET), whereby simple units assembled into groups that themselves became new units of biological organization (1–4). The popularization of these HET [also known as transitions in individuality (2, 5)] as part of the “major transitions in evolution” by Maynard Smith and Szathmáry (3), resulted in extensive research efforts—both empirical and theoretical—to understand how new units of biological organization can evolve. However, this endeavor has proved challenging, not least because a unique definition for what constitutes a unit of biological organization has eluded the field; instead, the literature abounds with definitions that differ in the minimal criteria for a group to be considered a unit of biological organization (SI Appendix, Text S1, Fig. S1, and Table S1). There seem to be only two points of general agreement: (i) a necessary criterion, common to all definitions, for a group to be a unit of biological organization is that the group must be a unit of selection (i.e., it can undergo evolutionary change by natural selection) (SI Appendix, Text S1); and (ii) there are certain entities that are unambiguously units of biological organization (e.g., animals, plants, eusocial colonies). This has engendered a “top-down” approach for the study of HET that starts with such paradigmatic examples of biological units, identifies their properties (e.g., high level of cooperation, reduced conflict, differentiated types, metabolic specialization) (SI Appendix, Text S1), and explores how a group could have evolved each of these properties. While this approach has revealed a wealth of valuable insights, we argue that it is insufficient to understand the origin and evolution of HET.
This type of top-down approach to the study of HET runs into two critical problems. First, by focusing on properties of groups that qualify as paradigmatic examples of biological units, studies largely ignore the ancestor, including its internal organization and properties, the ecological context, and the mechanisms that gave rise to the primitive instantiations of those groups (6–8). As a consequence, it often remains unclear how the organization of the group—including the properties of interest—originated from that of the ancestor, making it impossible to fully unravel the evolutionary trajectory from the solitary ancestor to a new unit of biological organization (9–12): Which organizing principles and properties (e.g., differentiation, conflict suppression, metabolic specialization, cooperation) evolved de novo and which appeared as by-products due to strong interdependencies? What was the order in which organizing principles evolved? How did the organization at one point in time constrain or potentiate the evolution of new organizing principles? What is the relative importance of various factors (e.g., ecological context, conflict avoidance, development/physiology/life history traits) for the evolution of new organizing principles? What types of organizing complexity can emerge from different ancestral properties and evolutionary trajectories?
Second, in addition to ignoring the ancestral properties, by fixating on certain properties common to the known paradigmatic examples of HET, the top-down approach fails to explore the full potential of evolutionary trajectories and transitions, not only the paradigmatic but also the peripheral, and not only the actual (i.e., realized) but also the possible (13). This likely paints an incomplete picture of HET and precludes a valuable comparison across potential evolutionary transitions: only by comparing their full spectrum can we determine the causal factors that explain why certain trajectories did result in new units of biological organization and others did not (14).
Here we identify six questions, Q1–Q6, that, regardless of the definition for what constitutes a new unit of biological organization, need to be addressed in a bottom-up approach to the study of HET:
Q1: When/how does a group originate that has the potential to undergo a HET?
Q2: What emergent properties do these groups have? (For example, in the case of multicellular groups: group size, composition, shape, and the interactions of cells inside the group, including cooperative interactions.)
Q3: How does selection act on these properties?
Q4: How does this affect the ancestral developmental program(s) and change group properties? Selection is only effective when group properties emerge from a heritable developmental program. In the case of newly formed groups, the developmental program is that of the solitary ancestor(s) that make up the group. Selection will therefore exert its effect by affecting the ancestral developmental program(s).
Q5: When/how does this lead to novel organizing/developmental principles within the new unit? (For example, in the case of multicellular groups: differential adhesion, pattern formation and cell signaling.)
Q6: What kinds of organizing complexity can evolve?
These questions separate the origination of the first group and group properties (Q1–Q2) from the selective pressures that underlie the conservation and further evolution of the group (Q3–Q6). This conceptual distinction helps disentangle the causal factors underlying HET; yet, importantly, it does not imply that these processes occur sequentially, since groups can have an instantaneous selective benefit upon their origination. Guided by these six questions, in this Perspective we propose a bottom-up approach to study the dynamics underlying HET, which builds on and integrates knowledge from existing research programs on biological construction: phylogenetic (12, 15–18), empirical (e.g., experimental evolution, developmental biology, sociobiology) (10, 19–22), and theoretical (e.g., on multilevel-selection, cooperation, self-organization) (4, 14, 23–29) (see also SI Appendix, Text S2). We illustrate this approach by focusing on the transition to multicellularity, but we showcase its wide applicability by briefly discussing the evolution of animal sociality in Other HET, below, and SI Appendix, Text S4.
Bottom-Up Approach
Through his work on multicellularity, John T. Bonner was one of the first to study evolutionary transitions in biological organization (1, 30). Bonner focused in particular on the role of the life cycle in the HET to multicellularity (30). He argued that the life cycle encapsulates all properties needed for the potential to evolve by natural selection (1) (i.e., reproduction and heritable variation) and considered the life cycle, and not the organism, to be the unit of biology (30) (SI Appendix, Text S3). With this view, biological entities (including groups) have the potential to be a unit of selection if and only if they are part of a life cycle. For example, if a cell acquires a mutation that makes it stick to its daughters after division (e.g., ref. 31), a group life cycle arises, in which cells form clumps that occasionally might break and give rise to new clumps. Over evolutionary time, these clumps could evolve new properties. Groups could also arise as part of the ancestral life cycle. In fact, an increasing number of studies show that groups are often expressed as facultative life stages—triggered by specific environmental conditions—in life cycles of otherwise solitary organisms (32). For example, Chlamydomonas reinhardtii, a close relative (i.e., sharing a recent common ancestor) of the multicellular volvocine green algae (33, 34), lives as a unicellular organism, but can induce stickiness and form groups in response to its natural predator Peranema trichophorum (35). Similarly, Capsaspora owczarzaki, a close relative of the metazoans, can form facultative aggregates in response to environmental stress (36). Even in endosymbioses, facultative associations between the symbiotic partners are hypothesized to have preceded obligate relationships (37).
Following these arguments, henceforth we will define a group as having the potential to undergo a HET (i.e., the potential to be a unit of selection) only when it is part of a life cycle, either as a reproducible life stage in the life cycle of the solitary precursor or as part of a life cycle in which the solitary life stage is effectively absent (i.e., groups that propagate by fragmentation) (see also refs. 38 and 39 and SI Appendix, Text S1). The reproducibility requirement pertains strictly to the act of group formation; for all other group properties, such as composition, size, or shape, we allow for potentially low or no reproducibility (for the purpose of this Perspective we distinguish between heritable material and reproducible properties; see Table 1). Therefore, according to this definition, one cannot establish a group’s potential to undergo a HET by examining its properties at a given moment in time; instead, one has to trace the group and its descendants over time to determine the reproducibility of group formation. Furthermore, we do not require the group to be formed in every successive instantiation of the life cycle (henceforth generation), only that it is formed sufficiently frequently for selection to potentially act on the group stage. For example, a group could be expressed as a facultative life stage only in response to certain recurrent environmental conditions, as in the examples above.
Table 1.
Definitions as used in this Perspective
| Term | Definition |
| Unit of biological organization | Multiple definitions (see SI Appendix, Text S1, Fig. S1 and Table S1). |
| Life cycle | The cycle of phenotypic properties that reoccurs every generation [not all properties need to reoccur (see SI Appendix, Text S3)]. |
| Group with potential to undergo a HET | Group that is part of a life cycle, such that the act of group formation is reproducible across subsequent instantiations of the life cycle. |
| Development | The intrinsic processes underlying an organism´s temporal and spatial organization. (Not confined to a particular life stage; encapsulates all processes underlying an organism’s life cycle, including solitary and potential group life stages). |
| Ecology | Biotic (e.g., competitors, predators) and abiotic environment (e.g., temperature, nutrient availability). |
| Emergent properties | Higher-level (e.g., group) properties that result from interactions between lower-level components (e.g., group members). |
| Heritable material | Material transmitted from parent to offspring as a direct continuation (e.g., DNA, developmental program). |
| Reproducible properties | Properties reconstructed in subsequent generations, as the product of the inherited material and the ecology. |
| Cooperation | Expression of a costly phenotype that is beneficial to others (e.g., public-good production). |
| Conflict | Expression of a beneficial phenotype that is costly to others (e.g., toxin production, social free-riding, parasitism, competition). |
To determine if groups are part of a life cycle, one needs to determine what constitutes the life cycle (SI Appendix, Text S3). This might seem a trivial task when thinking of the paradigmatic examples of biological organization (animals and plants), but it can be surprisingly difficult in general. Soil-dwelling unicellular organisms are a case in point: in the absence of information about their environment, the life cycle of single cells could be described by their division cycle; but many soil organisms are exposed to fluctuating environmental conditions, such as feast–famine cycles, where short periods of food availability are alternated with long periods of starvation. One could therefore argue that the feast-and-famine cycle, not the division cycle, determines the life cycle of these unicellular organisms. Thus, the feedback between the ecological context (biotic and abiotic interactions; also referred to below as the ecology) and development gives rise to the recurrent trait appearances that characterize the life cycle. Consequently, one can only evaluate life cycles accurately in the appropriate ecological context.
(Q1) Origination of a Group with the Potential to Undergo a HET.
Starting from the above definition of what constitutes a group with the potential to undergo a HET, we can examine the conditions necessary for its origination: first, something should trigger group formation; second, group formation should be reproducible across generations, either as an obligatory or as a facultative life stage. We discriminate between two scenarios that could trigger the appearance of the first group stage within a life cycle (SI Appendix, Fig. S2): (i) the ecology-first scenario, in which an ecological change results in the origination of the first group; and (ii) the mutation-first scenario, in which a genetic change results in the origination of the first group. Both scenarios pertain only to the mechanism that underlies the origination of the first groups, not to the selection pressures that might favor or oppose such groups.
In the ecology-first scenario, an ecological change (either biotic or abiotic) acts on preexisting cellular properties to lead to the formation of a group (19, 40). This can happen in many ways. For example, cells might be exposed to an atypical ecological condition that results in the overexpression (via regulatory induction) of a set of proteins. Many proteins carry promiscuous functions (41), such as weak adhesive properties [e.g., proteins involved in phagocytosis (16, 42)]; the overexpression of such proteins could lead to enhanced adhesion that would enable cell-to-cell attachment resulting in group formation. Thus, in this scenario, an ecological change is responsible for triggering group formation by acting on the preexisting plastic response of the solitary ancestor. Crucially, the ecological change should persist or reoccur sufficiently often to support the reproducibility of group formation across generations. It is important to note that here the role of ecology is distinct from the one typically considered in studies on HET: while most studies only consider the ecology when it comes to the selection pressures that favor group formation (e.g., ecological benefits) (see ref. 43), we emphasize that the ecology can also play a critical role in triggering and supporting the origination of the first group life cycles. We also consider the selective (dis)advantages of group formation, but we do so later, in Q3. As noted above, this conceptual separation is not meant to imply that the selective benefits only arise after the origination of the group, since groups can carry instantaneous benefits upon their origination; rather, it is done with the explicit purpose of highlighting the largely ignored, nonselective role that the ecology can play in group origination.
In the mutation-first scenario, a genetic change triggers group formation in a preexisting ecological context. This can also occur in many ways. For example, a genetic mutation could block the expression of an enzyme necessary for hydrolyzing the cell wall at the end of cytokinesis [e.g., a mutation in CTS1, a gene encoding for a chitinase that mediates cell separation in Saccharomyces cerevisiae (44)]. Then cells would remain attached after cell division and give rise to cell clumps. These clumps could grow and fragment under mechanical stress, thereby giving rise to a group life cycle (31). If the mutation is conditional on the environmental context, in that it only blocks the expression of the hydrolyzing enzyme under certain conditions [e.g., the conditional repression of autolysins in Bacillus subtilis (45)], environmental fluctuations might support a life cycle that alternates between a solitary life stage and a group life stage. Thus, although in this scenario ecological changes are not the primary cause for the origination of group formation, they can still play an important role in the emergent group life cycle and the reproducibility of the group stage.
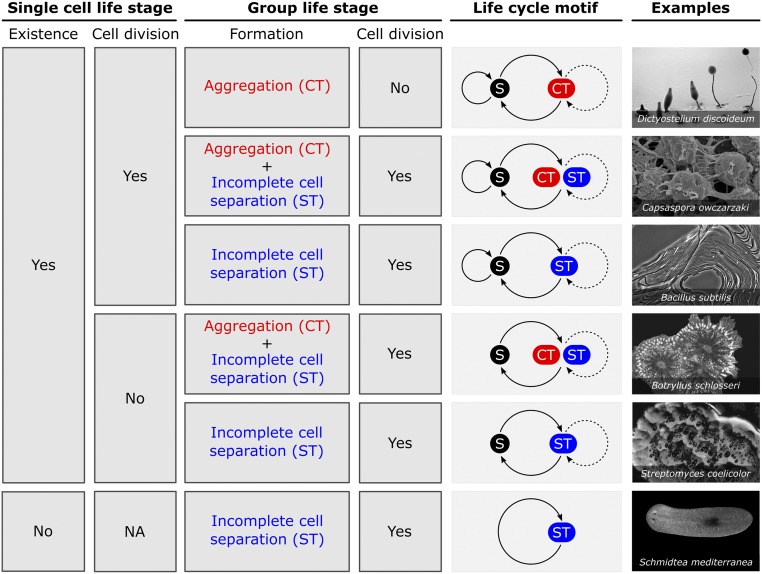
Fig. 1 gives an overview of the life-cycle motifs that could emerge upon origination of a group life stage (triggered by either ecological or genetic changes). These motifs represent the simplest possible life cycles (which could be part of more complex ones; see ref. 46) and they are categorized based on a few criteria (compare with figure S2 in ref. 4): (i) the presence/absence of the solitary life stage, (ii) the mechanism by which groups are formed, and (iii) the life stage at which cell division occurs (necessary to support the propagation of the life cycle). These criteria can be further extended to specify, for example, whether the group life stage is obligatory or facultatively expressed; how transitions between life stages take place (e.g., dispersal, sexual reproduction); or whether the solitary and group life stages coexist in time and space [e.g., when grouping is triggered by a change in ecological conditions, some cells might remain solitary; see Dictyostelium discoideum (47)].
Fig. 1.
Potential multicellular life cycles that could emerge upon the formation of the first multicellular groups. Categorization based on (i) existence of single cell (S), (ii) mechanism of group formation (CT/ST), and (iii) life stage where cell division occurs. Two life cycles have a group life stage formed by both CT and ST; here, aggregated cells divide inside the group. Arrows indicate cell division in solitary life stage, transition between solitary and group life stages, and potential fragmentation of the group (dotted line). Images show examples of species with a life cycle comparable to each life cycle motif. (Top to Bottom) D. discoideum, image courtesy of MJ Grimson and RL Blanton (17, 90), C. owczarzaki, image adapted from ref. 36, B. subtilis (107, 108), Botryllus schlosseri, reprinted from ref. 109 with permission from Elsevier, Streptomyces coelicolor, image courtesy of VM Zacharia and MF Traxler (52, 107), Schmidtea mediterranea asexual biotype CIW4, image adapted from ref. 110.
Consistent with Bonner (48), we discriminate between two grouping mechanisms (see also SI Appendix, Text S2): cells can either stay together (ST) due to incomplete cell separation after cell division (i.e., clonal development), or they can come together (CT) by means of aggregation (i.e., aggregative development) (4, 26, 49, 50). ST can take many forms: for example, cells could have incomplete cytokinesis, in which the cell walls at the division plane remain fused (44); a daughter cell could be engulfed by the mother cell during cell division (51); coenocytic filamentous cells could cellularize through septa formation (52); cells could undergo complete cell division, but remain attached due to adhesive molecules (53); and so forth. Similarly, CT can also take many forms: for example, cells can aggregate via chemotaxis (54), by binding a common surface (55), or by binding each other (35). Most forms of aggregation are mediated by soluble or membrane-bound adhesive molecules, such as extracellular polysaccharides, protein fibers, and adhesion receptors. ST and CT mechanisms can also be combined: for example, cells (clonal or mixed) could aggregate on a surface to form a group and subsequently undergo cell division without cell separation (22, 55).
Cells in groups formed via ST are necessarily “similar” (since they are clonal), while those in groups formed via CT can be similar (e.g., same or related genotypes) or “different” (e.g., different species). HET, in which group members are similar, are referred to as fraternal transitions, while those in which group members are different are referred to as egalitarian transitions (26). The bottom-up approach we outline can be employed to study both cases but, for simplicity of exposition, below we will focus on the fraternal case; thus, in the case of CT, the aggregating cells will be either clonal or at most of different genotypes of the same species. Bonner (48) pointed out that all aquatic origins of multicellularity arose via ST, while most terrestrial origins arose via CT. This shows that the physics of the environment—for example, a relative lack of surfaces that could support aggregative multicellularity in aquatic systems—can constrain the possible grouping mechanisms, reemphasizing the diverse and critical roles of ecology in the origination of groups.
(Q2) Emergent Group Properties.
The origination of a group with the potential to undergo a HET leads to the spontaneous emergence of group properties that fall into three categories (SI Appendix, Fig. S2).
Group formation.
Multiple properties characterize group formation, such as the rate at which a group forms, its timing relative to other events (e.g., the environmental fluctuations involved in triggering group formation), its location in physical space, or its efficiency. For example, a group that is triggered in response to starvation could form more or less quickly depending on the plastic response of individual cells to starvation (which could be different due to both phenotypic and genotypic variability); it could form in the same place where the cells starved, or elsewhere if cells first migrate to more appropriate conditions; and it could form more or less efficiently in terms of its inherent cohesion, depending on the level of adhesiveness of each cell.
Group features.
Group features emerge from the interactions between member cells and depend on cell properties. There can be many emergent group features, but here we briefly focus on group size, group composition, within-group interactions, and group shape. Group size is determined by the strength with which cells adhere to each other: stronger adhesion results in less fragmentation and hence bigger groups (56). Group composition is determined by members that make up the group, which could be clonal or nonclonal (different genotypes). Within the group, spontaneous interactions could emerge between cells. For example, in groups consisting of multiple genotypes, cells might spontaneously engage in antagonistic interactions via the production of toxins, but they might also engage in metabolic interactions, such as cross-feeding, whereby they exchange metabolites that improve growth (57). Such mutualistic interactions could further influence the organization of the group by promoting genotypic intermixing (58). In clonal groups (i.e., consisting of a single genotype), cells could spontaneously engage in a variety of interactions as well (25), some of which could be cooperative (31). Clonal groups could also spontaneously express phenotypic heterogeneity [e.g., via cell responses to local environmental gradients (see ref. 59)]. This capacity of cells to express phenotypic differences inside the group is in most cases already latently present in the ancestor (60). Solitary cells face a multitude of ecological challenges, which they overcome by adjusting their phenotype: for example, cells can express different metabolic pathways in response to the available resources, become motile in search for food, or induce dormancy to survive stress. The phenotypic states that the ancestor expresses in time can become expressed in space when cells form a group (61). Thus, the plasticity of the ancestor in response to its environment will likely influence the propensity of cells to vary inside the group. This phenotypic variability could even result in pattern formation if cells respond to each other through extracellular signals (62). Finally, group shape can also be affected by member cells. Models and experiments have shown that when cells differ in their adhesive properties, simple morphogenic processes could emerge (e.g., cell sorting, engulfment, folding) that can influence group shape (27, 63, 64). Differential adhesion is relevant to both clonal and nonclonal groups.
Propagation.
As part of a life cycle, groups need to propagate (SI Appendix, Text S3) to prevent the life cycle from ending with the group stage. Propagation can take many forms: groups might release single cells, they might shed fragments or fission, or they might dissolve altogether. The mode and rate of propagule production depend on the viscoelastic properties of the group as well as on the environmental conditions (65). For example, when groups are exposed to stronger shear stresses, they are expected to shed more propagules. The processes of propagule production and group formation are antagonistic (21, 66, 67): whereas the latter requires the attachment of cells, the former relies on their separation. This was experimentally illustrated in Vibrio cholerae (68): constitutive production of extracellular matrix enhanced group formation and growth due to cells firmly sticking together, but dramatically reduced propagule production. The trade-off between group formation and propagule production is just one of the many possible interdependencies that might characterize the first groups.
(Q3 and Q4) Selection and New Emergent Properties.
Selection could act on any of the emergent group properties and, due to interdependencies, indirectly affect others. For example, when there is selection for bigger group sizes, cells that produce more adhesive molecules might be favored, which strengthens their cohesion (56). This increased adhesiveness is likely to affect the group composition as well: for example, cells might start to sort based on their adhesive properties (69) or they might bind to nonadhesive cells in the environment. Increased adhesiveness can also change the group shape: for instance, adhesive molecules might alter the growth dynamics of the group (70) or change its viscoelastic properties (71), thereby changing the group response to external mechanical forces (e.g., shear stress). Finally, as mentioned above, increased adhesiveness can also influence propagule production: for example, adhesive molecules might decrease the rate of propagule production (68) and increase propagule size (65). Thus, selection for one property—group size—is likely to have consequences for many other group properties as well, some of which could be deleterious (e.g., reduced propagule production). Such interdependencies make it difficult to discriminate a posteriori between properties that were favored by selection and those that emerged as side-effects. For instance, in the study of HET, it is often claimed that the single-cell bottleneck evolved because it results in strict genetic homogeneity and, thereby, prevents conflict. However, the single-cell bottleneck might just as well be conserved because it can promote reproduction (72), improve dispersal (20, 21), support reliable development (73), or because it is simply associated with one of the ancestral life stages (e.g., syngamy) (74); this would lead to strict genetic homogeneity as an inevitable side-effect, even when it is not strictly required to prevent within-group conflict (75).
If trade-offs between group properties are deleterious to the life cycle, such as the one between group formation and propagule production, selection could favor mutations that overcome these trade-offs. This was demonstrated experimentally in Pseudomonas aeruginosa (21) by exposing it to a life-cycle regime in which cells had to alternate between two life stages: one in which group formation (i.e., adhesive cells) was favored and one in which propagule production (i.e., nonadhesive cells) was favored. Under this selection regime, cells evolved a surprising molecular trick to overcome the trade-off between group formation and propagule production. They increased mutation rates that—via frameshift mutations in a specific genomic region—facilitated the alternation between adhesive and nonadhesive phenotypic states. Consequently, groups always produced nonadhesive propagules, while a fraction of the propagules always reverted to group formation. Another solution to overcome this group formation–propagule production trade-off is regulation, as is the case in many strains of V. cholerae. These strains regulate matrix production based on nutrient availability (76): cells stimulate matrix production and stick together in good conditions, but inhibit matrix production and secrete enzymes that digest the remaining matrix to allow dispersal when conditions deteriorate.
The properties of the first groups are not only expected to be interdependent, but also to vary considerably across generations (4, 63, 77). In the relative absence of developmental control, groups are likely to be sensitive to small environmental perturbations. For example, a small change in the shear stress could affect the group size, group shape, and rate of propagule production. An important selective target might therefore be the reproducibility of group properties (4): selection in favor of developmental mechanisms that improve the reproducibility of beneficial group properties across generations (77). Selection for reproducibility is, in effect, selection for developmental control, since reproducible properties can evolve only to the extent that group formation is under the control of a heritable developmental program (78), whether it be encoded by a single or multiple genomes. Importantly, our bottom-up approach emphasizes that reproducibility of group properties can evolve after the origin of group formation, which only requires the act of group formation, and not the group properties, to be reproducible across generations (see discussion of Q1). Beneficial properties that might first be triggered by specific ecological conditions (i.e., facultatively expressed), can—via the evolution of new developmental mechanisms—become part of the developmental program, and therefore be expressed under a much wider range of conditions (i.e., genetic assimilation) (79, 80). For example, selection might favor groups that produce stress-resistant propagules. Initially, these might only be produced under starvation, which triggers sporulation as part of the ancestral developmental program. However, additional mutations might allow for quorum-sensing signaling (81), which could facilitate sporulation to also be triggered by high cell densities (i.e., bigger groups), even in the relative absence of starvation signals [in some colony-forming bacteria sporulation indeed depends on quorum-sensing signals (for example, see refs. 82 and 83)]. In the end, any developmental mechanism that facilitates the robust expression of a beneficial group property, over a large range of ecological conditions, is a mechanism that improves reproducibility via a form of developmental canalization.
Developmental mechanisms that promote reproducibility can also evolve in the presence of genetic diversity. For example, in the case of symbiosis, a group property (e.g., cross-feeding) might rely on the presence of two symbiotic partners, but might be difficult to reproduce if these partners dissociate after group formation and cannot re-establish a new group. Developmental mechanisms that prevent genotypes from dissociating (e.g., mechanisms that promote vertical transmission of the symbiotic partners) or promote their reestablishing a new group (e.g., partner-choice mechanisms) could improve the reproducibility of group properties (37). There might also be selection against the association of some genotypes. For example, if cooperation gives rise to a group property, noncooperative genotypes could reduce the fidelity with which this property is propagated across generations (e.g., noncooperating cells could undermine the development of the group property by exploiting cooperating cells). In that case, selection might favor developmental mechanisms that prevent noncooperative cells from joining the group [e.g., assortment mechanisms, such as kin discrimination and bottlenecks (84)]. The extent to which within-group conflict leads to reproducibility issues depends on the grouping mechanism and the ecological context in which groups are formed (28, 50). For example, groups formed by ST are less prone to internal conflict than those formed by CT (49), because in ST conflicts can only arise through mutations. Importantly, even if noncooperative cells might occasionally join a CT group, strong spatial assortment, if present in the environment, could still prevent those cells from parasitizing other groups, and therefore from reducing the reproducibility of group properties in the population.
Since within-group conflict is just one of many factors that could reduce the reproducibility of group properties, a lack of conflict does not guarantee accurate reproducibility of group properties. Conversely, the presence of within-group conflict does not automatically reduce reproducibility either, since there might be mechanisms that suppress within-group selection; for example, it could be physically impossible for the noncooperative cells to spread within the group, as is the case for cancerous tissues in plants (85). Thus, mechanisms that prevent within-group conflict (i.e., assortment mechanisms) and inhibit within-group selection [i.e., individuating mechanisms (see SI Appendix, Text S2) (86)] are merely a subset of the many developmental mechanisms that could influence the reproducibility of group properties.
(Q5 and Q6) New Organizing Principles and Organizing Complexity.
New organizing principles are those that underlie the organization of a group but that were not present in the ancestor. In the previous section we already alluded to some of these principles (e.g., quorum-sensing signaling). There are many organizing principles, which act at different spatial scales, ranging from the organization of single cells to that of organs. Some of these organizing principles are shared across a wide-range of multicellular organisms: for example, cell differentiation, cell-to-cell communication, pattern formation, lateral inhibition, induction, determination, regional differentiation, differential adhesion, segmentation, germ–soma differentiation, boundary formation, and tissue formation (10, 27, 64, 87). However, not all of these organizing principles are unique to multicellular groups: for example, in some cases, the solitary ancestor might already express cell differentiation or communication. Only when organizing principles evolved after the origin of the first groups do we consider them to be new organizing principles of the group.
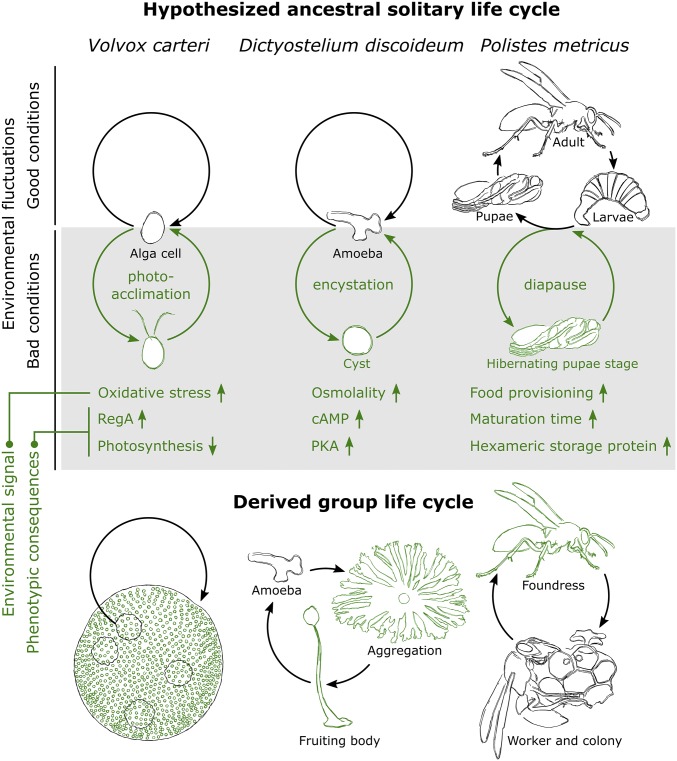
We have relatively little understanding of the origin of most organizing principles (e.g., germ–soma differentiation, tissue formation, pattern formation). However, there is accumulating evidence for the important role of the ancestor in the evolution of new organizing principles (12). For example, the aquatic and colonial green alga Volvox carteri exhibits germ–soma differentiation, with biflagellated somatic cells at the periphery of the spherical colony and dividing germ cells in the interior (Fig. 2) (34). Differentiation of somatic cells is regulated by RegA, a protein that suppresses photosynthesis and thereby prevents division (88). Interestingly, phylogenetic studies revealed that a close homolog of RegA is involved in photoacclimation, a plastic response that can be triggered by light deprivation (39, 89). In the unicellular ancestor, photoacclimation was likely required for cells to adjust to the diurnal light cycle: inhibiting photosynthesis during light limitation prevents oxidative stress. Thus, the regulatory protein involved in a switch between life stages in the solitary ancestor was co-opted for germ–soma differentiation in its multicellular descendant (Fig. 2). An even more striking case of co-option is found in the phagocytic and soil-dwelling amoeba Dictyostelium discoideum (90). This social amoeba is exposed to feast–famine cycles resulting from fluctuating resource levels in the soil. Upon starvation, cells aggregate into fruiting bodies that mediate spore dispersal. Cell aggregation, fruiting body formation, and sporulation depend on cAMP, which exerts its effect by activating cAMP receptors and the cAMP-dependent protein kinase (PKA) (54, 90). Interestingly, in species of solitary amoebae, encystation—which can be triggered by osmotic stress (e.g., due to soil dehydration)—also relies on cAMP-mediated activation of PKA (91). The disruption of cAMP receptors in the social amoeba Polysphondylium pallidum—a relative of D. discoideum—results in malformed fruiting bodies that are filled with cysts instead of spores (92). P. pallidum normally only forms cysts in the unicellular life stage (by comparison, D. discoideum never forms cysts). Supported by phylogenetic studies, these results indicate that the developmental program underlying fruiting body formation is derived from the encystation program (Fig. 2). In fact, one could argue that cysts in solitary amoebae and fruiting bodies in D. discoideum are distant homologies, in much the same way as fins and arms are homologies (93): they are different functional realizations of a (partly) conserved developmental program.
Fig. 2.
Relationship between life stages in hypothesized life cycles of solitary ancestors and group formation in derived group life cycles. (Upper) Simplified depiction of hypothesized ancestral solitary life cycles of V. carteri (33, 88, 89), D. discoideum (90), and Polistes metricus (103–105). Life cycles here consist of a life stage expressed under good conditions (black) and a life stage expressed under adverse conditions (green). For the latter life stage, we show an environmental signal that might trigger it and some phenotypic consequences. For P. metricus, high food provisioning at the end of the breeding season is hypothesized to be a cue for the upcoming winter season. (Lower) Simplified depiction of group life cycles of: V. carteri, corresponding to fifth life cycle in Fig. 1 (ST group and nondividing unicellular life stage; zygote, not shown); D. discoideum, corresponding to first life cycle in Fig. 1 (CT group and dividing unicellular life stage); and P. metricus, corresponding to seventh life cycle in SI Appendix, Fig. S3 (ST group and nonreproducing solitary life stage). Developmental program underlying life stages in solitary ancestor is co-opted for group formation (shown in green): differentiation of somatic cells (V. carteri), fruiting body formation (D. discoideum), and appearance of foundress phenotype (P. metricus).
This mounting evidence for the importance of the ancestral developmental program to the emergence of new organizing principles in its multicellular descendants (see also refs. 12, 36, 94, and 95) also emphasizes the need for caution when referring to HET in multicellularity as transitions in complexity. Many solitary organisms have intricate regulatory pathways—such as the encystation program in solitary amoebae—that could potentially support multicellular organization. In fact, multiple phylogenetic studies have shown that the regulatory complexity of solitary organisms, when focusing on specific regulatory pathways, can be comparable to that of their multicellular relatives. For example, the choanoflagellate Monosiga brevicollis, the closest unicellular relative of the metazoans, has a repertoire of phosphotyrosine signaling comparable to that of metazoans (96–98). This is particularly striking since phosphotyrosine signaling—involved in cell differentiation, adhesion, and the control of cell proliferation in metazoans (99)—was long considered to be unique to metazoan development. Along similar lines, Clarke et al. (100) showed that the solitary amoeba, Acanthamoeba castellanii, displays a rich repertoire of sensory receptors, transcription factors, and phosphotyrosine signaling, comparable to that of D. discoideum. The regulatory complexity in these solitary organisms likely reflects the complex ecology to which they are exposed—cells have to find food, avoid predation, and withstand many environmental changes (16, 19)—and therefore reveals that the life cycle of the solitary organisms can in many ways be more complex than that of their multicellular relatives. Hence, the full complexity of an organism cannot be adequately captured by measuring group properties alone (e.g., group size, number of differentiated cell types); one must also account for the properties of its life cycle (12, 101).
Even though we focus in our bottom-up approach largely on questions underlying the very origin of HET (Q1–Q4), we believe that this approach nevertheless can provide a valuable starting point toward understanding the kinds of organizational complexity that can emerge subsequently, which constitutes an important research challenge (Q5 and Q6). We are surrounded by an incredible diversity of multicellular organization, from filamentous algae to metazoan development, but it remains unclear what determines the organizational outcomes of these HET. Even though we have some intuitive understanding (e.g., filamentous organisms might be unlikely to evolve 3D structures), there are no theoretical or empirical studies yet that systematically approach this question. This is problematic, because intuition often fails. A salient example is the assumption that organizing principles arise in a certain intuitive order, from less to more complex, which has been disproven by phylogenetic studies in both volvocine green algae (102) and social amoebae (17). Traditional classifications based on phenotypic complexity do not match phylogenetic history; species that are phenotypically alike (i.e., similar complexity) are often far apart on the phylogenetic tree, while species that are phenotypically different are often closely related. Just as counterintuitively, many species with a relatively simple organization (e.g., small group sizes, few cell types, simple morphology) are derived from ones with more complex organization [e.g., the Acytosteliums, social amoebae that lack stalk cells, are derived from an ancestor with stalk cells (17)]. These phylogenetic studies further reveal that many organizing principles are invented multiple times (e.g., germ-soma differentiation) (102), which suggests that the developmental program underlying group formation strongly potentiates the evolution of some organizing principles more than others. A systematic, bottom-up approach to the study of HET could reveal what is possible, not only what seems intuitively probable. And by understanding how the earliest organizing principles came about, we could identify questions that help us understand the evolution of more advanced ones.
Other HET.
Although here we focused on the transition to multicellularity, the above questions can also be applied to other HET, both fraternal and egalitarian. Each HET has its own peculiarities that need to be accounted for. For example, in the case of animal sociality, a group cannot be defined in the same way as for multicellularity (SI Appendix, Text S4 and Fig. S3). However, despite these differences, the six questions we outline here help to identify commonalities and parallels among the various HET. For example, as for multicellularity, there is strong evidence that the ancestral life cycle plays an important role in the emergence of animal groups. This is exemplified in Polistes wasps, for which the bivoltine life cycle of the solitary ancestor was hypothesized to constitute a stepping stone to eusociality and caste differentiation (103–105). Wasps with a bivoltine life cycle have two reproductive broods a year (Fig. 2): the first brood occurs at the start of the breeding season and undergoes normal development; the second brood occurs in the summer and intercedes development by a diapause stage to survive winter. The phenotypic differences between the spring and summer brood result from a developmental switch, in which larvae can follow one of two possible developmental trajectories depending on the cues they experience (i.e., food provisioning). Substantiated by empirical evidence (105), the diapause ground-plan hypothesis (103, 104) states that this developmental switch is co-opted for caste differentiation in Polistes, in the same way that photoacclimation in the green algae and encystation in the amoebae were co-opted in the transition to multicellularity (89, 90) (Fig. 2). Recent work has further suggested that the bivoltine life cycle might also facilitate the transition to eusociality by allowing for the joint evolution of sex ratios and helping (106). Taken together, these studies highlight the paramount importance of the ancestral life cycle in the HET to animal sociality and reinforce the similarity across HET.
Conclusion
In this report, we proposed an integrative, bottom-up approach to study the dynamics underlying HET in biological organization. Starting from the solitary ancestor and its life cycle, we discussed how the first life cycles with a group life stage could originate (Q1); what properties characterize the first groups (Q2); how selection could act on those properties (Q3) and subsequently alter the organization of the groups (Q4); and, finally, how new organizing principles could evolve (Q5) and influence future organizational complexity (Q6). We argue that only by starting with the solitary ancestor and its life cycle, and studying these six questions, can we derive an understanding of the causal factors underlying HET. Then, by comparing different instantiations of the same transition (e.g., the multiple origins and transitions to multicellularity), we can determine whether the same causal factors underlie different transitions and which causal factors explain the different organizational outcomes of those transitions.
Supplementary Material
Acknowledgments
We thank John T. Bonner, whose pioneering work on multicellularity has been an inspiration to us and who provided invaluable feedback on this manuscript; Eörs Szathmáry and two anonymous reviewers for insightful comments and criticism; and Rob Pringle and the C.E.T. laboratory for discussion. J.v.G. received support from The Netherlands Organization for Scientific Research Rubicon Grant 2015-2. C.E.T. received support from the Alfred P. Sloan Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704631114/-/DCSupplemental.
References
- 1.Bonner JT. On Development: The Biology of Form. Harvard Univ Press; Cambridge, MA: 1974. [Google Scholar]
- 2.Buss LW. The Evolution of Individuality. Princeton Univ Press; Princeton, NJ: 1987. [Google Scholar]
- 3.Maynard Smith J, Szathmáry E. The Major Transitions in Evolution. Freeman; Oxford: 1995. [Google Scholar]
- 4.Szathmáry E. Toward major evolutionary transitions theory 2.0. Proc Natl Acad Sci USA. 2015;112:10104–10111. doi: 10.1073/pnas.1421398112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michod RE. Evolution of individuality during the transition from unicellular to multicellular life. Proc Natl Acad Sci USA. 2007;104:8613–8618. doi: 10.1073/pnas.0701489104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griesemer J. The units of evolutionary transition. Selection. 2000;1:67–80. [Google Scholar]
- 7.Clarke E. Origins of evolutionary transitions. J Biosci. 2014;39:303–317. doi: 10.1007/s12038-013-9375-y. [DOI] [PubMed] [Google Scholar]
- 8.De Monte S, Rainey PB. Nascent multicellular life and the emergence of individuality. J Biosci. 2014;39:237–248. doi: 10.1007/s12038-014-9420-5. [DOI] [PubMed] [Google Scholar]
- 9.Fontana W, Buss LW. The arrival of the fittest: Toward a theory of biological organization. Bull Math Biol. 1994;56:1–64. [Google Scholar]
- 10.Gerhart J, Kirschner M. Cells, Embryos, and Evolution: Toward a Cellular and Developmental Understanding of Phenotypic Variation and Evolutionary Adaptability. Blackwell; Oxford: 1997. [Google Scholar]
- 11.Szathmáry E. The origin of the human language faculty: The language amoeba hypothesis. In: Trabant J, Ward S, editors. New Essays on the Origin of Language. Mouton de Gruyter; Berlin: 2001. pp. 55–81. [Google Scholar]
- 12.Sebé-Pedrós A, Degnan BM, Ruiz-Trillo I. The origin of Metazoa: A unicellular perspective. Nat Rev Genet. 2017;18:498–512. doi: 10.1038/nrg.2017.21. [DOI] [PubMed] [Google Scholar]
- 13.Jacob F. The Possible and the Actual. Washington Univ Press; Seattle, WA: 1982. [Google Scholar]
- 14.Hogeweg P. Shapes in the shadow: Evolutionary dynamics of morphogenesis. Artif Life. 2000;6:85–101. doi: 10.1162/106454600568339. [DOI] [PubMed] [Google Scholar]
- 15.King N, Carroll SB. A receptor tyrosine kinase from choanoflagellates: Molecular insights into early animal evolution. Proc Natl Acad Sci USA. 2001;98:15032–15037. doi: 10.1073/pnas.261477698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King N. The unicellular ancestry of animal development. Dev Cell. 2004;7:313–325. doi: 10.1016/j.devcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Schaap P, et al. Molecular phylogeny and evolution of morphology in the social amoebas. Science. 2006;314:661–663. doi: 10.1126/science.1130670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rokas A. The molecular origins of multicellular transitions. Curr Opin Genet Dev. 2008;18:472–478. doi: 10.1016/j.gde.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Wolpert L. 1994 The evolutionary origin of development: Cycles, patterning, privilege and continuity Dev (Suppl):79–84. [Google Scholar]
- 20.Ratcliff WC, et al. Experimental evolution of an alternating uni- and multicellular life cycle in Chlamydomonas reinhardtii. Nat Commun. 2013;4:2742. doi: 10.1038/ncomms3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammerschmidt K, Rose CJ, Kerr B, Rainey PB. Life cycles, fitness decoupling and the evolution of multicellularity. Nature. 2014;515:75–79. doi: 10.1038/nature13884. [DOI] [PubMed] [Google Scholar]
- 22.Nadell CD, Drescher K, Foster KR. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol. 2016;14:589–600. doi: 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- 23.Wilson DS. A theory of group selection. Proc Natl Acad Sci USA. 1975;72:143–146. doi: 10.1073/pnas.72.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michod RE. Cooperation and conflict in the evolution of individuality. I. Multilevel selection of the organism. Am Nat. 1997;149:607–645. [Google Scholar]
- 25.Furusawa C, Kaneko K. Emergence of multicellular organisms with dynamic differentiation and spatial pattern. Artif Life. 1998;4:79–93. doi: 10.1162/106454698568459. [DOI] [PubMed] [Google Scholar]
- 26.Queller DC. Relatedness and the fraternal major transitions. Philos Trans R Soc Lond B Biol Sci. 2000;355:1647–1655. doi: 10.1098/rstb.2000.0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forgacs G, Newman SA. Biological Physics of the Developing Embryo. Cambridge Univ Press; Cambridge, UK: 2005. [Google Scholar]
- 28.Nowak MA. Five rules for the evolution of cooperation. Science. 2006;314:1560–1563. doi: 10.1126/science.1133755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fletcher JA, Doebeli M. A simple and general explanation for the evolution of altruism. Proc Biol Sci. 2009;276:13–19. doi: 10.1098/rspb.2008.0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonner JT. Size and Cycle: An Essay on the Structure of Biology. Princeton Univ Press; Princeton, NJ: 1965. [Google Scholar]
- 31.Koschwanez JH, Foster KR, Murray AW. Improved use of a public good selects for the evolution of undifferentiated multicellularity. eLife. 2013;2:e00367. doi: 10.7554/eLife.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alegado RA, et al. A bacterial sulfonolipid triggers multicellular development in the closest living relatives of animals. eLife. 2012;1:e00013. doi: 10.7554/eLife.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirk DL. Evolution of multicellularity in the volvocine algae. Curr Opin Plant Biol. 1999;2:496–501. doi: 10.1016/s1369-5266(99)00019-9. [DOI] [PubMed] [Google Scholar]
- 34.Kirk DL. A twelve-step program for evolving multicellularity and a division of labor. BioEssays. 2005;27:299–310. doi: 10.1002/bies.20197. [DOI] [PubMed] [Google Scholar]
- 35.Sathe S, Durand PM. Cellular aggregation in Chlamydomonas (Chlorophyceae) is chimaeric and depends on traits like cell size and motility. Eur J Phycol. 2016;51:129–138. [Google Scholar]
- 36.Sebé-Pedrós A, et al. Regulated aggregative multicellularity in a close unicellular relative of metazoa. eLife. 2013;2:e01287. doi: 10.7554/eLife.01287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Estrela S, Kerr B, Morris JJ. Transitions in individuality through symbiosis. Curr Opin Microbiol. 2016;31:191–198. doi: 10.1016/j.mib.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 38.Libby E, B Rainey P. A conceptual framework for the evolutionary origins of multicellularity. Phys Biol. 2013;10:035001. doi: 10.1088/1478-3975/10/3/035001. [DOI] [PubMed] [Google Scholar]
- 39.Herron MD, Nedelcu AM. Volvocine algae: From simple to complex multicellularity. In: Ruiz-Trillo I, Nedelcu AM, editors. Evolutionary Transitions to Multicellular Life. Springer; Amsterdam: 2015. pp. 129–152. [Google Scholar]
- 40.West-Eberhard MJ. Phenotypic plasticity and the origins of diversity. Annu Rev Ecol Syst. 1989;20:249–278. [Google Scholar]
- 41.Khersonsky O, Tawfik DS. Enzyme promiscuity: A mechanistic and evolutionary perspective. Annu Rev Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 42.Sakaguchi M, Murakami H, Suzaki T. Involvement of a 40-kDa glycoprotein in food recognition, prey capture, and induction of phagocytosis in the protozoon Actinophrys sol. Protist. 2001;152:33–41. doi: 10.1078/1434-4610-00041. [DOI] [PubMed] [Google Scholar]
- 43.West SA, Fisher RM, Gardner A, Kiers ET. Major evolutionary transitions in individuality. Proc Natl Acad Sci USA. 2015;112:10112–10119. doi: 10.1073/pnas.1421402112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuranda MJ, Robbins PW. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J Biol Chem. 1991;266:19758–19767. [PubMed] [Google Scholar]
- 45.Chai Y, Norman T, Kolter R, Losick R. An epigenetic switch governing daughter cell separation in Bacillus subtilis. Genes Dev. 2010;24:754–765. doi: 10.1101/gad.1915010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herron MD, Rashidi A, Shelton DE, Driscoll WW. Cellular differentiation and individuality in the ‘minor’ multicellular taxa. Biol Rev Camb Philos Soc. 2013;88:844–861. doi: 10.1111/brv.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarnita CE, Washburne A, Martinez-Garcia R, Sgro AE, Levin SA. Fitness tradeoffs between spores and nonaggregating cells can explain the coexistence of diverse genotypes in cellular slime molds. Proc Natl Acad Sci USA. 2015;112:2776–2781. doi: 10.1073/pnas.1424242112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonner JT. The origins of multicellularity. Integr Biol. 1998;1:27–36. [Google Scholar]
- 49.Grosberg RK, Strathmann RR. The evolution of multicellularity: A minor major transition? Annu Rev Ecol Evol Syst. 2007;38:621–654. [Google Scholar]
- 50.Tarnita CE, Taubes CH, Nowak MA. Evolutionary construction by staying together and coming together. J Theor Biol. 2013;320:10–22. doi: 10.1016/j.jtbi.2012.11.022. [DOI] [PubMed] [Google Scholar]
- 51.Angert ER. Alternatives to binary fission in bacteria. Nat Rev Microbiol. 2005;3:214–224. doi: 10.1038/nrmicro1096. [DOI] [PubMed] [Google Scholar]
- 52.Flärdh K, Buttner MJ. Streptomyces morphogenetics: Dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol. 2009;7:36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- 53.Berk V, et al. Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science. 2012;337:236–239. doi: 10.1126/science.1222981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loomis WF. Cell signaling during development of Dictyostelium. Dev Biol. 2014;391:1–16. doi: 10.1016/j.ydbio.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hölscher T, et al. Motility, chemotaxis and aerotaxis contribute to competitiveness during bacterial pellicle biofilm development. J Mol Biol. 2015;427:3695–3708. doi: 10.1016/j.jmb.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duran-Nebreda S, Solé R. Emergence of multicellularity in a model of cell growth, death and aggregation under size-dependent selection. J R Soc Interface. 2015;12:20140982. doi: 10.1098/rsif.2014.0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pande S, et al. Metabolic cross-feeding via intercellular nanotubes among bacteria. Nat Commun. 2015;6:6238. doi: 10.1038/ncomms7238. [DOI] [PubMed] [Google Scholar]
- 58.Momeni B, Brileya KA, Fields MW, Shou W. Strong inter-population cooperation leads to partner intermixing in microbial communities. eLife. 2013;2:e00230. doi: 10.7554/eLife.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 60.Schlichting CD. Origins of differentiation via phenotypic plasticity. Evol Dev. 2003;5:98–105. doi: 10.1046/j.1525-142x.2003.03015.x. [DOI] [PubMed] [Google Scholar]
- 61.Mikhailov KV, et al. The origin of Metazoa: A transition from temporal to spatial cell differentiation. BioEssays. 2009;31:758–768. doi: 10.1002/bies.200800214. [DOI] [PubMed] [Google Scholar]
- 62.Gierer A, Meinhardt H. A theory of biological pattern formation. Kybernetik. 1972;12:30–39. doi: 10.1007/BF00289234. [DOI] [PubMed] [Google Scholar]
- 63.Newman SA, Forgacs G, Muller GB. Before programs: The physical origination of multicellular forms. Int J Dev Biol. 2006;50:289–299. doi: 10.1387/ijdb.052049sn. [DOI] [PubMed] [Google Scholar]
- 64.Newman SA, Bhat R. Dynamical patterning modules: A “pattern language” for development and evolution of multicellular form. Int J Dev Biol. 2009;53:693–705. doi: 10.1387/ijdb.072481sn. [DOI] [PubMed] [Google Scholar]
- 65.Alpkvist E, Klapper I. Description of mechanical response including detachment using a novel particle model of biofilm/flow interaction. Water Sci Technol. 2007;55:265–273. doi: 10.2166/wst.2007.267. [DOI] [PubMed] [Google Scholar]
- 66.Rainey PB, Kerr B. Cheats as first propagules: A new hypothesis for the evolution of individuality during the transition from single cells to multicellularity. BioEssays. 2010;32:872–880. doi: 10.1002/bies.201000039. [DOI] [PubMed] [Google Scholar]
- 67.van Gestel J, Nowak MA. Phenotypic heterogeneity and the evolution of bacterial life cycles. PLoS Comput Biol. 2016;12:e1004764. doi: 10.1371/journal.pcbi.1004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nadell CD, Bassler BL. A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc Natl Acad Sci USA. 2011;108:14181–14185. doi: 10.1073/pnas.1111147108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garcia T, De Monte S. Group formation and the evolution of sociality. Evolution. 2013;67:131–141. doi: 10.1111/j.1558-5646.2012.01739.x. [DOI] [PubMed] [Google Scholar]
- 70.Ghosh P, Mondal J, Ben-Jacob E, Levine H. Mechanically-driven phase separation in a growing bacterial colony. Proc Natl Acad Sci USA. 2015;112:E2166–E2173. doi: 10.1073/pnas.1504948112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Serra DO, Richter AM, Hengge R. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J Bacteriol. 2013;195:5540–5554. doi: 10.1128/JB.00946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pichugin Y, Pena J, Rainey P, Traulsen A. 2017 Fragmentation modes and the evolution of life cycles. bioRxiv, https://doi.org/10.1101/120097.
- 73.Wolpert L, Szathmáry E. Multicellularity: Evolution and the egg. Nature. 2002;420:745. doi: 10.1038/420745a. [DOI] [PubMed] [Google Scholar]
- 74.Grosberg RK, Strathmann RR. One cell, two cell, red cell, blue cell: The persistence of a unicellular stage in multicellular life histories. Trends Ecol Evol. 1998;13:112–116. doi: 10.1016/S0169-5347(97)01313-X. [DOI] [PubMed] [Google Scholar]
- 75.Akçay E, Van Cleve J. Behavioral responses in structured populations pave the way to group optimality. Am Nat. 2012;179:257–269. doi: 10.1086/663691. [DOI] [PubMed] [Google Scholar]
- 76.Yan J, Nadell CD, Bassler BL. Environmental fluctuation governs selection for plasticity in biofilm production. ISME J. 2017;11:1569–1577. doi: 10.1038/ismej.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nanjundiah V. Cellular slime mold development as a paradigm for the transition from unicellular to multicellular life. In: Niklas KJ, Newman SA, editors. Multicellularity: Origins and Evolution. MIT Press; Cambridge, MA: 2016. pp. 105–130. [Google Scholar]
- 78.Griesemer J. Development, culture, and the units of inheritance. Philos Sci. 2000;67:S348–S368. [Google Scholar]
- 79.Waddington CH. Selection of the genetic basis for an acquired character. Nature. 1952;169:278. doi: 10.1038/169278a0. [DOI] [PubMed] [Google Scholar]
- 80.Pigliucci M, Murren CJ, Schlichting CD. Phenotypic plasticity and evolution by genetic assimilation. J Exp Biol. 2006;209:2362–2367. doi: 10.1242/jeb.02070. [DOI] [PubMed] [Google Scholar]
- 81.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 82.Lazazzera BA. Quorum sensing and starvation: Signals for entry into stationary phase. Curr Opin Microbiol. 2000;3:177–182. doi: 10.1016/s1369-5274(00)00072-2. [DOI] [PubMed] [Google Scholar]
- 83.van Gestel J, Nowak MA, Tarnita CE. The evolution of cell-to-cell communication in a sporulating bacterium. PLoS Comput Biol. 2012;8:e1002818. doi: 10.1371/journal.pcbi.1002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Powers ST, Penn AS, Watson RA. The concurrent evolution of cooperation and the population structures that support it. Evolution. 2011;65:1527–1543. doi: 10.1111/j.1558-5646.2011.01250.x. [DOI] [PubMed] [Google Scholar]
- 85.Sussex IM. Do concepts of animal development apply to plant systems. Brookhaven Symp Biol. 1973;25:145–151. [Google Scholar]
- 86.Clarke E. The multiple realizability of biological individuals. J Philos. 2013;110:413–435. [Google Scholar]
- 87.Niklas KJ, Newman SA. The origins of multicellular organisms. Evol Dev. 2013;15:41–52. doi: 10.1111/ede.12013. [DOI] [PubMed] [Google Scholar]
- 88.Kirk MM, et al. regA, a Volvox gene that plays a central role in germ-soma differentiation, encodes a novel regulatory protein. Development. 1999;126:639–647. doi: 10.1242/dev.126.4.639. [DOI] [PubMed] [Google Scholar]
- 89.Nedelcu AM, Michod RE. The evolutionary origin of an altruistic gene. Mol Biol Evol. 2006;23:1460–1464. doi: 10.1093/molbev/msl016. [DOI] [PubMed] [Google Scholar]
- 90.Schaap P. Evolutionary crossroads in developmental biology: Dictyostelium discoideum. Development. 2011;138:387–396. doi: 10.1242/dev.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ritchie AV, van Es S, Fouquet C, Schaap P. From drought sensing to developmental control: Evolution of cyclic AMP signaling in social amoebas. Mol Biol Evol. 2008;25:2109–2118. doi: 10.1093/molbev/msn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kawabe Y, et al. Activated cAMP receptors switch encystation into sporulation. Proc Natl Acad Sci USA. 2009;106:7089–7094. doi: 10.1073/pnas.0901617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shubin N, Tabin C, Carroll S. Fossils, genes and the evolution of animal limbs. Nature. 1997;388:639–648. doi: 10.1038/41710. [DOI] [PubMed] [Google Scholar]
- 94.Lee JH, Lin H, Joo S, Goodenough U. Early sexual origins of homeoprotein heterodimerization and evolution of the plant KNOX/BELL family. Cell. 2008;133:829–840. doi: 10.1016/j.cell.2008.04.028. [DOI] [PubMed] [Google Scholar]
- 95.Hanschen ER, et al. The Gonium pectorale genome demonstrates co-option of cell cycle regulation during the evolution of multicellularity. Nat Commun. 2016;7:11370. doi: 10.1038/ncomms11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.King N, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Manning G, Young SL, Miller WT, Zhai Y. The protist, Monosiga brevicollis, has a tyrosine kinase signaling network more elaborate and diverse than found in any known metazoan. Proc Natl Acad Sci USA. 2008;105:9674–9679. doi: 10.1073/pnas.0801314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pincus D, Letunic I, Bork P, Lim WA. Evolution of the phospho-tyrosine signaling machinery in premetazoan lineages. Proc Natl Acad Sci USA. 2008;105:9680–9684. doi: 10.1073/pnas.0803161105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hunter T. Tyrosine phosphorylation: Thirty years and counting. Curr Opin Cell Biol. 2009;21:140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Clarke M, et al. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol. 2013;14:R11. doi: 10.1186/gb-2013-14-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bell G, Koufopanou V. The architecture of the life cycle in small organisms. Philos Trans R Soc Lond B Biol Sci. 1991;332:81–89. [Google Scholar]
- 102.Herron MD, Michod RE. Evolution of complexity in the volvocine algae: Transitions in individuality through Darwin’s eye. Evolution. 2008;62:436–451. doi: 10.1111/j.1558-5646.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 103.Hunt JH. A conceptual model for the origin of worker behaviour and adaptation of eusociality. J Evol Biol. 2012;25:1–19. doi: 10.1111/j.1420-9101.2011.02421.x. [DOI] [PubMed] [Google Scholar]
- 104.Hunt JH, Amdam GV. Bivoltinism as an antecedent to eusociality in the paper wasp genus Polistes. Science. 2005;308:264–267. doi: 10.1126/science.1109724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hunt JH, et al. A diapause pathway underlies the gyne phenotype in Polistes wasps, revealing an evolutionary route to caste-containing insect societies. Proc Natl Acad Sci USA. 2007;104:14020–14025. doi: 10.1073/pnas.0705660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Quiñones AE, Pen I. A unified model of Hymenopteran preadaptations that trigger the evolutionary transition to eusociality. Nat Commun. 2017;8:15920. doi: 10.1038/ncomms15920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Claessen D, Rozen DE, Kuipers OP, Søgaard-Andersen L, van Wezel GP. Bacterial solutions to multicellularity: A tale of biofilms, filaments and fruiting bodies. Nat Rev Microbiol. 2014;12:115–124. doi: 10.1038/nrmicro3178. [DOI] [PubMed] [Google Scholar]
- 108.van Gestel J, Vlamakis H, Kolter R. From cell differentiation to cell collectives: Bacillus subtilis uses division of labor to migrate. PLoS Biol. 2015;13:e1002141. doi: 10.1371/journal.pbio.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Litman GW, Dishaw LJ. Histocompatibility: Clarifying fusion confusion. Curr Biol. 2013;23:R934–R935. doi: 10.1016/j.cub.2013.08.048. [DOI] [PubMed] [Google Scholar]
- 110.Adler CE, Seidel CW, McKinney SA, Alvarado AS. Selective amputation of the pharynx identifies a FoxA-dependent regeneration program in planaria. eLife. 2014;3:e02238. doi: 10.7554/eLife.02238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.