Significance
Thogotoviruses belong to Orthomyxoviridae family and infect a variety of mammalian hosts, including humans. The emergence of these viruses poses great threats to public health and the economy. In this work, we performed structural and phylogenic analyses on the fusogenic glycoproteins of Thogoto and Dhori viruses, two representatives of the Thogotovirus genus that cause severe human infections. Previous studies have shown that thogotovirus glycoproteins share ∼28% sequence identity with baculovirus Gp64s. Our structural analysis confirmed their homology in evolution and identified them as class III viral fusogens, in contrast to class I members of influenza viruses. Our studies provide structural evidence to help us to understand the evolution of these viruses and indicate a potential target for antiviral drug design.
Keywords: Thogotovirus, Othomyxoviridae, crystal structure, fusion protein, evolution
Abstract
Thogotoviruses are emerging tick-borne zoonotic orthomyxoviruses infecting both humans and domestic animals with severe clinical consequences. These viruses utilize a single-envelope glycoprotein (Gp) to facilitate their entry into host cells. Here, we present the Gp structures of Thogoto and Dhori viruses, both of which are members of the Thogotovirus genus in the family Orthomyxoviridae. These structures, determined in the postfusion conformation, identified them as class III viral fusion proteins. It is intriguing that the Gp structures are similar to the envelope protein of baculovirus, although sharing a low sequence identity of ∼28%. Detailed structural and phylogenic analyses demonstrated that these Gps originated from a common ancestor. Among the structures, domain I is the most conserved region, particularly the fusion loops. Domain II showed the highest variability among different viruses, which might be related to their distinct host tropism. These findings increase our understanding of the divergent evolution processes of various orthomyxoviruses and indicate potential targets for developing antiviral therapeutics by intercepting virus entry.
The Orthomyxoviridae family includes seven genera—four types of influenza viruses (Influenza virus A, B, C, and D), Thogotovirus, Quaranjavirus, and Isavirus—all of which are segmented negative-strand RNA viruses (1, 2). Many members in this family are important animal or human pathogens, as exemplified by the epidemic or pandemic influenza A viruses that have caused hundreds of thousands of human deaths in the past century and are still discretely circulating, posing a great threat to human health and the economy.
In contrast to influenza viruses, thogotoviruses (viruses in the Thogotovirus genus) are transmitted mainly through tick vectors and thus are also called “tick-borne viruses” (3, 4). These viruses mainly circulate in domestic animals, such as sheep, cattle, and camels, causing neural diseases and abortion (5). Among these viruses, two main species, Thogoto virus [Thogotovirus thogoto (THOV)] and Dhori virus [Thogotovirus dhori (DHOV)], have been reported to infect humans and even cause human deaths (6, 7). Antibodies against THOV and DHOV have been identified in humans living in parts of Europe, Asia, and Africa (5, 8). Although all these viruses are transmitted mainly by tick vectors, it has been reported that DHOV caused human infections in a vector-free manner, possibly by an aerosol route (7). This further complicates its transmission route among different hosts, indicating the potential to cause epidemics among humans. In 2014, a farmer in the United States was killed by a new virus called “Bourbon virus” (BOUV), which was then identified as an unknown type of thogotovirus (9). Therefore, characterization of thogotoviruses is of great importance for public health.
There is only one glycoprotein (Gp) embedded in the envelope of thogotoviruses, which is responsible for the entire process of virus entry, including attachment, entry, and fusion (10). By bioinformatics analysis, thogotovirus Gp does not show obvious similarity with the glycoproteins of influenza viruses or isavirus, indicating a distinct mechanism for entering host cells. The closest orthomyxovirus relative of thogotovirus is quaranjavirus, whose envelope Gp shares a sequence identity of ∼26% with thogotovirus Gps (11). Thus far the receptor of thogotoviruses has not been identified, and their entry pathway is unclear. The only evidence is that the BOUV viral particles could be observed in the endosomal compartment of infected cells (9), indicating that thogotoviruses might enter host cells by endocytosis and that the membrane fusion process might occur in the endosome.
To elucidate the mechanism of thogotovirus entry and pathogenicity, we biochemically and structurally characterized the envelope Gps of THOV and DHOV. We found that both THOV and DHOV Gps undergo pH-sensitive conformational changes, which supports the hypothesis that thogotoviruses enter cells by endocytosis pathway. In addition, we determined the crystal structures of the extracellular domains of THOV and DHOV Gps in the postfusion conformation, which display features of defined class III viral fusion proteins (12). These two Gps have high structural similarities with the Gp of Autographa californica multiple nucleopolyhedrosis virus (AcMNPV Gp64), which is a baculovirus with a double-stranded DNA genome that infects insect cells (13). It has been established that all class III viral fusion proteins are structural homologs with the same origin in evolution (12–14). The structures of thogotovirus Gps and baculovirus Gp64s indicate that they diverged much later than vesicular stomatitis virus (VSV) Gp and herpes simplex virus (HSV) glycoprotein B (gB). Combined with phylogenic analysis, we found that thogotovirus Gps and baculovirus Gp64s diverged from a common origin and that thogotoviruses further evolved in two different directions, the THOV and DHOV clades. These Gps exhibit different surface electrostatic potential patterns that might result from divergent evolution. Compared with THOV Gp and AcMNPV Gp64, the DHOV Gp has a more positively charged surface, indicating it is more diverged from the common ancestor. Our findings provide important clues to understand the evolution of thogotoviruses and may help in the development of antiviral therapeutics targeting the fusion proteins.
Results
Overall Structures of THOV and DHOV Gps.
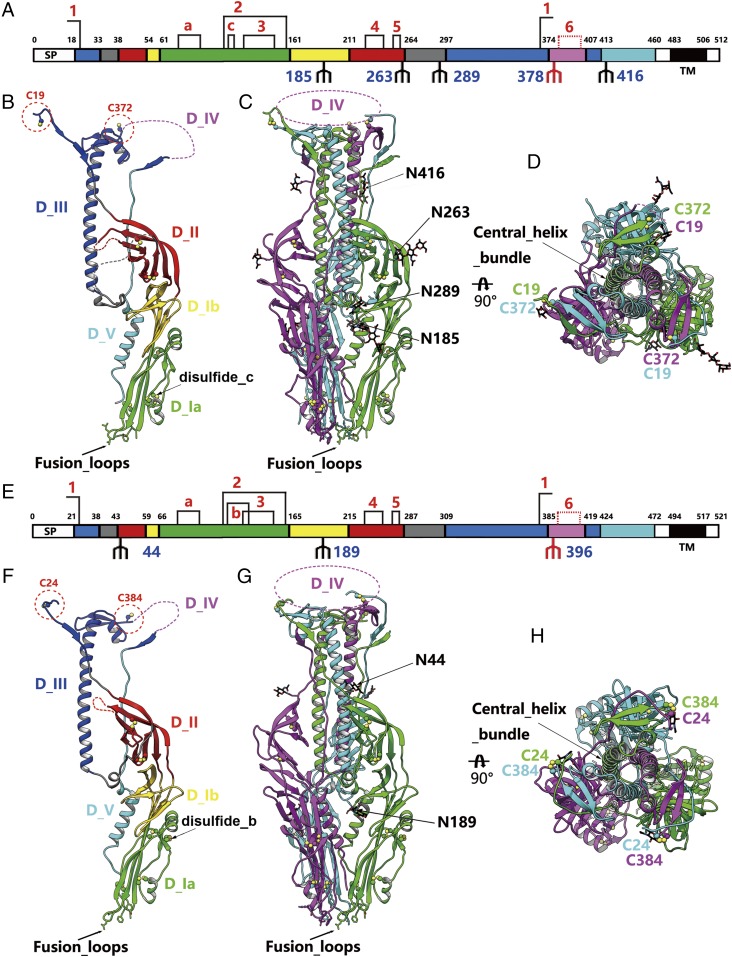
The ectodomains of THOV Gp (residues 18–483) and DHOV Gp (residues 21–494) were expressed using the baculovirus expression system. Initially, size-exclusion chromatography was performed to characterize the oligomerization state of THOV and DHOV Gps in solution. Judged by elution volumes, both proteins were estimated to be trimers of ∼180 kDa (Fig. S1). In addition, interchain disulfides were observed, as shown by SDS/PAGE under reducing and nonreducing conditions (Fig. S1). The crystal structures of THOV and DHOV Gps were determined at a resolution of 2.1 Å and 2.5 Å, respectively. Consistent with biochemical assays, both structures were composed of three identical chains that form an elongated bottle-shaped structure with a central helix bundle stabilizing the interchain interactions (Fig. 1). Three interchain disulfides occur at the peripheral loop regions to maintain Gps as trimers during the membrane-fusion process (Fig. 1).
Fig. S1.
Size-exclusion chromatograms and SDS/PAGE profiles of the THOV Gp (A) and DHOV Gp (B) ectodomains. The majority of the proteins are well ordered, as shown by a monodispersed peak, corresponding to ∼180 kDa, indicating that the proteins are trimeric in solution. SDS/PAGE under both reducing and nonreducing conditions indicates the presence of interchain disulfides.
Fig. 1.
Overall structure of the THOV and DHOV Gp ectodomains. (A and E) Schematic diagrams of THOV and DHOV Gp organization, colored by domains. The regions colored in gray are disordered loops not resolved in the structure. The disulfide bonds are connected by black lines. Disulfides 1–6 are conserved for all members of Gp64 protein family, but disulfide 6 is invisible in the structures. Disulfide a is shared by THOV Gp and DHOV Gp. Disulfides b and c are unique in DHOV Gp and THOV Gp, respectively. The observed glycans are shown as black branches in the diagram; red branches represent putative glycans not resolved in the structure. SP, signal peptide; TM, transmembrane region. (B and F) Ribbon representations of the THOV and DHOV Gp protomer, colored by domains as in A and E. The key residues at the putative fusion loops are shown as sticks. The unresolved disordered regions are represented by dashed lines, and disulfides are shown as spheres. Disulfides b and c are indicated by arrows. (C, D, G, and H) Two views of THOV (C and D) and DHOV (G and H) Gp trimers, colored by chains. The glycans are shown as black sticks, and the glycosylated residues are labeled. The major portion of domain IV is unresolved. The cysteines involved in interchain disulfides are labeled in the top view (D and H).
The two Gp structures are very similar, with a sequence identity of 35%. Both could be divided into five domains (Fig. 1 A, B, E, and F), reminiscent of the general class III viral fusion protein structures (12, 13, 15, 16). Domain I could be further divided into subdomains Ia and Ib; subdomain Ia is located at the end near the viral envelope and contains the conserved fusion loops (Fig. 1 B and F). Domain II is a typical pleckstrin homology (PH) domain composed of antiparallel β-strands that might undergo significant conformational changes during the membrane-fusion process. Domain III is mainly composed of a helix stalk in the center of the Gp trimer, which is the main interface for interprotomer interactions (Fig. 1 B, C, F, and G). The trimerization interfaces are rich in charged residues and form extensive charged interaction networks, making the trimers sensitive to environmental pH changes (Fig. S2). There are several histidines in this interface, which might serve as pH sensors by protonization or deprotonization in response to pH changes and thus trigger conformational changes to initiate membrane fusion (17). Domain IV is located at the viral membrane-distal end and is composed of disordered loops, which are largely invisible in the density map (Fig. 1 B, C, F, and G). Domain V links the membrane-distal portion (domains III and IV) and membrane-proximal portion (domain I), further guiding the protein to stretch to the transmembrane domain. Four and two N-linked glycosylation sites can be observed in the structures of THOV and DHOV Gp, respectively. Another putative glycosylation site in both Gps is located in domain IV but was not observed in the structure due to the disorder of this region (Fig. 1 A, C, E, and G).
Fig. S2.
Key pH-sensitive interfaces around the trimeric central helix bundle. A and B represent the interface closer to the membrane-distal side (interface_1), and C and D represent the interface closer to the membrane-proximal side (interface_2) of Gps; these interfaces are located at similar positions in THOV (A and C) and DHOV (B and D) Gps. The figures are presented with the extracellular views. All structures are colored by chains, and key charged or polar residues are shown as sticks. Polar contacts (hydrogen bonds and salt bridges) are shown as dashed lines.
All these features are similar to the well-defined class III viral fusion proteins in the postfusion conformation, such as HSV gB (15), VSV Gp (16), and AcMNPV Gp64 (13), indicating that thogotovirus Gps belong to class III viral fusion proteins and that the structures we obtained are in the postfusion conformation.
pH-Dependent Conformational Changes of Thogotovirus Gps.
It is interesting to note that both THOV and DHOV Gp were determined in the postfusion conformation although crystallized under acidic or neutral pH conditions (see details in SI Materials and Methods). As members of class III viral fusion proteins, both VSV Gp and AcMNPV Gp64 were reported to undergo reversible conformational changes in response to environmental pH values (13, 16, 18). This reversibility might be necessary for recovery to a high-pH conformation after passing through the acidic Golgi compartments during Gp maturation in the cell (16). As BOUV viral particles could be observed in the endosomal compartment (9), we assumed that thogotoviruses might enter cells via the endosomal pathway and that their Gps would also experience such pH-dependent structural changes during expression and infection.
By circular dichroism (CD) analysis, we observed a significant conformation shift of both THOV and DHOV Gps under different pH conditions, which was consistent with the observed pH-sensitive interfaces in Gp structures and indicated pH as the trigger that initiates member fusion (Fig. S3). Similarly, the cleaved H5 HA protein (HA1 and HA2), which is known to undergo conformational changes upon low-pH treatment, also showed different CD profiles at different pH conditions (19, 20), whereas the antibody fragment (Fab), as the negative control, showed no obvious changes in CD spectra under different pH conditions. Thus, the pH-sensitive CD profiles indicated that both THOV and DHOV Gps are expressed in the prefusion conformation but crystallized as postfusion forms. In addition, these observations also support the hypothesis that thogotoviruses might adopt the endosomal pathway to infect host cells.
Fig. S3.
CD spectra profile of THOV Gp and DHOV Gp ectodomains at different pH conditions. (A) CD spectra of a Fab molecule, a typical β-strand structure insensitive to pH conditions (negative control). (B) CD spectra profile of cleaved avian influenza virus H5 (HA1, HA2) protein, which is thought to undergo conformational changes upon pH shift (positive control). (C and D) CD spectra of soluble THOV Gp (C) and DHOV Gp (D). The CD spectra profiles at pH 8.5 and pH 5.0 are colored in black and red, respectively. The typical peak valleys for the β-strand and α-helix are indicated by black arrows.
Thogotovirus Gps Are Structurally Related to baculovirus Gp64s.
Previous studies have reported that AcMNPV Gp64 shares a low sequence identity (∼28%) with thogotovirus Gps and indicated that they are members of the same protein family (Gp64 family) (13). Our structural analysis revealed that their overall folds are very similar. The three structures could be superimposed with a certain degree of domain shifts (Fig. 2A). The individual domains of the three structures are highly similar, with domain Ia and domain III showing the highest similarity (Fig. 2 B–D). Notably, the putative fusion loops, which are located at the membrane-proximal end of domain Ia, are highly conserved among all proteins of the putative Gp64 family (Fig. 3 A and C). Domain IV of AcMNPV Gp64 is better resolved, with more loop density observed, but basically all three structures are disordered in this region. The most striking differences among these structures are located in domain II (Fig. 2A). It displays the poorest alignment, as obvious domain movement was observed relative to the rest of the protein, and it is the most variable region among all proteins in Gp64 family (Figs. 2A and 3A and Table S1). When we fixed domain Ia and compared the relative orientations of the other domains, we observed a striking difference among the three structures. With THOV Gp as the reference, domain Ib and domain II of DHOV Gp and AcMNPV Gp64 are rotated in opposite directions relative to domain Ia, thus leading to the poor alignment of this region (Fig. 2 B, E, and F). Overall, however, baculovirus Gp64 and thogotovirus Gps are closely related to each other in structure.
Fig. 2.
Structural comparison of THOV Gp, DHOV Gp, and AcMNPV Gp64. (A) Superimposition of THOV Gp, DHOV Gp, and AcMNPV Gp64 protomers, which are colored in cyan, magenta, and green, respectively. The disordered domain IV is labeled by a red dashed ellipsoid. The poorly superimposed domain II is highlighted by a black dashed ellipsoid. (B) Superimposition of domain I of THOV Gp, DHOV Gp, and AcMNPV Gp64 with the domain Ia fixed revealed that domain Ib shows different orientations among these structures. Domain Ia is colored in gray; domain Ib and domain II are colored as in A. (C and D) Superimposition of domain II and domain III of THOV Gp, DHOV Gp, and AcMNPV Gp64, individually. (E and F) With domain Ia fixed, domains Ib and domain II move in different orientations relative to domain I. Domain Ia is colored in gray; domains Ib and II are colored as in A. The pairwise Cα rmsd values are labeled below each structure comparison figure.
Fig. 3.
Conservation analysis of thogotovirus Gps among all members of Gp64 protein family. (A–C) Side (A), top (B), and bottom (C) views of the conservation map. Conserved regions are colored in purple, and variable regions are cyan. (D) Close-up view of the well-resolved basic loop density of DHOV Gp. The 2Fo-Fc electron density map at a contour level of 1.0 σ is shown as a mesh, and the loop residues are represented as sticks. The ribbon diagrams are colored by domains as in Fig. 2B.
Table S1.
Thogotovirus Gp-related protein sequences used for conservation analysis
| NCBI accession code | Annotations |
| YP_145808.1 | Gp75 (Thogoto virus) |
| BAQ22396.1 | Gp 75 (Thogoto virus) |
| AHB34057.1 | Gp (Upolu virus) |
| AHB34063.1 | Gp (Aransas Bay virus) |
| AKR14107.1 | Major budded virus envelope Gp (Dasychira pudibunda Nucleopolyhedrovirus) |
| AGR56918.1 | Gp64 (Choristoneura occidentalis Alphabaculovirus) |
| YP_008992122.1 | Gp64 (Choristoneura murinana Alphabaculovirus) |
| NP_848430.1 | Major budded virus envelope Gp (Choristoneura fumiferana multiple nucleopolyhedroviruses) |
| NP_932732.1 | Major budded virus envelope Gp (Choristoneura fumiferana DEF multiple nucleopolyhedroviruses) |
| AFY62837.1 | Gp64 (Philosamia cynthia ricini Nucleopolyhedrovirus) |
| ABQ12258.1 | Major budded virus envelope Gp (Antheraea pernyi Nucleopolyhedrovirus) |
| YP_611000.1 | Gp64 (Antheraea pernyi Nucleopolyhedrovirus) |
| AAW47929.1 | Envelope Gp (Antheraea pernyi Nucleopolyhedrovirus) |
| APP91608.1 | Gp (Sinu virus) |
| YP_008378382.1 | Gp64 (Choristoneura rosaceana Alphabaculovirus) |
| AED98372.1 | Gp (Jos virus) |
| APG77909.1 | Hemagglutinin (Hubei orthoptera virus 6) |
| ALR71098.1 | Major budded virus envelope Gp (Anticarsia gemmatalis multiple nucleopolyhedroviruses) |
| AAM82816.1 | Gp64 (Anticarsia gemmatalis Nucleopolyhedrovirus) |
| ALR71414.1 | Major budded virus envelope Gp (Anticarsia gemmatalis multiple nucleopolyhedroviruses) |
| ALR69841.1 | Major budded virus envelope Gp (Anticarsia gemmatalis multiple nucleopolyhedroviruses) |
| AFS51902.1 | DekiORF24 (Dendrolimus kikuchii Nucleopolyhedrovirus) |
| BAF32568.1 | Gp64 (Bombyx mori Nucleopolyhedrovirus) |
| AFN09034.1 | GpP64/67 (Bombyx mori Nucleopolyhedrovirus) |
| NP_203281.1 | Gp64 (Epiphyas postvittana Nucleopolyhedrovirus) |
| AMN92169.1 | Envelope Gp (Bourbon virus) |
| NP_046282.1 | Gp64 (Orgyia pseudotsugata multiple nucleopolyhedroviruses) |
| YP_803518.2 | Major budded virus envelope Gp (Anticarsia gemmatalis Nucleopolyhedrovirus) |
| YP_009316051.1 | Major budded virus envelope Gp (Anticarsia gemmatalis multiple nucleopolyhedroviruses) |
| ALR72199.1 | Major budded virus envelope Gp (Anticarsia gemmatalis multiple nucleopolyhedroviruses) |
| YP_473216.1 | Gp64 (Hyphantria cunea Nucleopolyhedrovirus) |
| AAF09194.1 | Envelope fusion Gp64 precursor (Hyphantria cunea Nucleopolyhedrovirus) |
| YP_009255283.1 | Gp64 (Catopsilia pomona Nucleopolyhedrovirus) |
| YP_009118511.1 | Major budded virus envelope Gp (Condylorrhiza vestigialis MNPV) |
| AAB53360.1 | Envelope Gp (Anagrapha falcifera MNPV) |
| AAN28029.1 | Gp67 (Rachiplusia ou MNPV) |
| YP_007250533.1 | Gp64 protein (Thysanoplusia orichalcea Nucleopolyhedrovirus) |
| BAS05039.1 | GP64/67 (Bombyx mori Nucleopolyhedrovirus) |
| YP_950827.1 | Gp64 (Maruca vitrata Nucleopolyhedrovirus) |
| AFN21223.1 | Gp64/67 (Bombyx mori Nucleopolyhedrovirus) |
| NP_047525.1 | Gp64/67 (Bombyx mori Nucleopolyhedrovirus) |
| AFN21083.1 | Gp64/67 (Bombyx mori Nucleopolyhedrovirus) |
| AGX01332.1 | Gp64/67 (Bombyx mori Nucleopolyhedrovirus) |
| ACQ57301.1 | Gp64/67 (Bombyx mandarina Nucleopolyhedrovirus) |
| AGX01204.1 | Gp64/67 (Bombyx mori Nucleopolyhedrovirus) |
| AAA72760.1 | Gp67 gene product (Autographa californica Nucleopolyhedrovirus) |
| AAA72759.1 | Envelope surface Gp (Autographa californica Nucleopolyhedrovirus) |
| NP_054158.1 | Major budded virus envelope Gp (Autographa californica Nucleopolyhedrovirus) |
| ABE68511.1 | Major budded virus envelope Gp (Plutella xylostella multiple nucleopolyhedroviruses) |
| AIU56980.1 | Ac-gp64 (Autographa californica Nucleopolyhedrovirus) |
| AFO10080.1 | Gp64 (Bombyx mandarina Nucleopolyhedrovirus S2) |
| AHX03161.1 | Envelope Gp (Batken virus) |
| AKN80958.1 | Gp64 (Lonomia obliqua multiple nucleopolyhedroviruses) |
| P27427.1 | RecName: full-envelope Gp; Flags precursor |
| ADF56027.1 | Gp (Dhori virus) |
| YP_009182316.1 | Gp64 (Diatraea saccharalis Granulovirus) |
| ACY56281.1 | Hemagglutinin (Quaranfil virus) |
| YP_009110688.1 | Hemagglutinin protein (Wellfleet Bay virus) |
| APG77868.1 | Hemagglutinin (Beihai orthomyxo-like virus 1) |
| AHY82907.1 | Envelope Gp (Tjuloc virus) |
| AFN73046.1 | Hemagglutinin (Tjuloc virus) |
| APG77884.1 | Hemagglutinin (Hubei earwig virus 1) |
Distinct Electrostatic Patterns of THOV Gp, DHOV Gp, and AcMNPV Gp64.
It has been shown that baculovirus can enter several types of mammalian cells using Gp64 as the fusogen, and the positive charges of Gp64 favor its entry (21–23). A disordered loop connecting domain II and domain III, rich in basic amino acid residues, plays a key role in this process. Increasing the number of positively charged amino acids of this basic loop would enhance the efficiency of baculovirus entry into mammalian cells (23). Although the precise mechanism of this phenomenon is unknown, it is clear that the positive charges of Gp64 have a great impact on the ability of the virus to infect mammalian cells.
The basic loop in both the AcMNPV Gp64 and THOV Gp structures was largely invisible but was well resolved in the structure of DHOV Gp (Figs. 1 A and E, 3C, and 4 A–D). Comparing the sequences, we found that AcMNPV Gp64 and THOV Gp harbor a longer basic loop than DHOV Gp (Fig. S4). As the loop of DHOV Gp is shorter, it packs much more closely to domain II and thus is less flexible. This feature also indicated that THOV Gp is much more similar to AcMNPV Gp64 than to DHOV Gp in structure.
Fig. 4.
Electrostatic potential maps of AcMNPV Gp64 (A, D, and G), THOV Gp (B, E, and H), and DHOV Gp (C, F, I) in side (A–C), top (D–F), and bottom (G–I) views. The surface potential is shown from −60 KT/e in red to +60 KT/e in blue. The most variable domain II and a surface pocket (pocket_A) are highlighted by black dashed ellipsoids. The basic loops are shown as green ribbons with dashed lines representing the unresolved parts.
Fig. S4.
Sequence alignment of THOV Gp, DHOV Gp, BOUV Gp, and AcMNPV Gp64. The secondary structures of THOV Gp, DHOV Gp, and AcMNPV Gp64 are denoted in green, blue, and black, respectively. Six conserved disulfides are labeled by numbers 1–6. Disulfide a is shared by THOV Gp, DHOV Gp, and BOUV Gp. Disulfides b, c, and d are unique for DHOV Gp, THOV Gp, and AcMNPV Gp64, respectively. The basic loop sequences are highlighted by red rectangles.
We next compared the electrostatic potential patterns of the THOV Gp, DHOV Gp, and AcMNPV Gp64 structures. Interestingly, the three proteins displayed significantly different patterns of surface charge distribution (Fig. 4). On the whole, THOV and DHOV Gps have more positively charged surfaces than AcMNPV Gp64, and DHOV Gp is the richest among them. Most of the positive-charge surfaces are focused on domain II, which shows the greatest variation among all proteins in Gp64 family (Figs. 3A and 4 A–C). Notably, a pocket formed in domain II (pocket A) is mainly negatively charged in AcMNPV Gp64 and THOV Gp, while it is highly positively charged in DHOV Gp (Fig. 4 A–C).
Accordingly, the conserved domain I shows a similar charge pattern among all three structures, while the top region of the domain III stalk (crown) also displays an increase in positive charge in the THOV and DHOV Gps compared with AcMNPV Gp64 (Fig. 4 D–I). These structural features indicate that AcMNPV Gp64 and DHOV Gp likely represent two evolutionary directions in favor of different hosts. In addition, the evolving route of THOV Gp is different from that of DHOV Gp, as it showed some obvious features similar to AcMNPV Gp64 rather than DHOV Gp.
Phylogenic Analysis of Thogotovirus Gps.
With the hypotheses generated by structural analysis, we then compared all the gene sequences of thogotovirus Gp homologs in the National Center for Biotechnology Information (NCBI) database, which involved all available thogotovirus and quaranjavirus Gp sequences. A phylogenic tree of Gps was built with the sequences of baculovirus Gp64s as the outgroup. All sequences clustered into three main clades, which are represented by THOV (clade A), DHOV (clade B), and Quaranfil virus (clade C, representing the Quaranjavirus genus) (Fig. 5). Among orthomyxovirus homologs, thogotoviruses diverged early from the quaranjavirus clade and later diverged into the THOV and DHOV clades.
Fig. 5.
Phylogenic tree of thogotovirus Gps and homologs. Two baculovirus Gp64 sequences were selected as the outgroup. Bootstrap values are shown at relevant nodes. Each taxon is labeled with the species name and the GenBank accession number. Evolutionary distances are expressed as the number of base substitutions per site. The three clade nodes A, B, and C correspond to the THOV clade, DHOV clade, and quaranjavirus clade, respectively. The DHOV clade is highlighted by a yellow rectangle. The taxons for the structures discussed in this study are highlighted in cyan (AcMNPV), green (THOV), and blue (DHOV. The newly identified BOUV is colored red. Quaranfil virus, the type species of the Quaranjavirus genus, is magenta.
Three unclassified thogotoviruses, Jos virus, Upolu virus, and Aransas virus, fell into clade A, indicating much closer relation to THOV. It is interesting that Sinu virus, an unclassified orthomyxovirus newly isolated from mosquitoes (24), clustered much more closely to baculoviruses than to the other three orthomyxovirus clades. The newly identified BOUV clustered with DHOV. The Gps of DHOV and BOUV share a sequence identity of ∼57%, and the basic loops are very similar to each other, i.e., shorter than that of THOV Gp and AcMNPV Gp64 (Fig. S4).
Although the orthomyxovirus homologs diverged from baculovirus Gp64 before they diverged from each other, the structure of THOV Gp showed some features more similar to baculovirus Gp64 than to DHOV Gp. This indicates that the evolutionary route of the THOV clade enabled these Gps to retain certain properties of the baculovirus clade. The DHOV clade, however, evolved via a route that was substantially distinct from that of the THOV and baculovirus clades. To further test the consistency of structural features with their phylogenic relationships, we conducted structural predictions for the representative Gps of each clade. In line with this hypothesis, the predicted structure models of BOUV, Quaranfil virus, and Sinu virus Gps displayed a similar trend of evolving electrostatic potential patterns (Fig. S5). The BOUV Gp, closest to DHOV Gp, shows a dominant positively charged surface profile. In contrast, Sinu virus Gp is rich in negative charges, similar to the pattern of AcMNPV Gp64. The Gp of Quaranfil virus, the type species of the Quaranjavirus genus, maintains some features of both BOUV and Sinu virus Gps and harbors moderate positively charged surfaces (Fig. S5).
Fig. S5.
Electrostatic surface patterns of BOUV, Quaranfil virus, and Sinu virus Gp models predicted by homology modeling. The structures of BOUV, Quaranfil virus, and Sinu virus Gps are predicted by homology modeling using different Gp structures as the input template to avoid model bias. The Top, Middle, and Bottom Rows represent models predicted based on AcMNPV Gp64, THOV Gp, and DHOV Gp, respectively. The best predicted models, based on the template structure with the highest sequence identity, are indicated by black rectangles. The most variable domain II and the pocket A discussed above are shown by dashed ellipsoids. The surface potential is shown from −60 KT/e in red to +60 KT/e in blue.
SI Materials and Methods
Protein Expression and Purification.
The coding sequences of ectodomains of THOV Gp (GenBank accession code: NC006506, residues 18–483) and DHOV Gp (GenBank accession code: GU969310, residues 21–494) were synthesized and subcloned into the baculovirus transfer vector pFastbac1 (Invitrogen) with an N-terminal gp67 signal peptide, a C-terminal thrombin cleavage site followed by a T4 fibritin trimerization domain, and a hexahistidine tag to facilitate purification. Both proteins were expressed with the Bac-to-Bac baculovirus expression system (Invitrogen). The soluble Gp ectodomains were captured by IMAC with a HisTrap HP column (GE Healthcare). The target protein was eluted with a buffer containing 20 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, and 100 mM imidazole. The eluted product was pooled and further purified by size-exclusion chromatography using a Superdex 200 10/300 GL column (GE Healthcare) equilibrated with a buffer containing 20 mM Tris⋅HCl (pH 8.0) and 150 mM NaCl. The eluted fractions were analyzed by SDS/PAGE under both reducing and nonreducing conditions, which showed a purity of more than 90%. The fractions were pooled and cleaved with thrombin at 4 °C for 12 h to remove the C-terminal trimerization domain and hexahistidine tag. The cleaved product was subjected to another round of size-exclusion chromatography for final purification. The eluted product was concentrated to ∼10 mg/mL for crystallization.
Crystallization and Data Collection.
Both THOV and DHOV Gps were crystallized by the sitting-drop vapor-diffusion method at 18 °C with 1 μL protein solution mixed with 1 μL reservoir buffer. For THOV Gp, initial crystals grew in 1 mo in a buffer containing 5% (vol/vol) (+/−)-2-Methyl-2,4-pentanediol, 0.1 M Hepes (pH 7.0), 20% (wt/vol) PEG 10,000, which yielded poor diffraction patterns. Matrixed optimization was performed to improve the quality of crystals. High-quality crystals were obtained in a buffer containing 5% (vol/vol) (+/−)-2-Methyl-2,4-pentanediol, 0.1 M Hepes (pH 7.5) and 10% (wt/vol) PEG 10,000, which were used for data collection. For DHOV Gp, small crystals appeared within a week in a buffer containing 6% (vol/vol) ethylene glycol, 0.1 M citric acid (pH 3.5), and 10% (wt/vol) PEG 6,000. After two rounds of tandem microseeding with the same crystallization buffer conditions, full-size crystals were obtained for data collection. Diffraction data were collected with cryoprotected crystals [in a reservoir solution containing 20% (vol/vol) glycerol] at SSRF beamline BL17U (36). All datasets were processed with HKL2000 software (37). The datasets of THOV and DHOV Gps were refined to 2.1 and 2.5 Å, respectively.
Structure Determination.
Due to the lack of a strong indication for available homologous structure, it was impossible to get a complete structure template for molecular replacement. As we searched the database, the AcMNPV Gp64 (PDB ID code: 3DUZ) was found to share ∼28% sequence identity with both THOV and DHOV Gps. By aligning the sequences, the most conserved region was mapped to the N-terminal 160 amino acids. With this partial model as search input, the initial phase of THOV Gp was obtained by molecular replacement using Phaser (38) from the CCP4 program suite (44). Aided by several rounds of density modification with noncrystallographic symmetry restraints applied using DM (39) in CCP4 (44) and automatic model building using Autobuild in Phenix (45), the atomic model of the major part was obtained. After several rounds of iterative refinement using Phenix (45) with secondary structure restraints imposed and manual model building in Coot (46), a nearly complete atomic model of THOV Gp was built successfully. The structure of DHOV Gp was determined by molecular replacement using THOV Gp with the loop removed as the search model. The model was completed by iterative refinement in Phenix (45) and manual model building in Coot (46). The final models of THOV and DHOV Gps were refined to an Rfree of 20.9 and 25.7%, respectively. The stereochemical quality of the final models was assessed with MolProbity (47). Data collection, processing, and refinement statistics are summarized in Table S2. All structural figures were generated with either Chimera (40) or PyMOL (41).
Table S2.
Crystallographic data collection and refinement statistics
| THOV Gp | DHOV Gp | |
| Data collection | ||
| Space group | P1 21 1 | P3 |
| Cell dimensions | ||
| a, b, c, Å | 106.20, 66.77, 120.74 | 106.73, 106.73, 134.44 |
| α, β, γ, ° | 90.00, 103.31, 90.00 | 90.00, 90.00, 120.00 |
| Resolution*, Å | 50.00–2.10 (2.18–2.10) | 50.00–2.50 (2.59–2.50) |
| Unique reflections | 95,874 (8,850) | 59,514 (5,937) |
| Rmerge*,† | 0.089 (0.794) | 0.150 (2.113) |
| Rpim | 0.043 (0.363) | 0.054 (0.768) |
| CC1/2 | 0.996 (0.879) | 0.998 (0.772) |
| I/σ(I)* | 16.8 (2.8) | 14.5 (1.1) |
| Completeness*, % | 99.3 (99.3) | 99.9 (99.8) |
| Redundancy* | 5.5 (5.6) | 8.5 (8.4) |
| Refinement | ||
| Resolution, Å | 34.45–2.10 (2.17–2.10) | 34.32–2.50 (2.59–2.50) |
| No. reflections | 95,818 (8,836) | 43,039 (1,425) |
| Rwork/Rfree‡ | 0.172/0.209 | 0.231/0.257 |
| No. atoms | ||
| Protein | 9,417 | 9,344 |
| Ligand | 232 | 56 |
| Water | 972 | 201 |
| B-factors, Å2 | ||
| Protein | 53.4 | 73.4 |
| Ligand | 91.3 | 102.6 |
| Water | 59.0 | 62.8 |
| Rmsd | ||
| Bond lengths, Å | 0.008 | 0.003 |
| Bond angles, ° | 0.93 | 0.71 |
| Ramachandran plot§, % | ||
| Favored | 96 | 94 |
| Allowed | 3.9 | 5.6 |
| Outliers | 0.1 | 0.4 |
Values for the outmost resolution shell are given in parentheses.
Rmerge = ΣiΣhkl| Ii < I> | /ΣiΣhklIi, where Ii is the observed intensity and <I> is the average intensity from multiple measurements.
Rwork = Σ | | Fo| | Fc | | /Σ | Fo|, where Fo and Fc are the structure-factor amplitudes from the data and the model, respectively. Rfree is the R factor for a subset (5%) of reflections that was selected before refinement calculations and was not included in the refinement.
Ramachandran plots were generated by using the program MolProbity.
CD Assay.
The CD spectra of soluble THOV and DHOV Gps in different pH conditions were measured. Initially, the proteins were buffer exchanged into a buffer containing either 20 mM Tris⋅HCl (pH 8.5) and 50 mM NaCl or 20 mM sodium citrate (pH 5.0) and 50 mM NaCl by dialysis. After the samples were loaded, the CD spectra at 200–260 nm were recorded on a Chirascan spectrometer (Applied Photophysics) using a thermostatically controlled cuvette at 25 °C. The concentration of the samples used for data collection was adjusted to 0.2 mg/mL accordingly. The curves were generated by nonlinear fitting with the OriginPro 8.0 program (OriginLab).
Conservation Mapping of THOV Gp-Related Proteins.
The amino acid sequences of all THOV Gp-related proteins available in the NCBI database were downloaded and aligned with Clustal Omega (48). The resultant alignment output was used as the input to the ConSurf (49) server and mapped to the structure of DHOV Gp, which has fewer unresolved loops than THOV Gp. The final output was used to prepare figures using the PyMOL (41) program. All THOV Gp-related sequences used in this analysis are listed in Table S1.
Structure-Based Sequence Alignment.
The structures of THOV Gp, DHOV Gp, and AcMNPV Gp64 were aligned with the MUSTANG (50) server. The resulting structure-based sequence alignment was used as the reference for aligning full-length sequences. The sequence of BOUV Gp was used as the query in the BLAST (51) server to search in the database; this search identified DHOV Gp, with a sequence identity of ∼57%, as the best match. Therefore the sequence of BOUV Gp was further aligned with the other three sequences with DHOV Gp as the reference. The alignment figure was generated with ESPript (52).
Phylogenic Analysis.
All Gp coding sequences of all annotated and unclassified thogotovirus and quaranjavirus species available in the NCBI database were analyzed. Two sequences of baculovirus Gp64, AcMNPV and BmNPV, were selected as the outgroup. The sequences were aligned using MUSCLE (42), and all poorly or ambiguously aligned regions were removed. The phylogenetic tree was inferred using the ME method (43) available in the MEGA (53) package, with bootstrap values estimated from 1,000 replicate trees. The evolutionary distances were computed using the maximum composite likelihood method (54) and are expressed as the number of base substitutions per site. Finally, the resulting phylogenetic trees were displayed and annotated with FigTree.
Homology Modeling.
The Gp structures of BOUV, Quaranfil virus, and Sinu virus were predicted by homology modeling using the SWISS-MODEL (55) server. To avoid model bias, each inquiry sequence was modeled based on three different template structures, AcMNPV Gp64, THOV Gp, and DHOV Gp. The outputs based on three different input template models were quite consistent. The disordered domain IV and the missing loops in domain II were not modeled.
Discussion
Among the seven genera in the Othomyxoviridae family, five (Influenza virus A, B, C, and D and Isavirus) possess class I fusogenic proteins to mediate membrane fusion and utilize sialic acid-related receptors for attachment to the host cell surface and subsequent entry (25–30). Meanwhile, they also harbor receptor-destroying enzymes to facilitate the release of progeny virions (31, 32). Precisely, influenza A and B viruses utilize HA proteins to bind to α2–3–, α2–6–, or α2–8–linked sialic acid receptors and mediate membrane fusion in the endosome, and they use neuraminidase proteins for receptor cleavage (25). For influenza C and D viruses, all three processes (i.e., attachment, fusion, and release) are mediated by a single protein, hemagglutinin-esterase-fusion (HEF) protein, and they recognize 9-O-acetylated sialic acid receptors (28–30, 33). In isaviruses, however, two envelope proteins are involved, the HE (hemagglutinin-esterase) and F (fusion) proteins, which are responsible for binding and destroying 4-O-acetylated sialic acid receptors and mediating membrane fusion, respectively (26, 27, 32). It is intriguing that thogotoviruses, as a genus in the Othomyxoviridae family, utilize a single-envelope Gp for entry into host cells, which is strikingly different from all the proteins mentioned above. To date, the receptors governing thogotoviruses’ entry have not been identified. Previous studies have shown that Gp can induce hemagglutination but lacks neuraminidase or esterase activities (10). All these unique features demonstrate that thogotoviruses represent a type of virus significantly distinct from other orthomyxoviruses.
Interestingly, our current study indicates that the closest homolog of thogotovirus Gps is AcMNPV Gp64, a class III viral fusogen (13). Although sharing a low sequence identity (∼28%) (Fig. S3), the overall folds of THOV/DHOV Gps and AcMNPV Gp64 are quite similar. Their individual domains can be superimposed well, but their relative orientations vary among different species (Fig. 2 A–F). Because of this variation, the electrostatic potential patterns of these proteins also show obvious diversity. The most variable domain, domain II, is obviously the result of the divergent evolution of the different viruses, which might be related to their distinctions in host tropism.
Baculoviruses can enter several mammalian cells using Gp64 as the fusion machine but cannot replicate (21, 22), which is reminiscent of the bitropism of thogotoviruses in mammalian hosts and tick vectors. Our data further provide strong support for the functional correlation between thogotovirus Gps and baculovirus Gp64s. Our structural and phylogenic analyses indicated that the AcMNPV Gp64 and DHOV Gp represent two evolving directions that favor insect and mammal hosts, respectively. Although THOV is closer to DHOV than to baculovirus, its Gp evolved in a direction different from that of DHOV and retains obvious similarities to baculovirus Gp64.
Among all orthomyxoviruses using thogotovirus Gp homologs for cell entry, only members of the Thogotovirus genus were reported to infect humans (5–7, 9). In particular, DHOV is reported to be capable of transmitting through an aerosol route and causing severe infections (7), implying the potential to cause human epidemics. Notably, the newly identified BOUV clustered within the DHOV clade. These observations indicate that the DHOV clade likely represents a group of well-adapted and dangerous mammalian-tropic viruses that pose great threats to human health. Thus, special care should be taken for the surveillance of potential epidemics of DHOV clade viruses in humans. Our study here indicates that structural information could also provide important clues for early warnings of potential human infections of emerging pathogens.
Although thogotovirus Gps and baculovirus Gp64s are clearly of the same origin, it is difficult to tell which is more similar to the common ancestor, i.e., whether the Gp64 of baculoviruses evolved from a thogotovirus-like ancestor or the Gp of thogotoviruses originated from a baculovirus-like prototype. We noticed that the group I baculoviruses using Gp64 as the fusogen appear later in evolution than those in group II utilizing an F protein. There is only one Gp, F protein, in the envelope of group II baculoviruses that facilitates their entry (34). Group I baculoviruses, however, have two envelop Gps, Gp64 and a vestigial F homolog that is incapable of mediating virus infection (34, 35). Thus, group I baculoviruses evolved from a group II-like ancestor that obtained the prototype of Gp64, which gradually replaced the function of F protein. Because thogotoviruses have only one envelope Gp, the most likely scenario might be that an ancient group II-like baculovirus obtained a Gp64 prototype from a thogotovirus-like ancestor and evolved into group I baculoviruses. The Gp64 prototype is conceivably quite similar to THOV Gp, which possesses the features of both baculovirus Gp64 and DHOV Gp.
The structural and phylogenic analyses in this study provide important insight into the evolution of viruses from different families: Orthomyxoviridae (segmented negative-strand RNA viruses) and Baculoviridae (double-stranded DNA viruses). Our findings are of great significance for understanding the origin and evolving process of orthomyxoviruses, many of which, including some of the thogotoviruses discussed above, are multitropism pathogens that infect both humans and animals.
In addition, the two Gp structures could potentially offer important guidance for antiviral drug design. Based on our analysis, the fusion loops of Gps are highly conserved among all Gp64 family members. By analogy to VSV Gp, domain I, where the fusion loop is located, would retain the same fold before and after membrane fusion (16, 18). Therefore, it would be a suitable target for the development of broad-spectrum antiviral drugs that block virus entry by interfering with the membrane-fusion process.
Despite these advances, some important questions remain to be answered to understand the mechanism of thogotovirus entry. It is noteworthy that all three structures were determined in the postfusion conformation, although they were crystallized under different pH conditions varying from 3.5 to 7.5 (see details in SI Materials and Methods). Both AcMNPV Gp64 and DHOV Gp were crystallized at acidic pHs, 5.0 and 3.5, respectively (13). However, THOV Gp crystals were obtained at nearly neutral conditions, pH 7.0–7.5, where Gps are thought to exist in the prefusion conformation. As AcMNPV Gp64 undergoes a pH-induced conformational change (13) and THOV and DHOV Gps also displayed significant conformational shifts at different pH conditions (Fig. S2), we infer that the prefusion structures of thogotovirus Gps are metastable and unfavorable for crystallization. In neutral pH solutions, Gp can also adopt a postfusion conformation, but at a low rate, thereby offering the opportunity for crystallization by displacing the equilibrium between different conformations. A similar phenomenon was also observed for the Gp of VSV, which could be crystallized in the postfusion conformation at pH 6.0–7.0 (16, 18). Thus far, the receptors for both baculoviruses and thogotoviruses are unknown. To fully uncover the mechanism of thogotovirus entry, more efforts will be required to resolve the Gp structures at the prefusion state and to identify their potential receptors.
Materials and Methods
Protein Expression, Purification, and Crystallization.
The ectodomains of THOV Gp (GenBank accession code: NC006506, residues 18–483) and DHOV Gp (GenBank accession code: GU969310, residues 21–494) fused with a C-terminal T4 fibritin trimerization domain and a 6× histidine affinity tag were expressed using the Bac-to-Bac baculovirus expression system (SI Materials and Methods). The soluble Gp ectodomains were purified by tandem immobilized metal affinity chromatography (IMAC) and size-exclusion chromatography (SI Materials and Methods). The eluted products in the buffer containing 20 mM Tris⋅HCl (pH 8.0) and 150 mM NaCl were concentrated to ∼10 mg/mL for crystallization. Both THOV and DHOV Gps were crystallized by the sitting-drop vapor-diffusion method at 18 °C (SI Materials and Methods).
Diffraction Data Collection and Structure Determination.
Diffraction data were collected with cryoprotected [in a reservoir solution containing 20% (vol/vol) glycerol] crystals at the Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U (36). All datasets were processed with HKL2000 software (37). The THOV and DHOV Gps datasets were refined to 2.1 and 2.5 Å, respectively. Both structures were solved by the molecular-replacement method (38). The initial phase of THOV Gp was obtained using the N-terminal 160-amino acid partial model of baculovirus Gp64 [Protein Data Bank (PDB) ID: 3DUZ] as the search input. Density modification (39) was performed with noncrystallographic symmetry restraints to improve the density of other parts. Iterative autobuilding and manual adjustments were conducted to complete the model (SI Materials and Methods). The DHOV Gp structure was solved with the THOV Gp structure as the search input. The final structures of the THOV and DHOV Gps were refined to an Rfree of 20.9% and 25.7%, respectively (SI Materials and Methods). Data collection, processing, and refinement statistics are summarized in Table S2. All structural figures were generated with either Chimera (40) or PyMOL (41).
Phylogenic Analysis.
The Gp coding sequences of thogotoviruses and quaranjaviruses (all annotated and unclassified species in the two genera available in the NCBI database) were analyzed. Two sequences of baculovirus Gp64, AcMNPV and Bombyx mori nucleopolyhedrosis virus (BmNPV), were selected as the outgroup. The sequences were aligned using MUSCLE (42), and the phylogenetic tree was inferred using the Minimum Evolution (ME) method (SI Materials and Methods) (43). The resulting phylogenetic trees were displayed and annotated with FigTree.
Data Availability.
The atomic coordinates of the THOV and DHOV Gp extracellular domains have been deposited in the PDB under the ID codes 5XEA and 5XEB, respectively.
Supplementary Material
Acknowledgments
We thank Haixia Xiao for excellent suggestions regarding this study and comments on the manuscript and the staff at the SSRF beamline 17U for assistance in diffraction data collection. This work was supported by Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDPB03, China Ministry of Science and Technology National 973 Project Grants 2013CB531502 and 2014CB542503, National Key Plan for Scientific Research and Development of China Grant 2016YFD0500305, and China National Science and Technology Special Project Grant 2017ZX10303403 of the National Natural Science Foundation of China (NSFC). Y.S. is supported by NSFC Excellent Young Scientist Program Grant 81622031 and Youth Innovation Promotion Association of the Chinese Academy of Sciences Grant 2015078. G.F.G. is partly supported as a leading principal investigator of the NSFC Innovative Research Group supported by NSFC Grant 81621091.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates of Thogoto virus and Dhori virus Gp extracellular domains have been deposited in the Protein Data Bank (PDB) database (PDB ID codes 5XEB and 5XEB, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706125114/-/DCSupplemental.
References
- 1.Kawaoka Y, Palese P. Family Orthomyxoviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball L, editors. Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic; San Diego: 2005. pp. 681–693. [Google Scholar]
- 2.Perez D, et al. Orthomyxoviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy. Elsevier; Oxford: 2011. pp. 749–762. [Google Scholar]
- 3.Haig DA, Woodall JP, Danskin D. Thogoto virus: A hitherto underscribed agent isolated from ticks in Kenya. J Gen Microbiol. 1965;38:389–394. doi: 10.1099/00221287-38-3-389. [DOI] [PubMed] [Google Scholar]
- 4.Anderson CR, Casals J. Dhori virus, a new agent isolated from Hyalomma dromedarii in India. Indian J Med Res. 1973;61:1416–1420. [PubMed] [Google Scholar]
- 5.Hubálek Z, Rudolf I. Tick-borne viruses in Europe. Parasitol Res. 2012;111:9–36. doi: 10.1007/s00436-012-2910-1. [DOI] [PubMed] [Google Scholar]
- 6.Moore DL, et al. Arthropod-borne viral infections of man in Nigeria, 1964-1970. Ann Trop Med Parasitol. 1975;69:49–64. doi: 10.1080/00034983.1975.11686983. [DOI] [PubMed] [Google Scholar]
- 7.Butenko AM, Leshchinskaia EV, Semashko IV, Donets MA, Mart’ianova LI. [Dhori virus–A causative agent of human disease. 5 cases of laboratory infection] Vopr Virusol. 1987;32:724–729. Russian. [PubMed] [Google Scholar]
- 8.Filipe AR, Calisher CH, Lazuick J. Antibodies to Congo-Crimean haemorrhagic fever, Dhori, Thogoto and Bhanja viruses in southern Portugal. Acta Virol. 1985;29:324–328. [PubMed] [Google Scholar]
- 9.Kosoy OI, et al. Novel thogotovirus associated with febrile illness and death, United States, 2014. Emerg Infect Dis. 2015;21:760–764. doi: 10.3201/eid2105.150150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Portela A, Jones LD, Nuttall P. Identification of viral structural polypeptides of Thogoto virus (a tick-borne orthomyxo-like virus) and functions associated with the glycoprotein. J Gen Virol. 1992;73:2823–2830. doi: 10.1099/0022-1317-73-11-2823. [DOI] [PubMed] [Google Scholar]
- 11.Presti RM, et al. Quaranfil, Johnston Atoll, and Lake Chad viruses are novel members of the family Orthomyxoviridae. J Virol. 2009;83:11599–11606. doi: 10.1128/JVI.00677-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Backovic M, Jardetzky TS. Class III viral membrane fusion proteins. Curr Opin Struct Biol. 2009;19:189–196. doi: 10.1016/j.sbi.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadlec J, Loureiro S, Abrescia NG, Stuart DI, Jones IM. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat Struct Mol Biol. 2008;15:1024–1030. doi: 10.1038/nsmb.1484. [DOI] [PubMed] [Google Scholar]
- 14.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heldwein EE, et al. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 16.Roche S, Bressanelli S, Rey FA, Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 17.Kampmann T, Mueller DS, Mark AE, Young PR, Kobe B. The role of histidine residues in low-pH-mediated viral membrane fusion. Structure. 2006;14:1481–1487. doi: 10.1016/j.str.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 18.Roche S, Rey FA, Gaudin Y, Bressanelli S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 2007;315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- 19.Lu X, et al. Structure and receptor-binding properties of an airborne transmissible avian influenza A virus hemagglutinin H5 (VN1203mut) Protein Cell. 2013;4:502–511. doi: 10.1007/s13238-013-3906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W, et al. An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level. Science. 2013;340:1463–1467. doi: 10.1126/science.1236787. [DOI] [PubMed] [Google Scholar]
- 21.Ho YC, et al. Baculovirus transduction of human mesenchymal stem cell-derived progenitor cells: Variation of transgene expression with cellular differentiation states. Gene Ther. 2006;13:1471–1479. doi: 10.1038/sj.gt.3302796. [DOI] [PubMed] [Google Scholar]
- 22.Zeng J, Du J, Zhao Y, Palanisamy N, Wang S. Baculoviral vector-mediated transient and stable transgene expression in human embryonic stem cells. Stem Cells. 2007;25:1055–1061. doi: 10.1634/stemcells.2006-0616. [DOI] [PubMed] [Google Scholar]
- 23.O’Flynn NM, Patel A, Kadlec J, Jones IM. Improving promiscuous mammalian cell entry by the baculovirus Autographa californica multiple nuclear polyhedrosis virus. Biosci Rep. 2012;33:23–36. doi: 10.1042/BSR20120093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Contreras-Gutiérrez MA, et al. Sinu virus, a novel and divergent orthomyxovirus related to members of the genus Thogotovirus isolated from mosquitoes in Colombia. Virology. 2017;501:166–175. doi: 10.1016/j.virol.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouvier NM, Palese P. The biology of influenza viruses. Vaccine. 2008;26:D49–D53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aspehaug V, Mikalsen AB, Snow M, Biering E, Villoing S. Characterization of the infectious salmon anemia virus fusion protein. J Virol. 2005;79:12544–12553. doi: 10.1128/JVI.79.19.12544-12553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rimstad E, Mjaaland S, Snow M, Mikalsen AB, Cunningham CO. Characterization of the infectious salmon anemia virus genomic segment that encodes the putative hemagglutinin. J Virol. 2001;75:5352–5356. doi: 10.1128/JVI.75.11.5352-5356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muraki Y, Hongo S. The molecular virology and reverse genetics of influenza C virus. Jpn J Infect Dis. 2010;63:157–165. [PubMed] [Google Scholar]
- 29.Song H, et al. An open receptor-binding cavity of hemagglutinin-esterase-fusion glycoprotein from newly-identified influenza D virus: Basis for its broad cell tropism. PLoS Pathog. 2016;12:e1005411. doi: 10.1371/journal.ppat.1005411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hause BM, et al. Characterization of a novel influenza virus in cattle and swine: Proposal for a new genus in the Orthomyxoviridae family. MBio. 2014;5:e00031-14. doi: 10.1128/mBio.00031-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen M, et al. Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase. Virol J. 2013;10:321. doi: 10.1186/1743-422X-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kristiansen M, Frøystad MK, Rishovd AL, Gjøen T. Characterization of the receptor-destroying enzyme activity from infectious salmon anaemia virus. J Gen Virol. 2002;83:2693–2697. doi: 10.1099/0022-1317-83-11-2693. [DOI] [PubMed] [Google Scholar]
- 33.Muchmore EA, Varki A. Selective inactivation of influenza C esterase: A probe for detecting 9-O-acetylated sialic acids. Science. 1987;236:1293–1295. doi: 10.1126/science.3589663. [DOI] [PubMed] [Google Scholar]
- 34.Lung O, Westenberg M, Vlak JM, Zuidema D, Blissard GW. Pseudotyping Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV): F proteins from group II NPVs are functionally analogous to AcMNPV GP64. J Virol. 2002;76:5729–5736. doi: 10.1128/JVI.76.11.5729-5736.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lung OY, Cruz-Alvarez M, Blissard GW. Ac23, an envelope fusion protein homolog in the baculovirus Autographa californica multicapsid nucleopolyhedrovirus, is a viral pathogenicity factor. J Virol. 2003;77:328–339. doi: 10.1128/JVI.77.1.328-339.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Q, et al. The macromolecular crystallography beamline of SSRF. Nucl Sci Tech. 2015;26:10102. [Google Scholar]
- 37.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 39.Cowtan K. Joint CCP4 and ESF-EACBM newsletter on protein. Crystallography. 1994;31:34–38. [Google Scholar]
- 40.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 41.DeLano WL. 2002 PyMOL molecular graphics system. Available at www.pymol.org. Accessed January 15, 2017.
- 42.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rzhetsky A, Nei M. A simple method for estimating and testing minimum-evolution trees. Mol Biol Evol. 1992;9:945–967. [Google Scholar]
- 44.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 45.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Landau M, et al. ConSurf 2005: The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: A multiple structural alignment algorithm. Proteins. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- 51.Johnson M, et al. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The atomic coordinates of the THOV and DHOV Gp extracellular domains have been deposited in the PDB under the ID codes 5XEA and 5XEB, respectively.