Significance
A mutation in myocilin is the most common known genetic cause of primary open-angle glaucoma (POAG). These mutations, which are dominant in nature, affect trabecular meshwork (TM) health and/or function and cause elevated intraocular pressure. Using in vitro human trabecular meshwork cells, an in vivo mouse model, and ex vivo human eyes, our study demonstrates the potential of clustered regularly interspaced short palindromic repeats (CRISPR)-mediated genome editing in human myocilin-associated POAG. By disrupting the mutant myocilin gene and its function using CRISPR-Cas9, we were able to reduce associated endoplasmic reticulum stress, lower intraocular pressure, and prevent further glaucomatous damage in mouse eyes. We also show the feasibility of using the CRISPR-Cas9 system in cultured human eyes.
Keywords: myocilin, CRISPR, glaucoma, trabecular meshwork, genome editing
Abstract
Primary open-angle glaucoma (POAG) is a leading cause of irreversible vision loss worldwide, with elevated intraocular pressure (IOP) a major risk factor. Myocilin (MYOC) dominant gain-of-function mutations have been reported in ∼4% of POAG cases. MYOC mutations result in protein misfolding, leading to endoplasmic reticulum (ER) stress in the trabecular meshwork (TM), the tissue that regulates IOP. We use CRISPR-Cas9–mediated genome editing in cultured human TM cells and in a MYOC mouse model of POAG to knock down expression of mutant MYOC, resulting in relief of ER stress. In vivo genome editing results in lower IOP and prevents further glaucomatous damage. Importantly, using an ex vivo human organ culture system, we demonstrate the feasibility of human genome editing in the eye for this important disease.
Glaucoma is a multifactorial disease, with increased intraocular pressure (IOP) as a major risk factor (1, 2). With the advent of efficient genome editing tools, it is now reasonable to think that the genomic component of glaucoma can be targeted for therapeutic and prophylactic purposes. Mutations in the myocilin gene (MYOC) have been shown to cause juvenile open-angle glaucoma, an early-onset form of primary open-angle glaucoma (POAG), as well as adult-onset POAG, and exhibit autosomal dominant inheritance (3–8). Almost 100 different pathogenic MYOC mutations have been identified, most of which are clustered in exon 3 encoding the olfactomedin domain (9). Myocilin mRNA and/or protein are expressed in eye structures including the retina, ciliary body, and trabecular meshwork (10–12), as well as in skeletal muscles, heart, brain, and testes (13–15), but the secreted protein does not appear to be necessary for ocular health, as both knockout animals and humans with homozygous, likely null mutations are not associated with glaucoma phenotypes (16–19).
Various knockin and knockout mouse models indicate that MYOC mutations are gain-of-function mutations (16, 20). Molecular and biochemical studies have shown that mutant myocilin accumulates inside cells instead of being secreted, resulting in activation of the unfolded protein response (UPR) cascade and ER stress (21–23). The UPR usually results in decreased translation and increased chaperone levels to promote proper folding and secretion of proteins (24); however, under a high burden of misfolding, cells undergo apoptosis. Trabecular meshwork (TM) cells are believed to be sensitive to chronic ER stress, ultimately dying, leading to elevated IOP and glaucoma in patients with MYOC mutations (22, 25, 26).
Novel approaches to treatment based on molecular mechanisms have been directed at the use of chemical chaperones to reduce protein misfolding and increase mutant myocilin secretion (27–30). These strategies have been successful in animal models of myocilin-associated glaucoma, and if delivered to human patients might provide short-term relief of ER stress, which likely will require chronic treatment. Another way to approach this treatment, especially given the gain-of-function nature of MYOC mutations, is by targeting the MYOC mRNA or protein itself (transcription or translational inhibition). While siRNA and shRNA provide potentially viable treatment options (31), we elected to directly target the MYOC gene using gene editing with clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 technology to treat myocilin-associated glaucoma.
Originally part of the prokaryotic adaptive immune system, the CRISPR-Cas9 system has been adapted as a genome-editing tool, in which the Cas9 endonuclease is directed by a guide RNA (gRNA) to cleave a 23-bp DNA sequence that contains a target-specific 20-bp sequence plus an adjacent NGG nucleotide motif, protospacer-adjacent motif (PAM) (32). DNA double-strand breaks (DSBs) induced by Cas9 nuclease can be repaired by two endogenous cellular repair processes: nonhomologous end-joining (NHEJ) and homology-directed repair (HDR). NHEJ is usually an error-prone process causing small insertions and deletions (indels) resulting from DNA ligation at the break sites. These indels can cause frame shift mutations in coding regions, resulting in an early stop codon leading to premature termination (32). In contrast, HDR is a less error-prone process relying on high sequence homology between the mutant and intact donor DNA strands. For gene therapy purposes, donor DNA or repair template is usually a wild-type (WT) copy of the gene with homology arms starting at the Cas9-induced DSB. As can be imagined, NHEJ has better efficiency than HDR with the currently available tools.
The simplicity of its design and flexibility in targeting candidate genes with gRNA sequences makes CRISPR-Cas9 promising as an in vivo genome-editing tool. We sought to use the CRISPR-Cas9 system to disrupt the MYOC gene and its aberrant function in human and mouse TM cells and in a mouse model of primary open glaucoma (Tg-MYOCY437H) that has been previously characterized in our laboratories (30). We aimed to design our gRNA near the start codon of human MYOC (exon1) to introduce an early frame shift mutation, resulting in termination of the protein. This proof-of-principle study of disruption of gene function should be equally effective for various MYOC mutations. Easy access to the TM of the eye, the dominant gain-of-function nature of MYOC mutations, and better efficiency of NHEJ compared with HDR also make this approach appealing for the treatment of myocilin-associated glaucoma.
Results
CRISPR-Mediated Gene Editing of MYOC in Vitro.
We generated NTM5 (normal trabecular meshwork 5) cell lines stably expressing wild-type and mutant MYOC containing the Y437H mutation (one of the most common MYOC mutations leading to early-onset POAG) (Fig. S1). Both WT and mutant myocilin were DsRed-tagged. Both WT and mutant cell lines express higher levels of myocilin compared with naïve control cell lines (Fig. S1). While mutant myocilin accumulates in the cell, WT myocilin is secreted into the conditioned medium. Both WT and mutant myocilin are posttranslationally modified by cleavage of the N-terminal signal sequence (shorter isoform) and/or glycosylation and thus sometimes appear as two bands on a Western blot (33, 34). Accumulation of mutant myocilin, visible as DsRed on immunocytochemistry (ICC) in Fig. S1D, leads to up-regulation of ER stress mRNA and proteins including BiP, calnexin, PDI, ATF4, and CHOP, as demonstrated by qPCR, Western blot analysis, and ICC (Fig. S1). Similar levels of both WT and mutant MYOC mRNA indicate that the changes in protein handling are posttranscriptional. These data confirm previous findings indicating that mutant myocilin accumulates inside glaucomatous TM cells and TM cells from transgenic mouse models expressing a human MYOC mutation (22, 30).
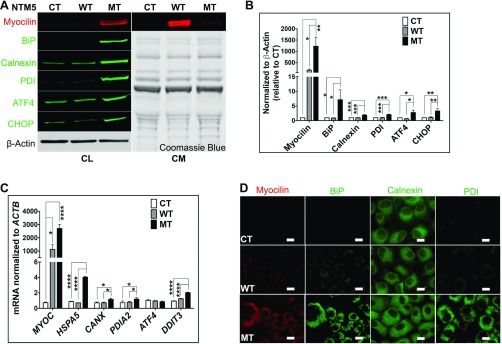
Fig. S1.
Mutant myocilin accumulates inside TM cells. (A) Stably transfected NTM5 cell lines overexpress WT and mutant (MT) forms of human MYOC compared with naïve untransfected control (CT) cells (n = 4). While mutated myocilin accumulates in cells, as demonstrated by analysis of the cell lysate (Left; CL), the WT form is efficiently secreted into the conditioned medium (Right; CM). The accumulation of mutant myocilin causes ER stress, as shown by increased expression of BiP, Calnexin, PDI, ATF4, and CHOP proteins in MT cells. β-actin served as a loading control for cell lysate, and Coomassie blue gel stain was used to test equal loading of secreted proteins in conditioned medium. (B) Densitometry showing significantly higher levels of myocilin, BiP, Calnexin, PDI, ATF4, and CHOP in NTM5 cells overexpressing mutant MYOC (MT) (n = 4) compared with CT or WT cells. (C) qRT-PCR showing overexpression of MYOC mRNA in WT and MT cells and significantly increased expression of ER stress markers mRNA in MT cells (n = 4). (D) An NTM5 cell line overexpressing MT myocilin showing ER accumulation of MYOC (DsRed), BiP (green), Calnexin (green), and PDI (green) compared with CT or NTM5 cells overexpressing WT myocilin. (Scale bar: 100 µm.) Error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, one-way ANOVA.
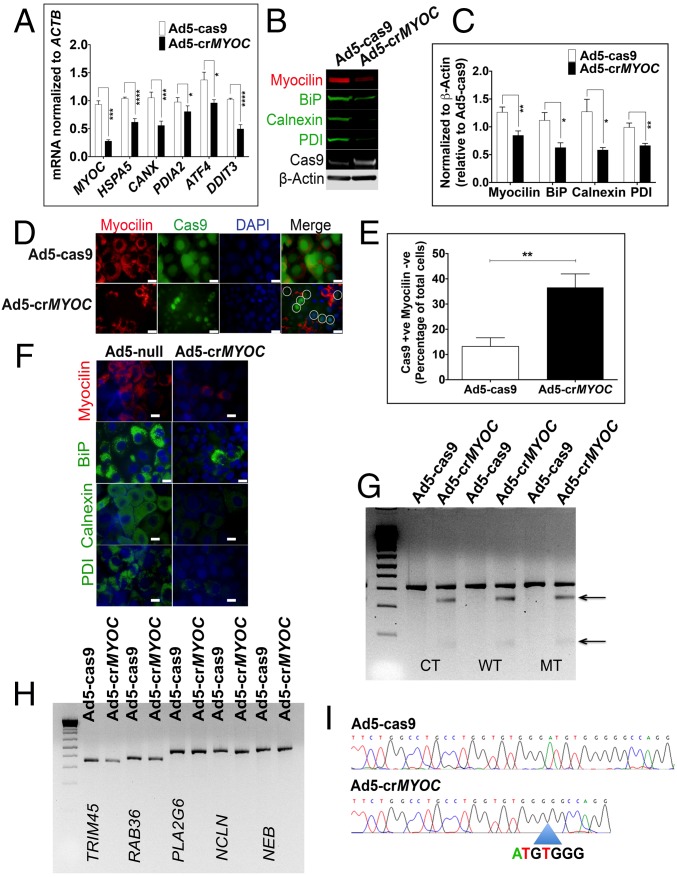
Adenovirus with CRISPR assembly (Ad5-crMYOC)-mediated transduction of NTM5 cell lines stably expressing mutant myocilin reduces levels of myocilin and associated key UPR markers, such as BiP, Calnexin, and PDI mRNA (Fig. 1A), as well as protein levels (Fig. 1B). Ad5-cas9 control does not contain any gRNA and expresses only Cas9. β-actin and Cas9 were used as internal loading controls. Densitometric analysis showed statistically significant differences in myocilin, BiP, Calnexin, and PDI protein levels between Ad5-cas9– and Ad5-crMYOC–treated samples (Fig. 1C). We observed a decrease in myocilin-positive cells with Ad5-crMYOC treatment (marked by circles in Fig. 1D). Quantification of Cas9-positive and myocilin-negative cells showed a significant increase in Ad5-crMYOC–treated samples compared with Ad5-cas9–treated samples (Fig. 1E). Since Cas9 from Ad5-cas9 control virus is GFP-tagged, we found a more robust staining pattern compared with that of Cas9 from the Ad5-crMYOC virus, but both showed nuclear staining. Assuming that every transduced cell expresses Cas9, we found ∼60–70% transduction efficiency using Ad5-cas9 and/or Ad5-crMYOC by quantifying Cas9-GFP–expressing cells. ICC showed decreased BiP, Calnexin, and PDI expression with Ad5-crMYOC treatment (Fig. 1F). A GeneArt genomic cleavage assay (described in Materials and Methods) revealed a cleavage product in Ad5-crMYOC–treated DNA samples, but not in Ad5-cas9–treated DNA samples (Fig. 1G), indicating that our gRNA targeted the MYOC gene. The absence of cleavage products in predicted off-target sites (TRIM45, RAB36, PLA2G6, NCLN, and NEB) also demonstrates that the gRNA is specific to MYOC (Fig. 1H). DNA sequencing of treated cells revealed that the most common change observed with the MYOC-specific gRNA is a 7-bp deletion (3 bp upstream of PAM sequence) in MYOC (Fig. 1I).
Fig. 1.
MYOC disruption by CRISPR-Cas9 in vitro. (A) Ad5-crMYOC treatment of stably transfected NTM5 cell lines overexpressing a mutant (MT) form of human MYOC decreases myocilin (MYOC), BiP (HSP5), Calnexin (CANX), PDI (PDIA2), ATF4 (ATF4), and CHOP (DDIT3) mRNA expression (n = 5). (B) Representative Western blot showing that Ad5-crMYOC decreases mutant myocilin and associated BiP, Calnexin, and PDI (n = 5). Cas9 and β-actin served as loading controls. (C) Densitometry showing significant decreases in myocilin, BiP, Calnexin, and PDI. (D) ICC showing that Ad5-crMYOC treatment decreases myocilin (n = 5). Most of the Cas9 (green)-positive cells are MYOC (red)-negative (denoted by circles in the last panel). (Scale bar: 20 µm.) (E) Quantification of Cas9-positive but myocilin-negative cells showing a significant increase in numbers after Ad5-crMYOC treatment. (F) Ad5-crMYOC–mediated decrease in myocilin (red) and associated BiP, Calnexin, and PDI levels (green; ICC). (Scale bar: 100 µm.) (G) GeneArt Genomic Cleavage assay showing cleavage bands from the MYOC PCR product (arrows) in Ad5-crMYOC–treated DNA in all three cell lines: control (CT), WT, and mutant (MT). (H) GeneArt genomic cleavage assay showing no cleavage bands from predicated off-target sites in PLA2G6, NCLN, and NEB, confirming gRNA specificity for the MYOC gene. (I) Clonal sequencing of the PCR product from Ad5-crMYOC treated samples revealing the most common 7-bp (ATGTGGG) deletion. The error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, one-way ANOVA or paired Student’s t test.
CRISPR-Cas9 Lowers IOP and Prevents Further Glaucomatous Damage in Vivo.
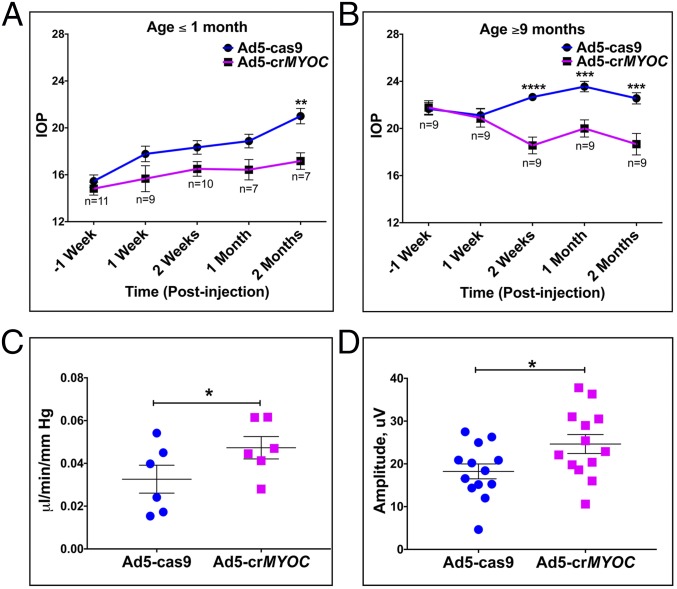
Our laboratory has previously developed and characterized a mouse model of POAG (Tg-MYOCY437H) due to expression of a human MYOC mutation (Y437H) in a human MYOC transgene (30). Tg-MYOCY437H mice exhibit elevated IOP, retinal ganglion cell (RGC) death and dysfunction, and optic nerve axonal degeneration, matching the phenotype seen in patients with POAG. We also showed that mutant myocilin is not secreted into the aqueous humor but rather accumulates inside TM cells, inducing ER stress in Tg-MYOCY437H mice. In the present study, we specifically targeted human mutant myocilin transgene with our Ad5-crMYOC virus injected intravitreally (2 × 107 pfu/eye). We treated a cohort of young Tg-MYOCY437H mice (age ≤1 mo) before the onset of elevated IOP with either Ad5-crMYOC or Ad5-cas9 (control) to determine whether this treatment could prevent IOP elevation. The results indicate that Ad5-crMYOC treatment prevents IOP elevation compared with Ad5-cas9–treated eyes, in which IOP increases with age (Fig. 2A). IOP values in Ad5-cas9 virus-injected eyes coincide with previously reported values (in a similar age group) in this model (30), suggesting that Ad5-cas9 does not resolve or worsen the phenotype and serves as a true control. We also injected a small cohort of 1- to 2-mo-old WT mice eyes with Ad5-cas9 and/or Ad5-crMYOC virus and did not observe any significant IOP changes. Mean nighttime IOP was 16 mm Hg and 15 mm Hg in 2-mo-old Ad5-cas9– and Ad5-crMYOC–injected eyes, respectively (n = 5). The mean nighttime IOP in untreated Tg-MYOCY437H mice (n = 4; ∼4 mo old) was 20 mm Hg, similar to values reported in previous studies (29, 30).
Fig. 2.
CRISPR-Cas9 lowers IOP and prevents further glaucomatous damage. (A) Ad5-crMYOC (intravitreal injections; 2 × 107 pfu/eye) prevents IOP elevation in ≤1-mo-old Tg-MYOCY437H mice (n = 7–11). (B) Ad5-crMYOC (intravitreal injections; 2 × 107 pfu/eye) lowers IOP in older (age ≥9 mo) Tg-MYOCY437H mice (n = 9). (C) Improved or preserved outflow facility in eyes of 4- to 5-mo-old Tg-MYOCY437H mice at 1 mo after Ad5-crMYOC treatment. The mean facility in these eyes is 0.0468, compared with 0.0309 μL/min/mmHg in Ad5-cas9–treated eyes (n = 7). (D) Improved retinal ganglion cell function measured by pERG in eyes of 4- to 5-mo-old Tg-MYOCY437H mice at 1 mo after Ad5-crMYOC treatment (mean amplitude, 24.644 μV vs. 18.239 μV in Ad5-cas9–treated eyes; n = 13). Error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, ANOVA with Bonferroni correction or paired Student’s t test.
Targeting myocilin with Ad5-crMYOC also lowered high IOP in a cohort of older Tg-MYOCY437H mice (age ≥9 mo) as early as 2 wk postinjection (Fig. 2B). The IOP in Ad5-crMYOC–treated eyes remained significantly lower out to 2 mo posttreatment (Fig. 3B). This suggests the presence of a pool of stressed TM cells that stay viable but poorly functional for long periods and, when corrected, have better proliferation ability and are able to repopulate the damaged TM. It is known that TM health and/or function is compromised in Tg-MYOCY437H mice, as shown by the reduced outflow facility in these mice (35). A significant increase in outflow facility at 1 mo after Ad5-crMYOC treatment in a separate cohort of 4- to 5-mo-old Tg-MYOCY437H mice (Fig. 2C) indicates that Ad5-crMYOC treatment improves TM cell function in vivo by disrupting mutant MYOC. The improved TM health and rescue in the IOP phenotype in Ad5-crMYOC–treated eyes correlates with the significantly improved RGC function at 1 mo after Ad5-crMYOC treatment, as measured by pattern electroretinography (pERG) in a cohort of 4- to 5-mo-old Tg-MYOCY437H mice with a mean amplitude of 24.644 μV vs. 18.239 μV in Ad5-cas9–treated eyes (n = 13) (Fig. 2D). Mean pERG values were 26.723 μV and 28.417 μV at 1 mo after Ad5-cas9 and Ad5-crMYOC injection, respectively, into eyes of 4- to 5-mo-old WT mice, a non-statistically significant difference (n = 2). Although there was mild self-limited anterior segment inflammation due to Ad5 injection (even with the Ad5-null vector) (36), this did not affect the iridocorneal angle (Fig. S2), IOP, or aqueous outflow facility.
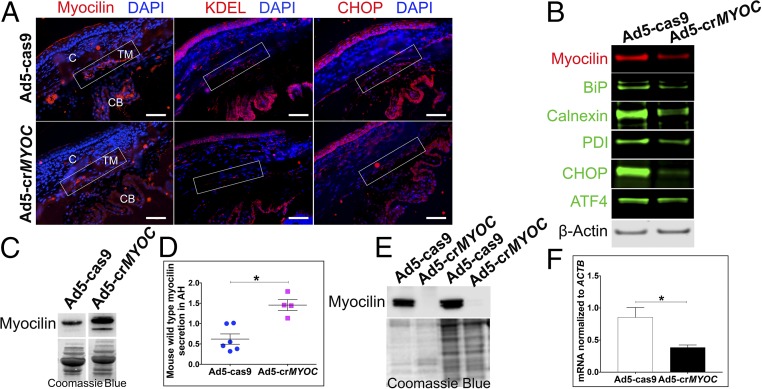
Fig. 3.
CRISPR efficiency in vivo in mice and ex vivo in human eyes. (A) Representative image showing that Ad5-crMYOC treatment reduces myocilin (red), KDEL (red), and CHOP (red) levels in the TM (rectangular box) of Tg-MYOCY437H mice (n = 3). C, cornea; CB, ciliary body. (Scale bar: 50 µm.) (B) Representative image of WB samples from a TM ring showing that Ad5-crMYOC treatment decreases myocilin levels and associated BiP, Calnexin, PDI, ATF4, and CHOP protein levels (n = 9). (C) Representative image showing increased secretion of mouse WT myocilin into the aqueous humor of Ad5-crMYOC–treated eyes compared with Ad5-cas9–treated eyes. (D) Densitometry showing a significant increase in the levels of secreted mouse WT myocilin in Ad5-crMYOC–treated eyes of Tg-MYOCY437H mice (n = 4). (E) inhibition of myocilin secretion by Ad5-crMYOC–mediated myocilin knockdown in a human ex vivo anterior segment perfusion culture (n = 2). Coomassie blue staining was performed to ensure relatively equal loading. (F) Decreased myocilin (MYOC) mRNA expression in human ex vivo anterior segment perfusion culture by Ad5-crMYOC treatment. Error bars represent SEM. *P < 0.05.
Fig. S2.
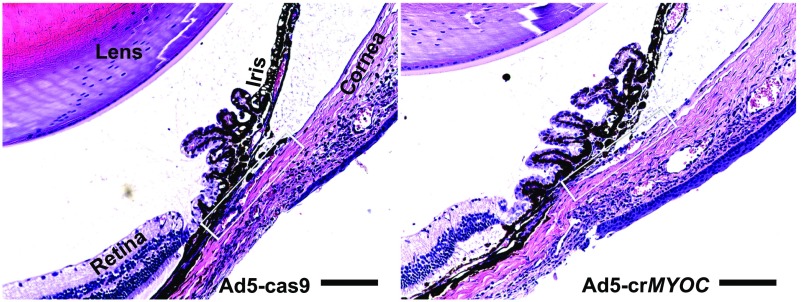
H&E staining of anterior segment tissues of Ad5-crMYOC–injected mice. H&E staining was used to examine inflammation and gross abnormalities in the anterior segment due to adenovirus 5 (Ad) expression of Cas9. The left eyes of Tg-MYOCY437H mice were injected intravitreally with Ad5-cas9, and the contralateral eyes were injected with Ad5-crMYOC (n = 2 mice). Eyes were enucleated at 1 mo postinjection, and paraffin-embedded anterior segment sections were stained with H&E. The H&E staining reveals an open iridocorneal angle, with no ocular abnormalities associated with Ad5-cas9 or Ad5-crMYOC. We have previously shown that Ad5 injections cause mild inflammation in the anterior segment (36). (Left) Mild inflammation was associated with control Ad5-cas9 injection, as evidenced by a few inflammatory cells. (Right) Interestingly, Ad5-crMYOC produced less inflammation in the anterior segment tissues (n = 2). The rectangular box indicates the TM. (Scale bar: 100 µm.)
CRISPR-Cas9 Efficiency in Vivo and ex Vivo.
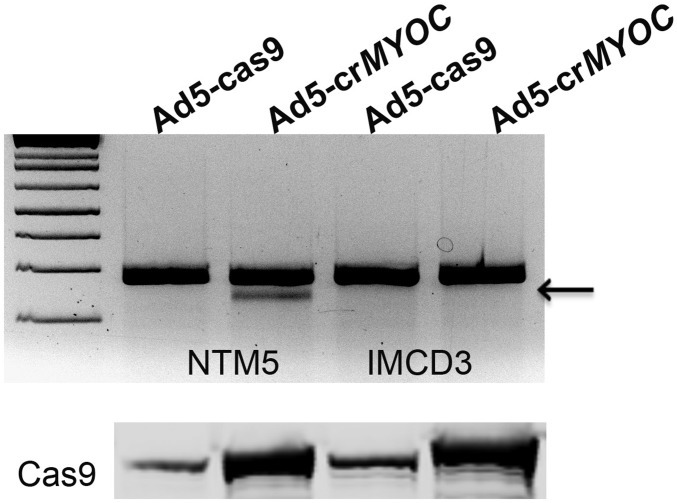
Having shown that our CRISPR gRNA (for MYOC) is both efficient and selective in vitro (Fig. 1), we then tested the efficiency of the same Ad5-crMYOC in vivo. Eyes from 4- to 5-mo-old mice were fixed for myocilin and ER stress marker immunolabeling, or protein was isolated from TM tissue (with some iris, corneal endothelium, and sclera, referred to as the “TM ring”) from the eyes at 1 mo after Ad5-cas9 or Ad5-crMYOC treatment to test CRISPR efficiency in vivo. TM tissue sections from Ad5-crMYOC–treated eyes showed reduced myocilin, KDEL, and CHOP labeling, indicating that the gRNA is targeting MYOC in vivo (Fig. 3A). The residual myocilin labeling (red) could be endogenous mouse myocilin that is not targeted by our gRNA. By quantifying the number of corrected TM cells in vivo, we found an estimated transduction efficiency of at least ∼60–70% using Ad5-cas9 and/or Ad5-crMYOC. Of note, we also found Cas9 expression in the cornea endothelium and parts of the iris and ciliary body, similar to previous studies with adenovirus transduction (36–38). Western blot analysis data also showed reductions in myocilin, BiP, Calnexin, PDI, ATF4, and CHOP levels, confirming our ICC data indicating gRNA targeting in Tg-MYOCY437H mice (Fig. 3B). Previous work has shown that mutant myocilin is able to form homodimers and tetramers as well as heterodimers with WT myocilin (23). This prevents the normal secretion of the WT isoform, initiates UPR, and causes glaucoma. Our Ad5-crMYOC treatment specifically targets human mutant MYOC in Tg-MYOCY437H mice. The decrease in mutant human protein in TM tissue lysate correlated with the increased secretion of mouse WT myocilin in aqueous humor samples of Ad5-crMYOC–treated eyes (Fig. 3C). Testing this exclusively was difficult owing to a lack of specific anti-mouse myocilin antibody. The light myocilin band in the Ad5-cas9 transgenic mice is likely due to a small amount of secreted WT mouse myocilin. Quantification of secreted protein showed significantly increased levels of WT mouse myocilin in the aqueous humor of Ad5-crMYOC–treated eyes (Fig. 3D). These findings are also supported by the fact that our gRNA does not target mouse WT Myoc in the mouse kidney cell line IMCD3 and is highly specific in targeting the human MYOC human trabecular meshwork cell line NTM5 (Fig. S3).
Fig. S3.
Ad5-crMYOC specifically targets human MYOC. The GeneArt genomic cleavage assay shows a cleavage band (from MYOC qPCR product; arrow) in Ad5-crMYOC–treated DNA in all NTM5 cell lines, but not in mouse kidney cell line IMCD3, confirming the gRNA specificity for the human MYOC gene. Cas9 expression appears similar in both Ad5-crMYOC–treated NTM5 and IMCD3 cells.
We further assessed the feasibility of using genome editing to treat glaucoma in human TM tissues using a human ex vivo anterior segment perfusion culture system (Materials and Methods). Ad5-crMYOC reduced myocilin mRNA (Fig. 3F) and myocilin secretion into the perfusate medium (medium in which eyes were cultured, collected at the end of the experiment) obtained from the TM outflow pathway compared with the contralateral fellow eye treated with Ad5-cas9 (Fig. 3E), further supporting the ex vivo efficiency of Ad5-crMYOC in perfusion-cultured human eyes.
Discussion
Although it has been almost 20 y since the discovery of MYOC as the first glaucoma gene (7), the physiological role of the normal MYOC protein remains unknown. The fact that most MYOC mutations result in early-onset glaucoma has a much greater impact on quality of life than typical late-onset POAG. The lack of a discernable phenotype in Myoc−/− mice and humans with MYOC deletions suggests that MYOC mutations are gain-of-negative function mutations (16, 17, 19, 20). We previously reported the development of a mouse model (Tg-MYOCY437H) that replicates the POAG phenotype (30). The Tg-MYOCY437H mouse has contributed to our understanding of the in vivo pathophysiology of myocilin-associated glaucoma. Unlike WT, mutant myocilin accumulates inside TM cells, causing ER stress. This chronic ER stress affects the normal function and/or survival of TM cells, resulting in increased aqueous humor outflow resistance and leading to elevated IOP.
We have previously shown that a chemical chaperone can be used to treat Tg-MYOCY437H mice (30). Administration of phenylbutyric acid (PBA; 20 mM) in drinking water for 5 wk prevents IOP elevation and also reduces IOP in Tg-MYOCY437H mice. Although PBA is effective in the MYOC mouse model and has Food and Drug Administration approval as an oral agent for urea cycle disorders, the continuous production of mutant myocilin requires chronic administration of PBA to relieve glaucoma symptoms. In the present study, we sought a more permanent solution for treatment by using genome editing. Given the dominant nature of MYOC mutations, we used the CRISPR-Cas9 system to disrupt the MYOC gene both in vitro and in vivo in mice. Our construct also works well in cultured human TM cells and skin fibroblasts in vitro, as well as in human ex vivo perfusion-cultured eyes, suggesting a feasible translational application of this technology to humans. Our data show a genome editing phenotypic response similar to that seen with the chronic use of PBA in young Tg-MYOCY437H mice (22, 23).
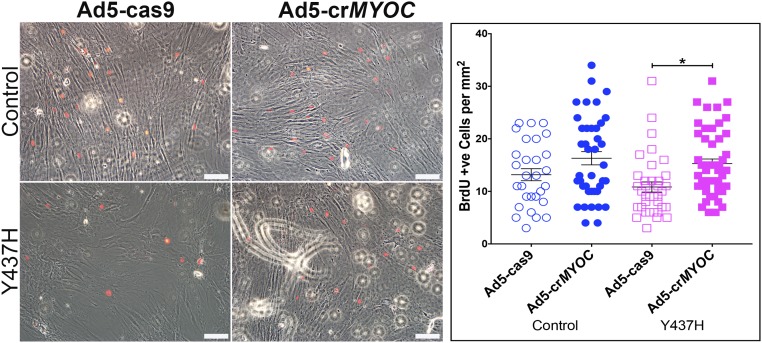
Our findings indicate that genome editing can prevent IOP elevation and the resulting development of glaucoma, and also effectively lower IOP in mutant MYOC ocular hypertensive mice. Using a gRNA specifically directed against the human MYOC gene, we were able to disrupt the gain of function of mutant myocilin, lower the misfolded protein load in TM cells, and rescue ER stress in vitro. Using the same gRNA and CRISPR-Cas9 assembly in vivo, we were able to prevent the development of glaucoma in young Tg-MYOCY437H mice before the onset of symptoms (i.e., elevated IOP). In addition, we were able to use genome editing to treat glaucoma in older Tg-MYOCY437H mice that had elevated IOP for 9 mo before treatment. Although we would expect genome editing to permanently eliminate the expression of the mutant myocilin protein, our present data do not address whether the genome-edited TM cells persist indefinitely. The reduction of elevated IOP in older animals indicates that TM cells tolerate ER stress without dying for a substantial period, potentially providing a broad therapeutic time frame. It is likely that a pool of stressed TM cells, when corrected, have better proliferation ability and are able to repopulate the damaged TM. This idea is supported by the increased proliferation in human skin fibroblasts from patients with myocilin-associated glaucoma after genome editing (Fig. S4). Ad5-crMYOC–mediated disruption of MYOC in these fibroblasts improved their proliferative ability, as demonstrated by increased incorporation of 5-bromo-2′-deoxyuridine (BrdU).
Fig. S4.
BrdU incorporation in skin fibroblasts. Skin fibroblasts from a control group (n = 3 cell strains) show inclusion of BrdU (red) in nuclei. There are relatively fewer BrdU-positive cells in fibroblasts from myocilin (Y437H) glaucoma patients (n = 3 cell strains). Quantification of BrdU- positive cells (per mm2 in ∼12 images per coverslip) reveals that Ad5-crMYOC treatment significantly increases the number of BrdU-positive cells in the Y437H group, suggesting better proliferation after mutant MYOC gene disruption. (Scale bar: 100 µm.) *P < 0.05.
It would have been nice to quantify changes in TM cellularity in these rescued mice after Ad5-crMYOC treatment, but the lack of an organized TM structure as well as a TM cell-specific marker in mice makes this quantification difficult and nonspecific, although we would expect to see an increase in viable and functional TM cells after disruption of the mutant transgene. While we were able to preserve RGC function in younger mice, the reduction of IOP in older mice might not allow significant RGC recovery owing to the high sensitivity to pressure variation and irreversible RGC loss. Nonetheless, lowering IOP should halt any further loss of RGCs, as is achieved by successful drug therapy in human patients.
It should be noted that in this study, we targeted the first exon, a target that should be equally effective for numerous different MYOC mutations. We also could have targeted the promoter or, alternatively, attempted to use homologous recombination to correct specific mutations in a given patient. However, the use of an identical gRNA in patients with different disease-causing variants may help overcome one potential problem with genome editing, the off-target effects. One concern associated with the use of NHEJ-based gene disruption is the possible introduction of additional mutations that might lead to longer or shorter protein isoforms; however, we did not observe other myocilin protein isoforms after Ad5-crMYOC treatment. Importantly, we have demonstrated the feasibility of CRISPR-Cas9 genome editing in a human organ. We successfully targeted human MYOC in ex vivo perfusion-cultured human eyes and reduced the secreted myocilin protein load. Such ex vivo systems also may be used to explore and eliminate potential off-target genome alterations, as well as to further explore Cas9 modifications with greater on-target efficiency.
Previously reported in vivo uses of CRISPR-Cas9 include correction of a dominant loss-of-function cataract-causing mutation in mouse embryos (39), HDR in a mouse model of type I tyrosinemina (40), correction of retinal dystrophy in a rat model of autosomal dominant retinitis pigmentosa (41), NHEJ-mediated gene disruption of PCSK9 in the mouse liver (42), and intrahepatic hepatitis B virus template clearance (43). Our present study demonstrates the potential of CRISPR-mediated genome editing in human myocilin-associated POAG, as well as in other human disorders resulting from gain-of-function mutations.
Materials and Methods
TM Cell Culture.
Human TM cells were isolated from carefully dissected human TM tissue explants derived from patients with glaucoma or from normal donors and characterized as described previously (44). All donor tissues were obtained from regional eye banks after receipt of informed consent from the donors’ families and were managed according to the guidelines in the Declaration of Helsinki on research involving human tissue. Isolated TM cells were grown in DMEM (Invitrogen) containing 0.292 mg/mL l-glutamine (Life Technologies), 100 U/mL penicillin-0.1 mg/mL streptomycin (Life Technologies), and 10% FBS (Life Technologies). The stably transformed human TM cell line NTM5 (45) was also used and cultured in the same medium.
Generation and Characterization of Stable Cell Lines.
NTM5 cell lines were transfected with pDsRedN1-MYOC (WT or Y437H) using LyoVec (InvivoGen). These plasmids express DsRed at the C terminus of myocilin protein. Once confluent, the cells were treated with 1.5 µg/mL puromycin (Thermo Fisher Scientific), and at 3–4 wk posttreatment, individual colonies were picked and expanded. Colonies were characterized using Western blot analysis and Sanger sequencing; colonies with the highest myocilin expression were used for subsequent experiments. Nontransfected naïve NTM5 cells served as a control.
Design of Human MYOC-Specific gRNA.
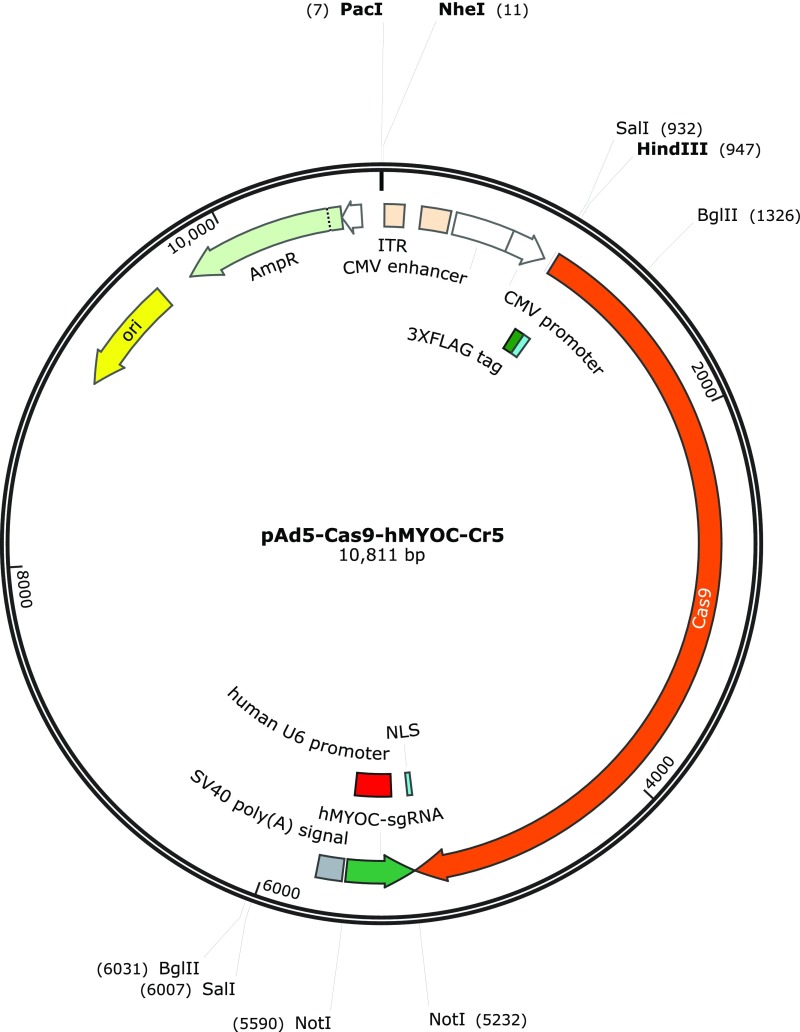
The human MYOC cDNA sequence at ≈300 bp from the start codon was loaded onto the Genome-Engineering website (tools.genome-engineering.org). (Note: We did not target the MYOC genome sequence, because human MYOC cDNA was the transgene inserted into Tg-MYOCY437H mice.) Several of the most suitable gRNAs, based on computationally predicted off-target sites for each intended target, were selected, cloned into px330 (with spCas9) vectors, and tested for efficiency and selectivity for human MYOC over mouse Myoc. gRNA (GGCCTGCCTGGTGTGGGATG) with the greatest efficiency in disrupting MYOC was cloned into a shuttle vector and subsequently into Ad5 vectors for use in our study. Ad5-cas9 control virus was purchased from Vector Biolabs (catalog no. 1901). Ad5-cas9 control virus does not contain any gRNA and expresses only GFP-tagged Cas9. Maps of the shuttle vectors used to generate these viruses are provided in Figs. S5 and S6.
Fig. S5.
Map of the shuttle vector used to generate the Ad5-cas9 virus.
Fig. S6.
Map of the shuttle vector used to generate the Ad5-crMYOC virus.
TM Cell Treatment.
Primary human TM cells were grown to 100% confluence in serum-containing medium. TM cells were incubated with medium containing either Ad5-cas9 (tagged with GFP) or Ad5-crMYOC for 24 h, followed by incubation in fresh medium for another 24 h.
Mouse Husbandry.
Tg-MYOCY437H mice (on a mixed C57BL/6 and A/J background) were housed and bred at the University of Iowa’s animal research facility. The mice were maintained on a 4%-fat NIH 31 diet provided ad libitum and housed in cages containing dry bedding (Soft-Zorb Enrichment Blend; Northeastern Products). A temperature of 21 °C and a 12-h light/12-h dark cycle were maintained. All animal procedures in this study were performed in compliance with the Association for Research in Vision and Ophthalmology’s Statement for the Use of Animals in Ophthalmic and Vision Research and were approved by both the University of Iowa’s Animal Care and Use Committee and the University of North Texas Health Science Center’s Institutional Animal Care and Use Committee. Power calculations were performed to determine the number of mice needed for each experiment.
Intravitreal Injections.
Mice were anesthetized with a mouse anesthesia mixture (ketamine + xylazine). One eye was injected with Ad5-crMYOC (2 × 107 pfu/eye in a volume of 2 μL of PBS) or Ad5-cas9 through the sclera into the vitreous chamber using a Hamilton microsyringe fitted with a sterile 33-gauge needle. The contralateral eye served as a control. The few mice that developed eye abnormalities were excluded from further study.
IOP Measurements.
IOP was measured with a TonoLab rebound tonometer (Colonial Medical Supply) in a masked manner (i.e., the researcher taking the reading was unaware of the type of treatment until the very end of the experiment) in two independent cohorts of mice at the University of North Texas Health Science Center’s and University of Iowa’s animal research facilities. Mice were anesthetized using 2.5% isoflurane plus 100% oxygen. Daytime IOP was measured between 8 and 10 AM, and nighttime IOP was measured in the dark between 9 and 11 AM. The data shown here are for nighttime IOPs.
Outflow Facility Measurements.
Aqueous humor outflow facility was measured after 1 mo of intravitreal virus injections in both eyes of each animal (age 4–5 mo) by constant flow infusion as described previously (46). In brief, anesthetized eyes were infused at a flow rate of 0.1 μL/min. Once the pressure stabilized, pressure measurements were recorded, and flow rate was increased sequentially to 0.2, 0.3, 0.4, and 0.5 μL/min. Three stabilized pressures (spaced 5 min apart) at each flow rate were recorded. Mean stabilized pressures were plotted with flow rates, and outflow facility in each eye was calculated as the reciprocal of the slope of the resulting line.
pERGs.
After 1 mo of intravitreal virus injections, retinal ganglion cell functional analysis was performed using a pERG system (Intelligent Hearing Systems) that records in both eyes simultaneously using snout electrodes as described previously (47). In brief, 4- to 5-mo-old mice were anesthetized (ketamine 100 mg/kg; xylazine 10 mg/kg i.p.), placed on a holder to allow unobstructed vision, and then moved at a fixed distance (10 cm) from the LED monitors. A constant body temperature of 37 °C was maintained using a rectal probe and feedback-controlled heating pad. A small amount of sterile PBS was applied topically to prevent corneal dryness. Visual stimuli consisted of contrast-reversing bars generated on two LED screens. pERG readings from each eye were generated simultaneously using an s.c. stainless steel needle placed in the snout. The reference and ground electrodes were placed medially on the back of the head and at the root of the tail, respectively. pERG waveforms were generated automatically, and average amplitude was represented graphically using GraphPad Prism.
Perfusion Anterior Segment Culture of Human Eyes.
Human donor eyes were obtained from the Lions Eye Institute for Transplantation and Research (Tampa, FL) after receipt of informed consent from the donors’ families. All donor tissues were managed in accordance with Declaration of Helsinki guidelines for research involving human tissue. The anterior segments of paired donor eyes were prepared for ex vivo perfusion culture as described previously (48, 49). Both Ad5-cas9 and Ad5-crMYOC (1010 pfu) were introduced as a bolus into each eye, followed by perfusion at a constant flow rate of 2.5 μL/min. Perfusate medium was collected over 4 d of culture and then subjected to Western blot analysis for myocilin expression. The SDS-polyacrylamide gels were also stained with Coomassie blue for total proteins to ensure equal protein loading. TM tissues were isolated, and DNA/RNA was extracted using the Qiagen AllPrep Kit for further analysis.
RNA Isolation and QRT-PCR.
Total RNA was collected using an RNA isolation kit (IBI Scientific), and 2 µg of RNA was converted into cDNA using iScript (Bio-Rad). cDNA samples were diluted in iQ SYBR Green Supermix (Bio-Rad) with the relevant PCR primers (Table S1) and run on a Bio-Rad CFX96 qPCR system. Data were analyzed with the system software.
Table S1.
PCR primers
| Gene name | Forward primer | Reverse primer |
| MYOC | ACCTCCTCCGAGACAAGTCA | TAGTTCTCCACATCCGGTGTCT |
| HSPA5 | GAACGTCTGATTGGCGATGC | TCAACCACCTTGAACGGCAA |
| CANX | CGCGGGGCAAGATCATGGA | TCCACCCTGACAGAGTTCCT |
| PDIA2 | CCAGGTCGGAAGGTGATTGA | GACCCCATAGTGGAGTTGGC |
| ATF4 | ATGGGTTCTCCAGCGACAAG | GAAGGCATCCTCCTTGCTGT |
| DDIT3 | ACCTCCTGGAAATGAAGAGGAAG | CAGTCAGCCAAGCCAGAGAA |
| ACTB | GCCGCCAGCTCACCAT | AATCCTTCTGACCCATGCCC |
Western Blot Analysis.
Cells were lysed with 1% Triton X-100 (containing protease inhibitors). For in vivo CRISPR efficiency determination studies, anterior segment or iridocorneal angle tissues were carefully dissected and lysed in tandom affinity purification lysis buffer. TM tissues were carefully dissected from perfusion cultures human anterior segments in the ex vivo model. In addition, aqueous humor (2–3 μL) was collected from each eye using a pulled glass capillary and mixed with 20 µL of 1× sodium dodecyl sulfate-PAGE buffer with reducing agent. Proteins were separated on denaturing polyacrylamide gels and then transferred to a nitrocellulose membrane by electrophoresis. Dried blots were blocked in Odyssey Blocking Buffer (LI-COR Biosciences), followed by overnight incubation with specific primary antibodies (Table S2). The membranes were washed with PBS/Tween buffer (PBST) and then incubated with corresponding IRdye secondary antibodies (LI-COR Biosciences). The proteins were then visualized using the Odyssey CLx Infrared Imaging System (LI-COR Biosciences), and quantitation was performed with ImageStudio software (LI-COR Biosciences).
Table S2.
Antibodies for Western blot analysis and immunofluorescence microscopy
| Protein name | Source | Dilution |
| Myocilin | sc-21243 (Santa Cruz Biotechnology) | WB: 1:500 |
| ICC: 1:200 | ||
| BiP | ab108615 (Abcam) | WB: 1:1,000 |
| ICC: 1:500 | ||
| Calnexin | ab133615 (Abcam) | WB: 1:1,000 |
| ICC: 1:500 | ||
| PDI | 2446 (Cell Signaling Technology) | WB: 1:1,000 |
| ICC: 1:500 | ||
| CHOP | NB600-1335 (Novus Biologicals) | WB: 1:1,000 |
| ICC: 1:500 | ||
| ATF4 | 11815 (Cell Signaling Technology) | WB: 1:1,000 |
| KDEL | ab12223 (Abcam) | IHC: 1:500 |
| Cas9 | 7A9-3A3 (Cell Signaling Technology) | WB: 1:2,000 |
| ICC: 1:500 | ||
| β-actin | 926–42212 (LI-COR Biosciences) | WB: 1:5,000 |
WB, Western blot.
Immunofluorescence Microscopy.
TM cells were grown on coverslips in 24-well plates. At 48 hours after Ad5-crMYOC treatment, the cells were fixed, permeabilized, and blocked with SuperBlock (Thermo Fisher Scientific). Alternatively, enucleated mouse eyes were fixed in 4% formaldehyde and embedded in cryoembedding medium, and 10-μm sections were cut using a Leica CM3050S cryostat. The sections were then blocked with 5% normal goat serum. Slides/coverslips were incubated overnight with the appropriate primary antibodies (Table S2) and then washed with PBS, followed by a 1-h incubation with the appropriate Alexa Fluor secondary antibodies. Nuclei were counterstained with Vectashield containing DAPI (Vector Laboratories). Images were captured with a Leica TCS SP8 confocal imaging system.
GeneArt Genomic Cleavage Assay.
Genomic DNA was extracted from cells treated with Ad5-crMYOC or Ad5-cas9. Loci in which the gene-specific double-strand breaks occurred were amplified by PCR (Table S3). The PCR product was denatured and reannealed so that mismatches were generated as strands with an indel reannealed to strands with no indel or a different indel. The mismatches were subsequently detected and cleaved by detection enzymes (Thermo Fisher Scientific), and the resulting bands were analyzed by gel electrophoresis.
Table S3.
Primers for the GeneArt genomic cleavage assay
| Gene name | Forward primer | Reverse primer |
| MYOC | GTTCTTCTGTGCACGTTGC | CTGGTCCAAGGTCAATTGGT |
| Myoc | ATGCCAGCTCTCCATCTGCT | TCGGCCCAAGGTCATCTGGT |
| TRIM45 | GAGAGCTAGACCTGGAAGCG | TCCCAAGTGCAGTCCCACTA |
| RAB36 | TCGCCCCTCTGGACTCACA | AGCCTCGTGTGTGTGTTTCATT |
| PLA2G6 | GAGTGTGCTGGTTGGGTTTG | GCTGACTTCATCACATCGCC |
| NCLN | TTTGGCCACCAGTTCTTCGG | GGAGGAACATGAACCCCCAA |
| NEB | CTACACTCTCCTGCCCGATG | AGCTGGGAGGGGGAGAATAAT |
| NEB | CTACACTCTCCTGCCCGATG | AGCTGGGAGGGGGAGAATAAT |
BrdU Assay.
All experiments were conducted with the approval and supervision of the University of Iowa’s Internal Review Board (application no. 200202022) in accordance with the Declaration of Helsinki. After receipt of informed consent, skin biopsy specimens were collected from patients for use in the generation of fibroblasts as described previously (50–52). Skin fibroblasts from patients with myocilin-associated glaucoma and age-matched controls (grown on coverslips in 24-well plates) were subjected to Ad5-cas9 or Ad5-crMYOC treatment. At 48 h after treatment, BrdU (0.03 μg/mL) was added to the medium for 1–2 h. Cells were fixed, permeabilized with 1 N HCL, blocked with 0.1% serum albumin, and incubated with anti-BrdU primary antibody (ab6326; Abcam) at 4 °C overnight. After three washes with PBS, Alexa Fluor 568 secondary antibody was added, followed by incubation for 1 h at room temperature. Coverslips were mounted using Vectashield containing DAPI (Vector Laboratories). Images were captured using a Leica TCS SP8 confocal imaging system.
Hematoxylin and Eosin Staining.
Formalin-fixed enucleated mouse eyes were washed in PBS, dehydrated with ethanol, and embedded in paraffin. The eyes were subsequently sectioned at 5 μm thickness and then stained with hematoxylin and eosin (H&E).
Statistics.
All data are presented as mean ± SEM. Comparisons between two groups were conducted using the unpaired one-tailed Student’s t test, and comparisons among three or more groups were done using one-way ANOVA with Bonferroni correction. A P value <0.05 was considered significant.
Study Approval.
All animal procedures performed in this study were conducted in compliance with the Association for Research in Vision and Ophthalmology’s Statement for the Use of Animals in Ophthalmic and Vision Research and were approved by the University of Iowa’s Animal Care and Use Committee. All experiments involving human skin biopsy specimens were conducted with the approval of and under the supervision of the University of Iowa’s Internal Review Board (application no. 200202022) and were consistent with the principles specified in the Declaration of Helsinki.
Acknowledgments
This project was supported by National Institutes of Health Grants R01 EY024259, R01 EY026177, and R00 EY022077, funding from the Howard Hughes Medical Institute, and from the Roy J. Carver Charitable Trust. Adenoviral vectors were obtained from the Gene Transfer Vector Core at the University of Iowa.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.J.Z. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706193114/-/DCSupplemental.
References
- 1.Hollands H, et al. Do findings on routine examination identify patients at risk for primary open-angle glaucoma? The rational clinical examination systematic review. JAMA. 2013;309:2035–2042. doi: 10.1001/jama.2013.5099. [DOI] [PubMed] [Google Scholar]
- 2.Weinreb RN, Aung T, Medeiros FA. The pathophysiology and treatment of glaucoma: A review. JAMA. 2014;311:1901–1911. doi: 10.1001/jama.2014.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alward WL, et al. Clinical features associated with mutations in the chromosome 1 open-angle glaucoma gene (GLC1A) N Engl J Med. 1998;338:1022–1027. doi: 10.1056/NEJM199804093381503. [DOI] [PubMed] [Google Scholar]
- 4.Meyer A, et al. [Linkage between juvenile glaucoma and chromosome 1q in 2 French families] C R Acad Sci III. 1994;317:565–570. French. [PubMed] [Google Scholar]
- 5.Richards JE, et al. Mapping of a gene for autosomal dominant juvenile-onset open-angle glaucoma to chromosome Iq. Am J Hum Genet. 1994;54:62–70. [PMC free article] [PubMed] [Google Scholar]
- 6.Sheffield VC, et al. Genetic linkage of familial open-angle glaucoma to chromosome 1q21-q31. Nat Genet. 1993;4:47–50. doi: 10.1038/ng0593-47. [DOI] [PubMed] [Google Scholar]
- 7.Stone EM, et al. Identification of a gene that causes primary open-angle glaucoma. Science. 1997;275:668–670. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- 8.Wiggs JL, et al. Genetic linkage of autosomal dominant juvenile glaucoma to 1q21-q31 in three affected pedigrees. Genomics. 1994;21:299–303. doi: 10.1006/geno.1994.1269. [DOI] [PubMed] [Google Scholar]
- 9.Fingert JH, Stone EM, Sheffield VC, Alward WL. Myocilin glaucoma. Surv Ophthalmol. 2002;47:547–561. doi: 10.1016/s0039-6257(02)00353-3. [DOI] [PubMed] [Google Scholar]
- 10.Karali A, Russell P, Stefani FH, Tamm ER. Localization of myocilin/trabecular meshwork–inducible glucocorticoid response protein in the human eye. Invest Ophthalmol Vis Sci. 2000;41:729–740. [PubMed] [Google Scholar]
- 11.Kubota R, et al. A novel myosin-like protein (myocilin) expressed in the connecting cilium of the photoreceptor: Molecular cloning, tissue expression, and chromosomal mapping. Genomics. 1997;41:360–369. doi: 10.1006/geno.1997.4682. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi H, et al. Mouse myocilin (Myoc) gene expression in ocular tissues. Biochem Biophys Res Commun. 1998;248:104–109. doi: 10.1006/bbrc.1998.8917. [DOI] [PubMed] [Google Scholar]
- 13.Abderrahim H, Jaramillo-Babb VL, Zhou Z, Vollrath D. Characterization of the murine TIGR/myocilin gene. Mamm Genome. 1998;9:673–675. doi: 10.1007/s003359900844. [DOI] [PubMed] [Google Scholar]
- 14.Swiderski RE, et al. Expression pattern and in situ localization of the mouse homologue of the human MYOC (GLC1A) gene in adult brain. Brain Res Mol Brain Res. 1999;68:64–72. doi: 10.1016/s0169-328x(99)00085-6. [DOI] [PubMed] [Google Scholar]
- 15.Tomarev SI, Tamm ER, Chang B. Characterization of the mouse Myoc/Tigr gene. Biochem Biophys Res Commun. 1998;245:887–893. doi: 10.1006/bbrc.1998.8541. [DOI] [PubMed] [Google Scholar]
- 16.Kim BS, et al. Targeted disruption of the myocilin gene (Myoc) suggests that human glaucoma-causing mutations are gain of function. Mol Cell Biol. 2001;21:7707–7713. doi: 10.1128/MCB.21.22.7707-7713.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morissette J, et al. Homozygotes carrying an autosomal dominant TIGR mutation do not manifest glaucoma. Nat Genet. 1998;19:319–321. doi: 10.1038/1203. [DOI] [PubMed] [Google Scholar]
- 18.Pang CP, et al. TIGR/MYOC gene sequence alterations in individuals with and without primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2002;43:3231–3235. [PubMed] [Google Scholar]
- 19.Wiggs JL, Vollrath D. Molecular and clinical evaluation of a patient hemizygous for TIGR/MYOC. Arch Ophthalmol. 2001;119:1674–1678. doi: 10.1001/archopht.119.11.1674. [DOI] [PubMed] [Google Scholar]
- 20.Gould DB, et al. Genetically increasing Myoc expression supports a necessary pathologic role of abnormal proteins in glaucoma. Mol Cell Biol. 2004;24:9019–9025. doi: 10.1128/MCB.24.20.9019-9025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carbone MA, et al. Overexpression of myocilin in the Drosophila eye activates the unfolded protein response: Implications for glaucoma. PLoS One. 2009;4:e4216. doi: 10.1371/journal.pone.0004216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joe MK, et al. Accumulation of mutant myocilins in ER leads to ER stress and potential cytotoxicity in human trabecular meshwork cells. Biochem Biophys Res Commun. 2003;312:592–600. doi: 10.1016/j.bbrc.2003.10.162. [DOI] [PubMed] [Google Scholar]
- 23.Jacobson N, et al. Non-secretion of mutant proteins of the glaucoma gene myocilin in cultured trabecular meshwork cells and in aqueous humor. Hum Mol Genet. 2001;10:117–125. doi: 10.1093/hmg/10.2.117. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida H. ER stress and diseases. FEBS J. 2007;274:630–658. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Vollrath D. Reversal of mutant myocilin non-secretion and cell killing: Implications for glaucoma. Hum Mol Genet. 2004;13:1193–1204. doi: 10.1093/hmg/ddh128. [DOI] [PubMed] [Google Scholar]
- 26.Yam GH, Gaplovska-Kysela K, Zuber C, Roth J. Aggregated myocilin induces Russell bodies and causes apoptosis: Implications for the pathogenesis of myocilin-caused primary open-angle glaucoma. Am J Pathol. 2007;170:100–109. doi: 10.2353/ajpath.2007.060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burns JN, et al. Rescue of glaucoma-causing mutant myocilin thermal stability by chemical chaperones. ACS Chem Biol. 2010;5:477–487. doi: 10.1021/cb900282e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yam GH, Gaplovska-Kysela K, Zuber C, Roth J. Sodium 4-phenylbutyrate acts as a chemical chaperone on misfolded myocilin to rescue cells from endoplasmic reticulum stress and apoptosis. Invest Ophthalmol Vis Sci. 2007;48:1683–1690. doi: 10.1167/iovs.06-0943. [DOI] [PubMed] [Google Scholar]
- 29.Zode GS, et al. Topical ocular sodium 4-phenylbutyrate rescues glaucoma in a myocilin mouse model of primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2012;53:1557–1565. doi: 10.1167/iovs.11-8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zode GS, et al. Reduction of ER stress via a chemical chaperone prevents disease phenotypes in a mouse model of primary open-angle glaucoma. J Clin Invest. 2011;121:3542–3553. doi: 10.1172/JCI58183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan C, Zins EJ, Clark AF, Huang AJ. Suppression of keratoepithelin and myocilin by small interfering RNAs (siRNA) in vitro. Mol Vis. 2007;13:2083–2095. [PubMed] [Google Scholar]
- 32.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark AF, et al. Glucocorticoid induction of the glaucoma gene MYOC in human and monkey trabecular meshwork cells and tissues. Invest Ophthalmol Vis Sci. 2001;42:1769–1780. [PubMed] [Google Scholar]
- 34.Shepard AR, et al. Characterization of rabbit myocilin: Implications for human myocilin glycosylation and signal peptide usage. BMC Genet. 2003;4:5. doi: 10.1186/1471-2156-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kasetti RB, Phan TN, Millar JC, Zode GS. Expression of mutant myocilin induces abnormal intracellular accumulation of selected extracellular matrix proteins in the trabecular meshwork. Invest Ophthalmol Vis Sci. 2016;57:6058–6069. doi: 10.1167/iovs.16-19610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pang IH, Millar JC, Clark AF. Elevation of intraocular pressure in rodents using viral vectors targeting the trabecular meshwork. Exp Eye Res. 2015;141:33–41. doi: 10.1016/j.exer.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueyama K, et al. Ocular localization and transduction by adenoviral vectors are serotype-dependent and can be modified by inclusion of RGD fiber modifications. PLoS One. 2014;9:e108071. doi: 10.1371/journal.pone.0108071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Millar JC, Pang IH, Wang WH, Wang Y, Clark AF. Effect of immunomodulation with anti-CD40L antibody on adenoviral-mediated transgene expression in mouse anterior segment. Mol Vis. 2008;14:10–19. [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Y, et al. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013;13:659–662. doi: 10.1016/j.stem.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 40.Yin H, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bakondi B, et al. In vivo CRISPR/Cas9 gene editing corrects retinal dystrophy in the S334ter-3 rat model of autosomal dominant retinitis pigmentosa. Mol Ther. 2016;24:556–563. doi: 10.1038/mt.2015.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding Q, et al. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin SR, et al. The CRISPR/Cas9 system facilitates clearance of the intrahepatic HBV templates in vivo. Mol Ther Nucleic Acids. 2014;3:e186. doi: 10.1038/mtna.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stamer WD, Clark AF. The many faces of the trabecular meshwork cell. Exp Eye Res. 2017;158:112–123. doi: 10.1016/j.exer.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pang IH, Shade DL, Clark AF, Steely HT, DeSantis L. Preliminary characterization of a transformed cell strain derived from human trabecular meshwork. Curr Eye Res. 1994;13:51–63. doi: 10.3109/02713689409042398. [DOI] [PubMed] [Google Scholar]
- 46.Millar JC, Clark AF, Pang IH. Assessment of aqueous humor dynamics in the mouse by a novel method of constant-flow infusion. Invest Ophthalmol Vis Sci. 2011;52:685–694. doi: 10.1167/iovs.10-6069. [DOI] [PubMed] [Google Scholar]
- 47.Chou TH, Bohorquez J, Toft-Nielsen J, Ozdamar O, Porciatti V. Robust mouse pattern electroretinograms derived simultaneously from each eye using a common snout electrode. Invest Ophthalmol Vis Sci. 2014;55:2469–2475. doi: 10.1167/iovs.14-13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pang IH, McCartney MD, Steely HT, Clark AF. Human ocular perfusion organ culture: A versatile ex vivo model for glaucoma research. J Glaucoma. 2000;9:468–479. doi: 10.1097/00061198-200012000-00009. [DOI] [PubMed] [Google Scholar]
- 49.Clark AF, Wilson K, de Kater AW, Allingham RR, McCartney MD. Dexamethasone-induced ocular hypertension in perfusion-cultured human eyes. Invest Ophthalmol Vis Sci. 1995;36:478–489. [PubMed] [Google Scholar]
- 50.Tucker BA, et al. Transplantation of adult mouse iPS cell-derived photoreceptor precursors restores retinal structure and function in degenerative mice. PLoS One. 2011;6:e18992. doi: 10.1371/journal.pone.0018992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grinnell KL, Bickenbach JR. Skin keratinocytes pre-treated with embryonic stem cell-conditioned medium or BMP4 can be directed to an alternative cell lineage. Cell Prolif. 2007;40:685–705. doi: 10.1111/j.1365-2184.2007.00464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bickenbach JR. Isolation, characterization, and culture of epithelial stem cells. Methods Mol Biol. 2005;289:97–102. doi: 10.1385/1-59259-830-7:097. [DOI] [PubMed] [Google Scholar]