Significance
The circadian oscillator is a cell-autonomous biological timer driving daily physiological rhythms to ensure fitness and health. Regulatory mechanisms of the oscillator are complex and not fully understood. We previously generated two circadian reporter mouse lines that differ only in the 3′-UTR region of the core clock gene Per2. Interestingly, substitution of the endogenous Per2 3′-UTR with an SV40 late poly(A) signal led to a lengthened period, enhanced PER2 protein level, and more robust oscillatory amplitude. PER2 also displayed a positive role in circadian transcription. Molecular and genetic studies showed the microRNA miR-24 binds to the Per2 3′-UTR to attenuate rhythmic PER2 accumulation. These results identified an important posttranscriptional regulatory mechanism of PER2 expression required for normal circadian timekeeping.
Keywords: Per2 gene, circadian, miR-24, microRNA, 3′-UTR regulation
Abstract
We previously created two PER2::LUCIFERASE (PER2::LUC) circadian reporter knockin mice that differ only in the Per2 3′-UTR region: Per2::Luc, which retains the endogenous Per2 3′-UTR and Per2::LucSV, where the endogenous Per2 3′-UTR was replaced by an SV40 late poly(A) signal. To delineate the in vivo functions of Per2 3′-UTR, we analyzed circadian rhythms of Per2::LucSV mice. Interestingly, Per2::LucSV mice displayed more than threefold stronger amplitude in bioluminescence rhythms than Per2::Luc mice, and also exhibited lengthened free-running periods (∼24.0 h), greater phase delays following light pulse, and enhanced temperature compensation relative to Per2::Luc. Analysis of the Per2 3′-UTR sequence revealed that miR-24, and to a lesser degree miR-30, suppressed PER2 protein translation, and the reversal of this inhibition in Per2::LucSV augmented PER2::LUC protein level and oscillatory amplitude. Interestingly, Bmal1 mRNA and protein oscillatory amplitude as well as CRY1 protein oscillation were increased in Per2::LucSV mice, suggesting rhythmic overexpression of PER2 enhances expression of Per2 and other core clock genes. Together, these studies provide important mechanistic insights into the regulatory roles of Per2 3′-UTR, miR-24, and PER2 in Per2 expression and core clock function.
Virtually all living organisms have evolved an intrinsic timing system, the circadian clock, to anticipate and exploit daily environmental changes. Circadian clocks commonly display three cardinal features, namely, a circadian free-running period, entrainment to environmental cues, and temperature compensation of circadian periodicity (1). These features allow the clocks to run on a consistent daily schedule, but on the other hand also confer adaptation to various environmental cues such as light, food, and temperature to achieve biological homeostasis and well-being (2, 3). In mammals, the clock system is organized in a hierarchical manner, with the central pacemaker in the hypothalamic suprachiasmatic nuclei (SCN) coordinating peripheral tissue clocks to perform essential physiological functions (4).
The mammalian cell-autonomous molecular oscillator consists of interlocked feedback loops (5). In the core loop, the positive transcription factors CLOCK and BMAL1 form heterodimers to drive the E-box enhancer-dependent transcription of Cryptochrome (Cry) and Period (Per) genes. The CRY and PER proteins in turn heterodimerize and translocate to the nucleus to inhibit CLOCK/BMAL1 activity. PER proteins are generally believed to play an auxiliary role in transcriptional repression (6), required for nuclear translocation of the primary repressor CRY (7–10). However, accumulating evidence also supports a coactivator function of PER in circadian transcription (11–14), suggesting complex molecular mechanisms. In the secondary loop, BMAL1 also drives the nuclear receptor Rev-Erb expression, and reciprocally Bmal1 is tightly regulated by REV-ERBs and their opposing nuclear receptor RORs (15, 16). These nuclear receptors are thought to compete for binding to the shared element RORE and thus play important roles in circadian amplitude regulation (17). More recent studies have also shown amplitude regulatory mechanisms distinct from the competitive binding model (18, 19), suggesting an integrated control of circadian amplitude by diverse pathways.
Expression and activity of core clock factors are subject to posttranscriptional and posttranslational regulation (20–26). One emerging regulatory mechanism is microRNA (miRNA)-mediated mRNA degradation or translational repression (27). miRNAs are small noncoding RNAs that bind to target 3′ untranslated regions (3′-UTRs) of mRNAs via sequence-specific interaction, regulating mRNA stability and/or translational efficiency (28). Together, miRNAs target a majority of protein-coding genes involved in various physiological processes. In particular, global knockout of Dicer, encoding the enzyme responsible for cleaving pre-miRNA to generate miRNAs, significantly shortened the circadian activity period (29), providing genetic evidence that miRNAs collectively play a regulatory role in clock function. Several other studies have also investigated specific miRNAs that regulate expression of core clock components (27, 30–32). For example (33), miR-132 and miR-219 were previously identified as enriched in mouse SCN neurons, displaying light-inducible and circadian expression requiring CREB and CLOCK/BMAL1, respectively. Functional analysis further revealed their respective activity to diminish light-responsive phase delay and shorten circadian period length, at least in part involving Per1 activation. These studies together suggest a pervasive miRNA regulation of the clock that requires further mechanistic understanding.
We previously generated Per2::Luc reporter mice where the luciferase coding sequence was inserted before the endogenous Per2 stop codon such that expression of PER2::LUC fusion proteins is driven by the natural Per2 promoter (34, 35). This reporter mouse line has proven highly versatile, allowing direct visualization of robust molecular rhythms of PER2 as a state variable of the circadian clock. Consistent with a normal circadian function of PER2::LUC, the reporter line showed indistinguishable behavioral and molecular characteristics compared with the wild-type (WT) mice (34). Using these mice, we showed that peripheral tissues are self-sustained, SCN-independent oscillators, and the SCN coordinates the phases of peripheral oscillators (34). We produced a modified version of reporter mice, Per2::LucSV, that harbors a replacement of the endogenous Per2 3′-UTR with an SV40 poly(A) signal, and fibroblast cells derived from these mice were previously reported (36, 37). To investigate the regulation and function of Per2 in vivo, particularly its 3′-UTR, we conducted in-depth molecular and behavioral studies of Per2::LucSV mice. Our results revealed a regulatory role of miR-24 and Per2 3′-UTR in Per2 expression and provided in vivo evidence for a positive transcription function of PER2 in the circadian oscillator leading to amplitude enhancement.
Results
Generation and Circadian Characterization of Per2::LucSV Knockin Mice.
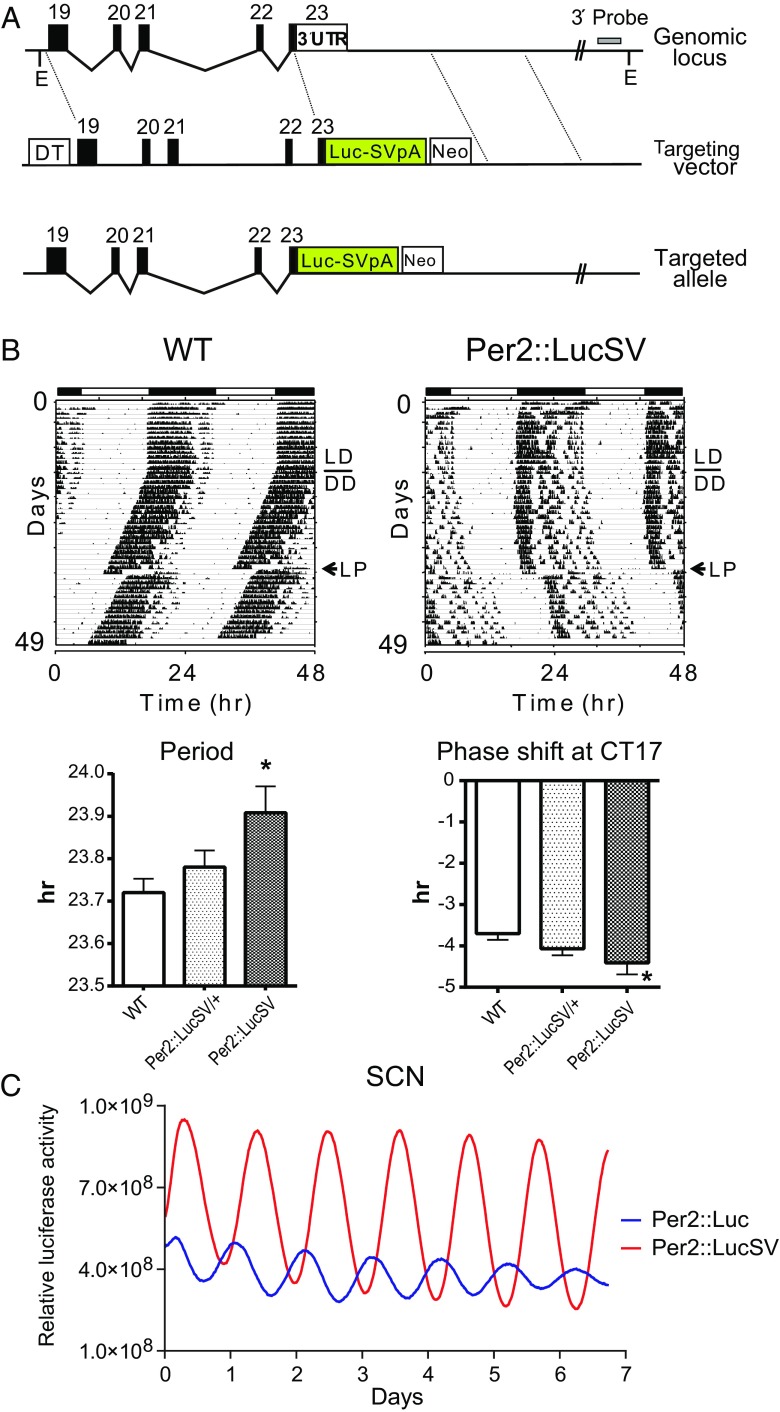
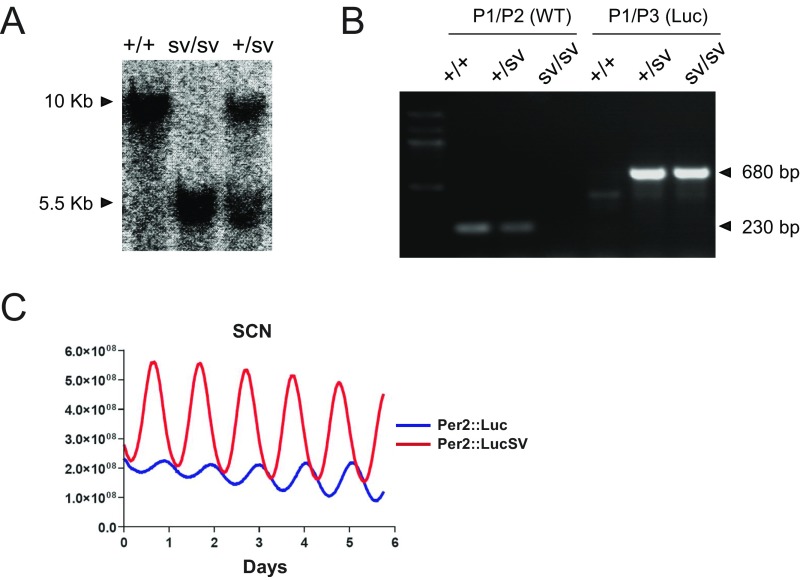
We previously generated the Per2::Luc reporter knockin mouse line with an intact endogenous Per2 3′-UTR (34). To investigate the role of Per2 3′-UTR in mRNA half-life and translational regulation, we created a modified knockin version expressing PER2::LUC. Specifically, the endogenous 3′-UTR of the original Per2::Luc knockin targeting vector (34) was replaced with an SV40 poly(A) signal sequence. The short arm for homologous recombination from the original vector was also changed to a 3.1-kb genomic DNA region 3 kb downstream from the original short arm used for Per2::Luc knockin (Fig. 1A). The presence of the correctly targeted Per2::LucSV allele was confirmed by Southern blotting analysis and PCR (Fig. S1 A and B). Intercrosses between heterozygous (C57BL/6J × 129S6SvEvTac) F1 offspring produced WT, heterozygous Per2::LucSV knockin, and homozygous F2 animals at the expected 1:2:1 Mendelian ratio. Mice heterozygous and homozygous for the targeted allele were developmentally and morphologically indistinguishable from WT littermates.
Fig. 1.
Generation and circadian characterization of Per2::LucSV knockin mice. (A) Diagram of the mouse Per2 locus, targeting vector, and targeted knockin allele. Exons are indicated by filled blocks with numbers. DT, diphtheria toxin A chain; E, EcoRI; Neo, neomycin resistance gene; triangle, loxP site. (B) Representative locomotor activity records from WT and Per2::LucSV homozygous knockin mice (WT, n = 10; Per2::LucSV, n = 12). Animals were maintained on LD12:12 for the first 14 d, indicated by the filled and empty bars above the records, before being transferred to DD to measure free-running periods. The free-running period of Per2::LucSV homozygous knockin mice was significantly longer than that of WT mice (t test: *P < 0.0001). On day 20 in DD conditions, a 6-h light pulse (LP; arrow) was administered at circadian time 17. Per2::LucSV homozygous knockin mice showed greater phase delay compared with WT mice (t test: *P < 0.001). WT, n = 10; Per2::LucSV/+, n = 15; Per2::LucSV, n = 12. (C) SCN explants were placed on an inverted microscope (DMI6000B, Leica) within an environmentally controlled Lucite chamber (Solent Scientific) maintained at 36 °C. Cultures were placed on a 36 °C Peltier heated stage within the chamber and tissues were imaged using a camera cooled to −20 °C (Stanford Photonics). Images were collected at 15 fps and threshold was applied to eliminate artifactual signals in real time. Images were subsequently integrated and analyzed by using the CellCycle program (Actimetrics), which can track single-cell images and quantify bioluminescence signals. Representative imaging data from Per2::Luc and Per2::LucSV heterozygous mouse SCN cultures with subtracted baseline are indicated as blue and red traces, respectively.
Fig. S1.
Generation of Per2::LucSV knockin mice. (A) Southern blot of tail genomic DNA from F2 mice. After XbaI digestion, 3′ external probe detect 10-kb WT fragment and 5.5-kb targeted fragment. (B) PCR genotyping of F2 mice. Agarose gel electrophoresis shows the presence of a 230-bp WT allele and a 680-bp knockin allele. (C) Additional bioluminescence recording from SCN explants as described in Fig. 1C. Representative imaging data from Per2::Luc and Per2::LucSV heterozygous mouse SCN cultures with subtracted base line are indicated as blue and red traces, respectively.
To begin to determine the effect of the Per2::LucSV allele on the circadian clock, we first analyzed the circadian behavior of Per2::LucSV mice. Unlike the previous Per2::Luc reporter mice (34), which showed normal behavioral periodicity compared with WT (23.69 ± 0.07), Per2::LucSV mice displayed longer free-running periods in constant darkness (DD) (23.91 ± 0.06, n = 12, t test: *P < 0.0001) (Fig. 1B). Furthermore, they also exhibited greater phase delays than WT when light pulse was administrated at CT17 (Fig. 1B). Together, these results suggest a functional role of the Per2 3′-UTR in the circadian clock.
A Regulatory Role of 3′-UTR in PER2 Level and Oscillatory Amplitude.
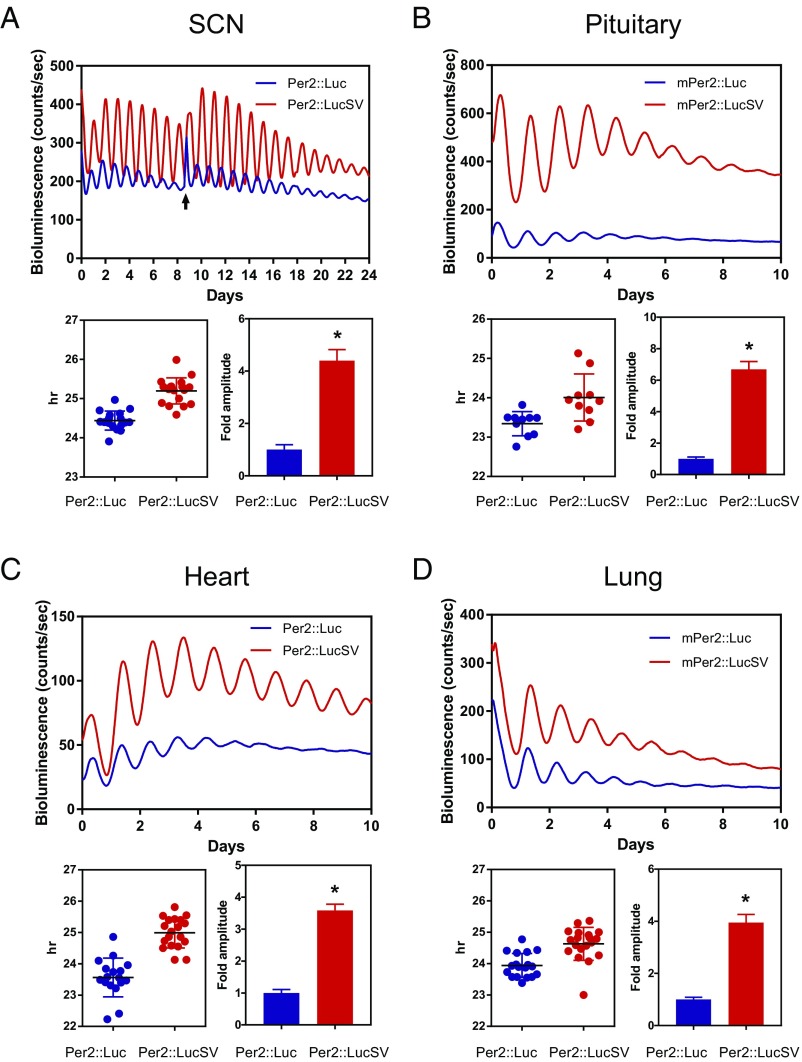
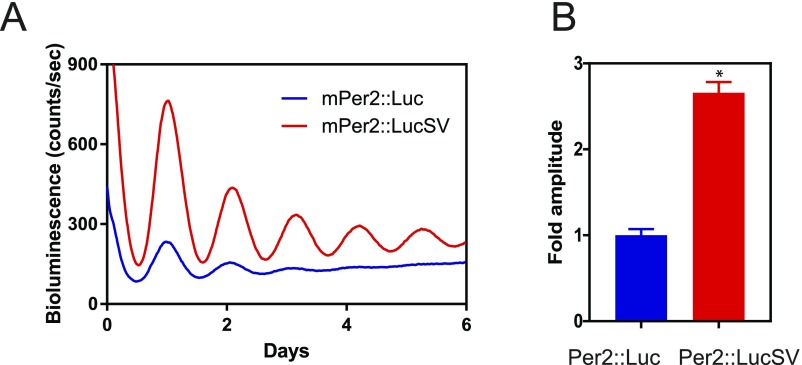
To delineate the circadian function of Per2 3′-UTR, we compared circadian rhythms between Per2::Luc and Per2::LucSV via real-time recording of bioluminescence from the SCN explants. Per2::Luc and Per2::LucSV mice were entrained to 12:12 light/dark (LD) cycles and SCN tissues were isolated 1 h before lights off. Whole-field bioluminescence from SCN slices was captured by camera and image stacks were then analyzed using the CellCycle program (Actimetrics). Interestingly, bioluminescence level and oscillatory amplitude in Per2::LucSV SCN explants were higher than those from Per2::Luc (Fig. 1C and Fig. S1C and Movie S1). Next we compared circadian bioluminescence rhythms in SCN and various peripheral tissues via photomultiplier tubes (PMTs) as previously described (34). All tissue explants from Per2::LucSV showed more robust and sustained circadian bioluminescence rhythms compared with those from Per2::Luc explants, characterized by three- to four-fold amplitude enhancement and elevated baseline levels. Per2::LucSV also showed tissue-specific period lengths that are significantly longer than in Per2::Luc in all tissues tested (Fig. 2). We derived mouse embryonic fibroblasts (MEFs) from both mouse lines, and reporter bioluminescence from MEFs also showed patterns consistent with the tissue explants. The robust and sustained bioluminescence in Per2::LucSV MEFs (Fig. S2) has been previously exploited for sensitive single-cell imaging and high-throughput screening (36, 37).
Fig. 2.
Enhanced amplitude and baseline level of real-time bioluminescence rhythms in Per2::LucSV compared with Per2::Luc. Representative PER2::LUC bioluminescence recording of SCN (A), pituitary (B), heart (C), and lung (D) from Per2::Luc and Per2::LucSV homozygous mice. Mice were kept in a light/dark cycle before dissection. Tissue sections were harvested just before lights off and immediately cultured for recording. Arrow indicates a medium change (A). Recordings are neither normalized nor corrected for baseline drift. Blue and red traces represent PER2 rhythm from Per2::Luc and Per2::LucSV mice, respectively. For each tissue period and relative amplitude, comparisons are shown in the graphs Below. Relative fold amplitude of Per2::Luc and Per2::LucSV tissues were calculated by the LM fit (damped sin) method (lumicycle analysis, Actimetrics). In all four tissues, relative amplitude was significantly higher in Per2::LucSV (t test: *P < 0.0001). Error bars represent SD. Per2::Luc SCN mean circadian period: 24.44 ± 0.05885 (n = 17), Per2::LucSV SCN mean circadian period: 25.20 ± 0.07833 (n = 18). t test: P value < 0.0001 (A). Per2::Luc pituitary mean circadian period: 23.34 ± 0.09702 (n = 10), Per2::LucSV pituitary mean circadian period: 24.01 ± 0.1894 n = 10. t test: P value 0.0059 (B). Per2::Luc heart mean circadian period: 23.56 ± 0.1500 (n = 17), Per2::LucSV heart mean circadian period: 24.99 ± 0.1114 (n = 19). t test: P value < 0.0001 (C). Per2::Luc lung mean circadian period: 23.94 ± 0.09045 (n = 18), Per2::LucSV lung mean circadian period: 24.63 ± 0.1205 (n = 19). t test: P value < 0.0001 (D).
Fig. S2.
Enhanced amplitude of real-time bioluminescence rhythms in Per2::LucSV compared with Per2::Luc MEF. (A) Representative PER2::LUC bioluminescence recording of Per2::LucSV compared with Per2::Luc fibroblasts. Recordings are neither normalized nor corrected for baseline drift. (B) Relative fold amplitude of Per2::Luc and Per2::LucSV fibroblast luciferase activity (t test: *P < 0.0001).
Effects of the Per2 3′-UTR Replacement on Core Clock Gene Repression.
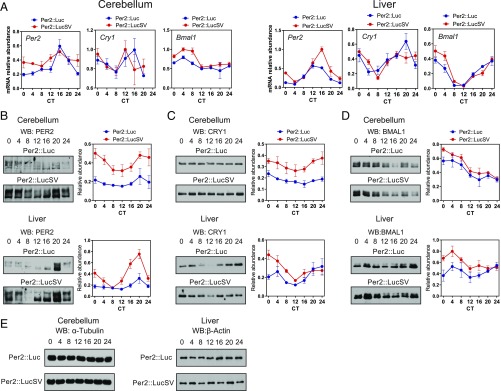
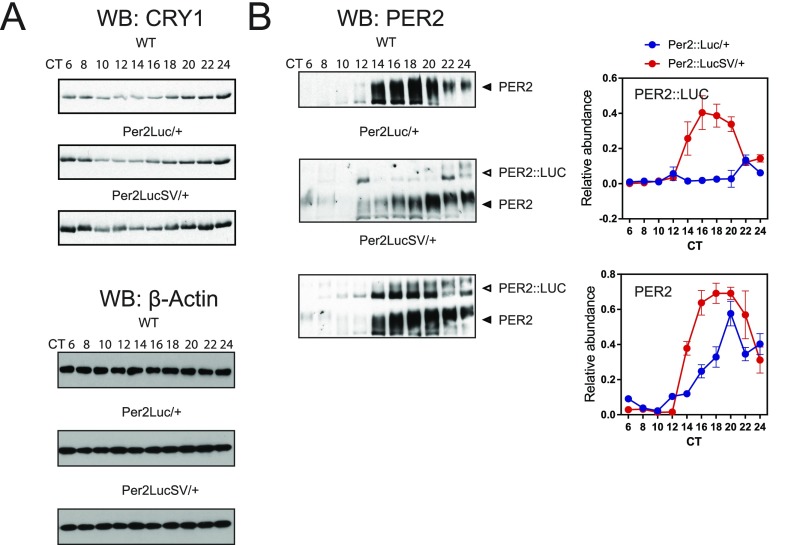
To explore how the Per2 3′-UTR substitution with the SV40 poly(A) affects clock gene expression, we determined both mRNA and protein expression patterns for several core clock genes in cerebellum and liver samples from Per2::Luc and Per2::LucSV mice maintained in constant darkness. Compared with the significantly enhanced bioluminescence, Per2 mRNA level increase in Per2::LucSV relative to Per2::Luc was more modest and temporally restricted (Fig. 3A). Moreover, whereas Cry1 mRNA oscillation was not significantly altered in cerebellum and liver of Per2::LucSV, Bmal1 mRNA level increased significantly during subjective daytime in Per2::LucSV cerebellum and liver compared with Per2::Luc (Fig. 3A). In accordance with bioluminescence measurements (Fig. 2), the Per2 3′-UTR replacement caused markedly elevated PER2::LUC protein level and circadian amplitude throughout the circadian cycle in both cerebellum and liver (Fig. 3B). Therefore, a posttranscriptional mechanism was primarily responsible for the PER2 protein accumulation to which transcriptional control may also contribute. Although Cry1 mRNA level was not significantly altered in Per2::LucSV (Fig. 3A), CRY1 protein level was increased in both tissues, suggesting that increased PER2::LUC protein levels promote CRY1/2 protein expression and/or stability (Fig. 3C and Fig. S3A). BMAL1 protein level increased during subjective daytime in Per2::LucSV cerebellum and liver compared with Per2::Luc (Fig. 3D). Rev-Erbα mRNA level was reduced in Per2::LucSV compared with Per2::Luc at CT4 (Fig. S3B), suggesting increased PER2::LUC protein level in Per2::LucSV at CT0 and CT4 may repress E-box–driven Rev-Erbα transcription, which in turn contributes to increased Bmal1 transcription. These results together suggested that enhanced circadian amplitude and baseline level of PER2 augment the mRNA and/or protein oscillation of core clock components, including Cry1 and Bmal1.
Fig. 3.
Circadian genes display enhanced amplitude of mRNA and protein oscillation in Per2::LucSV cerebellum and liver. (A) Real-time RT-PCR analysis of clock gene expression in Per2::Luc and Per2::LucSV homozygous mice. Blue and red circles represent Per2::Luc and Per2::LucSV mice, respectively. Error bars represent SEM for each time point from three independent homozygous repeats. Two-way ANOVA shows significant statistical differences between Per2::Luc and Per2::LucSV mice for Per2 (cerebellum and liver, P < 0.0001) Bmal1 (cerebellum, P < 0.0001, liver, P < 0.01). (B–D) PER, CRY1, and BMAL1 protein oscillation in cerebellum (Upper Left) and liver (Lower Left). Quantification from the three independent experiments is shown to the Right of each representative. Blue and red circles represent values from Per2::Luc and Per2::LucSV mice. Error bars represent SEM (n = 3). Two-way ANOVA shows significant statistical differences between Per2::Luc and Per2::LucSV mice for PER2 (cerebellum and liver, P < 0.0001), CRY1 (cerebellum, P < 0.0001; liver, P < 0.01), BMAL1 (cerebellum, P < 0.05; liver, P < 0.001). (E) Actin Western blot from cerebellum and liver of Per2::Luc and Per2::LucSV homozygous mice.
Fig. S3.
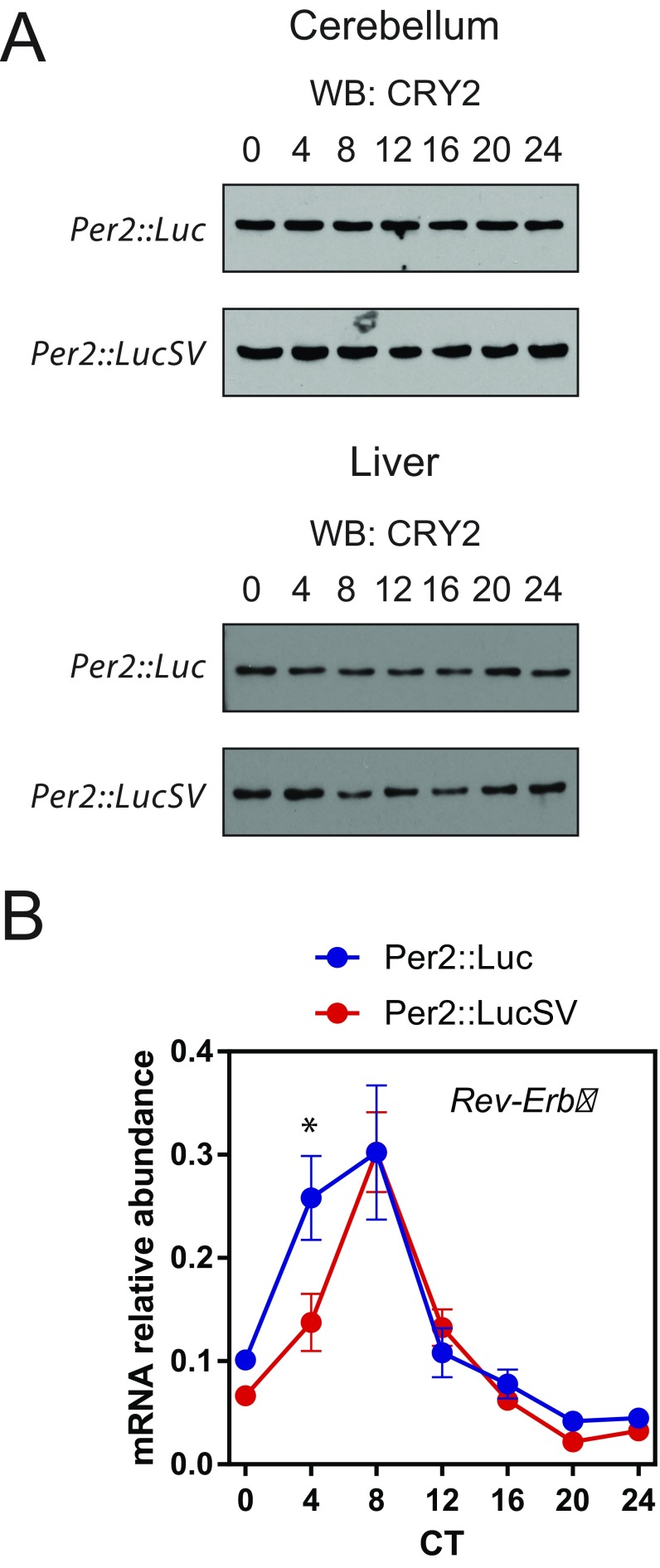
Per2::LucSV mice showed enhanced amplitude of clock protein oscillation in cerebellum and liver. (A) Western blotting shows CRY2 protein oscillation in cerebellum (Upper) and liver (Lower). (B) Rev-Erbα mRNA oscillation in liver. Blue and red circles represent values from Per2::Luc and Per2::LucSV mice. Error bars represent SEM for each time point from three independent homozygous repeats (two-way ANOVA, P < 0.01).
MicroRNA Regulation of PER Translation.
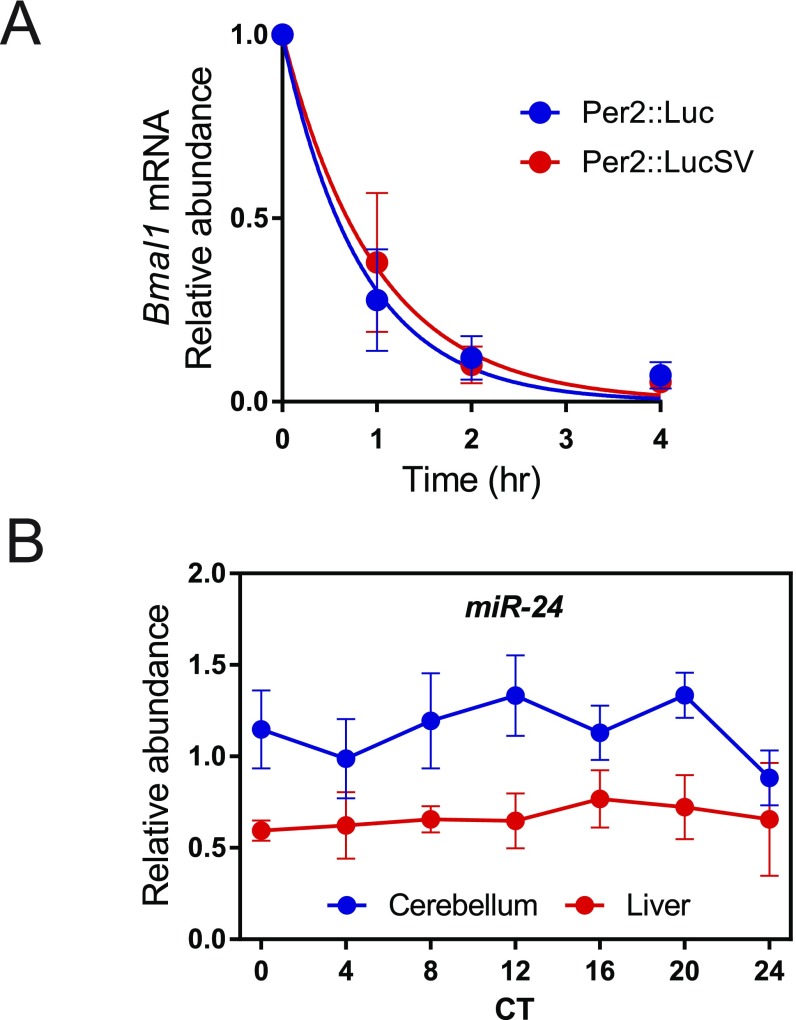
To delineate the underlying molecular mechanism for increased PER2::LUC protein levels in Per2::LucSV mice, we first measured Per2 mRNA half-life using actinomycin D-treated Per2::Luc and Per2::LucSV MEF cells. Per2::Luc mRNA half-life was estimated to be 0.45 h and 0.67 h in Per2::Luc and Per2::LucSV MEFs, respectively. This result indicated that replacement of the endogenous 3′-UTR with the SV40 poly(A) sequence moderately stabilized the mRNA (Fig. 4A). In addition, Bmal1 mRNA half-life was not affected by the Per2 3′-UTR change (Fig. S4A).
Fig. 4.
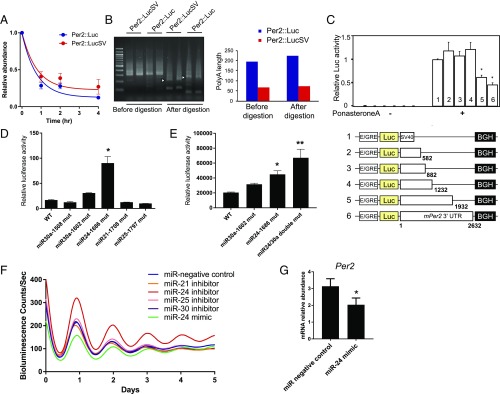
miR-24 regulates Per2 mRNA stability and protein translation. (A) Per2 mRNA half-life measurement. MEFs generated from mPer2::Luc and mPer2::LucSV homozygous mice were plated 2 d before treatment with 2 μg/mL actinomycin D for the indicated periods of time. Total RNA was isolated from the harvested cells, and quantitative PCR was performed using firefly luciferase primers. Blue and red circles represent values from Per2::Luc and Per2::LucSV MEFs. Error bars represent SEM for each time point from three independent repeats. The half-life of Per2::Luc mRNA from Per2::Luc and Per2::LucSV MEFs was determined to be 0.45 h (blue) and 0.67 h (red), respectively. The half-life parameter K (rate constant) is not significantly different; P = 0.314 based on model-based nonlinear one-phase exponential decay analysis (GraphPad Prism 7). (B) The poly(A) tail length of Per2 mRNA at ZT12 in the liver of Per2::Luc or Per2::LucSV mice. Ligation-mediated (LM)-PATs were performed and a representative gel image is shown. Each lane represents an individual mouse (n = 2). Arrowheads indicate the fragment derived after restriction enzyme treatment for poly(A) length measurement. Graph on the Right represents the poly(A) tail length calculated from the LM-PAT gel shown on the Left. (C) Serial truncation in the mouse Per2 3′-regulatory region were incorporated into a luciferase reporter construct containing an ecdysone-inducible expression system. BGH, BGH poly(A) signal; E/GRE, ecdyson/glucocorticoid responsive element; Luc, Luciferase; SV40, SV40 poly(A) signal. An equal amount of each reporter DNA construct was transiently transfected into NIH 3T3 cells with or without ponasterone A. Numbers at the Right of each construct indicate the end positions of the insert from the stop codon. Each value is the mean ± SEM of three replicates from three independent assays. The value of construct 1 was set as 1 for normalization. *P < 0.005 compared with the construct 1 (Student’s t test). (D) Putative microRNA binding sites (miR30a, miR-24, miR21, and miR25) located between constructs 4 and 5 (1,232–1,932 bp) were mutated individually from construct 5, and 1–1,932 bp 3′-UTR with mutant miRNA sites were used to replace the SV40 poly(A) signal sequence. Two hundred nanograms of each reporter construct was transfected into NIH 3T3 cells. Each value is the mean ± SEM of three replicates. The results shown are representative of three independent experiments. The miR-24 site mutation caused significant reporter activation (one-way ANOVA for all three independent experiments: *P < 0.0001). (E) The binding sites for miR-24 and miR-30a were mutated. Two hundred nanograms of each reporter construct was transfected into NIH 3T3 cells. Each value is the mean ± SEM of three replicates. The results shown are representative of three independent experiments. The miR-24/30a binding site double mutation showed an additive effect for reporter activation compared with WT (one-way ANOVA: *P < 0.001, **P < 0.0001 for the representative results, for two other experiments, miR24-1686 and miR24/30 double: P < 0.0001). (F) Amplitude of the circadian reporter rhythm was increased in Per2::Luc MEFs treated with miR-24 inhibitor oligonucleotides. Representative PER2::LUC bioluminescence recording from MEFs transfected with miRNA negative control (blue), miR-24 inhibitor (red) and miR-24 mimic (green), miR-25 mimic (pink), miR-30 mimic (purple), and miR-21 mimic (orange) are shown. (G) Per2 mRNA amount was reduced by miR-24 mimic transfection in Per2::Luc MEFs (t test: *P < 0.01).
Fig. S4.
(A) Per2 3′-UTR replacement did not affect Bmal1 mRNA half-life. Bmal1 mRNA half-life measurement. MEFs generated from Per2::Luc and Per2::LucSV homozygous mice were plated 2 d before treatment with 2 μg/mL actinomycin D for the indicated periods of time. Total RNA was isolated from the harvested cells, and quantitative PCR was performed using firefly luciferase primers. Blue and red circles represent values from Per2::Luc and Per2::LucSV MEFs. Error bars represent SEM for each time point from three independent repeats. The half-life of Bmal1 mRNA from Per2::Luc and Per2::LucSV MEFs was determined to be 0.44 h and 0.61 h, respectively, nonlinear one phase exponential decay analysis. The half-life parameter K (rate constant) is not significantly different (P = 0.41) based on model-based nonlinear one-phase exponential decay analysis (GraphPad Prism 7). (B) miR-24 amount was measured by real-time qRT-PCR analysis using brain and liver samples. Two-way ANOVA shows significant statistical differences between brain and liver for miR-24 expression (P < 0.0001).
As mentioned above, while the PER2::LUC protein level was highly elevated throughout the circadian cycle in Per2::LucSV cerebellum and liver compared with Per2::Luc, the increase in Per2::LucSV mRNA amount was more modest and limited to certain time points (Fig. 3A). To investigate the primary cause for the increased PER2::LUC protein accumulation, we measured the poly(A) length of Per2::Luc mRNA in Per2::Luc and Per2::LucSV mouse liver using a poly(A) tail-length (PAT) assay (38–40). To our surprise, the poly(A) length of Per2::Luc mRNA was longer in Per2::Luc than in Per2::LucSV, indicating that increased PER2::LUC protein accumulation in Per2::LucSV was not due to poly(A) length change (Fig. 4B).
Next, we investigated whether any cis-regulatory elements in the Per2 3′-UTR were altered in Per2::LucSV, which might be responsible for PER2::LUC protein accumulation. We generated five reporter constructs containing serial truncations in the Per2 3′-UTR. Luciferase activity was significantly increased when the 3′-UTR fragment +1,232 to +1,932 was deleted (Fig. 4C). We analyzed this sequence using the TargetScanMouse algorithm (41) and identified five putative miRNA binding sites. Individual seed sequences for miRNA binding sites were mutated in the no. 5 3′-UTR construct and reporter assays were performed by using NIH 3T3 cells. Compared with the WT control (WT-5), the miR-24 binding site mutant clone (miR24-1686 mut) displayed significantly enhanced luciferase activity (Fig. 4D), suggesting a regulatory role of miR-24 in Per2 expression. A mutation in the miR-30 site (miR30a-1602 mut) also induced luciferase activity as previously reported (29), albeit to a much lesser degree compared with the miR-24 mutation (Fig. 4D). When we introduced double mutations for both miR-24 and miR-30a binding sites in the 3′-UTR, an additive effect was observed in reporter assays (Fig. 4E).
We next transfected Per2::Luc MEF cells with either miRNA inhibitor or mimic oligonucleotides and monitored real-time PER2::LUC bioluminescence (Fig. 4F). MEF cells transfected with the miR-24 inhibitor oligonucleotide phenocopied Per2::LucSV and showed increased amplitude for the luciferase rhythm. Conversely, miR-24 mimic oligonucleotides decreased the reporter rhythm amplitude, together suggesting miR-24 plays an inhibitory role in PER2 protein expression. We further tested inhibitor oligonucleotides of several other candidate miRNAs, including miR-21, miR-25, and miR-30, and they did not potentiate the PER2::LUC rhythm in Per2::Luc MEFs. Next, we performed qPCR to determine the effect of miR-24 mimic on Per2 mRNA level (Fig. 4G). Relative to control oligonucleotides, transfection of miR-24 mimic significantly reduced Per2 mRNA level in Per2::Luc MEF cells, suggesting miR-24 alters PER2 protein level by destabilizing Per2 mRNA, at least in part. We also performed qPCR analysis to investigate whether miR-24 expression is under circadian control. miR-24 expression was not rhythmic in cerebellum and liver, and cerebellum showed significantly higher miR-24 expression than liver (Fig. S4B).
Positive Transactivation Role of PER2 for Its Own Transcription.
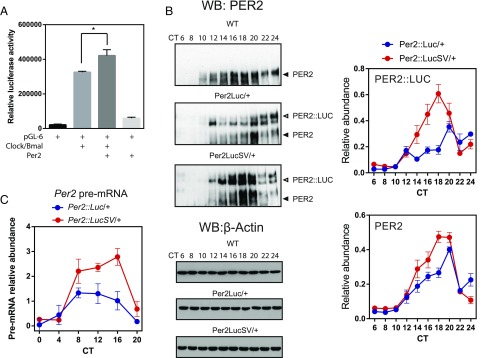
Somewhat surprisingly, based on the above analysis of E-box–driven transcription of Per2 and Cry1, increased PER2::LUC protein levels in Per2::LucSV mouse did not impose greater transcriptional inhibition in the core loop. Furthermore, Bmal1 transcription was augmented during the subjective daytime in Per2::LucSV tissues, suggesting a positive role of PER2::LUC protein in circadian transcription. Consistent with other reports indicating PER2 as a coactivator for its own transcription (11, 13, 42), reporter assays using a Per2 promoter construct showed potent stimulatory effects of Per2 cotransfection on CLOCK/BMAL1-dependent transcriptional induction (Fig. 5A). PER2 expression not only enhanced CLOCK/BMAL1 transactivation but also increased the baseline reporter activity in the absence of Clock/Bmal1 cotransfection (Fig. 5A).
Fig. 5.
PER2 functions as a positive regulator for Per2 transcription in vitro and in vivo. (A) The mouse Per2 promoter reporter construct pGL6 was transiently cotransfected into 293A cells with the indicated expression constructs. CLOCK/BMAL1-mediated transcriptional activation was significantly increased by PER2 coexpression (F test to compare variances, F = 40.74, P = 0.04). (B) Oscillation of PER2 (WT) and PER2::LUC fusion protein in liver. Western blotting was performed using liver protein extracts with the PER2 antibody. Both PER2 from the WT allele and PER2::LUC from the knockin allele were increased in the liver extract from Per2::LucSV heterozygous mice. Quantification for PER2::LUC (Upper) and PER2 (Lower) is shown at Right. Two-way ANOVA shows significant statistical differences in PER2::LUC and PER2 levels between Per2::Luc/+ and Per2::LucSV/+ mice; P < 0.0001. (C) Real-time RT-PCR analysis of Per2 pre-mRNA in liver from Per2::Luc heterozygous (Per2::Luc/+) and Per2::LucSV heterozygous (Per2::LucSV/+) mice. Error bars represent SEM for each time point from three independent repeats. Two-way ANOVA shows significant differences between Per2::Luc heterozygous and Per2::LucSV heterozygous (P < 0.0001).
To determine effects of knockin alleles on transcription and translation of the endogenous Per2 allele, we collected liver tissues at the indicated CT from Per2::Luc and Per2::LucSV heterozygous mice. Western blotting analysis showed that circadian expression of PER2::LUC protein was significantly enhanced from the Per2::LucSV allele relative to the Per2::Luc allele, due to the 3′-UTR replacement in the former and consequently removal of the endogenous miRNA binding sites (Fig. 5B, Left Middle and Lower panels of PER2 WB; quantification: Upper Right). Importantly, PER2 protein expression from the WT allele in Per2::LucSV heterozygote mouse liver was significantly elevated compared with that in Per2::Luc heterozygote mice (Fig. 5B, Left Middle and Lower panel of PER2 WB; quantification: Lower Right), and a similar pattern was observed for CRY1 in liver (Fig. S5A) and PER2 in kidney (Fig. S5B). These results strongly suggested that increased PER2::LUC protein expression from the Per2::LucSV allele may in turn activate Per2 transcription. We next determined transcription initiation rate by measuring Per2 pre-mRNA levels via qPCR analysis. From CT8 to CT20, the levels of Per2 pre-mRNA was significantly elevated in Per2::LucSV relative to Per2::Luc, indicating an activating role of PER2 protein in the transcription of the endogenous Per2 allele (Fig. 5C).
Fig. S5.
PER2 functions as a positive regulator for Per2 transcription in vitro and in vivo. Oscillation of CRY1 and PER2::LUC fusion protein in liver. (A) Western blotting was performed using liver protein extracts with the CRY1 antibody. CRY1 amount was increased in the kidney extract from Per2::LucSV heterozygous mice. (B) Oscillation of PER2 (WT) and PER2::LUC fusion protein in liver. Western blotting was performed using kidney protein extracts with the PER2 antibody. Both PER2 from the WT allele and PER2::LUC from the knockin allele were increased in the kidney extract from Per2::LucSV heterozygous mice. PER2::LUC quantification on the Upper Right, PER2 quantification on the Lower Right. Two-way ANOVA shows significant statistical differences between Per2::Luc/+ and Per2::LucSV/+ mice for PER2::LUC and PER2, P < 0.0001. Representative blots from two independent experiments are shown.
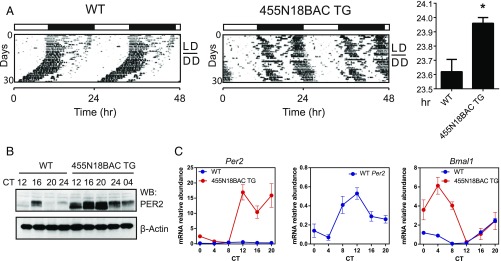
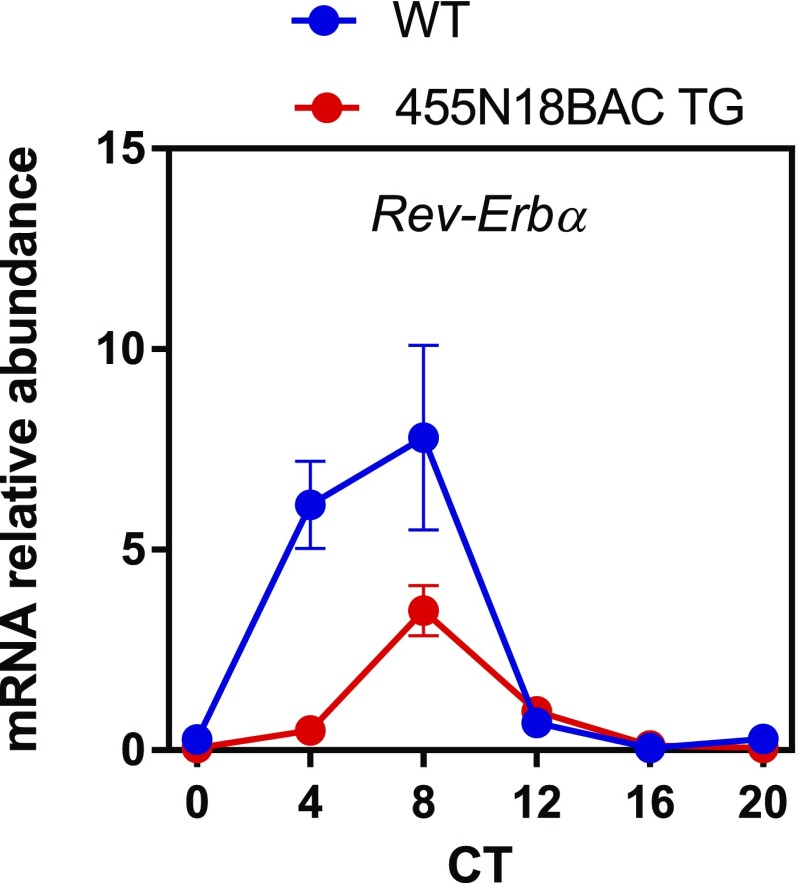
In a recent study (29), global Dicer knockout, as well as simultaneous knockdown of three PER1/2-targeting miRNAs including miR-24 and miR-30a, was found to shorten the circadian period lengths in cells and mice, contrary to the period-lengthening effect observed in Per2::LucSV mice. To delineate the role of PER2 and cognate miRNAs specifically, we analyzed an independent BAC transgenic (TG) mouse model of PER2 rhythmic overexpression, namely the 455N18BAC Per2 TG mice. Consistent with Per2::LucSV, we again observed lengthened circadian periods (Fig. 6A). Per2 mRNA and protein expression was highly elevated in the 455N18BAC TG mice compared with WT (Fig. 6 B and C). Bmal1 expression was also markedly enhanced in the TG mice compared with WT (Fig. 6C, Right), again consistent with what we previously observed from Per2::LucSV mice (Fig. 3C). Interestingly, Rev-Erbα level was significantly reduced in 455N18BAC TG mice at CT4 and CT8 compared with WT (Fig. S6). Consistent with results in Fig. S3B, this observation suggested that increased PER2 protein level potentiates Bmal1 transcription by repressing REV-ERBα transcription. These results further affirmed the effects of rhythmic PER2 overexpression on circadian behavior and gene expression.
Fig. 6.
Rhythmic PER2 overexpression in Per2 BAC transgenic (TG) mice lengthened circadian activity period and increased Bmal1 circadian amplitude. (A) Representative locomotor activity records from WT and 455N18BAC TG mice. Animals were maintained on LD12:12 for the first 12 d, indicated by the filled and empty bars Above the records, before transfer to DD to measure free-running period. The 455N18BAC TG mice exhibited significantly longer free-running periods than WT mice (23.96 ± 0.0400 vs. 23.62 ± 0.08602, n = 5, t test: P = 0.0071). (B) PER2 proteins are rhythmically overexpressed in 455N18BAC TG. Western blotting was performed using total protein extracts from liver with the PER2 antibody. (C) Real-time RT-PCR analysis of Per2 and Bmal1 expression in liver tissues from WT and 455N18BAC TG mice. (Left) Per2 mRNA level from 455N18BAC TG samples (red circles) were significantly elevated (two-way ANOVA, P < 0.0001) compared with the WT control (blue circles, Middle). (Right) Bmal1 mRNA level from 455N18BAC TG samples (red circles) were also significantly elevated (two-way ANOVA, P < 0.0001) compared with the WT control.
Fig. S6.
Real-time qRT-PCR analysis of Rev-Erbα in Per2::Luc and Per2::LucSV homozygous mice liver. Blue and red circles represent Per2::Luc and Per2::LucSV mice, respectively. Error bars represent SEM for each time point from three independent homozygous repeats. Two-way ANOVA shows significant statistical differences between Per2::Luc and Per2::LucSV mice for Rev-Erbα (P < 0.001).
Per2::LucSV Showed Enhanced Temperature Compensation in Tissue Clocks Compared with Per2::Luc.
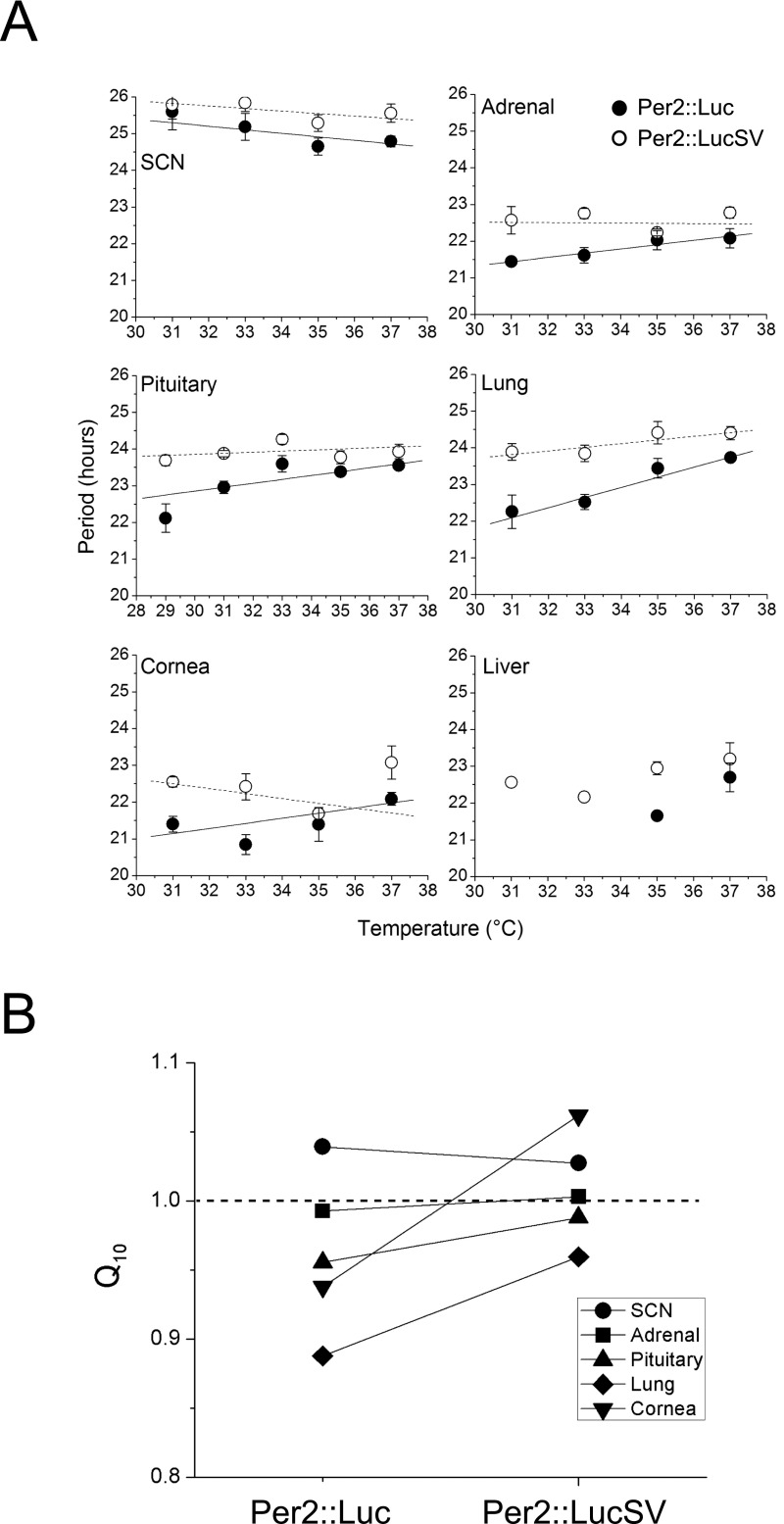
Temperature compensation is a defining feature of the circadian clock, evolved to maintain consistent periodicity over temperature fluctuation in daily and seasonal cycles (1). Previously, we employed Per2::Luc mice and showed that circadian rhythms in peripheral tissues are temperature compensated (43). To examine whether the enhanced central and peripheral circadian clocks in Per2::LucSV display varying temperature compensation relative to Per2::Luc, we harvested SCN and peripheral tissue explants from Per2::LucSV heterozygous mice, recorded reporter luminescence rhythms at temperatures ranging between 31 °C and 37 °C, and calculated the average periods for individual tissues. SCN and peripheral tissues from Per2::LucSV showed clear temperature compensation; however, compared with Per2::Luc (data adopted from ref. 43), Per2::LucSV tissues exhibited Q10 values closer to 1.0 than those of Per2::Luc, suggesting a more robust temperature compensation in the former (Fig. S7). The largest differences between the Q10 values of the two lines were observed in cornea and lung, while the other four tissues displayed more subtle differences.
Fig. S7.
Temperature compensation of central and peripheral oscillators in Per2::LucSV mice. (A) Average periods of central (SCN) and peripheral (adrenal, pituitary, lung, cornea, and liver) tissues are shown for both Per2::Luc and Per2::LucSV heterozygous mice within the range of temperatures tested (Per2::Luc data were adopted from ref. 43). All tissues show temperature compensation (Q10 value near 1.0). The periods of the Per2::LucSV oscillators are consistently longer (∼1 h) than those of Per2::Luc. Closed circle, Per2::Luc; open circle, Per2::LucSV. The y-error bars represent SE. (B) Comparison of Q10 values of five tissues from Per2::Luc and Per2::LucSV mice.
Discussion
Posttranscriptional 3′-UTR regulation of Per2 gene expression as revealed by the Per2::LucSV circadian reporter mouse showed lengthened circadian period lengths, greater phase responses to light pulse, and enhanced temperature compensation. Molecular analysis of Per2::LucSV revealed elevated abundance and oscillatory amplitude of PER2 as well as several other circadian components, suggesting a functional link between molecular amplitude enhancement and changes in circadian behavioral rhythms. In addition, we found a pivotal role of miR-24 in Per2 expression, providing a mechanistic explanation for the enhanced PER2 levels and circadian amplitude. Finally, we found that PER2 protein accumulation activates its own expression, supporting a positive role for PER2 in circadian transcription. These results highlight the multifunctional roles of PER2 within the circadian clock mechanism.
Our studies on the enhanced rhythms in Per2::LucSV revealed important miRNA regulation of PER2 expression and circadian rhythms. Specifically, mapping of the 3′-UTR identified a miR-24 binding site, and cellular functional assays showed a dominant role of miR-24 in PER2 expression. In comparison, miR-30 seems to play a less prominent role, as a mutation in the miR-30 site only weakly enhanced reporter expression. Previously, 248 mRNAs were found to be down-regulated by miR-24 overexpression, and Per2 is among the gene targets with a miR-24 seed sequence (44). On the other hand, no other miR-24 target genes identified therein are known to regulate PER2 protein level directly. A recent study (29) also implicated both miR-24 and miR-30 in PER2 expression; our results further define their relative contribution. Contrary to the period shortening observed in global Dicer knockout mice (29), both Per2::LucSV and 455N18BAC Per2 TG mice with rhythmic PER2 overexpression showed lengthened circadian periods, suggesting confounding effects of other miRNAs (in addition to miR-24 and miR-30a) on PER2 expression and circadian periodicity. Previously, the miR-192/194 cluster has been shown to coordinately repress expression of all three Per genes; similar to the role of miR-24 reported here (Fig. 4), expression of miR-192/194 in NIH 3T3 cells shortened the circadian period (45). Apart from circadian period, miRNAs have also been found to modulate oscillatory amplitude. For example, a miRNA overexpression screen identified miR-276a as a clock-related miRNA expressed in fly head (46). Deregulation of miR-276a expression, either up or down, in clock neurons attenuated circadian rhythmicity. The Timeless gene, playing an important role in the negative loop of Drosophila oscillator, was identified as a direct target of miR-276a and disruption of the cognate binding site on Timeless 3′-UTR elevated TIM protein levels. However, unlike PER2 overexpression in our study, TIM accumulation due to the miR-276a binding site mutation led to behavioral arrhythmicity. These studies together indicate important yet diverse roles of miRNAs in circadian clock regulation.
PER proteins are central circadian components with important roles in (patho)physiology (47, 48), yet their mechanistic functions are not fully understood. Previously, constitutive overexpression of PER2 was found to disrupt the clock in fibroblasts and tissues (10); in contrast, Per2::LucSV mice maintain high-amplitude oscillation of overexpressed PER2 and correspondingly show robust behavioral rhythms. These results suggest that rhythmic accumulation, rather than expression level per se, of PER2 is critical for circadian oscillation. Our results also provide in vivo evidence for a positive role of PER2 in the circadian oscillator. In a canonical model, PER proteins are believed to play a requisite role in nuclear translocation of CRYs as well as the subsequent transcriptional repression (2, 7). On the other hand, biochemical/molecular and in vivo evidence has also shown that PERs can function as positive factors to enhance E-box induction (11–14, 42). Complementing the mouse loss-of-function studies where Per knockout mice displayed reduced Per expression (48, 49), our current work provides an in vivo gain-of-function context highlighting a positive role of PER2 in circadian transcription. Our results showed reduced Rev-Erbα transcript levels in both Per2::LucSV and 455N18BAC TG, suggesting that PER2 may repress the E-box–driven Rev-Erbα transcription, thus relieving REV-ERB repression of circadian genes. Other mechanisms are also possible. It was recently shown that PER1/2 interfere with CRY interaction with CLOCK/BMAL1 immediately following translocation to the nucleus, causing delay of CRY-mediated repression (12). Consistent with the notion that such phase delay is required for the circadian period length (13), Per2::LucSV mice displayed longer period length (∼24.0 h) potentially attributable to heightened PER2 levels and activating activities. Alternative mechanisms for PER2 regulation of circadian periodicity are also possible. For example, prolonged degradation arising from the higher abundance of PER2 in Per2::LucSV mice could also contribute to circadian period lengthening. Moreover, in the aforementioned study (29), the period shortening in Dicer knockout mice was attributed to the ascending phase of the circadian cycle, specifically correlating with the accelerated cytoplasmic accumulation of PER2. Further studies are required to integrate PER2 expression, cellular localization, dual activity, and phosphorylation-coupled turnover in the circadian clock mechanism.
Per2::LucSV cells, displaying three- to fourfold enhancement in reporter bioluminescence over Per2::Luc, are ideally suited for high-sensitivity applications such as single-cell or microplate circadian monitoring (36, 37). Per::LucSV mice also represent an excellent in vivo model to understand regulation of circadian amplitude. While robust amplitude has clear health implications as chronic diseases and aging are known to correlate with damped circadian amplitude (50–52), the molecular mechanisms underlying circadian amplitude regulation are not well understood, mainly involving REV-ERBs and RORs in the secondary loop (17–19). The current study, on the other hand, suggests the core loop factor PER2 functions as a regulator of circadian amplitude. The positive role of PER2 in circadian transcription is in agreement with a functional plasticity of this pivotal core clock component (12). The ability to “buffer” or modulate the antagonism between CLOCK/BMAL1 and CRY may place PER2 in a unique position to modulate amplitude. The clock is inherently a self-limiting, rhythmic machinery, known as a “limit cycle” (53). Measures that perturb the positive–negative arm balance beyond a homeostatic range will conceivably damp the overall amplitude of the following cycles. This notion is supported by a stoichiometric ratio between the positive and negative core factors in fibroblast cells (54), as well as recent studies showing that a clock-enhancing compound only moderately enhances ROR activity and clock output gene expression (55–57). A possible dual role of PER proteins affords a built-in modulatory mechanism in the core loop, bridging the two competing arms and conferring functional dexterity to ensure clock robustness.
The circadian clock is highly sensitive to temperature changes (58), yet it is also imperative that the circadian period is temperature compensated such that constant periodicity is maintained despite environmental fluctuations (1). Temperature compensation has been described in various experimental systems (59–62), and several studies highlighted a mechanistic role of phosphorylation in temperature compensation. For example, casein kinase 1 (CK1), a key enzyme targeting PER proteins, has been shown to be relatively temperature insensitive (59). A recent study (63) employed both computational modeling and biochemical assays to demonstrate that PER2 protein stability, and therefore its level and activity, is regulated by a two-site phosphoswitch controlled by CK1 and a more temperature-sensitive priming kinase. The phosphoswitch was shown to regulate the mode of PER2 phosphorylation-dependent degradation, thereby enabling constant periodicity under varying temperatures. In the current study, Per2::LucSV mice showed a more robust temperature compensation compared with Per2::Luc (43), as evidenced by Q10 values from Per2::LucSV tissue clocks that were closer to 1. Future studies will examine whether the elevated PER2 levels directly impinge on temperature compensation through the phosphoswitch mechanism.
In conclusion, we generated a circadian reporter mouse line, Per2::LucSV, expressing high levels of PER2::LUC fusion proteins. Mechanistic studies revealed key regulatory functions of the Per2 3′-UTR and PER2 proteins in core circadian oscillators. The high-amplitude rhythms also render Per2::LucSV mice a useful research tool for future function and mechanism studies.
Materials and Methods
The Per2::LucSV knockin targeting vector was derived from the backbone of Per2::Luc targeting vector with modifications. The 455N18BAC Per2 TG mice were generated as previously described by using the 455N18BAC clone, which covers the Per2 genomic locus (64). Animal husbandry for all of the studies was carried out under Institutional Animal Care and Use Committee guidelines and the procedures were conducted as described in animal protocols approved by Northwestern University, Vanderbilt University, The University of Texas Southwestern Medical Center at Dallas, and The University of Texas Health Science Center at Houston (UTHSC-H). The ligation-mediated (LM)-PAT was performed as described previously (36) with slight modifications. Per2::Luc and Per2::LucSV MEFs were isolated from 12.5 d postcoitum (DPC) embryos and immortalized (23). Circadian gene expression (23, 65) and temperature compensation (39) experiments were performed as described. Data are presented as means ± SEM. P < 0.05 was considered statistically significant. For details regarding transgenic mice, tissue explant cultures, mRNA and protein analysis, plasmids and small hairpin RNAs, cell culture and reporter assays, please refer to SI Materials and Methods.
SI Materials and Methods
Per2::LucSV Knockin Mice and 455N18BAC Per2 TG Mice.
The Per2::LucSV knockin targeting vector was derived from the backbone of Per2::Luc targeting vector (34) with the modifications of including the SV40 poly(A) signal sequence from the pGL3 vector and replacement of the short arm localized 3 kb downstream of the original short arm region. Homologous recombinants were isolated after electroporation with 40 μg of the targeting construct into 2 × 107 W4 ES cells (129S6SvEvTac; provided by A. L. Joyner, New York University, New York). Following G418 selection (200 μg/mL), 500 surviving clones were screened by Southern analysis to detect homologous recombinants. Targeted ES cell clones were injected into C57BL/6J blastocysts and transferred to pseudopregnant C57BL/6J female recipients. Resulting male chimeras were bred with C57BL/6J females. Germ-line transmission was confirmed by agouti coat color and Southern blotting. Primers for PCR genotyping are the same as described previously (34). The 455N18BAC Per2 transgenic (TG) mice were generated as previously described by using the 455N18BAC clone, which covers the Per2 genomic locus (64).
Animals and Tissue Explant Cultures.
Animal husbandry for all of the studies except tissue explant experiments was carried out under Institutional Animal Care and Use Committee guidelines and the procedures were conducted as described in animal protocols approved by Northwestern University, Vanderbilt University, the University of Texas Southwestern Medical Center at Dallas, and the University of Texas Health Science Center at Houston (UTHSC-H). Per2::LucSV heterozygotes intercrossed to generate F2 or backcrossed to C57BL/6J. At 8–12 wk of age they were transferred into individual cages equipped with running wheels in LD12:12. After a minimum of 7 d of entrainment to LD12:12, animals were transferred to constant darkness (DD) for 3 wk. Mice were again subjected to LD12:12 for 3 wk, followed by another 3-wk DD exposure. Six-hour light pulses (fluorescent light, 300 lx) were given at circadian time 16 (4 h after activity onset) on day 21 during the second DD exposure. Animals were then returned to DD for 2 wk. WT, Per2::Luc mice or Per2::LucSV mice were killed between ZT11 and ZT12. SCN tissues were isolated from 300-μm coronal sections, and pituitary tissues were dissected and kept in chilled Hanks’ buffered salt solution (Invitrogen). All dissected tissues were cultured on Millicell culture membranes (PICMORG50, Millipore) and placed in 35-mm tissue culture dishes containing 2 mL DMEM media (Mediatech) supplemented with 352.5 μg/mL sodium bicarbonate, 10 mM Hepes (Invitrogen), 2 mM l-glutamine, 2% B-27 serum-free supplement (Invitrogen), 25 units/mL penicillin, 25 μg/mL streptomycin (Invitrogen), and 0.1 mM luciferin potassium salt (l-8240; Biosynth AG). Sealed dishes were placed in LumiCycle luminometers (Actimetrics) and bioluminescence was recorded continuously.
Real-Time PCR and Western Blotting.
RNA isolation, real-time PCR, and immunoblotting were performed as described previously (23, 34, 65). For Per2 qPCR, Per2-3173f: GCTTCTGGTCTGGACTGCAC, Per2-3257r: GAGTGTCTGAGG GCTCGTTG were used. Briefly, tissues were homogenized at 4 °C in 10 vol of lysis buffer (0.1 M NaC, 20 mM Hepes, pH 7.9, 1 mM EDTA, 1 mM DTT, 0.1% Triton X-100, 20 mM NaF, 1 mM Na3OV4) (Complete mini, Roche). Homogenates were cleared by centrifugation for 20 min at 15,600 × g. Twelve micrograms of protein samples were separated by 10% or 6% SDS-polyacrylamide gel and then transferred to a nitrocellulose membrane (NEN). Five percent nonfat dry milk in Tris-buffered saline containing 0.05% Tween 20 was used for blocking buffer. PER2 (epitope: amino acids 41–180), CRY1 (epitope: amino acids 496–606), CRY2 (epitope: amino acids 514–592), and BMAL1 antibodies (epitope: amino acids 1–220) were generated using guinea pig or chicken (Cocalico Biologicals) and serum was affinity purified using the same protein or peptide used to raise the antibody. β-Actin and α-tubulin from Santa Cruz were used as controls for immunoblotting.
LM-PATs.
LM-PATs were performed as described previously (40) with a slight modification. In brief, poly(A)-enriched RNAs (∼150 ng) were first incubated with 5′-phosphorylated oligo(dT)15 in the presence of T4 DNA ligase for 30 min at 42 °C to anneal with poly(A) tails of RNAs, followed by an excess amount of anchor primer with oligo(dT)12 (5′-GCGAGCTCCGCGGCCGCGTTTTTTTTTTTT-3′) to anneal at the end of poly(A) tails, and further incubated for 2 h at 12 °C to complete ligation between oligo(dT)s. These oligo(dT)-annealed RNAs were then subjected to reverse transcription reaction using SuperScript II (Invitrogen) for cDNA synthesis. Aliquots of this cDNA were used as templates for PCR reactions with Per2 3′ end-specific primers (sequences can be found below). Then, PCR products were digested by either XbaI (Per2::LucSV) or DraI (Per2::Luc) to confirm the PCR specificity. Resulting DNA fragments were visualized by Alpha Multiphotoimager II (AlphaInnotech), and poly(A) tail lengths were calculated by AlphaImager software (AlphaInnotech). The primer sequences for PAT_SV40 (Per2::LucSV) and PAT_Per2_DraI (Per2::Luc) are GTGGACGAAGTACCGAAAGGTC and GCCAAGCATCCAGCCCTGTTTTC, respectively.
Plasmids and Small Hairpin RNAs.
Serial truncations of the Per2 3′-UTR in the luciferase reporter construct containing an ecdysone-inducible expression system was generated by PCR mutagenesis. miRNA binding site mutant constructs (miR30a-1508 mut, miR30a-1602 mut, miR24-1686 mut, miR21-1708 mut, and miR25-1797 mut) were generated by DNA synthesis (GenScript) and subsequent insertion into the WT-5 UTR sequence. Site-directed mutagenesis was performed by using miR24-1686 as a template to generate the miR-24/-30 double mutant with the following primers: 5′-miR-30 mutF: AGGATAATTTTGTGAATCATGAGCGCAGATGCCAAGCATCCAGC and 5′-miR-30 mutR: GCTGGATGCTTGGCATCTGCGCTCATGATTCACAAAATTATCCT.
Cell Culture and Reporter Assay.
Per2::Luc and Per2::LucSV mouse embryo fibroblasts (MEFs) were isolated from 12.5 d postcoitum (DPC) embryos and immortalized (23). Per2::Luc MEF were transfected with miRIDIAN microRNA mimic or inhibitor for miR-21, miR-24, miR-25, and miR-30 (Dharmacon), and bioluminescence was measured in LumiCycle luminometers 24 h after transfection (Actimetrics). For the reporter assays, NIH 3T3 (2 × 105) cells, plated the day before were transfected with indicated vectors by using iMFectin (GenDEPOT). Thirty hours after transfection, cells were lysed and luminescence was measured (Promega) using a luminometer (Auto-Lumet Plus, Berthold Technologies).
Temperature Compensation.
Temperature compensation experiments were performed as described before (43). All animal studies were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee at Vanderbilt University (protocol M/03/153).
Statistical Analysis.
Data are presented as means ± SEM. Statistical significance was determined by t test, one-way ANOVA, and two-way ANOVA by using the GraphPad Prism software. P < 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank A. L. Joyner for providing W4 ES cells, L. Doglio for performing ES cell microinjections, I. Kornblum and J.-H. Choe for technical support, and other members of the S.-H.Y. and J.S.T. laboratories for advice and discussion. This work was supported in part by NIH/National Institute of General Medical Sciences (NIGMS) Grant R01GM114424 (to S.-H.Y.); The Welch Foundation, AU-1731 and NIH/National Institute on Aging Grants R01AG045828 and R01AG045828-04S1 (to Z.C.); NIH Grant NS099813 (to C.L.); NARSAD Young Investigator Grant 21267, the Sumitomo Foundation Grant for Basic Science Research Projects 150056, and the Tomizawa Jun-ichi & Keiko Fund of Molecular Biology Society of Japan for Young Scientists (to S.K.); Japan Society for the Promotion of Science (JSPS) KAKENHI JP26293048, the Uehara Memorial Foundation (to N.K.); JSPS 15H04683 and JSPS 16H01880 (to K.Y.); NIH/NINDS NS051278 (to S.Y.); NIH/NIGMS R01GM112991 and R01GM111387 (to C.B.G.); and NIH/National Institute of Mental Health (NIMH) Silvio O. Conte Center P50MH074924 and NIH/NIMH U01MH61915 (to J.S.T.). J.S.T. is an Investigator in the Howard Hughes Medical Institute, and a cofounder and Scientific Advisory Board member of Reset Therapeutics, Inc., a biotech company working on circadian rhythms and metabolism.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706611114/-/DCSupplemental.
References
- 1.Pittendrigh CS. Circadian rhythms and the circadian organization of living systems. Cold Spring Harb Symp Quant Biol. 1960;25:159–184. doi: 10.1101/sqb.1960.025.01.015. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardin PE, Panda S. Circadian timekeeping and output mechanisms in animals. Curr Opin Neurobiol. 2013;23:724–731. doi: 10.1016/j.conb.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herzog ED, Hermanstyne T, Smyllie NJ, Hastings MH. Regulating the suprachiasmatic nucleus (SCN) circadian clockwork: Interplay between cell-autonomous and circuit-level mechanisms. Cold Spring Harb Perspect Biol. 2017;9:a027706. doi: 10.1101/cshperspect.a027706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18:164–179. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kume K, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 7.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 8.Yagita K, et al. Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes Dev. 2000;14:1353–1363. [PMC free article] [PubMed] [Google Scholar]
- 9.Yagita K, et al. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 2002;21:1301–1314. doi: 10.1093/emboj/21.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen R, et al. Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol Cell. 2009;36:417–430. doi: 10.1016/j.molcel.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hampp G, et al. Regulation of monoamine oxidase A by circadian-clock components implies clock influence on mood. Curr Biol. 2008;18:678–683. doi: 10.1016/j.cub.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 12.Akashi M, et al. A positive role for PERIOD in mammalian circadian gene expression. Cell Rep. 2014;7:1056–1064. doi: 10.1016/j.celrep.2014.03.072. [DOI] [PubMed] [Google Scholar]
- 13.Ogawa Y, et al. Positive autoregulation delays the expression phase of mammalian clock gene Per2. PLoS One. 2011;6:e18663. doi: 10.1371/journal.pone.0018663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye R, Selby CP, Ozturk N, Annayev Y, Sancar A. Biochemical analysis of the canonical model for the mammalian circadian clock. J Biol Chem. 2011;286:25891–25902. doi: 10.1074/jbc.M111.254680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 16.Sato TK, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 17.Hogenesch JB, Herzog ED. Intracellular and intercellular processes determine robustness of the circadian clock. FEBS Lett. 2011;585:1427–1434. doi: 10.1016/j.febslet.2011.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, et al. Circadian amplitude regulation via FBXW7-targeted REV-ERBα degradation. Cell. 2016;165:1644–1657. doi: 10.1016/j.cell.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu B, et al. Coactivator-dependent oscillation of chromatin accessibility dictates circadian gene amplitude via REV-ERB loading. Mol Cell. 2015;60:769–783. doi: 10.1016/j.molcel.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 21.Kojima S, Shingle DL, Green CB. Post-transcriptional control of circadian rhythms. J Cell Sci. 2011;124:311–320. doi: 10.1242/jcs.065771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim C, Allada R. Emerging roles for post-transcriptional regulation in circadian clocks. Nat Neurosci. 2013;16:1544–1550. doi: 10.1038/nn.3543. [DOI] [PubMed] [Google Scholar]
- 23.Yoo SH, et al. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell. 2013;152:1091–1105. doi: 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J, et al. Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Mol Cell Biol. 2008;28:6056–6065. doi: 10.1128/MCB.00583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reischl S, Kramer A. Kinases and phosphatases in the mammalian circadian clock. FEBS Lett. 2011;585:1393–1399. doi: 10.1016/j.febslet.2011.02.038. [DOI] [PubMed] [Google Scholar]
- 26.Cardone L, et al. Circadian clock control by SUMOylation of BMAL1. Science. 2005;309:1390–1394. doi: 10.1126/science.1110689. [DOI] [PubMed] [Google Scholar]
- 27.Mehta N, Cheng HY. Micro-managing the circadian clock: The role of microRNAs in biological timekeeping. J Mol Biol. 2013;425:3609–3624. doi: 10.1016/j.jmb.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 28.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 29.Chen R, D’Alessandro M, Lee C. miRNAs are required for generating a time delay critical for the circadian oscillator. Curr Biol. 2013;23:1959–1968. doi: 10.1016/j.cub.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kadener S, et al. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 2009;23:2179–2191. doi: 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kojima S, Gatfield D, Esau CC, Green CB. MicroRNA-122 modulates the rhythmic expression profile of the circadian deadenylase Nocturnin in mouse liver. PLoS One. 2010;5:e11264. doi: 10.1371/journal.pone.0011264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo W, Sehgal A. Regulation of circadian behavioral output via a microRNA-JAK/STAT circuit. Cell. 2012;148:765–779. doi: 10.1016/j.cell.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng HY, et al. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoo SH, et al. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoo SH, et al. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc Natl Acad Sci USA. 2005;102:2608–2613. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Z, et al. Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc Natl Acad Sci USA. 2012;109:101–106. doi: 10.1073/pnas.1118034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sallés FJ, Strickland S. Analysis of poly(A) tail lengths by PCR: The PAT assay. Methods Mol Biol. 1999;118:441–448. doi: 10.1385/1-59259-676-2:441. [DOI] [PubMed] [Google Scholar]
- 39.Kojima S, Green CB. Analysis of circadian regulation of poly(A)-tail length. Methods Enzymol. 2015;551:387–403. doi: 10.1016/bs.mie.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kojima S, Sher-Chen EL, Green CB. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 2012;26:2724–2736. doi: 10.1101/gad.208306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 42.Miki T, et al. PML regulates PER2 nuclear localization and circadian function. EMBO J. 2012;31:1427–1439. doi: 10.1038/emboj.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reyes BA, Pendergast JS, Yamazaki S. Mammalian peripheral circadian oscillators are temperature compensated. J Biol Rhythms. 2008;23:95–98. doi: 10.1177/0748730407311855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lal A, et al. miR-24 inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagel R, Clijsters L, Agami R. The miRNA-192/194 cluster regulates the Period gene family and the circadian clock. FEBS J. 2009;276:5447–5455. doi: 10.1111/j.1742-4658.2009.07229.x. [DOI] [PubMed] [Google Scholar]
- 46.Chen X, Rosbash M. mir-276a strengthens Drosophila circadian rhythms by regulating timeless expression. Proc Natl Acad Sci USA. 2016;113:E2965–E2972. doi: 10.1073/pnas.1605837113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 48.Bae K, et al. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 49.Zheng B, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 50.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schroeder AM, Colwell CS. How to fix a broken clock. Trends Pharmacol Sci. 2013;34:605–619. doi: 10.1016/j.tips.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nohara K, Yoo SH, Chen ZJ. Manipulating the circadian and sleep cycles to protect against metabolic disease. Front Endocrinol (Lausanne) 2015;6:35. doi: 10.3389/fendo.2015.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schibler U, Naef F. Cellular oscillators: Rhythmic gene expression and metabolism. Curr Opin Cell Biol. 2005;17:223–229. doi: 10.1016/j.ceb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 54.Lee Y, Chen R, Lee HM, Lee C. Stoichiometric relationship among clock proteins determines robustness of circadian rhythms. J Biol Chem. 2011;286:7033–7042. doi: 10.1074/jbc.M110.207217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He B, et al. The small molecule nobiletin targets the molecular oscillator to enhance circadian rhythms and protect against metabolic syndrome. Cell Metab. 2016;23:610–621. doi: 10.1016/j.cmet.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nohara K, et al. Ammonia-lowering activities and carbamoyl phosphate synthetase 1 (Cps1) induction mechanism of a natural flavonoid. Nutr Metab (Lond) 2015;12:23. doi: 10.1186/s12986-015-0020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gloston GF, Yoo SH, Chen ZJ. Clock-enhancing small molecules and potential applications in chronic diseases and aging. Front Neurol. 2017;8:100. doi: 10.3389/fneur.2017.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330:379–385. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Isojima Y, et al. CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci USA. 2009;106:15744–15749. doi: 10.1073/pnas.0908733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Z, McKnight SL. A conserved DNA damage response pathway responsible for coupling the cell division cycle to the circadian and metabolic cycles. Cell Cycle. 2007;6:2906–2912. doi: 10.4161/cc.6.23.5041. [DOI] [PubMed] [Google Scholar]
- 61.Mehra A, et al. A role for casein kinase 2 in the mechanism underlying circadian temperature compensation. Cell. 2009;137:749–760. doi: 10.1016/j.cell.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Portolés S, Más P. The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet. 2010;6:e1001201. doi: 10.1371/journal.pgen.1001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou M, Kim JK, Eng GW, Forger DB, Virshup DM. A Period2 phosphoswitch regulates and temperature compensates circadian period. Mol Cell. 2015;60:77–88. doi: 10.1016/j.molcel.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 64.Antoch MP, et al. Functional identification of the mouse circadian clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jeong K, et al. Dual attenuation of proteasomal and autophagic BMAL1 degradation in Clock Δ19/+ mice contributes to improved glucose homeostasis. Sci Rep. 2015;5:12801. doi: 10.1038/srep12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.