Significance
Plants have the ability to live and grow for many thousands of years due to the activity of groups of cells called meristems. Meristems contain stem cells that can survive the entire life of the plant and ensure the continuous supply of new cells. Stem cells are thought to be qualitatively different compared with their neighboring daughter cells. Here we show that in the case of the proximal root meristem, there does not seem to be such an on-off type of organization. We show that the majority of transcripts, together with other cellular properties, gradually transition from stem cell activity to differentiation, by opposing gradients. This impacts our understanding of meristem organization and will determine the direction of future research.
Keywords: plant development, root meristem, Arabidopsis, transcriptional regulation
Abstract
In plants, apical meristems allow continuous growth along the body axis. Within the root apical meristem, a group of slowly dividing quiescent center cells is thought to limit stem cell activity to directly neighboring cells, thus endowing them with unique properties, distinct from displaced daughters. This binary identity of the stem cells stands in apparent contradiction to the more gradual changes in cell division potential and differentiation that occur as cells move further away from the quiescent center. To address this paradox and to infer molecular organization of the root meristem, we used a whole-genome approach to determine dominant transcriptional patterns along root ontogeny zones. We found that the prevalent patterns are expressed in two opposing gradients. One is characterized by genes associated with development, the other enriched in differentiation genes. We confirmed these transcript gradients, and demonstrate that these translate to gradients in protein accumulation and gradual changes in cellular properties. We also show that gradients are genetically controlled through multiple pathways. Based on these findings, we propose that cells in the Arabidopsis root meristem gradually transition from stem cell activity toward differentiation.
The root apical meristem (RAM) is responsible for generating the cells that are incorporated into existing cell files (Fig. 1A). Cells close to the root tip have stem cell properties: they generate daughters that are displaced from the tip and ones that remain close to the tip, and could act as long-term progenitors for the cell file (1). Embedded within the root meristem is a region of very low mitotic activity, the quiescent center (QC) (2). Ablation of the QC in Arabidopsis causes neighboring cells in the root cap to become differentiated (3). Thus, the QC has been proposed to maintain a stem cell state in neighboring cells. This property has only been unequivocally shown for the (distal) root cap (3), in which a single nondifferentiated cell layer separates the QC from morphologically differentiated cells (4). However, the concept of a single “stem cell” layer surrounding the QC has been transposed to other tissue initials. The cell directly adjacent to the QC is believed to be the stem cell for the cell file, and have unique properties. Its displaced daughters, similar to transit-amplifying cells, have increased division capacity, which is lost as cells are displaced into the elongation and differentiation zones (reviewed in refs. 5 and 6). In contrast to the binary definition of the stem cell and the nonstem cell daughters, which is anatomically evident in the root cap, the process of cell differentiation in the proximal meristem follows a gradual progression. Differentiation of xylem and phloem cell types (7–9), as well as endodermis (10) and hair-producing epidermal cells (11, 12), is progressive and involves several consecutive steps. A key question is whether the proximal meristem undergoes binary (on–off) cell fate transitions or more gradual differentiation steps. This would be reflected in on–off or gradual gene-expression patterns along the longitudinal axis of the root meristem. While several studies have described genome-wide patterns of gene activity in cell types or zones within the root meristem (13–15), these studies lack the resolution in the longitudinal axis to distinguish these two possibilities. Here we use a dedicated approach to isolate specific cell populations, within the proximal meristem, differing in their distance from the QC. From the transcriptome organization of these different cell populations we conclude that there is a gradual decrease in stem cell-related transcripts and an increase in differentiation-related transcripts with increasing distance from the QC.
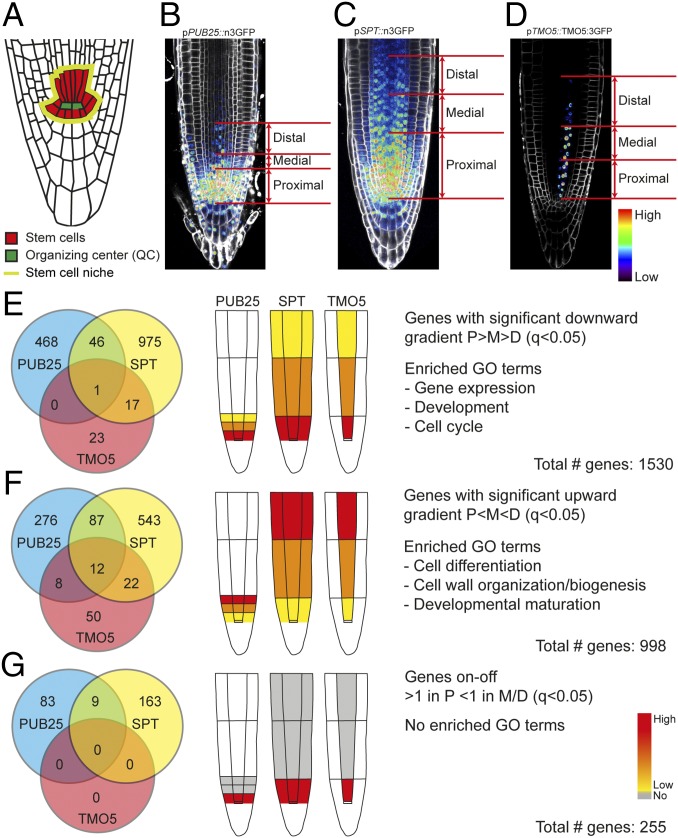
Fig. 1.
Experimental set-up and results of high-resolution datasets. (A) Schematic overview of organization of stem cells in the Arabidopsis RAM. The QC is depicted in green, stem cells in red, and stem cell niche is outlined in light green. (B and C) Schematic indication of isolated cell populations by cell sorting. Three lines were used: two with broad gradients, one steep (B) the other shallow (C), and a cell-type–specific line (D). (E) Genes identified (1,530 in total) in gradients emanating from proximal populations (P > M > D). Enriched GO terms include those associated with developmental processes. (F) Genes identified (998 in total) in P < M < D gradient. Enriched GO terms include those associated with differentiation processes. (G) Genes identified (255 in total) expressed only in proximal cell population. No GO-terms were enriched. Micrographs (B–D) were taken at the same magnification.
Results
Intensity-Based Cell-Sorting Can Separate Cell Populations Along Gradients.
To molecularly characterize the organization of the proximal meristem, we used FACS (16). By separating cells into different pools based on fluorescence intensity, we expected to separate the meristem into ontogenetic zones. As a proof-of-concept, we initially made use of the xylem-specific TARGET OF MONOPTEROS5 (TMO5) gene reporter (pTMO5::TMO5:3GFP) (17). In addition to being cell-type–specific, this gene shows high expression close to the QC, which gradually decreases shootward (SI Appendix, Fig. S1C). GFP+ cells were divided into two populations, one with high-GFP (i.e., cells close to the QC) and one with low-GFP expression (i.e., cells away from the QC) (SI Appendix, Fig. S2). Microarray analysis revealed a large number of genes with differential expression [1,227 genes with fold-change > 1.3 and significantly changed (q < 0.05) in high vs. low; including, HISTIDINE PHOSPHOTRANSFER PROTEIN6 (AHP6) (18), LONELY GUY3 (LOG3) (19, 20), several CYCLIN genes, and TMO5 itself] (Dataset S1). Gene Ontology (GO) enrichment analysis revealed significant enrichment of “developmental and cell cycle processes” in the “high” population (Dataset S2). Because these results indicate that it is possible to obtain informative cell-type–specific transcriptomic data from intensity-based sorted cells, we next exhaustively sampled cell populations along expression gradients. Sorting was performed to generate complementary datasets: one cell-type–specific xylem dataset and two “general” datasets (across cell types). The same TMO5 reporter line was used for the cell-type–specific dataset, while two general gradient lines with either a short/steep (pPLANT U-BOX25; pPUB25::n3GFP) (21) or a long/shallow (pSPATULA; pSPT::n3GFP) (21) gradient were used for the general datasets (Fig. 1 B–D and SI Appendix, Fig. S1). The shallow gradient in the SPT line should inform about expression patterns across the entire meristem, while the steep gradient in the PUB25 line should provide increased resolution in the region closest to the QC. We employed the same differential cell sorting method as described above, except that GFP+ cells were divided into three populations: proximal (P, with highest GFP signal), medial (M, with intermediate GFP signal), and distal to the QC (D, with lowest GFP signal), schematically represented in Fig. 1 B–D. It should be noted that expression in the columella can be observed in both pPUB25-n3GFP and pSPT-n3GFP lines, and these cells are likely sorted into one of the zones, along with cells from the proximal meristem. Given that there are between 24 and 30 columella cells, and that each zone in the proximal meristem conservatively consists of more than 300 cells, we do not expect that cosorting of root cap cells will strongly influence gene-expression profiles. Following differential sorting, transcript levels of PUB25 and SPT were evaluated in each sample by qPCR. These transcripts were found to be about 10-fold higher in the proximal population compared with the distal population (and intermediate in the medial population) in agreement with the observed GFP signal in the root (SI Appendix, Fig. S3). Thus, this validates the use of GFP levels as a proxy for transcript levels, and also demonstrates that differential cell sorting could successfully separate cell populations along gradients.
Transcript Profiling Reveals Dominant Opposing Transcriptional Gradients.
After cell sorting, we used total RNA for transcriptome profiling by RNA-sequencing. Differential gene expression was calculated through comparisons between each of the cell populations (SI Appendix, Table S1 and Dataset S3). Similar to our qPCR results, transcript levels of PUB25, SPT, and TMO5 were found to gradually decrease in the cell populations farther away from the QC (SI Appendix, Fig. S4 A–C). Further statistical analysis, using hierarchical clustering and principal component analysis, indicated that the cell-type–specific TMO5 dataset is distinct from the general datasets of PUB25 and SPT, and that sampling of similar cell populations (SPT proximal and three regions of PUB25) from different gradient lengths resulted in similar datasets. As predicted, all three PUB25 cell populations clustered with the proximal cell population derived from the SPT line (SI Appendix, Fig. S4 D and E).
Quality threshold-clustering (22) of all significantly differentially expressed genes (q < 0.05) resulted in six dominant expression profiles (SI Appendix, Fig. S5 A–C). When performing the same clustering after removing all genes expressed in the root cap (above an expression level of 5 in ref. 14), we identified identical dominant clusters for both SPT and PUB25 datasets (SI Appendix, Fig. S5 D and E), further supporting the idea that columella cosorting has negligible effects on the data, and suggesting that the sorted population indeed represent zones in the proximal meristem.
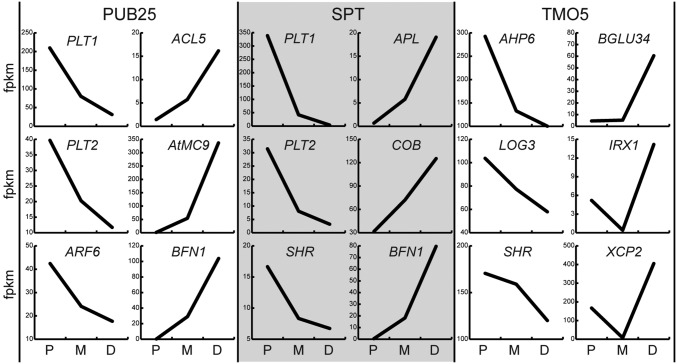
Interestingly, most of the genes (TMO5: 42%, PUB25: 60%, and SPT: 77%) showed a graded expression profile; either downward (P > M > D) or upward (P < M < D). Specifically, over 1,500 genes showed a graded expression profile that was higher in the proximal population and significantly reduced in the other populations (i.e., a downward gradient, P > M > D, q < 0.05) (Fig. 1E and Dataset S4) over all of the datasets. The enriched GO terms from this cluster were related to “gene expression,” “development,” and “cell cycle” (Fig. 1E and Dataset S2). Accordingly, expression profiles of genes known to be important for root meristem development and cell division [e.g., AHP6, AUXIN RESPONSE FACTOR6 (ARF6), ARF8, KINESIN-12B, LOG3, PIN-FORMED1 (PIN1), PIN4, PLETHORA1 (PLT1), PLT2, SHORT-ROOT, ZWILLE, and several CYCLIN and CYCLIN-DEPENDENT KINASE (CDK) genes] (18–20, 23–29) exhibited this graded profile (Fig. 2). Interestingly, analysis of the overlapping genes between the datasets revealed a set of 468 genes with a downward gradient, specific for the PUB25 dataset. When we examined the expression profile of these genes in the SPT dataset, we found that 217 (46%) were not significantly changed between any of the populations in SPT, while most of the remaining genes [146 (31%)] were expressed significantly lower in the SPT medial population compared with the proximal population, but did not change further shootward. This confirmed our expectation that the steeper gradient of PUB25 is capable of capturing expression dynamics at a higher resolution than SPT, increasing the overall resolution of our datasets. The clustering analysis also identified many genes with a graded expression profile in the opposite direction. Analysis of genes with low but detectable expression [>one fragment per kilobase of exon per million fragments (FPKM)], in the proximal population revealed around 1,000 genes with significantly increased expression in the more distal cell populations (i.e., an upward gradient, P < M < D, q < 0.05) (Fig. 1F and Dataset S4). GO terms for genes in this category associated mostly with “cell differentiation and maturation processes” (Fig. 1F and Dataset S2). These included ACAULIS 5, BIFUNCTIONAL NUCLEASE 1, COBRA, IRREGULAR XYLEM 1, and XYLEM CYSTEINE PEPTIDASE 2 genes (Fig. 2), known to control aspects of differentiation (30–34). This suggests that genes associated with differentiation are expressed in very young meristematic cells, and increase in expression level as cells progress toward differentiation.
Fig. 2.
Graphs depicting expression profiles of genes known for their involvement in development (Left) or differentiation (Right) in PUB25, SPT, and TMO5 datasets, showing opposite profiles. FPKM expression level in proximal (P), medial (M), or distal (D) population is shown.
Importantly, there were no dominant clusters resembling a binary expression profile (SI Appendix, Fig. S5). We tested if our analysis has the depth and analytical power to identify genes with such expression patterns by exploring the profiles of well-established markers for small cell populations in the QC or cells in its vicinity. The ACR4, CYCD6, NTT, and WIP4 genes all displayed the predicted binary profile. Hence, such genes are identified by our analysis. We next identified all genes showing this pattern and investigated what functions are associated (Methods, Fig. 1G, and Dataset S4). While this resulted in 255 genes (over all datasets; none were found in the TMO5 dataset), including the abovementioned NTT and WIP4, there were no significantly enriched GO-terms associated with these genes (Fig. 1G).
Thus, instead of discrete zones of stem cells, division, and differentiation, most genes in our datasets are expressed in opposing gradients of stem cell activity and differentiation potential.
Transcriptional Gradients Translate to Protein Gradients and Correlate with Gradual Differentiation in the RAM.
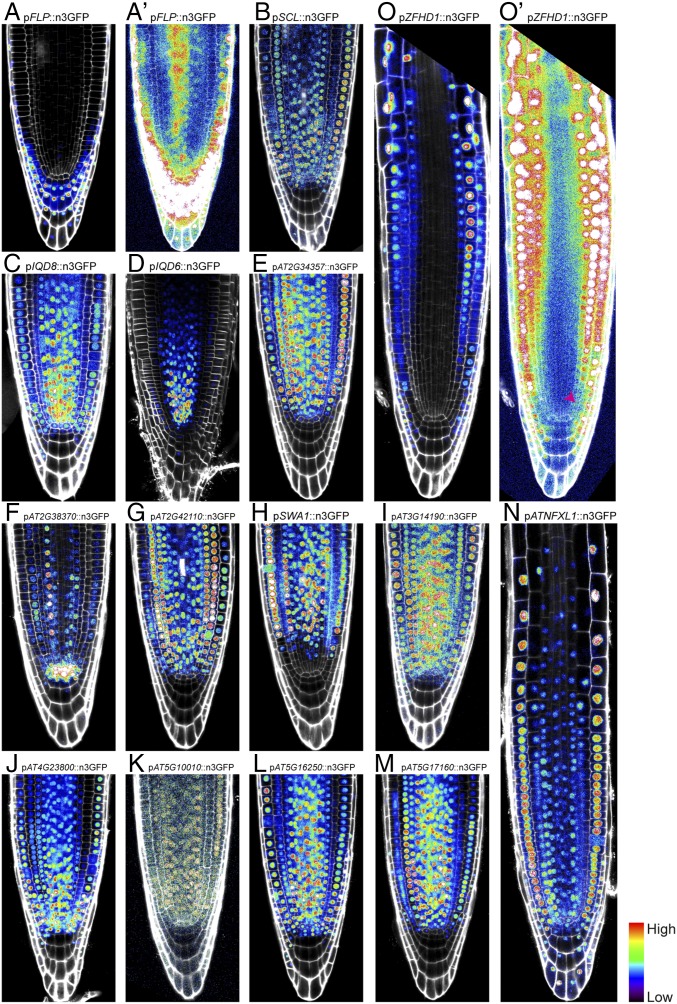
To determine if the transcript gradients observed in our RNA-sequencing datasets are a consequence of complex posttranscriptional regulation, or are generated by direct transcriptional control, we made reporter lines for 16 genes predicted to show a gradient of expression (SI Appendix, Fig. S6). Fifteen of the 16 lines showed the predicted expression gradient, and the remaining line could not be detected in the root (Fig. 3, Table 1, and SI Appendix, Fig. S7). Thus, finding that the vast majority of the sampled genes followed the predicted expression pattern indicated that the RNA-sequencing expression profiles can be highly predictive. Notably, lines 3 (IQD8), 4 (IQD6), 8 (AT2G42110), and 11 (AT4G23800) (Fig. 3 C, D, G, and J) showed very strong expression in the proximal region that gradually decreased. Expression of genes following this downward gradient (P > M > D) were more easily imaged than those following an upward gradient. This is likely due to detection limits of the microscope, as expression levels of genes in an opposite gradient are generally very low in the proximal region (Fig. 3O′). Overall, these data suggest that these expression profiles are likely generated by direct transcriptional output and promoter activity.
Fig. 3.
Overview of expression profile as visualized by promoter activity in confocal micrographs, of selected genes: all lines with visible expression in the root tip follow the RNA-sequencing-predicted graded expression profile. (A) 1: FLP; (A′) overexposed image of A; (B) 2: SCL; (C) 3: IQD8; (D) 4: IQD6; (E) 5: AT2G34357; (F) 7: AT2G38370; (G) 8: AT2G42110; (H) 9: SWA1; (I) 10: AT3G14190; (J) 11: AT4G23800; (K) 12: AT5G10010; (L) 13: AT5G16250; (M) 14: AT5G17160; (N) 16: ATNFXL1; (O) 17: ZFHD1; (O′) overexposed image of O shows very low signal in first meristematic cells (pink arrowhead). Micrographs were taken at the same magnification.
Table 1.
Expression values, fold-changes, and observed expression domain of selected genes
| No. | AGI | Short description | FC | FPKM | Observed expression pattern | |||||||||
| LR | TMO-P | TMO-M | TMO-D | PUB-P | PUB-M | PUB-D | SPT-P | SPT-M | SPT-D | Root | Regulated by PLT1/2* | |||
| 1 | AT1G14350 | FLP | 1.57 | 38.57 | 36.95 | 31.27 | 17.59 | 7.58 | 8.69 | 15.43 | 8.57 | 6.95 | P > D Gradient† (p) | No |
| 2 | AT1G63100 | SCL | 1.69 | 68.83 | 53.79 | 42.91 | 34.11 | 23.48 | 22.34 | 36.91 | 8.73 | 3.10 | P > D Gradient (p) | No |
| 3 | AT1G72670 | IQD8 | 2.69 | 25.74 | 19.30 | 13.33 | 11.69 | 8.03 | 8.48 | 7.76 | 2.82 | 0.62 | P > D Gradient | Yes+ |
| 4 | AT2G26180 | IQD6 | 2.05 | 73.89 | 45.14 | 35.85 | 11.70 | 7.76 | 14.20 | 14.51 | 3.74 | 1.79 | P > D Gradient (p + g) | Yes+ |
| 5 | AT2G34357 | ARM repeat protein | 1.57 | 48.74 | 43.09 | 34.31 | 17.73 | 16.78 | 24.41 | 22.73 | 9.11 | 6.95 | P > D Gradient (p) | Yes+ |
| 6 | AT2g38160 | Unknown protein | 1.84 | 41.02 | 24.75 | 16.82 | 23.64 | 11.75 | 1.10 | 18.25 | 7.41 | 2.44 | ND | No |
| 7 | AT2G38370 | Unknown protein | 1.86 | 44.81 | 34.09 | 20.85 | 11.29 | 8.77 | 10.66 | 14.30 | 5.33 | 2.14 | P > D Gradient (p) | Yes+ |
| 8 | AT2G42110 | Unknown protein | 2.20 | 834.66 | 389.55 | 333.09 | 82.49 | 92.37 | 62.94 | 153.11 | 80.45 | 0.00 | P > D Gradient (p) | No |
| 9 | AT2G47990 | SWA1 | 1.55 | 65.08 | 62.28 | 50.50 | 29.22 | 31.83 | 45.30 | 41.58 | 26.85 | 13.87 | P > D Gradient (p) | Yes+ |
| 10 | AT3G14190 | Unknown protein | 2.04 | 41.09 | 25.01 | 20.81 | 25.53 | 20.99 | 15.13 | 12.82 | 11.82 | 13.55 | P > D Gradient (p) | No |
| 11 | AT4G23800 | HMG family protein | 2.12 | 211.09 | 135.06 | 99.43 | 147.95 | 92.50 | 96.52 | 108.21 | 32.33 | 14.12 | P > D Gradient (p + g) | No |
| 12 | AT5G10010 | Unknown protein | 1.52 | 43.41 | 43.00 | 33.39 | 17.79 | 19.94 | 26.55 | 26.79 | 19.02 | 14.37 | P > D Gradient (p) | No |
| 13 | AT5G16250 | Unknown protein | 2.50 | 65.80 | 37.66 | 27.53 | 46.66 | 27.45 | 21.79 | 33.58 | 13.59 | 9.86 | P > D Gradient (p) | No |
| 14 | AT5G17160 | Unknown protein | 2.10 | 62.36 | 37.89 | 26.90 | 15.43 | 14.67 | 19.85 | 15.39 | 3.88 | 1.56 | P > D Gradient (p) | Yes+ |
| 15 | AT5G67390 | Unknown protein | 1.68 | 24.39 | 16.72 | 12.70 | 0.43 | 4.44 | 6.46 | 2.00 | 5.72 | 8.67 | ND | No |
| 16 | AT1G10170 | ATNFXL1 | −1.68 | 10.97 | 11.08 | 17.96 | 34.26 | 65.63 | 47.62 | 29.68 | 66.50 | 127.43 | P < D Gradient (p) | No |
| 17 | AT1G69600 | ZFHD1 | −1.83 | 5.23 | 11.64 | 14.48 | 17.90 | 24.29 | 18.87 | 11.06 | 38.05 | 50.68 | P < D Gradient (p) | No |
| 18 | AT2G43680 | IQD14 | −2.31 | 11.00 | 14.24 | 31.30 | 12.72 | 17.18 | 19.52 | 23.33 | 58.01 | 65.47 | No expression (p) | No |
| 19 | AT2G44840 | ERF13 | −2.45 | 27.84 | 31.76 | 51.55 | 373.37 | 212.88 | 183.69 | 162.44 | 254.96 | 169.86 | P < D Gradient (g) | No |
| 20 | AT3G58120 | BZIP61 | ND | 13.81 | 26.56 | 41.93 | 4.08 | 49.61 | 68.36 | 6.14 | 22.12 | 72.08 | P < D Gradient (g) | Yes− |
FC, fold-change high-GFP vs. low-GFP population; g, tested by translational (genomic) fusion; LR, low-resolution TMO5 dataset; ND, Not determined; p, tested by transcriptional (promoter) reporter; PUB-D, PUB25 distal dataset; PUB-M, PUB25 medial dataset; PUB-P, PUB25 proximal dataset; SPT-D, SPT distal dataset; SPT-M, SPT medial dataset; SPT-P, SPT proximal dataset; TMO-D, TMO5 Distal dataset; TMO-M, TMO5 medial dataset; TMO-P, TMO5 proximal dataset. Regulation by PLT1/2: Yes+, positive regulation; Yes−, negative regulation;
Santuari et al. (37).
Very weak expression in vascular cells showing P > D gradient, strong expression in root cap and columella.
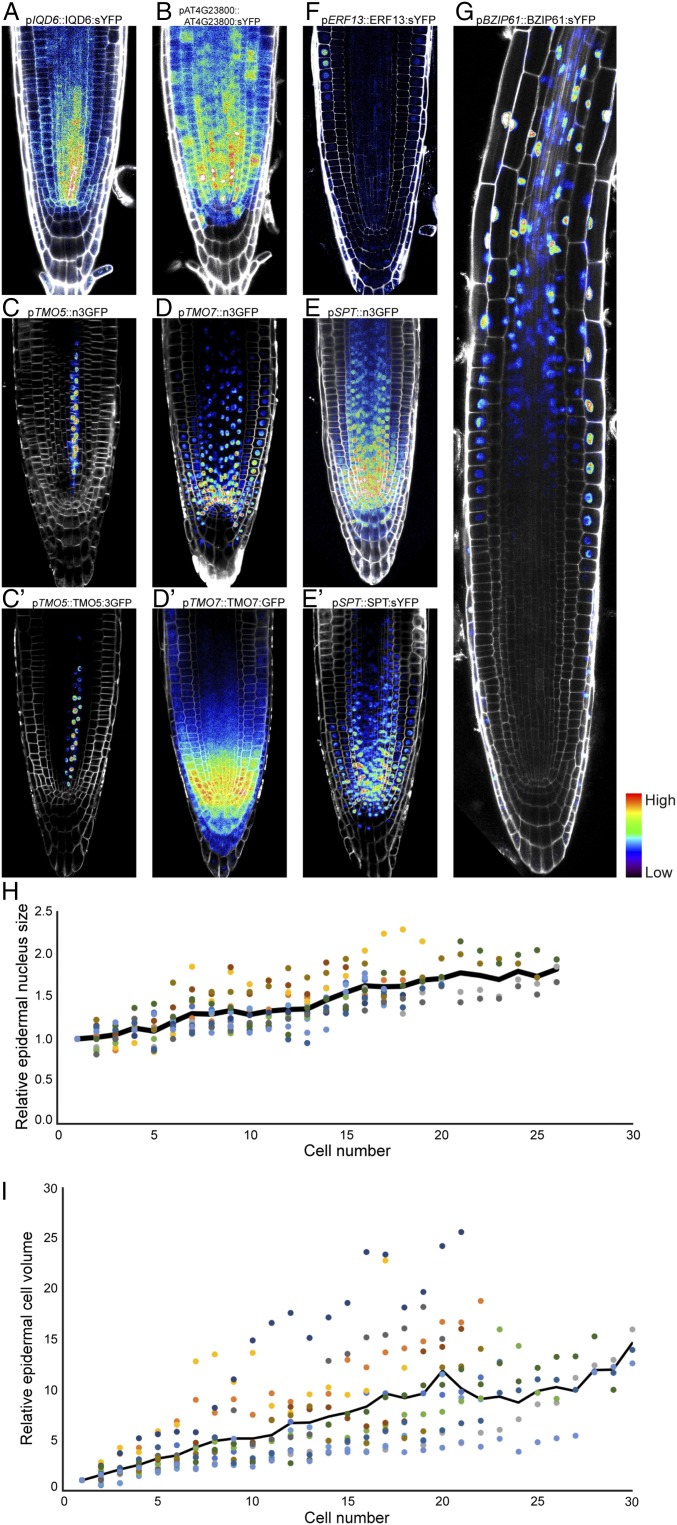
Next, we addressed whether posttranscriptional control influences the output of the observed transcriptional gradients. To this end, we analyzed translational fusion proteins of seven different genes whose expression formed a gradient. These included four genes identified here (4: IQD6, 11: AT4G23800, 19: ERF13, and 20: BZIP61), as well as previously identified TMO5 (17), TMO7 (17), and SPT (21). In all cases, the protein reporters were expressed in similar graded protein abundance, as seen for their transcripts (Table 1, SI Appendix, Fig. S4 B and C, and Dataset S3) and transcriptional fusion reporters (Figs. 3 D and J and 4 A–G). This indicates that transcriptional gradients can be directly translated into protein gradients.
Fig. 4.
Transcriptional gradients are translated to gradients in protein abundance and correlate with cellular properties. (A–G) Translational fusions of 4: IQD6 (A), 11: AT4G23800 (B), TMO5 (C and C′), TMO7 (D and D′), SPT (E and E′), 19: ERF13 (F), and 20: BZIP61 (G) indicate similar protein gradients as observed for their transcripts (compare with Figs. 3 D and J, panels C–E, and Table 1, respectively). (H and I) Quantification of epidermal nuclear size (H) and cell volume (I) relative to cell contacting QC reveals progressive increase of both cellular parameters. Black line indicates mean relative size/volume, of 13 (nuclear size) and 18 (cell volume) cell files. Micrographs (A–G) were taken at the same magnification.
Cellular properties are defined by the activities of their proteins. Therefore, a key question is whether the gradual molecular organization of the root meristem (by transcript and protein gradients) also translates into gradual cellular changes in the root meristem. In support of this hypothesis, xylem (8, 9), phloem (7), and endodermis (10) differentiation processes have been shown to involve several successive steps. However, each of these processes commences in cells that are some distance from the QC. It is unclear if gradual cellular changes can be observed in the youngest meristem cells. We therefore imaged cellular properties (nucleus size and cell volume) along the meristem, starting in the cells adjacent to the QC. We found that both epidermis nuclear size (Fig. 4H) and cell volume (Fig. 4I and SI Appendix, Fig. S8) did not abruptly increase along the meristem, but instead follow a graded pattern of increase. Thus, gradients of gene expression in the root meristem correlate with the existence of gradients of protein accumulation, as well as gradual cellular reorganization.
Expression Gradients Can Be Uncoupled from Meristem Size.
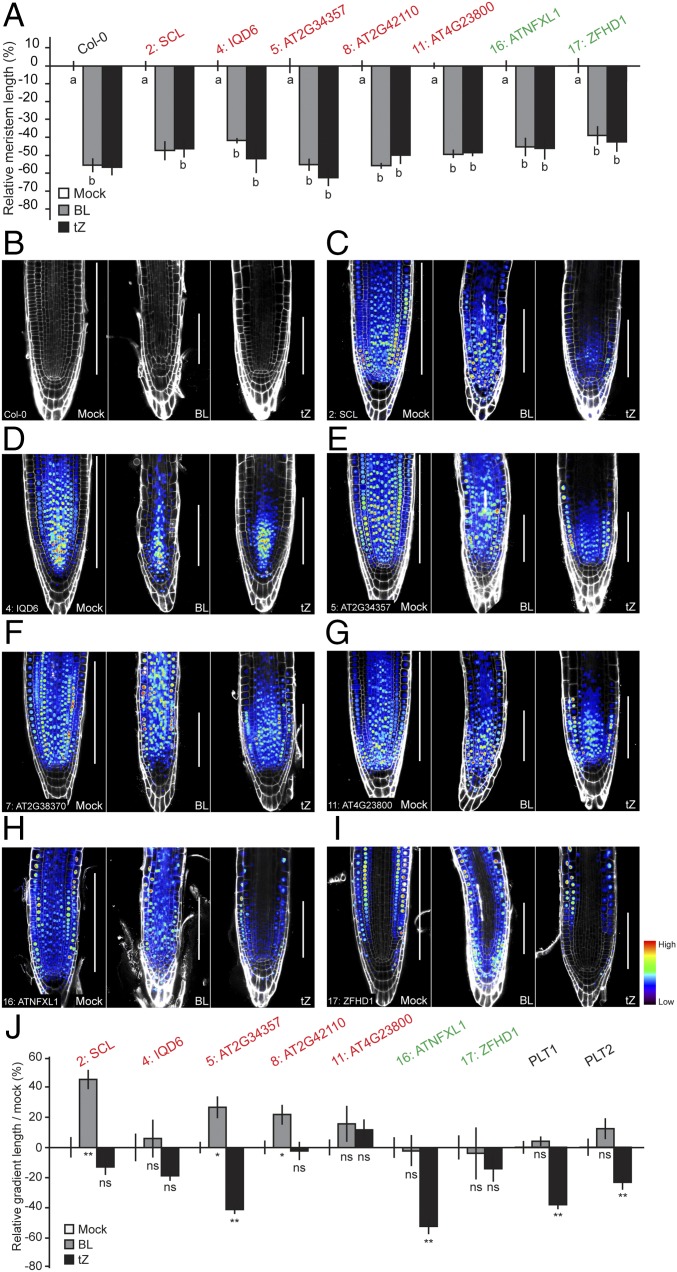
Although our data suggest that a large number of genes are expressed in gradients along the RAM, it remains unclear how and if these gradients are developmentally regulated. One possibility is that gradients form through inheritance of regulators during cell division. Alternatively, the gradient-forming mechanism may set gradient length independently of growth (SI Appendix, Fig. S9A). Therefore, we asked if the identified transcriptional gradients scale proportionally with meristem size. To address this question, we exogenously applied brassinolide (BL) and trans-zeatin (tZ). Treatments with these phytohormones decrease meristem length, which is easily quantifiable by measuring meristem length from QC to the first elongated cortical cell (Fig. 5 A and B). We used the fluorescent reporter lines of gradient-expressed genes isolated from the RNA-sequencing analysis to determine the extent of expression of these gradient-expressed genes in plants treated with BL or tZ (Methods and SI Appendix, Fig. S9B). We first established that all lines showed wild-type sensitivity to BL and tZ treatment (Fig. 5A). However, only two of these (lines 5: AT2G34357 and 16: ATNFXL1) and the known gradient-expressed PLT1 and PLT2 genes (23, 35, 36) showed a reduction of gradient length proportional to the reduction in meristem length upon tZ treatment (Fig. 5 E, H, and J and SI Appendix, Fig. S9 C–E). None of the other five tested lines showed this for either treatment (Fig. 5 C–J).
Fig. 5.
Gradient length can be uncoupled from growth. (A) Bar graph showing the relative meristem length after treatment with BL or tZ, as measured by the length from QC until the first elongating cortex cell. n = 3–10. This shows stable and easily measurable meristem length reduction after 3 d of treatment in Col-0 and reported lines. Response to hormones was found to be not significantly different between transgenic lines and Col-0 (P > 0.05; two-way ANOVA). (B–I) Confocal micrographs of Col-0 (B) and promoter lines 2: SCL (C), 4: IQD6 (D), 5: AT2G34357 (E), 7: AT2G38370 (F), 11: AT4G23800 (G), 16: ATNFXL1 (H), and 17: ZFHD1 (I) after 3 d mock treatment (Left) or with BL and tZ (Center and Right, respectively). White bars indicate meristem length. (J) Gradient length quantification of all analyzed lines after treatment. Mean relative gradient length is shown with bars indicating SE. ns, not significant; P > 0.05; *P < 0.05; **P < 0.001, as calculated by Student’s t test. Results show that gradient expression can be uncoupled from growth and PLT regulation. Genes indicated in red were idenfified in this study to show a downward gradient (P > M > D) and those in green an upward gradient (P < M < D). n = 10–19. Micrographs (B–I) were taken at the same magnification.
Given the relatively large overlap between genes with a graded expression profile and PLT1/2 target genes (for downward gradient dataset: TMO5: 30%, PUB25: 31%, and SPT: 34%; for upward gradient dataset: TMO5: 53%, PUB25: 43%, and SPT: 35%) (37) (SI Appendix, Table S2), there could be PLT-dependent regulation that is growth-dependent. However, the presumed PLT1/2 target 4: IQD6) (37) (Table 1), behaves differently from PLT1/2 expression upon perturbation of the meristem (Fig. 5J). This demonstrates that several of the tested expression profiles can be uncoupled from both growth and PLT regulation, and suggests a diverse genetic basis for transcriptional gradient formation.
Disrupting Transcription Gradients Leads to Growth Defects.
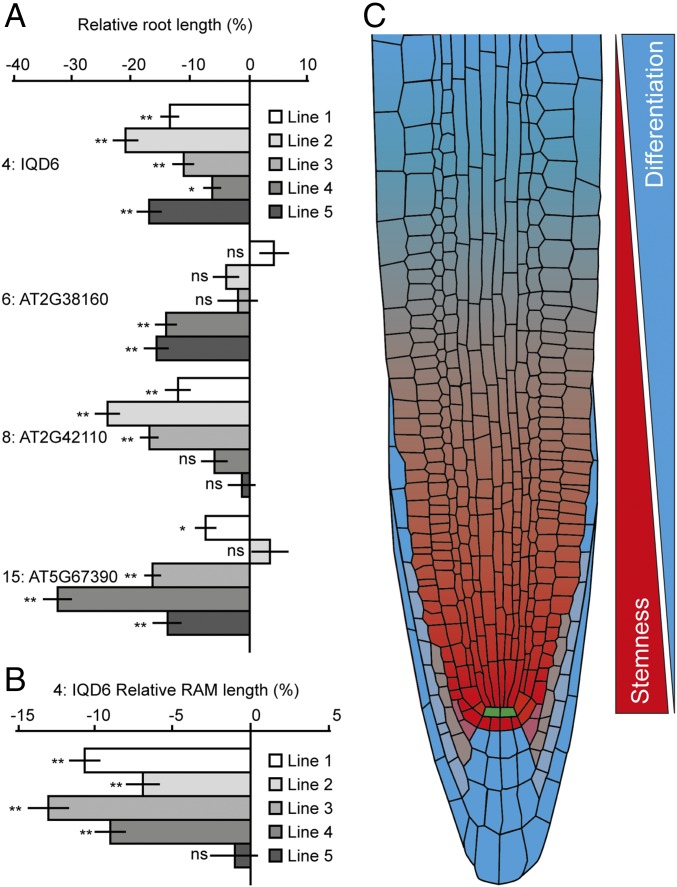
To determine if normal regulation of the observed gradients is relevant for normal growth and development, we aimed to override endogenous expression regulation. To this end, we expressed four of the gradient genes under control of the strong 35S promoter. All analyzed lines had stable overexpression of the respective gene of at least 10-fold the normal expression level (SI Appendix, Fig. S10). For each gene, multiple independent lines had disrupted root growth as a result of the overexpression (Fig. 6A). For example, in lines overexpressing gene 4: IQD6, all independent lines showed disturbed root growth and this coincided with reduced RAM length (Fig. 6B). These data indicate that disruption of a single transcriptional expression gradient can lead to reduced root growth, indicating that these gradients are important for normal growth and development.
Fig. 6.
Disruption of expression gradient can lead to reduced root growth. (A) Bar graph depicting relative root length in respective overexpression lines, relative to Col-0. For each gene, multiple independent lines show a significant reduction in root length as a result of overexpression. For 4: IQD6 all independent lines had reduced root length. Mean relative root length is shown with bars indicating SE. n = 45–77. (B) Bar graph showing relative RAM length in overexpression lines of 4: IQD6, relative to Col-0. n = 56–71. RAM length reduction correlates with root length reduction and overexpression of gene 4: IQD6. Mean relative RAM length is shown with bars indicating SE. ns, not significant; P > 0.05; *P < 0.05; **P < 0.001, as calculated by Student’s t test. (C) Proposed model for proximal meristem organization by opposing gradients of stemness and differentiation potential.
Discussion
In this study, we have established a framework for the molecular organization of the root meristem. We have characterized genes expressed in cell populations of varying ontogeny to critically assess the mode of organization. Our results indicate that there are few, if any, qualitative differences between the most distal cells, commonly considered stem cells, and more proximal cells. Rather, differences between the regions within the proximal meristem are quantitative and appear to be dynamic (Fig. 6C). We have identified large gene clusters expressed in opposing graded profiles along the meristem. There is a strong association with developmental and differentiation processes in these respective clusters. We found that the transcriptional gradients can be directly translated into protein gradients and are well correlated with cellular features (i.e., nuclear size and cell volume). While it is possible that direct contact of meristem cells to the QC confers unique properties, these properties do not appear to translate into a striking gene-expression difference between them and their more shootward progeny. In addition to the distinction between stem cells and their dividing daughters (often referred to as “transit amplifying cells”), additional zones have been distinguished, including a transition zone, elongation zone, and differentiation zone (5, 6). Given the dominant gradient transcript patterns, a distinct possibility is that the qualitative properties that have been assigned to different zones are in fact different outputs of the same quantitative core framework. In this scenario, thresholds of expression could result in differential outputs.
An open question is how these gradients are regulated. Our results indicate that expression regulation is not generic, as some gradients are affected by changes in meristem size, while others are not. Thus, gradients do not simply follow from a growth or dilution process, but seem to have diverse inputs. One such input could be the PLT transcription factors (23, 35, 36), as there is considerable overlap between gradient genes and PLT targets (37). Nevertheless, the majority of genes are not PLT targets and several of the gene-expression reporters had responses to root growth inhibition that are distinct from the changes in PLT expression, suggesting independent regulation. It will be interesting to determine the gene regulatory networks that generate gradients. Particularly, the growth-independent gradients are fascinating, as it is unknown at present how such patterns could be regulated. Another open question is whether the upward and downward gradients are independent of each other, or whether they interact, for example, by mutual inhibition of graded transcriptional regulators. A precedent for such a system comes from the analysis of PLT target genes. PLTs can activate or repress target genes in a dose-dependent manner (36, 37), and could thus influence both up- and downward gradient genes. While PLTs are indisputably important in this process, their action alone cannot explain all observed expression profiles and behavior upon perturbation. It will therefore be interesting to further investigate what PLT-independent mechanisms play a role in this and how they interact.
Methods
Plant Material and Growth Conditions.
Previously described reporter lines for TMO5 [pTMO5::n3GFP; pTMO5::TMO5:3xGFP (17)], TMO7 [pTMO7::n3GFP; pTMO7::TMO7:GFP; (17)], SPT [pSPT::n3GFP (21)], PUB25 [pPUB25::n3GFP (21)], PLT1 [pPLT1::PLT1:YFP (35)], and PLT2 [pPLT2::PLT2:YFP (35)] were used. Seeds were surface-sterilized and grown on MS plates under standard long-day growth conditions (22 °C, 16:8-h light/dark cycles) following a 1- to 4-d stratification at 4 °C. Arabidopsis ecotype Columbia-0 was used as wild-type control in all cases. Hormone treatments were performed for 3 d by transferring 2-d-old seedlings to plates containing either 5 nM of BL or 1 µM of tZ.
Cloning and Plant Transformation.
Promoter fragments (up to 3-kb upstream of the start codon) and coding sequences (CDS) from selected genes were amplified from genomic DNA (promoter) or root cDNA (CDS) using PCR and Phusion Flash master mix (Thermo Scientific) and the primers described in SI Appendix, Table S4. PCR products were cloned into the pPLV4_v2 (promoter) or pPLV26 (CDS) vectors using ligation-ndependent cloning (LIC) (38, 39). Translational fusion constructs (for 4, 11, 19, and SPT) were generated by cloning the whole genomic region, including up to 3-kb upstream promoter into the pPLV16 vector using LIC (38, 39) and primers listed in SI Appendix, Table S4. For meristematic tagging of the nuclear envelope, a nuclear targeting factor (NTF) (40) was cloned after the RPS5A promoter (41) using LIC and primers listed in SI Appendix, Table S4. All constructs were confirmed by sequencing, and subsequently transformed into Arabidopsis Col-0 wild-type plants through Agrobacterium-mediated transformation. At least three independent transformants were checked and representative pictures are shown.
Microscopic Analysis.
Confocal laser-scanning microscopy was performed as previously described (21) using a Leica SP5. Briefly, 5-d-old seedling roots were stained using propidium iodide (PI; Invitrogen) and visualized using the following wavelengths: 488-nm excitation and 500- to 535-nm detection for GFP and 600- to 700-nm detection for PI. Measurements were performed and brightness and contrast were adjusted using Leica Application Suite Advanced Fluorescence and ImageJ software. Nucleus size was measured using a marker for the nuclear envelop (pRPS5A::NTF:GFP) and measuring the width of the nucleus in each cell along 13 epidermal cell files in 5 independent roots. For cell volume determination, 5-d-old seedling roots were first stained with mPS-PI (42) and cell outlines were subsequently imaged using confocal microscopy. Cell volumes from 18 cell files were calculated from 3D z-stacks of 13 independent roots by 3D segmentation using MorphoGraphX software (43).
To measure gradient length, confocal images were taken and fluorescence intensity was normalized to show only the top 50%. The length of the remaining fluorescence was measured, comparing mock to treated root tips (SI Appendix, Fig. S9B).
For esthetic reasons, all micrographs were globally adjusted in brightness and contrast, rotated, and displayed on a matching background.
FACS.
FACS of plant protoplasts was performed as described in ref. 16. Briefly, root tips of 6-d-old seedlings were cut and incubated in protoplasting Solution B [1.5% (wt/vol) cellulysin and 0.1% (wt/vol) pectolyase in Solution A (600 mM mannitol, 2 mM MgCl2, 0.1% [wt/vol] BSA, 2 mM CaCl2, 2 mM Mes, 10 mM KCl, pH 5.5)] for 1 h at room temperature. Cells were spun down at 200 × g for 6 min and resuspended in Solution A. Cells were sorted on a MoFlo Astrios (Beckmann), based on strength of GFP signal at 488-nm excitation. Cells were collected in RLT buffer from a Qiagen RNeasy Micro kit and directly frozen.
RNA Isolation and RNA-Sequencing Library Preparation.
Total RNA from sorted cells was isolated using a Qiagen RNeasy Micro kit, RNA concentration was measured using a Life Technologies Qubit 2.0 fluorometer, and RNA integrity was measured using an Agilent 2100 bioanalyzer plant RNA 6000 pico kit, all following the manufacturers’ instructions. Ten nanograms of total RNA was used for amplification in an Ovation RNA-Seq System V2 (NuGEN). Approximaely 3 µg of amplified cDNA was fragmented to 200 bp on a Covaris Sonication system. Next, 100 ng was subsequently used to prepare libraries for Illumina RNA-sequencing, using Ovation Ultralow Library Systems (NuGEN), following the manufacturer’s instructions.
Quantitative RT-PCR and RNA-Sequencing.
qPCR was performed as previously described (44). Poly(dT) cDNA prepared from 10 ng of total RNA using an Invitrogen SuperScript III or Bio-Rad iScript kit and the manufacturer’s instructions, followed by qPCR using SYBR Green Master Mix (Applied Biosystems) or iQ SYBR green mix (Bio-Rad). All reactions were performed in triplicate, using primers designed in Beacon designer 8.0 (Premier Biosoft International) (SI Appendix, Table S4) and data were analyzed using the qBase program (45). Signals were normalized against expression of PP2A or ACTIN2 and EEF1α4 (46).
RNA-sequencing was performed using Illumina HiSEq. 50SE (Duke IGSP Genome Sequencing and Analysis Core Resource, Duke University, NC). Data were analyzed and visualized using the Tuxedo suite (47). Bowtie was used to create an Arabidopsis whole-genome index file based on TAIR10 Arabidopsis genome annotation (48). Next, TopHat was used to align sequencing reads to the genome and build a transcriptome index using the following parameters: minimal intron length = 35 bp; maximal intron length = 2,000 bp. An average of 16.9 million reads were mapped to the genome, accounting for about 85% of all reads (SI Appendix, Table S1). The mapped read counts were then used to calculate differential expression using Cufflinks and the following parameters: minimal isoform fraction = 0.05; pre-mRNA fraction = 0.05; minimal intron length = 35 bp; intron overhang tolerance = 8, comparing all cell populations to each other. A combination of R and FileMaker pro software was used to visualize and interpret the data.
Quality threshold-clustering was performed using MultiExperiment Viewer software (22) calculating a Pearson correlation with a maximum cluster diameter of 0.5 and minimum cluster population of 5.
Direct analysis of different expression profile clusters was performed with the following criteria:
For downward (P > M > D) gradient: genes were included only when the fold-change was significantly (q < 0.05) larger in the proximal population compared with the medial population and when the same was true between the medial and distal populations.
For upward (P < M < D) gradient: genes were only included when the fold-change was significantly (q < 0.05) larger in the distal population compared with the medial population, when the same was true between the medial and proximal populations, and when detectable expression (>1 FPKM) was observed in all populations.
For on–off profiles: genes were included when expression was detectable (>1 FPKM) in the proximal population, but not in the other populations (<1 FPKM), and where the difference between the proximal and medial populations was significant (q < 0.05).
GO-term enrichment analysis was performed using the Cytoscape software with BiNGO plugin (49, 50). Overrepresented biological process terms were calculated compared with the Arabidopsis genome using a hypergeometric test and a Benjamini–Hochberg false-discovery rate (P value) below 0.05.
Data Availability.
RNA-seq data has been deposited in Gene Expression Omnibus with accession no. GSE98097. In addition, for easy accessibility and visualization, these data are available at the AraBidopsis Embryonic and Root Transcriptome brOwser (AlBERTO; albertodb.org), an interactive gene expression database.
Supplementary Material
Acknowledgments
We thank Heather Belcher, Manuel Valdes, and Masashi Yamada for help with protoplasting and RNA-sequencing library preparations; the Duke Cancer Institute Flow Cytometry service for performing cell sorting; Peter Schaap for providing server capacity for RNA-sequencing data analysis; and the MicroSpectroscopy Centre Wageningen and Vlaams Instituut voor Biotechnologie Imaging Core for providing microscopy equipment and assistance. This work was supported by European Research Council Starting Grants CELLPATTERN 281573 (to D.W.) and TORPEDO 714055 (to B.D.R.); Netherlands Organization for Scientific Research Grant VIDI 864.13.00; (to B.D.R.); NIH Grant R01-GM043778 and Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation Grant GBMF3405 (to P.N.B.); and European Molecular Biology Organization Short-term Fellowships ASTF 427-2013 and ASTF 4-2012 (to J.R.W. and B.K.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE98097).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707400114/-/DCSupplemental.
References
- 1.Scheres B, et al. Embryonic origin of the Arabidopsis primary root and root meristem initials. Development. 1994;120:2475–2487. [Google Scholar]
- 2.Clowes FAL. Nucleic acids in root apical meristems of Zea. New Phytol. 1956;55:29–34. [Google Scholar]
- 3.van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature. 1997;390:287–289. doi: 10.1038/36856. [DOI] [PubMed] [Google Scholar]
- 4.Willemsen V, et al. The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Dev Cell. 2008;15:913–922. doi: 10.1016/j.devcel.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 5.Bennett T, Scheres B. Root development-two meristems for the price of one? Curr Top Dev Biol. 2010;91:67–102. doi: 10.1016/S0070-2153(10)91003-X. [DOI] [PubMed] [Google Scholar]
- 6.Heidstra R, Sabatini S. Plant and animal stem cells: Similar yet different. Nat Rev Mol Cell Biol. 2014;15:301–312. doi: 10.1038/nrm3790. [DOI] [PubMed] [Google Scholar]
- 7.Furuta KM, et al. Plant development. Arabidopsis NAC45/86 direct sieve element morphogenesis culminating in enucleation. Science. 2014;345:933–937. doi: 10.1126/science.1253736. [DOI] [PubMed] [Google Scholar]
- 8.Yamaguchi M, Kubo M, Fukuda H, Demura T. Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 2008;55:652–664. doi: 10.1111/j.1365-313X.2008.03533.x. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi M, et al. VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell. 2010;22:1249–1263. doi: 10.1105/tpc.108.064048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geldner N. The endodermis. Annu Rev Plant Biol. 2013;64:531–558. doi: 10.1146/annurev-arplant-050312-120050. [DOI] [PubMed] [Google Scholar]
- 11.Dolan L, Costa S. Evolution and genetics of root hair stripes in the root epidermis. J Exp Bot. 2001;52:413–417. doi: 10.1093/jexbot/52.suppl_1.413. [DOI] [PubMed] [Google Scholar]
- 12.Masucci JD, et al. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development. 1996;122:1253–1260. doi: 10.1242/dev.122.4.1253. [DOI] [PubMed] [Google Scholar]
- 13.Birnbaum K, et al. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- 14.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 15.Li S, Yamada M, Han X, Ohler U, Benfey PN. High-resolution expression map of the Arabidopsis root reveals alternative splicing and lincRNA regulation. Dev Cell. 2016;39:508–522. doi: 10.1016/j.devcel.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer-Pascuzzi AS, Benfey PN. Fluorescence-activated cell sorting in plant developmental biology. Methods Mol Biol. 2010;655:313–319. doi: 10.1007/978-1-60761-765-5_21. [DOI] [PubMed] [Google Scholar]
- 17.Schlereth A, et al. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature. 2010;464:913–916. doi: 10.1038/nature08836. [DOI] [PubMed] [Google Scholar]
- 18.Mähönen AP, et al. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science. 2006;311:94–98. doi: 10.1126/science.1118875. [DOI] [PubMed] [Google Scholar]
- 19.De Rybel B, et al. Plant development. Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science. 2014;345:1255215. doi: 10.1126/science.1255215. [DOI] [PubMed] [Google Scholar]
- 20.Ohashi-Ito K, et al. A bHLH complex activates vascular cell division via cytokinin action in root apical meristem. Curr Biol. 2014;24:2053–2058. doi: 10.1016/j.cub.2014.07.050. [DOI] [PubMed] [Google Scholar]
- 21.Wendrich JR, et al. A set of domain-specific markers in the Arabidopsis embryo. Plant Reprod. 2015;28:153–160. doi: 10.1007/s00497-015-0266-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heyer LJ, Kruglyak S, Yooseph S. Exploring expression data: Identification and analysis of coexpressed genes. Genome Res. 1999;9:1106–1115. doi: 10.1101/gr.9.11.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aida M, et al. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119:109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 24.Friml J, et al. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell. 2002;108:661–673. doi: 10.1016/s0092-8674(02)00656-6. [DOI] [PubMed] [Google Scholar]
- 25.Gälweiler L, et al. Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science. 1998;282:2226–2230. doi: 10.1126/science.282.5397.2226. [DOI] [PubMed] [Google Scholar]
- 26.Helariutta Y, et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101:555–567. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- 27.Lee YR, Li Y, Liu B. Two Arabidopsis phragmoplast-associated kinesins play a critical role in cytokinesis during male gametogenesis. Plant Cell. 2007;19:2595–2605. doi: 10.1105/tpc.107.050716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moussian B, Schoof H, Haecker A, Jürgens G, Laux T. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 1998;17:1799–1809. doi: 10.1093/emboj/17.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rademacher EH, et al. A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J. 2011;68:597–606. doi: 10.1111/j.1365-313X.2011.04710.x. [DOI] [PubMed] [Google Scholar]
- 30.Avci U, Petzold HE, Ismail IO, Beers EP, Haigler CH. Cysteine proteases XCP1 and XCP2 aid micro-autolysis within the intact central vacuole during xylogenesis in Arabidopsis roots. Plant J. 2008;56:303–315. doi: 10.1111/j.1365-313X.2008.03592.x. [DOI] [PubMed] [Google Scholar]
- 31.Clay NK, Nelson T. Arabidopsis thickvein mutation affects vein thickness and organ vascularization, and resides in a provascular cell-specific spermine synthase involved in vein definition and in polar auxin transport. Plant Physiol. 2005;138:767–777. doi: 10.1104/pp.104.055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pérez-Amador MA, et al. Identification of BFN1, a bifunctional nuclease induced during leaf and stem senescence in Arabidopsis. Plant Physiol. 2000;122:169–180. doi: 10.1104/pp.122.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roudier F, et al. COBRA, an Arabidopsis extracellular glycosyl-phosphatidyl inositol-anchored protein, specifically controls highly anisotropic expansion through its involvement in cellulose microfibril orientation. Plant Cell. 2005;17:1749–1763. doi: 10.1105/tpc.105.031732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor NG, Laurie S, Turner SR. Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell. 2000;12:2529–2540. doi: 10.1105/tpc.12.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galinha C, et al. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449:1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- 36.Mähönen AP, et al. PLETHORA gradient formation mechanism separates auxin responses. Nature. 2014;515:125–129. doi: 10.1038/nature13663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santuari L, et al. The PLETHORA gene regulatory network guides growth and cell differentiation in Arabidopsis roots. Plant Cell. 2016;28:2937–2951. doi: 10.1105/tpc.16.00656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Rybel B, et al. A versatile set of ligation-independent cloning vectors for functional studies in plants. Plant Physiol. 2011;156:1292–1299. doi: 10.1104/pp.111.177337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wendrich JR, Liao CY, van den Berg WA, De Rybel B, Weijers D. Ligation-independent cloning for plant research. Methods Mol Biol. 2015;1284:421–431. doi: 10.1007/978-1-4939-2444-8_21. [DOI] [PubMed] [Google Scholar]
- 40.Deal RB, Henikoff S. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev Cell. 2010;18:1030–1040. doi: 10.1016/j.devcel.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weijers D, et al. An Arabidopsis minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development. 2001;128:4289–4299. doi: 10.1242/dev.128.21.4289. [DOI] [PubMed] [Google Scholar]
- 42.Truernit E, et al. High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell. 2008;20:1494–1503. doi: 10.1105/tpc.107.056069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barbier de Reuille P, et al. MorphoGraphX: A platform for quantifying morphogenesis in 4D. eLife. 2015;4:05864. doi: 10.7554/eLife.05864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Rybel B, et al. A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev Cell. 2013;24:426–437. doi: 10.1016/j.devcel.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 45.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghosh S, Chan CK. Analysis of RNA-seq data using TopHat and Cufflinks. Methods Mol Biol. 2016;1374:339–361. doi: 10.1007/978-1-4939-3167-5_18. [DOI] [PubMed] [Google Scholar]
- 48.Lamesch P, et al. The Arabidopsis information resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 50.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data has been deposited in Gene Expression Omnibus with accession no. GSE98097. In addition, for easy accessibility and visualization, these data are available at the AraBidopsis Embryonic and Root Transcriptome brOwser (AlBERTO; albertodb.org), an interactive gene expression database.