Significance
RNase P is a tRNA-processing enzyme of unique architectural diversity: either a catalytic RNA plus one or more (up to 10) proteins, or one (or three) unrelated proteins only. We identified yet another enzyme form in the bacterium Aquifex aeolicus, a 23-kDa protein and the smallest known form of RNase P. Apparently, it was acquired by horizontal gene transfer from Archaea. In some other bacteria and many archaea, it is simultaneously present with the presumably more ancient RNA-based enzyme form. Bacteria with both activities may represent the missing link of RNase P evolution, a transition state that had also been once traversed by the Aquificaceae, which, however, later lost their RNA-based RNase P.
Keywords: protein-only RNase P, Aquifex aeolicus, tRNA processing, HARP
Abstract
RNase P is an essential tRNA-processing enzyme in all domains of life. We identified an unknown type of protein-only RNase P in the hyperthermophilic bacterium Aquifex aeolicus: Without an RNA subunit and the smallest of its kind, the 23-kDa polypeptide comprises a metallonuclease domain only. The protein has RNase P activity in vitro and rescued the growth of Escherichia coli and Saccharomyces cerevisiae strains with inactivations of their more complex and larger endogenous ribonucleoprotein RNase P. Homologs of Aquifex RNase P (HARP) were identified in many Archaea and some Bacteria, of which all Archaea and most Bacteria also encode an RNA-based RNase P; activity of both RNase P forms from the same bacterium or archaeon could be verified in two selected cases. Bioinformatic analyses suggest that A. aeolicus and related Aquificaceae likely acquired HARP by horizontal gene transfer from an archaeon.
The architectural diversity of RNase P enzymes is unique: In Bacteria, Archaea, and in the nuclei and organelles of many Eukarya, RNase P is a complex consisting of a catalytic RNA subunit and a varying number of proteins (one in Bacteria, at least four in Archaea, and up to 10 in Eukarya) (1, 2). A different type of RNase P was discovered more recently in human mitochondria (3) and, subsequently, in land plants and some protists (4, 5). This form, termed proteinaceous or protein-only RNase P (PRORP), lacks any RNA subunit and consists of one or three (animal mitochondria) protein subunit(s); it is found in most branches of the eukaryotic phylogenetic tree (6).
Bacterial RNase P enzymes identified so far are composed of a ∼400-nt-long catalytic RNA subunit (encoded by rnpB) and a small protein subunit of ∼14 kDa (encoded by rnpA) (7). However, no rnpA and rnpB genes were identified in the genome of Aquifex aeolicus or other Aquificaceae (8–12). The genetic organization of A. aeolicus tRNAs in tandem clusters and as part of ribosomal operons and the detection of tRNAs with canonical mature 5′-ends in total RNA extracts from A. aeolicus implied the existence of a tRNA 5′-maturation activity (9) that was indeed subsequently detected in cell lysates of A. aeolicus (11, 13). However, to date, the identity and biochemical composition of RNase P in A. aeolicus has remained enigmatic.
Results and Discussion
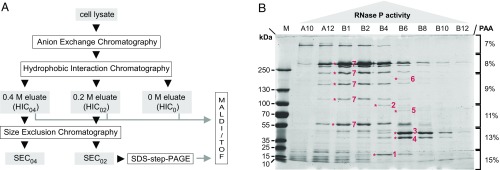
Here, we pursued a classical biochemical approach to identify the RNase P of A. aeolicus. The purification procedure consisted of three consecutive chromatographic steps: anion exchange, hydrophobic interaction, and size exclusion chromatography (AEC, HIC, and SEC, respectively; Fig. 1A and SI Appendix, Figs. S1–S8). RNase P activity was assayed at all purification steps. To identify putative protein components of the enzyme, fractions with low and high RNase P activity from different purification steps were comparatively analyzed by step-gradient SDS/PAGE, and protein bands correlating with activity (Fig. 1B) were subjected to mass spectrometry. An example is SEC (SI Appendix, Fig. S5) fraction B4 displaying maximum activity combined with an enrichment of protein bands 1 and 2 identified as the hypothetical protein Aq_880 and polynucleotide phosphorylase (PNPase), respectively (Fig. 1B). Several protein bands correlating less strongly with RNase P activity were identified as well. The most abundant proteins in fractions containing highest RNase P activity (SI Appendix, Figs. S9 and S10), as inferred from mass spectrometry, were in the order of decreasing peptide representation: (i) Aq_880, (ii) glutamine synthetase, (iii) ribosomal protein S2, (iv) PNPase, (v) N utilization substance protein B homolog (NusB), and (vi) Aq_707 (with similarity to an Escherichia coli tRNA binding protein of the MnmC family). Furthermore, Aq_880 was the only protein that was also found in an HIC fraction with low RNase P activity eluting at 0 M (NH4)2SO4 (SI Appendix, Figs. S3 and S10). The presence of ribosomal protein S2 combined with a previously described association of RNase P with 30S ribosomal subunits in Bacillus subtilis (14) suggested a possible association of A. aeolicus RNase P with the ribosome, which, however, could not be confirmed experimentally.
Fig. 1.
Partial purification and identification of A. aeolicus RNase P. (A) Schematic overview of the purification procedure. The applied chromatography steps and methods are shown as open boxes; the cell lysate and column fractions with RNase P activity are indicated as gray boxes. (B) SDS/PAGE analysis of “SEC0.2” fractions with RNase P activity eluting from a SEC column that had been loaded with a “HIC0.2” sample [material eluted from the hydrophobic interaction column at 0.2 M (NH4)2SO4]. Fractions with low (A10, B12), increasing (A12, B1, B2), maximum (B4), and decreasing (B6, B8, B10) RNase P activity were loaded in order of their elution from the column (SI Appendix, Fig. S5). Prominent protein bands correlating with RNase P activity were excised and identified by mass spectrometry as: 1, hypothetical protein Aq_880; 2, polynucleotide phosphorylase (PNPase); 3, uncharacterized protein homologous to RNA pseudouridine synthase from B. subtilis; 4, hypothetical protein Aq_707; 5, Aq_1754/Aq_707; 6, Aq_808/Aq_707; 7, glutamine synthetase; only two of these proteins (1 and 2) were most abundant in fraction B4, which displayed the highest RNase P activity; M, molecular mass marker.
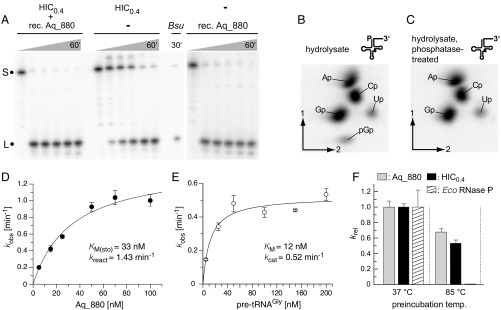
The 22.6-kDa protein Aq_880 was recombinantly expressed in E. coli and affinity-purified using a C-terminal His tag. Addition of affinity-purified Aq_880 protein to an active A. aeolicus HIC fraction not only boosted RNase P activity, but recombinant Aq_880 alone was able to process the precursor tRNAs specifically and efficiently at the canonical RNase P cleavage site without requiring any additional components (Fig. 2A). We further analyzed whether processing by recombinant Aq_880 generates tRNA with a 5′-phosphate end, as any other previously characterized RNase P. TLC indeed confirmed a 5′-phosphate at the 5′-terminal G+1 residue of tRNAGly processed by recombinant Aq_880 (Fig. 2 B and C). A contamination of the recombinant Aq_880 preparation with endogenous E. coli RNase P was excluded by RT-PCR analysis and micrococcal nuclease pretreatment (SI Appendix, Figs. S11 and S12). Since Aq_707 and PNPase (Aq_221) copurified with RNase P activity and peptides matching Aq_707 and Aq_880 were identified in band 6 (Fig. 1B) excised from SDS gels, we tested whether these recombinantly produced A. aeliocus proteins support RNase P activity (SI Appendix, Fig. S13). This was done by adding excess amounts of recombinant Aq_707 or Aq_221 to a HIC0.4 fraction and analyzing processing at a pretRNA concentration of 33 nM (SI Appendix, Fig. S13A). The rationale was that Aq_707 or Aq_221, if being part of the RNase P enzyme complex and contributing to activity, were likely present in substoichiometric amounts owing to depletion during purification, such that their exogenous addition might boost RNase P activity. However, neither Aq_707 nor PNPase showed intrinsic endonuclease activity nor did they stimulate the RNase P activity of Aq_880; PNPase caused some unspecific degradation of the pretRNA and the 5′-leader cleavage product, which we attribute to its 3′-exonuclease activity (SI Appendix, Fig. S13A). To further examine the possibility that Aq_707, identified together with Aq_880 in band 6 (Fig. 1B) by MS analysis, might support interaction of Aq_880 with pretRNA substrates, we analyzed processing under dilute multiple-turnover conditions where substrate binding may be rate-limiting, using 1 nM recombinant Aq_880, 5 nM pretRNA, and 1 nM recombinant Aq_707. However, the rate constant of cleavage was essentially identical in the presence and absence of Aq_707 (SI Appendix, Fig. S13B). These findings argue against a direct interaction of the two proteins. Band 6 in Fig. 1B migrated between the 130- and 250-kDa marker proteins. Protein monomers of Aq_880 and Aq_707 have sizes of 23 and 40 kDa, respectively, which left open the possibility that one or two other macromolecules mediate incorporation of the two proteins into a larger complex. However, MS analysis did not detect any other protein in band 6. Finally, addition of total RNA prepared from A. aeolicus did not affect the reaction catalyzed by recombinant Aq_880 (SI Appendix, Fig. S14). We thus concluded that Aq_880 is the principal constituent of A. aeolicus RNase P.
Fig. 2.
(A) Processing of Thermus thermophilus pretRNAGly by the “HIC0.4” fraction [material eluted from a HIC column at 0.4 M (NH4)2SO4; Center], by recombinant Aq_880 (final concentration 0.1 µg/µL) (Right), or by a mixture of both (Left); Bsu, B. subtilis RNase P as positive control; S, the 5′-[32P]-endlabeled pretRNAGly substrate; L, the 5′-leader cleavage product. (B and C) Analysis of the 5′-end of pretRNAGly as processed by recombinant Aq_880 in vitro. PretRNAGly was labeled by in vitro transcription in the presence of [α-32P]GTP and processed with recombinant Aq_880 under standard conditions to nearly completeness. The 5′-mature tRNA product was gel-purified, and a part of the eluate was treated with alkaline phosphatase. Untreated (B) and phosphatase-treated (C) RNAs were subjected to alkaline hydrolysis, and monophosphate and diphosphate nucleosides were resolved by 2D TLC. The pGp bisphosphate (B) is sensitive to phosphatase pretreatment (C) consistent with its origin from the tRNA’s 5′-end. Spot intensities correspond to the number of the respective nucleosides that are followed by a G in the tRNA sequence and, thereby, are radioactively labeled (A, 8; C, 8; G, 9; U, 1). (D) Single-turnover kinetics of recombinant Aq_880 for the processing of pretRNAGly (data points are based on three to six independent determinations for each concentration of Aq_880); error bars are SDs. Kinetic constants and the standard errors of the curve fit are kreact = 1.43 ± 0.14 min−1 and Km(sto) = 33 ± 8 nM. (E) Multiple-turnover kinetics of pretRNAGly processing using 1 nM Aq_880. Results are based on three independent determinations for each pretRNA concentration. Error bars are SDs of the mean. Kinetic constants and the standard errors of the curve fit are kcat = 0.52 ± 0.04 min−1 and Km = 12 ± 4 nM. (F) Thermostability of recombinant Aq_880 (gray bars) or a HIC0.4 fraction containing native RNase P (black bars) in comparison with the E. coli RNase P holoenzyme (white and hatched bars, respectively). RNase P activities were preincubated either at 37 °C or 85 °C before their catalytic activity was determined under single-turnover conditions at 37 °C. Error bars are standard deviations of the mean based on at least three experiments. Note that the activity of E. coli RNase P after preincubation at 85 °C was so low that the corresponding bar to the right of the black HIC0.4 bar does not rise above the baseline.
The single-turnover kinetic parameters of recombinant Aq_880 in a low-salt buffer with 4.5 mM Mg2+ at 37 °C (Fig. 2D) were determined as kreact = 1.43 min−1 and KM(sto) = 33 nM, thus similar to the values previously obtained for protein-only RNase P (PRORP3) from Arabidopsis thaliana [kreact = 1.7 min−1; KM(sto) ∼ 5 nM] (15) and similar to activities determined by others (16). The multiple-turnover kinetic parameters determined under the same conditions at 1 nM enzyme and 5–200 nM substrate were 0.5 min−1 for kcat and 12 nM for Km (Fig. 2E). The two- to threefold decrease of both parameters relative to those of the single turnover might be explained by product release limiting the rate of the multiple-turnover reaction. SEC indicated that native and recombinant Aq_880 elute as a ∼420-kDa complex (SI Appendix, Fig. S15), suggesting that the protein forms homooligomers (e.g., three hexamers or six trimers). This is in line with a weak band of ∼70 kDa observed for recombinant Aq_880 in SDS gels in addition to the monomer (∼23 kDa; Fig. 3B and SI Appendix, Fig. S16). Mass spectrometry identified this additional band at ∼70 kDa as Aq_880 (SI Appendix, Fig. S16), supporting the notion that Aq_880 has the potential to form stable homotrimers.
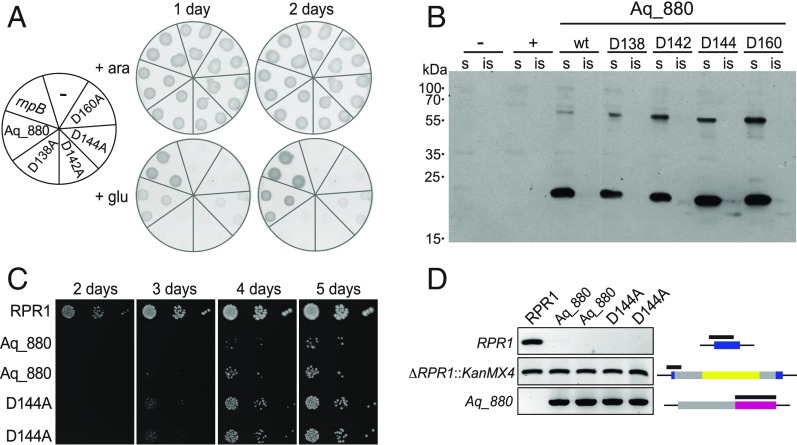
Fig. 3.
Complementation of E. coli and yeast RNase P by Aq_880. (A) A. aeolicus Aq_880 is able to functionally replace bacterial RNase P in vivo. E. coli BW cells transformed with expression plasmids for Aq_880 or its variants D138A, D142A, D144A, and D160A were grown at 37 °C under permissive (in the presence of arabinose; +ara) or nonpermissive (in the presence of glucose; +glu) conditions. Already after 1 d, cell growth was detectable in E. coli BW cells containing the expression vector for Aq_880; whereas no complementation was observed with the aspartate to alanine mutants D138A, D142A, and D160A, weak colony formation was seen with D144A. rnpB: E. coli RNase P as positive control. (−), the empty vector as negative control. (B) Expression and solubility of Aq_880 variants in E. coli BW. Soluble (s) and insoluble (is) protein fractions from E. coli BW expressing the Aq_880 variants were analyzed by Western blotting using a polyclonal antiserum to Aq_880. No signal was obtained when E. coli BW was transformed with the empty vector (−) or E. coli rnpB (+), respectively; molecular mass marker (in kilodaltons) indicated on the left. (C) Aq_880 and its variant D144A rescue growth upon deletion of the yeast nuclear RNase P RNA gene RPR1. Two colonies each obtained through rescue by Aq_880 or its variant D144A were applied as spots in 10-fold serial dilution to YPD plates, and the growth of the strains was monitored in parallel to a control (rescue by RPR1). Note the later appearance and smaller colony size of Aq_880 and D144A strains indicating slow growth. (D) Genotyping of Aq_880 and D144A colonies derived from plasmid shuffle. The analysis of a control (rpr1Δ::kanMX4 [RPR1]) and two complementation isolates each (Aq_880, genotype rpr1Δ::kanMX4 [Aq_880]; D144A, genotype rpr1Δ::kanMX4 [Aq_880D144A]) is shown. The deletion of RPR1, the integrity of the chromosomal gene disruption, and the presence of Aq_880 were verified by PCR. The part of the gene interrogated by the genotyping PCR is indicated by a black bar on top of the gene cartoon (RPR1, blue; kanMX4, yellow; Aq_880, magenta; promoter/terminator regions, gray) to the right of each agarose gel panel.
We further investigated the thermostability of recombinant Aq_880 as the hyperthermophile A. aeolicus naturally thrives at 85–95 °C. This was done by preincubating recombinant Aq_880 for 10 min at 85 °C before conducting the cleavage assay at 37 °C. The E. coli RNase P holoenzyme served as a control treated in the same way. Preincubation at 85 °C preserved the bulk of Aq_880 activity, but essentially inactivated E. coli RNase P (Fig. 2F, Left). Similar results were obtained when we used a native HIC0.4 fraction instead of recombinant Aq_880 (Fig. 2F, Right). These findings confirm the expected thermostability of Aq_880 and do not provide evidence for substantial differences in thermostabilty between the native and recombinant Aq_880.
Bioinformatic analyses predicted limited sequence and structure similarities between Aq_880 and the PIN (PilT N-terminal) ribonuclease domain. A PIN domain-like fold is also characteristic for the NYN domains of eukaryotic protein-only RNase P (PRORP) enzymes, for which a two metal-ion catalytic mechanism with metal ions coordinated by conserved aspartate residues was proposed (17). Limited similarity of Aq_880 to the metallonuclease domain of A. thaliana PRORP isoenzymes was evident from the alignment of the full-length sequences in SI Appendix, Fig. S17A. This alignment also suggested sporadic similarity to the PPR RNA binding and the central domains of PRORPs. Considering that an N-terminal truncation of 8 of the 11 helices in the PPR domain essentially abolished pretRNA-processing activity (17), the N-proximal part of Aq_880 may exert a corresponding function in pretRNA binding, although this is unclear at present and has to be addressed in future studies. The partial alignment of the major portion of the PRORP metallonuclease domain with Aq_880 (SI Appendix, Fig. S17B) suggested that aspartate residues D138, D142, and D160 of Aq_880 are functionally equivalent to D399, D475, and D493 in the catalytic center of PRORP1 from A. thaliana (SI Appendix, Fig. S18). We also considered a role for D144 of Aq_880 (corresponding to Y477 in PRORP1) since this residue, assumed to be in close proximity to the putative active site, was found to be strongly conserved (as D or E) in Aq_880 homologs identified by bioinformatic screens (SI Appendix, Figs. S23 and S24). We mutated the conserved aspartate residues in Aq_880 individually to alanines and analyzed activity of the resulting recombinant Aq_880 variants under single-turnover conditions at 37 °C (SI Appendix, Figs. S19 and S20). Mutations D138A, D142A, and D160A essentially abolished activity, while the D144A mutant was fully active.
Aq_880 (and less efficiently also the D144A variant) was also able to rescue the growth of the conditionally lethal E. coli RNase P mutant strain BW (18) (Fig. 3A), demonstrating that the small protein from A. aeolicus provides sufficient RNase P activity to support growth of the E. coli host, even if the growth rate was slower than upon complementation with the homologous RNase P RNA gene (rnpB). Bacteria harboring an empty plasmid or expressing the inactive variants D138A, D142A, and D160A were unable to grow under the same conditions. Western blotting confirmed expression of Aq_880 mainly in the soluble protein fraction of E. coli lysates (Fig. 3B).
Aq_880 was even able to replace the complex RNase P of Saccharomyces cerevisiae (Fig. 3C), a 400-kDa ribonucleoprotein (RNP) consisting of one RNA and nine protein subunits. For this analysis, we employed a previously established plasmid-shuffle protocol (19). Here, the otherwise lethal deletion of the yeast RNase P RNA gene RPR1 was rescued by the plasmid-based expression of A. aeolicus Aq_880, demonstrating that the highly dissimilar RNase P enzymes are basically exchangeable. The catalytically inactive variant D160A did not give rise to any viable colonies. Yeast cells dependent on Aq_880 nevertheless grew considerably slower than the parental wild-type strain, with only a slight improvement by the D144A variant (Fig. 3C); the deletion of the endogenous RPR1 gene (rpr1Δ::kanMX4) and the presence/absence of plasmid-encoded RPR1 and Aq_880 were verified by PCR (Fig. 3D). The poor growth contrasts with similar complementation experiments using eukaryal PRORPs, of which some, but not all, resulted in strains with wild-type–like growth properties (19). However, even strains harboring the less effectively complementing A. thaliana PRORP1 grew better than any of the Aq_880 strains. The slow growth could be due to problems in expressing sufficient bacterial protein in the eukaryal host, unfavorable subcellular localization, differences in substrate recognition between Aq_880 and yeast nuclear RNase P, or a combination of them leading to retarded biogenesis of mature tRNAs in general, or of a subset of tRNAs as observed for A. thaliana PRORP1 complementation in E. coli (20).
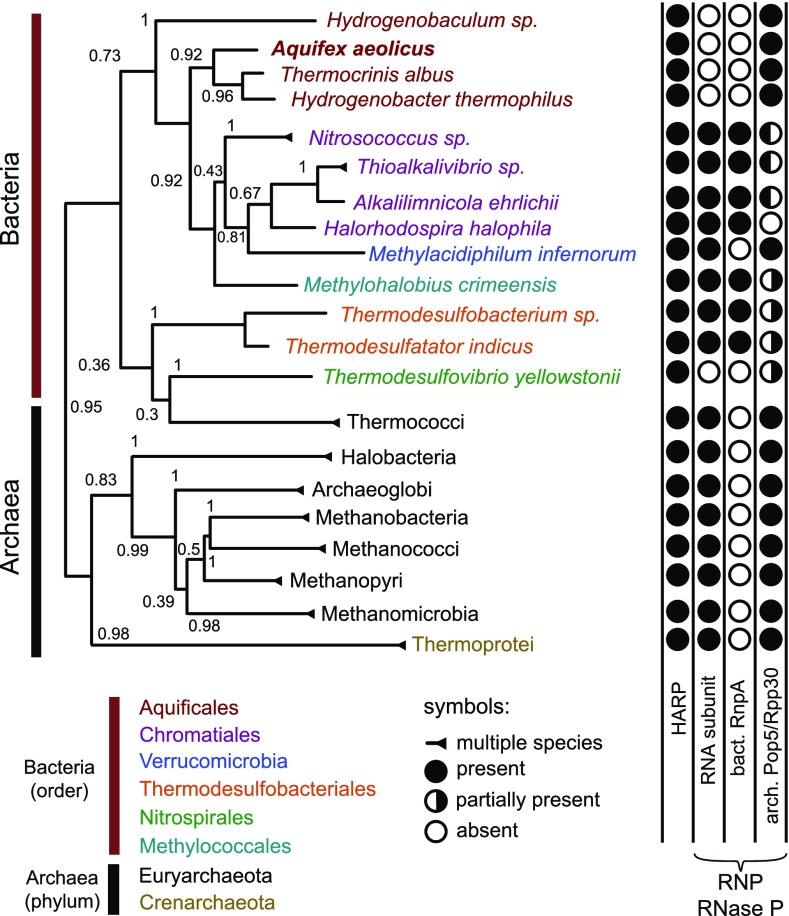
Bioinformatic screens identified numerous Aq_880 homologs in Archaea, few in Bacteria (Fig. 4 and SI Appendix, Figs. S22–S24 and Table S1), but none in Eukarya. We therefore propose to name this type of protein-only RNase P “HARP” for Homolog of Aquifex RNase P. The HARP proteins fall within the PIN_5 group of a recently published classification of the PIN domain-like superfamily (21). The relatively large group of archaeal organisms that encode HARP (SI Appendix, Table S1) have in common that they additionally encode an RNP RNase P. This suggests at least partially divergent functions of HARP and RNP enzymes in these Archaea, rather than reflecting evolutionary transition states where RNP RNase P is about to be replaced by HARP. In the latter scenario, one would have expected an irregular distribution of the two RNase P types among these archaeal species. The group of Bacteria-encoding HARPs was considerably smaller than the archaeal group (SI Appendix, Table S1). In the Aquificaceae, which all lack genes for a canonical bacterial RNase P, we identified homologs of the Archaea-specific RNase P protein subunits Pop5 and Rpp30, which bind to the catalytic domain of archaeal RNase P RNA (22). This suggests that the genes for HARP, Pop5, and Rpp30 were acquired by horizontal gene transfer from Archaea, in line with bioinformatic evidence that A. aeolicus has acquired at least 10% of its protein-coding genes by horizontal gene transfer from archaeal organisms (23). In the genomes of the Aquificaceae A. aeolicus and Hydrogenobacter thermophilus TK6, HARP and Pop5 genes are separated by only 12.5 and 3.1 kb, respectively, which may be evidence for their horizontal cotransfer. The findings may also suggest a functional link between HARP and Pop5/Rpp30. Thermodesulfovibrio yellowstonii seems to represent a situation similar to the Aquificaceae (no RNP RNase P), although a Pop5 homolog could not be identified (SI Appendix, Table S1). Remarkably, we identified a few bacteria that encode both an RNP RNase P and HARP. In these cases, either Rpp30 or Pop5 is not detectable (SI Appendix, Table S1), suggesting relaxed constraints to keep Rpp30 and Pop5 when RNA-based RNase P is expressed. All rnpA genes of this bacterial group with assumed dual RNase P activities encode regular RnpA proteins. Likewise, their rnpB genes encode RNase P RNAs that largely conform to canonical bacterial type A structures. This raises the question whether HARP and RNP RNase P both exert RNase P activity when coexpressed in the same organism. So far, evidence suggesting that both enzyme types might have RNase P activity within the same organism was obtained for one bacterium, Thermodesulfatator indicus, and one archaeon Methanothermobacter thermautotrophicus. T. indicus RNase P RNA has RNase P activity in vitro (Table 1), and the RNase P activity of the M. thermautotrophicus RNP enzyme has been thoroughly characterized (24–26). The HARP enzymes of both organisms were active in vitro and able to complement E. coli BW bacteria in vivo (Table 1 and SI Appendix, Fig. S21).
Fig. 4.
Phylogenetic distribution of HARP. Condensed phylogenetic tree of HARPs. Bacteria were condensed at order level, Archaea at phylum level. Bootstrap values are indicated at the branches (inferred from 1,000 replicates). Filled and empty circles indicate the presence or absence, respectively, of RNase P RNA (RNA subunit), the bacterial RNase P protein (RnpA), or the archaeal RNase P protein subunits Pop5/Rpp30; half-filled circles indicate that we identified for (at least some) bacterial species in this order only an Rpp30 (white/black circles) or Pop5 homolog (black/white circle), but not both.
Table 1.
Activity of RNP RNase P and HARP in selected bacteria and archaea
| Organism | RNP RNase P in vitro | HARP | |
| in vitro | in vivo | ||
| Bacteria | |||
| E. coli | +++ | — | — |
| A. aeolicus | — | +++ | +++ |
| T. indicus | + | ++ | +++ |
| Archaeon | |||
| M. thermautotrophicus | Active* | + | ++ |
RNP RNase P (RNA alone and holoenzyme) and HARP in vitro activities were tested as described in SI Appendix. —, enzyme not present; +(++), relative enzyme activities in processing assays or relative efficiencies in genetic complementation of strain BW; for example, in RNA-alone reactions, the kobs of pretRNAGly cleavage was 0.54 min−1 (+++) for E. coli and 0.11 min−1 (+) for T. indicus RNase P RNA; in complementation experiments (HARP in vivo), +++ was assigned to colony densities roughly corresponding to those obtained for BW bacteria complemented with aq_880 after overnight incubation at 37 °C.
We finally directed our attention to Archaea associated with deviations from canonical RNase P processing. The archaeal symbiont Nanoarchaeum equitans lacks RNase P and the need for tRNA 5′-end maturation is obviated by transcription of leaderless tRNAs (27). In the archaeon Ignicoccus hospitalis KIN4 I, which is the host for N. equitans (28), both HARP and RNA-based RNase P components were identified (SI Appendix, Table S1), although Pop5 and Rpp30 homologs were only found upon increasing the e value from 10−10 to 10−6 in genome searches. Pyrobaculum, Caldivirga, and Vulcanisaeta species encode shortened RNase P RNAs lacking the specificity domain (22). This suggested the possibility that these minimized enzymes have restricted functionality, which might be compensated by the presence of a HARP enzyme. However, even when increasing the e value from 10−10 to 10−4 in genome searches, no HARP candidates could be identified in any of these Archaea.
Conclusions
We discovered a minimal and RNA-free RNase P in the hyperthermophilic bacterium A. aeolicus, which was apparently acquired by horizontal gene transfer from an archaeon. Thus, we not only solved the last cold case in the RNase P field, but identified a previously unknown class of RNase P. Homologs were found in only a few Bacteria but many Archaea. At least for one archaeon and one bacterium analyzed so far we could provide evidence that both forms, RNA-based and protein-only RNase P, catalyze tRNA 5′-end maturation. As these findings are based on in vitro data and some complementation results in E. coli (Table 1), future studies will have to clarify if these homologs indeed function as genuine RNase P enzymes in vivo. Our findings have revealed yet another face of the unique architectural diversity of the RNase P enzyme family, comprised of diverse members of RNA and protein catalysts. RNA-based and protein-only RNase P enzymes have also been found in Eukarya (6), but appear to be mutually exclusive there, i.e., in no case they coexist within the same genetic compartment. With the identification of Bacteria that have protein- and RNA-based RNase P, we seem to have identified the kind of evolutionary transition state that was traversed by the Aquificaceae, which then, at some point in evolution, lost their “ancient” RNA-based enzyme. The situation in Archaea is evolutionarily and biologically intriguing as well, as the presence of this protein-based enzyme form seems to be linked to the presence of an RNA-based RNase P, suggesting that the two enzyme types functionally complement each other in tRNA processing, or one may exert another function, unrelated to tRNA processing, in Archaea. Finally, it will be challenging to understand why the Aquificaceae acquired and have retained the two archaeal RNase P RNA binding proteins Pop5 and Rpp30.
Materials and Methods
Growth of Bacterial Strains.
A. aeolicus VF5 cell pellets were purchased from M. Thomm and R. Huber (University of Regensburg, Germany). After growing for 1 d, cells were harvested as described (29) in the late exponential phase. E. coli strains were generally grown in liquid or on solid LB medium (10 g of tryptone, 5 g of yeast extract, 10 g of sodium chloride and, if required, 15 g of agar-agar per liter) at 37 °C under shaking (220 rpm; warm air incubation shaker, GFL 3033) in the case of liquid cultures. For the selection of strains expressing antibiotic resistance genes, growth media were supplemented with ampicillin (100 µg/mL), kanamycin (50 µg/mL), and/or chloramphenicol (34 µg/mL). E. coli DH5α was used as the host strain for cloning, E. coli Rosetta (DE3) for protein expression, and E. coli BW (18) for complementation studies. Liquid overnight cultures were usually incubated in 2–8 mL of LB medium containing the appropriate antibiotics and, if required, specific carbon sources.
Purification and Enrichment of RNase P Activity.
Purification of A. aeolicus RNase P by AEC, HIC, and SEC, as well as size exclusion chromatography to study the oligomeric state of Aq_880, was performed with an ÄKTA basic 10 (GE Healthcare) FPLC system at room temperature. Details are described in SI Appendix.
Step Gradient SDS/PAGE and Qualitative and Quantitative Mass Spectrometry Analysis.
The procedures are detailed in SI Appendix.
In Vitro Transcription of RNAs, 5′-[32P]-Endlabeling, RNase P Processing Assays.
This was done according to standard protocols (30) detailed in SI Appendix.
Pretreatment with Micrococcal Nuclease.
RNase P enzyme solutions were preincubated with micrococcal nuclease (Thermo Scientific) before addition of substrate and excess amounts of carrier RNA (6S-1 RNA from B. subtilis). The experimental details can be found in SI Appendix.
Cloning Procedures, Recombinant Protein Expression, and Complementation Studies in E. coli and Yeast.
This was performed by standard procedures; complementation analyses were carried out as described (18, 19). Experimental details are provided in SI Appendix.
Determination of Single and Multiple Turnover Kinetic Parameters for pretRNAGly Processing by Aq_880.
The experimental details are described in SI Appendix.
5′-End Group Analysis.
5′-end group analysis of mature tRNAGly derived from processing of pretRNAGly by Aq_880 was performed as described (31).
Bioinformatic Prediction of RNase P Components.
Based on the Aq_880 reference sequence, Aq_880 homologs were searched in two iterations in all bacterial and archaeal genomes provided at the National Center for Biotechnology Information (NCBI) (download 2015-10-06) using blastp+ at the proteome level and tblastn+ at the genome level with an e value of 10–10 (32, 33). Similarly, we used the reference sets provided in ref. 34 for Rpp29 (Pop4) and a self-assembled set of well-annotated reference sequences for RnpA homolog searches (the bacterial P protein, also termed C5). The latter comprised sequences from Avibacterium, Bibersteinia, Enterobacter, Escherichia, Herbaspirillum, Mycobacterium, and Serratia. The RNA subunit was predicted using Infernal (35) with the Rfam v12 models RF00010, RF00011, and RF00373 (bacterial RNase P class A, B, and archaeal RNase P, respectively) (36). In the case of multiple hits, only the best one was chosen. All results were aligned using Clustal Omega (37) and manually inspected for likely false positives (none identified). The alignment of Aq_880 homologs (SI Appendix, Fig. S23) and the reference sequences/IDs used for the RnpA search can be downloaded at bioinf.pharmazie.uni-marburg.de/supplements/harp_2016/. Based on the alignment region 19–253, a WebLogo (SI Appendix, Fig. S24) was generated (38). An overview of the results is given in SI Appendix, Table S1.
Phylogenetic Tree.
The phylogenetic tree of Aq_880 homologs was generated using all identified sequences (see above) with a RAxML (39) rapid bootstrap analysis with 1,000 bootstrap runs to search for the best-scoring maximum likelihood tree with respect to the GTR substitution model and the Gamma model of rate heterogeneity (40). For the sake of clarity, bacteria were condensed at order level, archaea at phylum level for Fig. 4. SI Appendix, Table S1 shows the results in more detail.
Alignment of Aq_880 and A. thaliana PRORP1-3.
The alignment of Aq_880 to the metallonuclease domains of A. thaliana PRORP1-3 was performed in a semiautomatic manner. The NYN domains were manually aligned based on the NYN WebLogo provided in ref. 4. The regions upstream and downstream of the NYN domain were aligned using Clustal Omega (37). NCBI accession codes were NP_850186 for A. thaliana PRORP1, NP_179256 for PRORP2, and NP_193921 for PRORP3.
Supplementary Material
Acknowledgments
We thank Diogo Monteiro for his help with the yeast complementation experiments and Dominik Helmecke for support with respect to processing assays, Johann Heider and Wolfgang Buckel for giving us access to the French press and ultracentrifuge, and Tina Krieg for technical assistance in sample preparation for mass spectrometry. The Superose 6 column was a loan from Peter Friedhoff, Justus-Liebig-Universität Gießen. A. aeolicus ribosomal subunits were kindly prepared in the laboratory of Ciarán Condon (Paris).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707862114/-/DCSupplemental.
References
- 1.Hartmann E, Hartmann RK. The enigma of ribonuclease P evolution. Trends Genet. 2003;19:561–569. doi: 10.1016/j.tig.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Klemm BP, et al. The diversity of ribonuclease P: Protein and RNA catalysts with analogous biological functions. Biomolecules. 2016;6:E27. doi: 10.3390/biom6020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holzmann J, et al. RNase P without RNA: Identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Gobert A, et al. A single Arabidopsis organellar protein has RNase P activity. Nat Struct Mol Biol. 2010;17:740–744. doi: 10.1038/nsmb.1812. [DOI] [PubMed] [Google Scholar]
- 5.Taschner A, et al. Nuclear RNase P of Trypanosoma brucei: A single protein in place of the multicomponent RNA-protein complex. Cell Rep. 2012;2:19–25. doi: 10.1016/j.celrep.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lechner M, et al. Distribution of ribonucleoprotein and protein-only RNase P in Eukarya. Mol Biol Evol. 2015;32:3186–3193. doi: 10.1093/molbev/msv187. [DOI] [PubMed] [Google Scholar]
- 7.Hartmann RK, Gössringer M, Späth B, Fischer S, Marchfelder A. The making of tRNAs and more - RNase P and tRNase Z. Prog Mol Biol Transl Sci. 2009;85:319–368. doi: 10.1016/S0079-6603(08)00808-8. [DOI] [PubMed] [Google Scholar]
- 8.Swanson RV. Genome of Aquifex aeolicus. Methods Enzymol. 2001;330:158–169. doi: 10.1016/s0076-6879(01)30373-7. [DOI] [PubMed] [Google Scholar]
- 9.Willkomm DK, Feltens R, Hartmann RK. tRNA maturation in Aquifex aeolicus. Biochimie. 2002;84:713–722. doi: 10.1016/s0300-9084(02)01447-5. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Altman S. In search of RNase P RNA from microbial genomes. RNA. 2004;10:1533–1540. doi: 10.1261/rna.7970404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marszalkowski M, Willkomm DK, Hartmann RK. 5′-end maturation of tRNA in aquifex aeolicus. Biol Chem. 2008;389:395–403. doi: 10.1515/BC.2008.042. [DOI] [PubMed] [Google Scholar]
- 12.Lechner M, et al. Genomewide comparison and novel ncRNAs of Aquificales. BMC Genomics. 2014;15:522. doi: 10.1186/1471-2164-15-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lombo TB, Kaberdin VR. RNA processing in Aquifex aeolicus involves RNase E/G and an RNase P-like activity. Biochem Biophys Res Commun. 2008;366:457–463. doi: 10.1016/j.bbrc.2007.11.165. [DOI] [PubMed] [Google Scholar]
- 14.Barrera A, Pan T. Interaction of the Bacillus subtilis RNase P with the 30S ribosomal subunit. RNA. 2004;10:482–492. doi: 10.1261/rna.5163104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brillante N, et al. Substrate recognition and cleavage-site selection by a single-subunit protein-only RNase P. Nucleic Acids Res. 2016;44:2323–2336. doi: 10.1093/nar/gkw080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howard MJ, et al. Differential substrate recognition by isozymes of plant protein-only Ribonuclease P. RNA. 2016;22:782–792. doi: 10.1261/rna.055541.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howard MJ, Lim WH, Fierke CA, Koutmos M. Mitochondrial ribonuclease P structure provides insight into the evolution of catalytic strategies for precursor-tRNA 5′ processing. Proc Natl Acad Sci USA. 2012;109:16149–16154. doi: 10.1073/pnas.1209062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wegscheid B, Hartmann RK. The precursor tRNA 3′-CCA interaction with Escherichia coli RNase P RNA is essential for catalysis by RNase P in vivo. RNA. 2006;12:2135–2148. doi: 10.1261/rna.188306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber C, Hartig A, Hartmann RK, Rossmanith W. Playing RNase P evolution: Swapping the RNA catalyst for a protein reveals functional uniformity of highly divergent enzyme forms. PLoS Genet. 2014;10:e1004506. doi: 10.1371/journal.pgen.1004506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gößringer M, et al. Protein-only RNase P function in Escherichia coli: Viability, processing defects and differences between PRORP isoenzymes. Nucleic Acids Res. 2017;45:7441–7454. doi: 10.1093/nar/gkx405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matelska D, Steczkiewicz K, Ginalski K. Comprehensive classification of the PIN domain-like superfamily. Nucleic Acids Res. 2017;45:6995–7020. doi: 10.1093/nar/gkx494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai LB, et al. Discovery of a minimal form of RNase P in Pyrobaculum. Proc Natl Acad Sci USA. 2010;107:22493–22498. doi: 10.1073/pnas.1013969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aravind L, Tatusov RL, Wolf YI, Walker DR, Koonin EV. Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet. 1998;14:442–444. doi: 10.1016/s0168-9525(98)01553-4. [DOI] [PubMed] [Google Scholar]
- 24.Andrews AJ, Hall TA, Brown JW. Characterization of RNase P holoenzymes from Methanococcus jannaschii and Methanothermobacter thermoautotrophicus. Biol Chem. 2001;382:1171–1177. doi: 10.1515/BC.2001.147. [DOI] [PubMed] [Google Scholar]
- 25.Hall TA, Brown JW. Archaeal RNase P has multiple protein subunits homologous to eukaryotic nuclear RNase P proteins. RNA. 2002;8:296–306. doi: 10.1017/s1355838202028492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall TA, Brown JW. Interactions between RNase P protein subunits in archaea. Archaea. 2004;1:247–254. doi: 10.1155/2004/743956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Randau L, Schröder I, Söll D. Life without RNase P. Nature. 2008;453:120–123. doi: 10.1038/nature06833. [DOI] [PubMed] [Google Scholar]
- 28.Paper W, et al. Ignicoccus hospitalis sp. nov., the host of ‘Nanoarchaeum equitans’. Int J Syst Evol Microbiol. 2007;57:803–808. doi: 10.1099/ijs.0.64721-0. [DOI] [PubMed] [Google Scholar]
- 29.Huber R, et al. Aquifex pyrophilus gen. nov. sp. nov., represents a novel group of marine hyperthermophilic hydrogen-oxidizing bacteria. Syst Appl Microbiol. 1992;15:340–351. [Google Scholar]
- 30.Busch S, Kirsebom LA, Notbohm H, Hartmann RK. Differential role of the intermolecular base-pairs G292-C(75) and G293-C(74) in the reaction catalyzed by Escherichia coli RNase P RNA. J Mol Biol. 2000;299:941–951. doi: 10.1006/jmbi.2000.3789. [DOI] [PubMed] [Google Scholar]
- 31.Rossmanith W, Tullo A, Potuschak T, Karwan R, Sbisà E. Human mitochondrial tRNA processing. J Biol Chem. 1995;270:12885–12891. doi: 10.1074/jbc.270.21.12885. [DOI] [PubMed] [Google Scholar]
- 32.Tatusova T, et al. Update on RefSeq microbial genomes resources. Nucleic Acids Res. 2015;43:D599–D605. doi: 10.1093/nar/gku1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenblad MA, López MD, Piccinelli P, Samuelsson T. Inventory and analysis of the protein subunits of the ribonucleases P and MRP provides further evidence of homology between the yeast and human enzymes. Nucleic Acids Res. 2006;34:5145–5156. doi: 10.1093/nar/gkl626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nawrocki EP, et al. Rfam 12.0: Updates to the RNA families database. Nucleic Acids Res. 2015;43:D130–D137. doi: 10.1093/nar/gku1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.