Significance
The large superfamily of cadherins serve essential roles in cell–cell interactions and guidance. The extracellular cadherin (EC) domains responsible for the biological functions are decorated with O-linked mannose glycans, but the functions of these O-glycans are poorly understood. Here we describe an O-mannosylation pathway orchestrated by four homologous TMTC1–4 genes that is dedicated selectively to the cadherin superfamily. Mutations in the TMTC3 gene cause cobblestone lissencephaly, demonstrating the importance of this type of O-mannosylation.
Keywords: glycoproteomics, O-glycosylation, glycosyltransferase, mass spectrometry, gene editing
Abstract
The cadherin (cdh) superfamily of adhesion molecules carry O-linked mannose (O-Man) glycans at highly conserved sites localized to specific β-strands of their extracellular cdh (EC) domains. These O-Man glycans do not appear to be elongated like O-Man glycans found on α-dystroglycan (α-DG), and we recently demonstrated that initiation of cdh/protocadherin (pcdh) O-Man glycosylation is not dependent on the evolutionary conserved POMT1/POMT2 enzymes that initiate O-Man glycosylation on α-DG. Here, we used a CRISPR/Cas9 genetic dissection strategy combined with sensitive and quantitative O-Man glycoproteomics to identify a homologous family of four putative protein O-mannosyltransferases encoded by the TMTC1–4 genes, which were found to be imperative for cdh and pcdh O-Man glycosylation. KO of all four TMTC genes in HEK293 cells resulted in specific loss of cdh and pcdh O-Man glycosylation, whereas combined KO of TMTC1 and TMTC3 resulted in selective loss of O-Man glycans on specific β-strands of EC domains, suggesting that each isoenzyme serves a different function. In addition, O-Man glycosylation of IPT/TIG domains of plexins and hepatocyte growth factor receptor was not affected in TMTC KO cells, suggesting the existence of yet another O-Man glycosylation machinery. Our study demonstrates that regulation of O-mannosylation in higher eukaryotes is more complex than envisioned, and the discovery of the functions of TMTCs provide insight into cobblestone lissencephaly caused by deficiency in TMTC3.
Protein O-mannosylation on select Ser/Thr residues is found in yeast and metazoans, and the genetic and biosynthetic basis for this type of glycosylation has long been thought to be an evolutionarily conserved family of protein O-mannosyltransferases with seven members in yeast (Pmt1-7) and two in higher eukaryotes (POMT1/POMT2) (1). O-linked mannose (O-Man) glycosylation is the only type of O-glycosylation found in yeast (2, 3), and O-Man glycans are widely found on proteins trafficking the secretory pathway, similar to O-GalNAc glycosylation in metazoans (4). In contrast, O-Man glycosylation in metazoans is found only on select proteins, with α-dystroglycan (α-DG) being the best characterized (5–7), but, with the advent of sensitive glycoproteomics strategies, we recently found O-Man glycans on the large superfamily of cadherins (cdhs) and protocadherins (pcdhs) as well as a family of IPT/TIG domain-carrying proteins, including the hepatocyte growth factor receptor (HGFR) (6). The O-Man glycans on cdhs and pcdhs are found in highly conserved positions in the β-strands of extracellular cdh (EC) domains, and these glycosites appear highly conserved in evolution, suggesting that they serve important biological functions (6).
Deficiencies in enzymes catalyzing the structurally complex and diverse O-Man glycans on α-DG, including the two human POMT1 and POMT2 genes, underlie a subgroup of congenital muscular dystrophies that have been designated α-dystroglycanopathies because deficient O-Man glycosylation of α-DG disrupts the interaction between the dystrophin glycoprotein complex and the ECM (7–9). Several studies have also implicated deficiency of POMT2 with E-cdh dysfunction (10–12), although direct evidence for a role in glycosylation of cdhs and pcdhs is missing. To explore the functions of O-Man glycans on cdhs and pcdhs, we previously used a combinatorial gene-targeting strategy in multiple cell lines and found that the two POMTs are essential for glycosylation of α-DG but not cdhs, pcdhs, and IPT/TIG domain-containing proteins (13). In contrast to α-DG, we and others (6, 10, 11, 13) also found that O-Man glycans on the latter proteins were not elongated, suggesting that the biosynthesis of these distinct classes of proteins were different. We therefore predicted the existence of a new type of O-Man glycosylation machinery in higher eukaryotes (13).
Here, we report the discovery of a homologous family of putative O-Man glycosyltransferases (GTs) encoded by the four transmembrane and tetra-trico-peptide repeat (TPR) repeat-containing protein (TMTC) genes (TMTC1–4). The TMTC proteins were previously shown to be located in the endoplasmic reticulum (ER) and predicted to serve in Ca2+ regulation and protein folding (14, 15). Moreover, biallelic mutations in TMTC3 were reported to cause cobblestone lissencephaly (16), intellectual disabilities, and epilepsy (17), and family linkage and association analysis points to a role of TMTC2 in hearing loss (18). In murine models, tmtc3-KO results in early neonatal death, and in vitro studies have demonstrated abnormal TGF-β signaling in embryonic fibroblasts from these mice (19).
We used combinatorial gene KO targeting in HEK293 cells combined with differential O-Man glycoproteomics to discover the roles of the TMTC genes in O-Man glycosylation of cdhs and pcdhs, and we validated this by analysis of recombinant-expressed secreted cdh construct in TMTC mutant cell lines. We present strong evidence that the TMTC genes encode distinct O-Man GTs that cooperatively glycosylate distinct regions in the EC domains of cdhs and pcdhs. We also present evidence that the TMTCs do not glycosylate the IPT/TIG domain-containing proteins, and therefore we predict the existence of yet another undiscovered O-Man glycosylation pathway dedicated to IPT/TIG domains. The severe phenotype associated with deficiency of TMTC3 (16, 17) indicates that O-Man glycans on the cdh superfamily serve important biological functions.
Results
Bioinformatic Identification of the TMTC1–4 Genes.
Our finding that the POMT1/2 genes were dispensable for O-mannosylation of many proteins (13) prompted a search for additional enzymes. We first tested the possibility that candidate enzyme(s) responsible for cdh/pcdh O-mannosylation were already classified as GTs in the Carbohydrate-Active enZYmes (CAZy) database, albeit without known function, and KO of six such genes in combinations predicted to cover potential redundant isoenzymes in individual GT families did not affect O-mannosylation (GLT1D1/GTDC1, GLT8D1/GLT8D2, and KDELC1/KDELC2; not shown). We next hypothesized that the candidate enzyme(s) would resemble existing protein mannosyltransferases in CAZy GT39 and GT98 families with respect to overall multitransmembrane domain structure but without significant sequence similarity. We performed a broad search considering criteria common for Pmts/POMTs and the DPY19L genes responsible for C-mannosylation, including ER localization, general topology, and domain structure features. We identified the TMTC genes based on a conserved domain from yeast Pmts. Yeast Pmts match the PMT_2 superfamily domain hmm model (cl21590), and specifically the PMT_2 PFAM model (pfam13231); however, searching for these models directly in the human proteome identified only POMTs. By using the Conserved Domain Architecture Retrieval Tool (20), the complete set of metazoan proteins in refseq that match PMT_2 were retrieved, and the TMTC genes with C-terminal TPR domain invariant presence of DUF1736 were identified. TPR domains are found in various proteins, where they serve as interaction scaffolds and multiprotein complex mediators (21). The TMTCs have been reported to be located in the ER (15), and we have identified GalNAc-type and O-Man O-glycans in CHO cells that support luminal orientation of the C-terminal TPR domain (13, 22) (https://glycodomain.glycomics.ku.dk). The PMT_2 fold belongs to a larger family of folds whose structure is similar to the ArnT arabinosyltransferase identified in bacteria classified in CAZy GT83 (23, 24). Structural alignments of TMTCs and ArnT yielded good models and indicated that the catalytic residues are conserved in the first ER lumen loop. By using human TMTC3 protein sequence without the TPR repeats as source in the iterative threading assembly refinement (I-TASSER) prediction tool (25), the best-aligned structure, with a TM score (a metric for measuring the similarity of two proteins) of 0.782, was for the ArnT GTs from Cupriavidus metallidurans, and the second best TM score of 0.675 was for the oligosaccharyltransferase from Archaeoglobus fulgidus. We therefore considered the TMTCs as good candidates and proceeded with a KO strategy to test this hypothesis (13).
KO of TMTC1–4 in HEK293 Selectively Impairs O-Man Glycosylation of cdhs/pcdhs.
We previously generated HEK293 cells with KO of COSMC and POMGNT1 (HEK293SC), which results in a double “SimpleCell” line with truncated O-Man and O-GalNAc glycans suitable for lectin weak affinity chromatography (LWAC) enrichment of O-Man glycopeptides with the ConA lectin as well as O-GalNAc with the VVA lectin (6, 13, 26) (Fig. 1A). α-DG contains a central mucin-like domain with O-Man glycans in the N-terminal region and O-GalNAc glycans in the C-terminal region, and is therefore a useful probe for detection of substitution of O-Man with O-GalNAc glycans and exploration of common substrates (6, 13, 22), but also serves as a sensitive control for expression and Golgi processing of α-DG in the panel of isogenic KO cells.
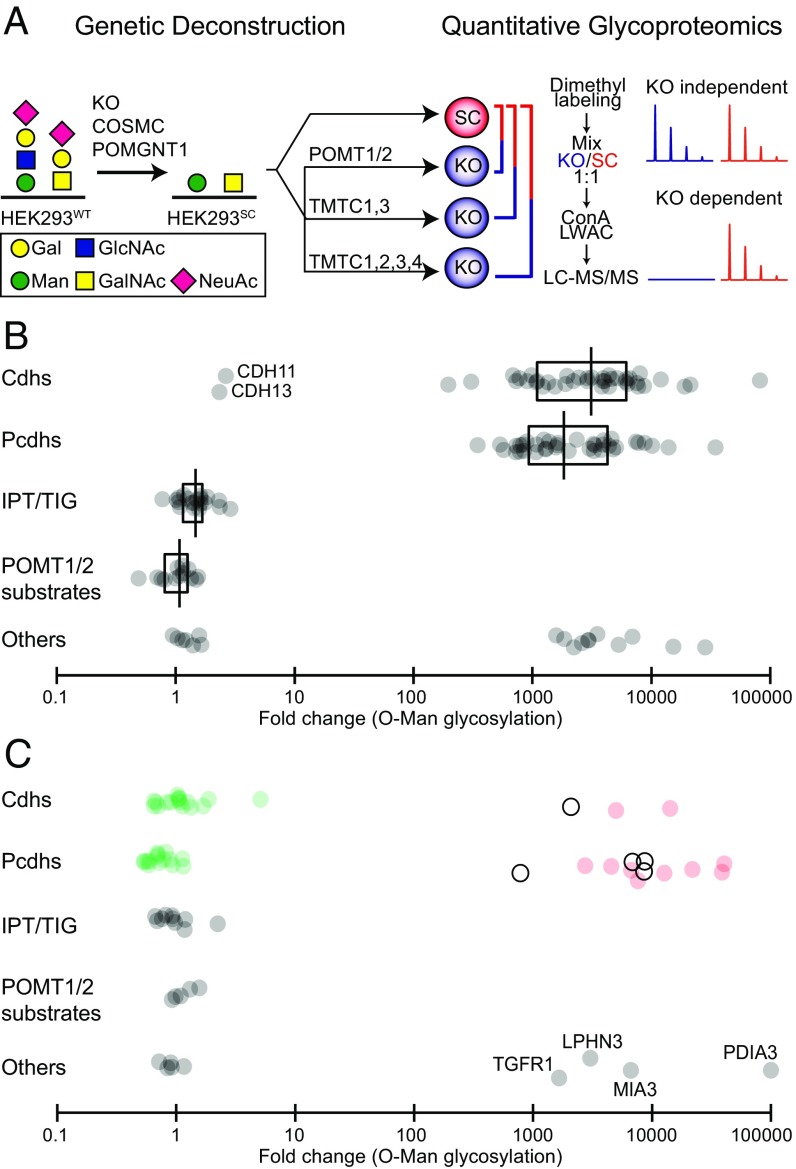
Fig. 1.
Genetic dissection and differential quantitative glycoproteome analysis identify TMTC1–4 as directing O-Man glycosylation of cdhs and pcdhs. (A) Graphical depiction of gene editing in HEK293 double SimpleCells (SC) with truncated O-Man and O-GalNAc glycans targeting TMTCs. For differential O-Man glycoproteome analyses, paired dimethyl labeled digests from HEK293SC and HEK293SC/TMTC1,2,3,4 or HEK293SC/TMTC1,3 cells were performed. We previously reported the differential O-Man glycoproteome analyses with HEK293SC/POMT1,2 (13). (B) Scatter plot of dimethyl labeled O-Man glycopeptide ratios (HEK293SC/HEK293SC/TMTC1,2,3,4) expressed on a log10 scale showing loss of O-Man glycopeptides (>100-fold change) derived from cdhs/pcdhs and other proteins, but not glycopeptides derived from IPT/TIG domains or previously identified POMT1/2 substrates (α-DG, KIAA1549, SUCO; <10-fold change) (13) in HEK293 cells with KO of TMTC1–4. Median ratios are plotted for unique O-Man glycopeptides. The box represents the interquartile range, and the vertical line represents the median value within each protein group. (C) Scatter plot of dimethyl labeled O-Man glycopeptide ratios (HEK293SC/HEK293SC/TMTC1,3) showing selective loss of O-Man glycopeptides derived from EC G-strands (red dots; >100-fold change) of cdhs and pcdhs, but not from B-strands (green dots; <10-fold change) in HEK293 cells with KO of TMTC1/3. White dots represent overlapping O-Man glycosylations on B-strand/loop regions. Additional proteins with TMTC1/3-dependent O-Man glycosylations are indicated at lower right: TGF-β receptor type-1 (TGFR1), Latrophilin-3 (LPHN3), Melanoma inhibitory activity protein 3 (MIA3) and protein disulfide-isomerase A3 (PDIA3).
Sequence analysis of the four TMTC-predicted protein products predicts that they have similar overall structures with N-terminal domains with multiple transmembrane spanning domains (predicted 9–12) and C-terminal domains with TPRs (predicted 10), but the sequence similarity and predicted domain structures do not enable segregation of the four TMTCs into potential subgroups. We therefore reasoned that the TMTCs would have similar and likely overlapping functions, and we designed a two-step KO strategy to obtain HEK293 cells without functional TMTC genes. In the HEK293SC background, we first simultaneously targeted TMTC1 and TMTC3 to obtain HEK293SC/TMTC1,3 cells by using the CRISPR/cas9 strategy and subsequently targeted TMTC2 and TMTC4 to generate HEK293SC/TMTC1,2,3,4 cells (Fig. 1A). The genetic engineering did not affect viability, growth including attachment, and gross morphology of the HEK293 cells. We used our previously established comparative quantitative O-glycoproteomics workflow based on differential labeling of tryptic digests from isogenic cells with stable dimethyl isotopes to probe global O-Man (27, 28). Total digests from isogenic HEK293SC and HEK293SC/TMTC1,2,3,4 cells were labeled with light and medium labels and mixed, and glycopeptides were enriched by LWAC. Following liquid chromatography (LC)/tandem MS (LC-MS/MS), medium/light (M/L) ratios were calculated for individual glycopeptides to quantify the relative changes of O-Man glycosylations between HEK293SC and HEK293SC/TMTC1,2,3,4 cells.
We identified and quantified O-Man glycopeptides derived from α-DG, 27 members of the cdh and pcdh family, five proteins containing IPT/TIG domains (plexins and HGFR), and 13 additional proteins (Table 1). The identified α-DG O-Man glycopeptides were equally present in HEK293SC and HEK293SC/TMTC1,2,3,4 cells (Fig. 1B and Dataset S1), which demonstrates that the POMT1/2-directed O-mannosylation is unaffected by KO of TMTC1–4. Unaffected O-Man glycosylation was also observed on glycopeptides derived from plexins, HGFR, and additional proteins. In striking contrast, nearly all O-Man glycopeptides derived from members of the cdh/pcdh family were absent in HEK293SC/TMTC1,2,3,4 cells. We quantified 79 unique O-Man glycopeptides from cdh/pcdhs, of which 77 were >100-fold less abundant in the HEK293SC/TMTC1,2,3,4 cell line compared with HEK293SC (Fig. 1B and Dataset S1). Representative members of classical/type I cdhs, desmosomal cdhs, clustered and nonclustered pcdhs, as well as unconventional/ungrouped cdhs were all identified with O-Man glycopeptides that were >100-fold less abundant. Interestingly, two quantified O-Man glycopeptides from cdh11 and cdh13 were found to be equally abundant. Nevertheless, these results clearly indicate that the TMTC1–4 genes play an important role for glycosylation of cdhs and pcdhs.
Table 1.
Summary of O-Man glycoproteins identified in KO HEK293 cells
| Protein | SC | SC TMTC1,2,3,4 | SC | SC TMTC1,3* |
| POMT1/T2 substrates | ||||
| Dystroglycan | 1 | 1 | 1 | 1 |
| SUCO | 1 | 1 | 1 | 1 |
| KIAA 1549 | 1 | 1 | 1 | 1 |
| Cdhs | ||||
| Classical type I | 4 | 1 | 4 | 4 |
| Atypical type II | 3 | 1 | 1 | 1 |
| Flamingo | 2 | 1 | 1 | |
| Desmocollin | 2 | 1 | 1 | |
| Desmoglein | 1 | |||
| Pcdhs | ||||
| Clustered | 7 | 5 | 2 | |
| Nonclustered | 5 | 5 | 3 | |
| FAT | 3 | 3 | 3 | |
| Plexins | 4 | 4 | 3 | 3 |
| Other | 12 | 7 | 8 | 4 |
| Total | 46 | 16 | 34 | 25 |
Specific sites are differentially O-mannosylated.
We next performed the same comparative analysis with HEK293SC/TMTC1,3 cells, and identified 34 O-Man glycoproteins, of which 20 were members of the cdh/pcdh family (Fig. 1C and Dataset S2). We quantified 47 unique O-Man glycopeptides from cdhs/pcdhs, of which 15 were >100-fold less abundant in the HEK293SC/TMTC1,3 cell line, and 32 peptides showed no substantial change compared with HEK293SC. Interestingly, essentially all of the identified O-Man glycosites that were affected by the TMTC1,3 KO were located on the G-strands of EC domains, whereas the unaffected glycosites were localized to B-strands in EC domains of desmocollin-2, E-, P-, N-, T-cdh, and cdh-11.
We identified two differentially regulated O-Man glycopeptides (four O-Man glycosites) >100-fold less abundant in HEK293SC/TMTC1,3 cells for classical/type I cdhs, all of which were localized to the EC3 G-strand of N-cdh (Fig. 1C and Dataset S2). A single differentially regulated O-Man glycosite, localized in the loop between EC3 B and C-strands, was identified for cdh EGF LAG seven-pass G-type receptor 2. The same trend was observed for pcdhs; eight glycopeptides were identified from pcdh10, 16, 17, α11, β2, and γB5, all of which had O-Man glycans on G-strands and were >100-fold less abundant in HEK293SC/TMTC1,3 cells. In contrast, all O-Man glycosites (15 glycopeptides; pcdh7, 9, 17, α11, γA11, and FAT1, 2, and 3) mapped to pcdh B-strands showed no change in relative abundance between in HEK293SC and in HEK293SC/TMTC1,3 cells. However, we note that four pcdh glycopeptides with overlapping O-Man glycosylations in B-strand/loop regions were identified as differentially regulated in HEK293SC/TMTC1,3 cells, indicating that the addition of O-Man glycans on loops adjacent to B-strands is dependent on TMTC1 and/or TMTC3 isoenzymes. Taken together, these results indicate that individual TMTC genes have distinct roles for glycosylation of the two opposite regions of EC domains (B- and G-strands) on cdhs and pcdhs.
The quantitative O-Man glycoproteomic analysis revealed additional proteins targeted for O-mannosylation by the TMTC1–4 family. Protein disulfide-isomerase A3 (PDIA3) was previously also identified with several O-Man glycans (6, 13), and all quantified PDIA3 O-Man glycopeptides were >100-fold less abundant in HEK293SC/TMTC1,2,3,4 and HEK293SC/TMTC1,3 cells compared with HEK293SC (Fig. 1C and Datasets S1 and S2). In addition, one O-Man glycopeptide from the extracellular region of TGF-β receptor type-1 was found to be differentially regulated in HEK293SC/TMTC1,3 cells, and this observation is further discussed later.
We previously demonstrated that O-mannosylation of IPT/TIG domain-containing proteins (plexins and HGFR) was not dependent on POMT1/2, and we predicted that these would then be controlled by the enzyme(s) dedicated to cdh/pcdh glycosylation. Surprisingly, the quantitative analysis of HEK293SC/TMTC1,2,3,4 and HEK293SC/TMTC1,3 cells undertaken here revealed unaffected O-Man glycosylation within the IPT/TIG domains of these proteins. HGFR was identified with five unique O-Man glycopeptides from IPT/TIG domains, all of which were equally abundant between HEK293SC and HEK293SC/TMTC1,2,3,4 cells (Fig. 1B and Dataset S1). We quantified 18 unique glycopeptides from plexin-A1, -A2, -A3, and -B2, of which 15 were localized within IPT/TIG domains. All O-Man glycans mapped to plexin IPT/TIG domains were equally abundant (Fig. 1B and Datasets S1 and S2) in HEK293SC and HEK293SC/TMTC1,2,3,4 cells.
Interestingly, we also identified three unique O-Man glycopeptides from plexin-B2, covering two O-Man glycosites localized outside the IPT/TIG domains that were >100-fold less abundant in HEK293SC/TMTC1,2,3,4 cells (Dataset S1). These O-Man glycosites in plexin-B2 were unaffected in the HEK293SC/TMTC1,3 cells (Dataset S2), indicating that TMTC2 and/or TMTC4 play selective roles for glycosylation of other proteins than cdhs and pcdhs.
TMTCs Selectively Control O-Mannosylation of Recombinant Expressed E-cdh.
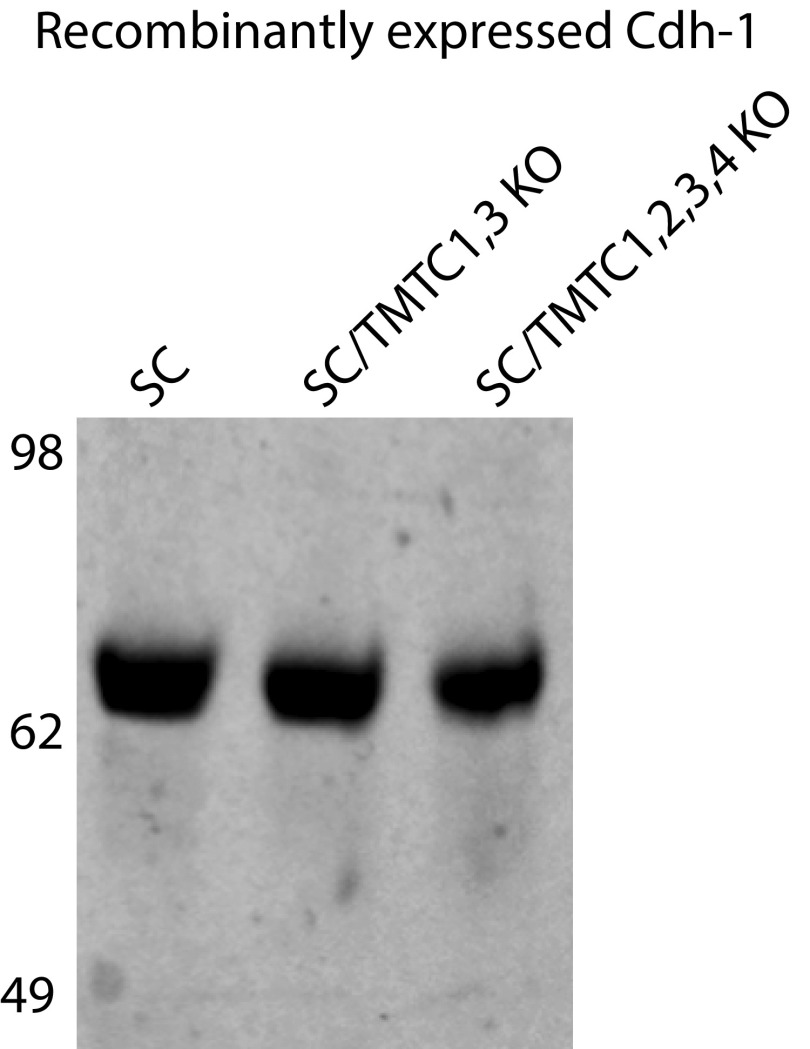
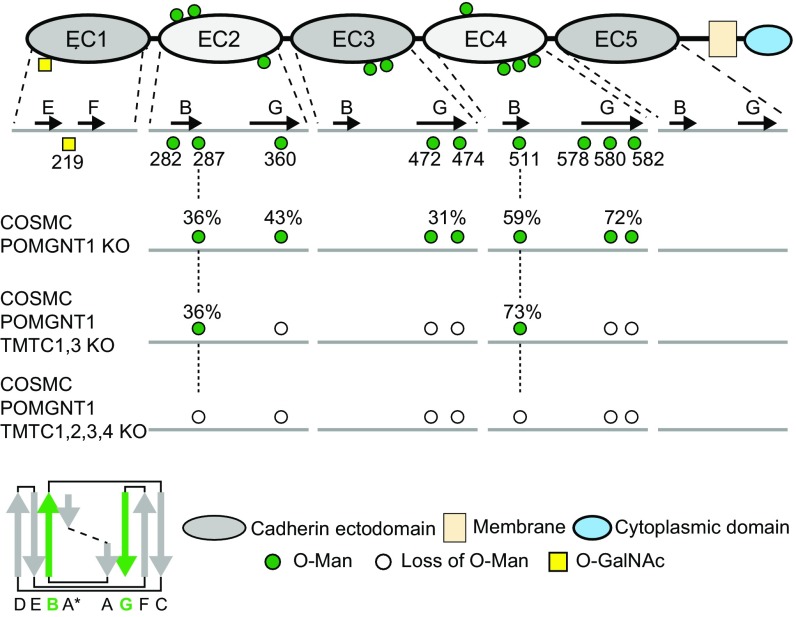
We previously demonstrated efficient expression and O-mannosylation of a secreted E-cdh ectodomain construct in HEK293 WT and POMT1/2 KO cells (13). Here, we expressed the same His-tagged E-cdh ectodomain fragment in HEK293SC, HEK293SC/TMTC1,3, and HEK293SC/TMTC1,2,3,4 cells to evaluate the role of TMTCs on E-cdh production, secretion, and O-mannosylation. E-cdh was expressed and secreted equally in all three cell lines and purified (Fig. S1). Label-free MS analysis of tryptic digests from HEK293SC revealed seven O-Man glycosites on EC2–EC4 with relatively high stoichiometry (30–70% site occupancy; Fig. 2). In contrast, E-cdh expressed in HEK293SC/TMTC1,2,3,4 produced essentially no detectable O-Man glycopeptides (within the signal-to-noise level). However, E-cdh expressed in HEK293SC/TMTC1,3 cells had normal O-Man glycans at two residues (Ser287 and Thr511) localized to the B-strand of EC2 and EC4 (Fig. 2), whereas the O-Man glycopeptides derived from the G-strands of EC2-EC4 were absent. This correlates well with the global analysis of O-Man cdh/pcdh glycoproteins in HEK293SC/TMTC1,3, and further confirms that the TMTC genes play different roles in glycosylation of the cdh/pcdh family.
Fig. S1.
SDS/PAGE analysis of recombinant expressed E-cdh. E-cdh (5 µg) purified from HEK293SC, HEK293SC/TMTC1,3, and HEK293SC/TMTC1,2,3,4 cells was separated on a NuPAGE 4–12% Bis-Tris protein gel and visualized by InstantBlue.
Fig. 2.
Summary of O-Man glycosites identified on recombinant E-cdh expressed in HEK293 mutant cell lines. Schematic representation of E-cdh domain organization (Top) with known O-Man glycosylation sites (green circles). Selected β-strands are indicated by black arrows for each EC domain, and all O-Man and O-GalNAc glycosites identified previously are shown with positions and numbering relative to EC β-strands and amino acids of E-cdh (UniProt Knowledgebase ID code P12830). Below the schematic representation are the O-Man glycosites identified on the recombinantly expressed E-cdh in HEK293SC, HEK293SC/TMTC1,3, and HEK293SC/TMTC1,2,3,4 cells, respectively. The stoichiometry (i.e., site occupancy) of O-Man glycans was calculated from the LC peak areas for each peptide (with and without O-Man glycans) and is indicated above each glycosylation site (as percentages). White circles represent loss of O-Man glycosylation sites (not detectable). (Bottom Left) Canonical EC β-strand arrangement, with B- and G-strands indicated in green.
Discussion
The present study presents evidence indicating that the four TMTC genes encode protein O-mannosyltransferases dedicated primarily to the cdh superfamily, and that the individual enzymes serve distinct roles in decorating the EC domains with O-Man glycans at specific regions. O-Man glycans on these proteins were only recently discovered (6, 10), and although our knowledge of the functions of the conserved glycans in cdhs and pcdhs is still limited, the recent findings that biallelic mutations of TMTC3 cause cobblestone lissencephaly (16), clearly indicates that this type of O-mannosylation has critical functions. This is further supported by the finding that mice deficient in tmtc3 suffer early postnatal death (19). TMTCs are predicted to serve distinct nonredundant functions, which is in excellent agreement with our finding that KO of TMTC1 and TMTC3 eliminated O-Man glycans found on G-strands but not B-strands of EC domains, whereas KO of all four was required to eliminate glycans completely. Clearly, further work is needed to confirm the roles of the TMTC genes, but the potential existence of a distinct gene family dedicated to O-mannosylation of cdhs and pcdhs was a surprising finding obtained through genetic dissection that now opens the possibility of wider studies of the mechanism and biological functions.
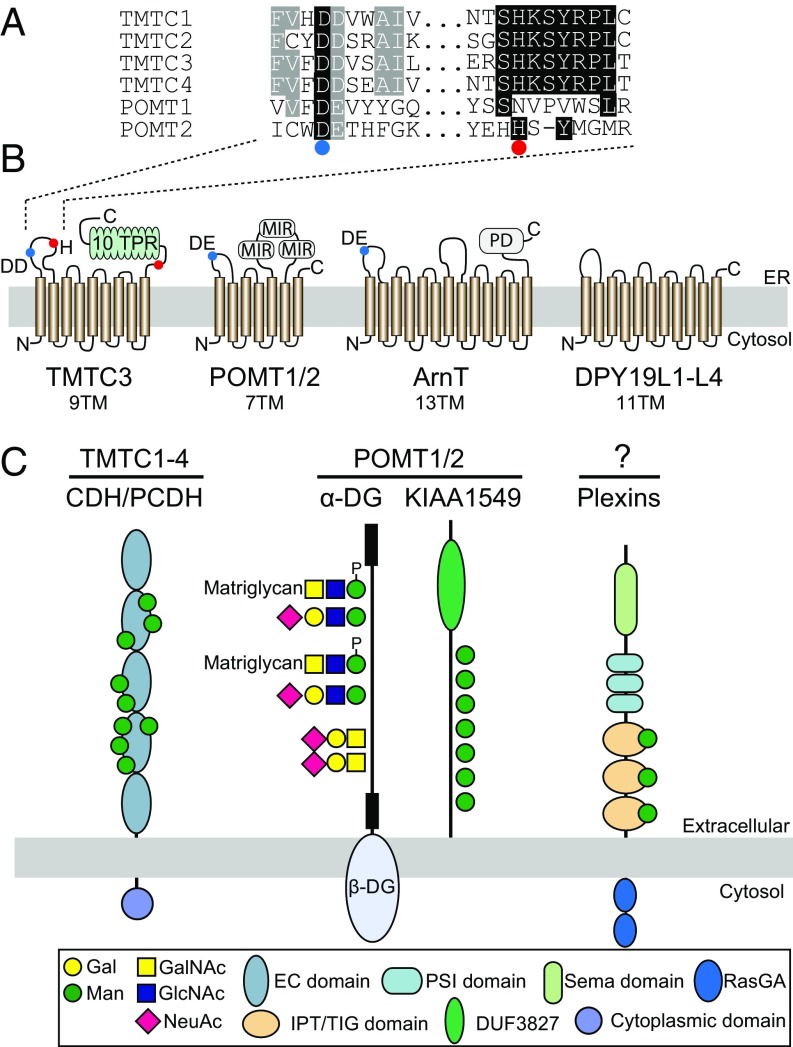
The present results demonstrate that the TMTC genes are required for O-mannosylation of cdhs and pcdhs, and indicate that they serve as O-mannosyltransferases predicted to use Dol-P-Man as donor substrate, similar to the POMTs and DPY19s isoenzymes (29–31). Direct demonstration of enzyme activity of multitransmembrane proteins is a considerable challenge. Several findings, however, support our genetic interpretation that TMTCs are enzymes. The TMTCs share predicted overall structure with the POMT, ArnT, and DPY19 proteins, and the first luminal loop contains an conserved acidic DD/DE motif essential for POMT/Pmt activity; however, the cross-conservation in this region between TMTCs and POMTs is quite limited (32) (Fig. 3). Moreover, a homozygous mutation of His67Asp close to the DD/DE motif in this loop of TMTC3 causes cobblestone lissencephaly (16) (Fig. 3). Deciphering the contributions of individual family members of GT is not straightforward, and such analyses have to include spatiotemporal context to deduce unique and potentially disease-causing functions (28). The level of difficulties faced is perhaps best illustrated by the limited understanding of the functions of the related yeast and mammalian O-mannosyltransferase isoenzymes (pmts and POMT1/POMT2) (2, 33, 34). Although we demonstrate here that the TMTC genes clearly have the capacity to differentially direct O-Man on cdh/pcdh EC domain G- and B-strands, we expect that the characterization of the individual contributions of each gene will require a complete combinatorial set of single and triple KOs of TMTC1–4. The TMTC genes are rather ubiquitously expressed in human tissues and cell lines as evaluated by RNA sequencing data, and they are highly evolutionary conserved, with apparent orthologs of all four genes present in fish, flies, and worms.
Fig. 3.
A proposed model for differential genetic regulation of protein O-mannosylation in higher eukaryotes. (A) Sequence alignment of a segment from the first luminal loop of human TMTCs and POMTs. A conserved acidic amino acid motif DD/DE in the TMTCs, POMTs (32), and ArnT is indicated by a blue circle. Single amino acid mutations in TMTC3 causing cobblestone lissencephaly are indicated by a red circle. (B) The transmembrane (TM) organization of TMTCs is based on the predicted structure of TMTC3 (16). POMT1/2 is adapted from ref. 44. The ArnT structure is based on the crystal structure from C. metallidurans (23), and the structure of DPY19Ls is adapted from the Caenorhabditis elegans DPY-19 (29). (C) O-mannosylation in higher eukaryotes is predicted to be controlled by at least three distinct enzyme families. The TMTCs control the cdh superfamily, whereas the classical POMT1/2 controls α-DG and KIAA1549. O-Man glycans on α-DG are elongated by different core structures (45), but it is currently unknown if the O-Man glycans on KIAA1549 are further elongated to form core M1–M3 and/or other glycan structures. The POMT1/2-independent O-mannosylation of cdhs, plexins, and other proteins does not appear to be further elongated (13). A novel O-Man glycosylation capacity dedicated to plexins and IPT/TIG domains is predicted to exist.
TMTC3 has previously been reported to be involved in ER stress response and interact with PDIA3 and pcdhs (15), and here we demonstrate that these are O-mannosylated by the TMTC-dependent pathway, although the function is still unclear (Fig. 1C and Dataset S2). In addition, two O-Man glycans previously localized on a specific β-strand of the TGF-β receptor type-1 (6) were also found to be differentially regulated in HEK293SC/TMTC1,3 cells. Abnormal TGF-β signaling has been linked to tmtc3 in a murine KO model (19), and it is possible that loss of O-mannosylation in mice deficient in tmtc3 may underlie the phenotype. Finally, TMTC1 and TMTC2 have been proposed to be ER-resident proteins involved in calcium homeostasis (15), and this may not be inconsistent with a role in glycosylation of cdhs and pcdhs, as these are major Ca2+-dependent adhesion molecules.
We did find two O-Man glycopeptides, one derived from cdh11 (EC2 G-strand) and the other from cdh13 (EC2 B-strand), that were not affected by KO of TMTCs (Fig. 1B). It is currently unclear why these appear to be O-mannosylated independently of the TMTCs in contrast to all other cdh/pcdh glycosites; however, both cdhs are less related in sequence to classical cdhs, with cdh11 classified as a type II cdh and cdh13 being truncated and a glycosylphosphatidylinositol-anchored protein. Moreover, the larger group of plexins, HGFR, and other proteins that we previously found to be O-mannosylated independently of POMT1/2 (13) were also found here to be O-mannosylated independently of the TMTC pathway. We were unable to identify specific characteristics for the proteins that appear to be O-mannosylated independently of the POMTs and the TMTCs. It is possible that the POMTs and TMTCs share some overlap in substrate specificities that will require KO of both groups of genes to elucidate, but we predict the existence of yet another novel O-mannosylation capacity selectively serving IPT/TIG domains and HGFR, and hence a total of three distinct pathways for O-mannosylation in higher eukaryotes (Fig. 3).
Several reports have implicated the POMTs in E-cdh glycosylation and function (10, 12). Although we do not have explanations for these seemingly contradictory results, we note that most of the data used to infer this relationship rely on antibodies generated to O-Man glycopeptides, and these appear to have highly restricted reactivity with O-glycopeptides (11). Further work is needed to confirm this, but our previous studies clearly demonstrate that POMTs are not required for cdh/pcdh O-mannosylation in HEK293 cells, and a truncated E-cdh construct can be expressed and secreted normally with O-Man glycans in HEK293 without POMT1 and/or POMT2 (13). The present results show that the TMTC genes are required for O-mannosylation of cdhs and pcdhs, and, interestingly, KO of TMTCs in HEK293 cells did not appear to affect expression, secretion, and immediate stability of recombinant expressed E-cdh (Fig. S1). We did not explore the N-glycans on these constructs, but a study has suggested that POMT-directed O-mannosylation interacted with N-glycosylation (12), and further studies may be needed to clarify this.
POMTs and Pmts are part of the family of GT-C fold enzymes classified in CAZy GT39, whereas DPY19s are classified in CAZy GT98. We propose that the TMTCs be classified as a novel CAZy GT105 family. The C-terminal TPR domains of TMTCs have been found on only one other GT, the nucleocytoplasmic O-GlcNAc-transferase, OGT (35), where it serves in protein–protein interactions and is proposed to function in guiding selection of substrates and kinetic efficiency (21, 35, 36). It is tempting to speculate that TMTC TPRs drive the distinct substrate specificity for the EC folds of cdhs and pcdh. In this respect truncating mutations in the TPR region of TMTC3 were also found to result in cobblestone lissencephaly (16), but future studies need to address the role of the TPRs in detail.
Cobblestone lissencephaly is a severe brain malformation with cortical dysplasia, irregular borders between white and gray matter, brainstem hypoplasia, and cystic cerebellar dysplasia (37). It is found associated with mutations in a large number of genes, of which most are involved in the elaborate POMT1/2 O-mannosylation pathway of α-DG (38), including POMT1, POMT2, POMGNT1, POMGNT2, B4GAT1, B3GALNT2, ISPD, TMEM5, LARGE, FKRP, and FKTN, or associated with the basement membrane, including LAMB1, LAMC3, and COL4A1 (16). The reported cases of cobblestone lissencephaly caused by deficiency in TMTC3 appear to differ from deficiencies in the genes involved in the classical α-DG O-mannosylation pathway by not being characterized by eye and muscle phenotypes. In contrast, mutations in LAMB1 have also been reported to cause cobblestone lissencephaly without eye and muscle involvement (39).
In conclusion, we present evidence suggesting the existence of an O-mannosylation pathway dedicated to cdhs/pcdhs orchestrated by the four TMTC1–4 genes. Preliminary evidence moreover suggests that the individual TMTCs serve distinct functions in glycosylation of different sites in cdh and pcdh EC domains, indicating that glycosylation of these important cell-adhesion proteins may be differentially regulated by expression of different TMTC repertoires. The TMTC-directed O-mannosylation is clearly biologically important, and our findings now open the possibility of more detailed studies.
Experimental Procedures
Detailed information on experimental procedures is included in SI Experimental Procedures.
Precise Gene Targeting of Glycogenes in HEK293 Cells.
HEK293SC cells with double KO of COSMC and POMGNT1 (6) were maintained in DMEM supplemented with 10% FBS and 2 mM l-glutamine. Gene targeting was performed as previously described (40). KO clones with frame-shift mutations were identified by indel detection by amplicon analysis (IDAA) (41) with gene-specific primers (Tables S1–S4). All selected clones were confirmed by Sanger sequencing of 200–300 bp of the target regions.
Table S1.
List of gRNAs
| Genes | Exons | Sequence |
| TMTC1 | 6/18 | 5′-GACCTGCCAGTCATAGCACA GGG-3′ |
| TMTC2 | 2/12 | 5′-GCCTGAACCATGCCATTGGA GGG-3′ |
| TMTC3 | 5/13 | 5′-ACTGCTGGACAGTTTCTCCG TGG-3′ |
| TMTC4 | 1/17 | 5′-GCTGCGTGCAAAACACACAA TGG-3′ |
Table S4.
Sequencing results for HEK293SC/TMTC1,2,3,4 KO
| Genes | Alleles | Indels | Sequence |
| TMTC1 | WT | CTGCCGACCTGCCAGTCATAGC–ACAgggTCACGGGTGCAAGCAGAAGCCACAC | |
| Allele 1 | −5 bp | CTGCCGACCTGCCAGTC––––––ACAgggTCACGGGTGCAAGCAGAAGCCACAC | |
| Allele 2 | +1 bp | CTGCCGACCTGCCAGTCATAGCCACAgggTCACGGGTGCAAGCAGAAGCCACAC | |
| TMTC2 | WT | TTTTCGCCTGAACCATGCCATT–GGAgggTTGAATCCCTGGAGCTACCATCTTG | |
| Allele 1 | −1 bp | TTTTCGCCTGAACCATGCCAT––GGAgggTTGAATCCCTGGAGCTACCATCTTG | |
| Allele 2 | +1 bp | TTTTCGCCTGAACCATGCCATTTGGAgggTTGAATCCCTGGAGCTACCATCTTG | |
| TMTC3 | WT | GTACTACTGCTGGACAGTTTCT–CCGtggAAAGGGTAGCATTCCATTTTCTATG | |
| Allele 1 | −7 bp | GTACTACTGCTGGACAGTTTCT––––––––AAGGGTAGCATTCCATTTTCTATG | |
| Allele 2 | +1 bp | GTACTACTGCTGGACAGTTTCTTCCGtggAAAGGGTAGCATTCCATTTTCTATG | |
| TMTC4 | WT | TCATAGCTGCGTGCAAAACACA––CAAtggCAACCGATCCCACTACTAACTTAGC | |
| Allele 1 | −7 bp | TCATAGCTGCGTGCAAAACACA–––––––––AACCGATCCCACTACTAACTTAGC | |
| Allele 2 | +2 bp | TCATAGCTGCGTGCAAAACACATCCAAtggCAACCGATCCCACTACTAACTTAGC |
Table S2.
List of primers
| Genes | Forward primer | Reverse primer |
| TMTC1 | 5′-GGTGACTGAACGCTTACAGAATG-3′ | 5′-AGATGGTGGCTAAGTTCCGC-3′ |
| TMTC2 | 5′-CTCCATGGACGCACATTTTC-3′ | 5′-TGGCTGAGTAGCCTCTTGTAG-3′ |
| TMTC3 | 5′-GAATCATTAGGCAGCTTGG-3′ | 5′-CCTGGTGAATACTGGAAGTTGG-3′ |
| TMTC4 | 5′-TGCAGAAACGGTCCTACCAG-3′ | 5′-ACTGAGACACAGTTGGGCAG-3′ |
Table S3.
Sequencing results for HEK293SC/TMTC1,3 KO
| Genes | Alleles | Indels | Sequence |
| TMTC1 | WT | CTGCCGACCTGCCAGTCATAGC–ACAgggTCACGGGTGCAAGCAGAAGCCACAC | |
| Allele 1 | −5 bp | CTGCCGACCTGCCAGTC––––––ACAgggTCACGGGTGCAAGCAGAAGCCACAC | |
| Allele 2 | +1 bp | CTGCCGACCTGCCAGTCATAGCCACAgggTCACGGGTGCAAGCAGAAGCCACAC | |
| TMTC3 | WT | GTACTACTGCTGGACAGTTTCT–CCGtggAAAGGGTAGCATTCCATTTTCTATG | |
| Allele 1 | −7 bp | GTACTACTGCTGGACAGTTTCT––––––––AAGGGTAGCATTCCATTTTCTATG | |
| Allele 2 | +1 bp | GTACTACTGCTGGACAGTTTCTTCCGtggAAAGGGTAGCATTCCATTTTCTATG |
Sample Preparation for Differential Glycoproteomics.
Packed cell pellets (0.5 mL) were lysed and digested with trypsin or chymotrypsin and subsequently labeled with dimethyl stable isotopes as described previously (13, 27). Following N-glycan removal by PNGase F (Roche), O-Man glycopeptides were separated from nonglycosylated peptides by LWAC by using a 2.8-m column packed in-house with ConA-conjugated agarose beads as previously described (13).
Recombinant Expression.
A secreted His-tagged E-cdh construct was transiently expressed by using PEI and conditioned media collected 3 d posttransfection as previously described (13). Secreted E-cdh was purified from 20 mL on 200 µL of Ni-NTA Sepharose beads as described previously (13), analyzed by NuPAGE, and subjected to trypsin digestions (10 μg E-cdh) and LC-MS/MS.
Nano–LC-MS/MS Analyses.
Samples were analyzed essentially as previously described (13).
Data Analyses.
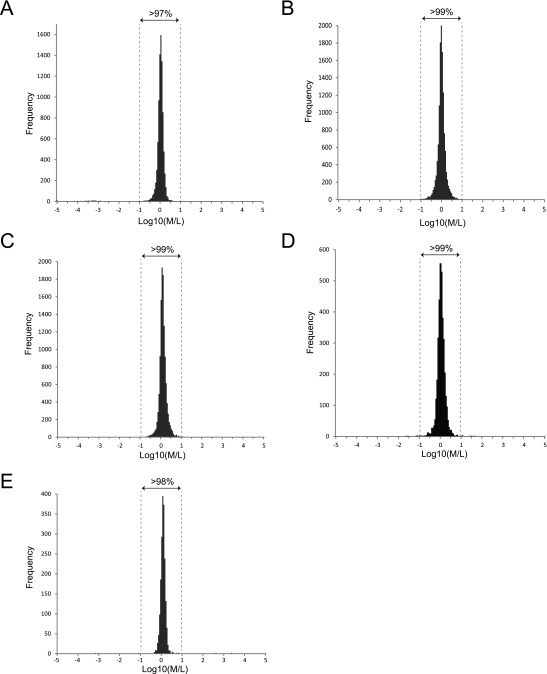
Data processing used Proteome Discoverer 1.4 software (Thermo Fisher Scientific) with minor modifications as outlined as follows (13). Raw data files (.raw) were processed by using the SequestHT or MS Amanda (42) node and searched against the human proteome (January 2013) downloaded from the UniProt Knowledgebase (UniProtKB) database (www.uniprot.org/). In all cases, the precursor mass tolerance was set to 5 ppm and fragment ion mass tolerance to 0.02 Da. Carbamidomethylation on Cys was used as a fixed modification, and oxidation of Met and hexose modification of Ser and Thr residues were used as variable modifications. Enzyme specificity (full or semi-) was trypsin or chymotrypsin; a maximum of two missed cleavage sites were tolerated. Final results were filtered for high-confidence (P < 0.01) identifications only. All O-Man glycopeptides identified by the aforementioned stringent constraints were further manually validated to confirm the accuracy of ion assignments as previously described (6, 13, 43). Relative quantification on the total proteolytic digests from HEK293SC/HEK293SC/TMTC1,2,3,4 and HEK293SC/HEK293SC/TMTC1,3 was performed to determine the technical/biological variability of the assay. A ±10-fold change was estimated for the assay, as >97% of the quantified peptides showed <10-fold relative change in abundance (Fig. S2). We therefore chose to use >100-fold change as a stringent cutoff value when categorizing differentially regulated glycopeptides in this assay.
Fig. S2.
MS-based relative quantification of the total proteolytic digest from paired isogenic KO cells using stable dimethyl isotopes. The technical/biological variability of the assay was estimated by comparing the proteome-level fold change of peptide M/L ratios. (A–C) Total tryptic digests of HEK293SC and HEK293SC/TMTC1,2,3,4 KO cells; (D) total chymotryptic digest of HEK293SC and HEK293SC/TMTC1,2,3,4 KO cells, and (E) total tryptic digest of HEK293SC and HEK293SC/TMTC1,3 KO cells. The bar-chart plots demonstrate that >97% of the quantified peptides in all datasets show <10-fold change. The data are centered around log10(M/L) = 0, which further shows that the proteolytic digest from paired isogenic KO cells have been accurately mixed in 1:1 ratios.
SI Experimental Procedures
Precise Gene Targeting of Glycogenes in HEK293 Cells.
HEK293SC cells with double KO of COSMC and POMGNT1 (6) were maintained in DMEM supplemented with 10% FBS and 2 mM l-glutamine. Gene targeting was performed as previously described (40): briefly, 20-nt guide sequences targeting human TMTC1–4 were designed by using a Desktop Genetics algorithm (Horizon Discovery). The single-guide RNAs were cotransfected with the GFP-tagged Cas9 plasmid by using Lipofectamine 3000 according to the manufacturer’s instructions (Thermo Fisher Scientific). Twenty-four hours after transfection, GFP-positive cells were enriched by FACS, and, following 1 wk of culture, cells were single-sorted in 96-well plates. KO clones with frame-shift mutations were identified by IDAA (41) with gene-specific primers (Tables S1–S4). Clones were selected with frame-shift mutations that result in premature stop codons. All genes were targeted in the center of an exon, and clones were selected with small introduced indels limited to the particular exon. All selected clones were confirmed by Sanger sequencing of 200–300 bp of the target regions.
Sample Preparation for Differential Glycoproteomics.
Packed cell pellets (0.5 mL) were lysed and digested with trypsin or chymotrypsin and subsequently labeled with dimethyl stable isotopes as described previously (13, 27). Briefly, proteolytic digests were labeled by NaBH3CN and formaldehyde (COH2; light label) or NaBH3CN and deuterated formaldehyde (COD2; medium label) and finally mixed at a 1:1 ratio. Following N-glycan removal by PNGase F (Roche), the differentially labeled peptides were resuspended in ConA loading buffer (20 mM Tris⋅HCl, pH 7.4, 300 mM NaCl, 1 mM CaCl2/MgCl2/MnCl2/ZnCl2). O-Man glycopeptides were separated from nonglycosylated peptides by LWAC using a 2.8-m column packed in-house with ConA-conjugated agarose beads as previously described (13). The glycopeptide-containing fractions were purified by in-house packed Stage tips (Empore disk-C18; 3M) before LC-MS/MS analysis.
Recombinant Expression.
A secreted His-tagged E-cdh construct was transiently expressed by using PEI and conditioned media collected 3 d posttransfection as previously described (13). Secreted E-cdh was purified from 20 mL on 200 µL of Ni-NTA Sepharose beads as described previously (13), analyzed by NuPAGE, and subjected to trypsin digestions (10 µg E-cdh) and LC-MS/MS.
Nano–LC-MS/MS Analyses.
Samples were analyzed essentially as previously described (13). Briefly, a setup composed of an EASY-nLC 1000 system (Thermo Fisher Scientific) was interfaced via a nanoSpray Flex ion source to an LTQ-Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific). The EASY-nLC 1000 system was operated by using a single analytical column setup packed in-house with Reprosil-Pure-AQ C18 phase (Dr. Maisch). The LC gradient duration was 120 min at 200 nL/min. The mobile phases were composed of solvent A (H2O) and solvent B (acetonitrile), with both solvents containing 0.1% formic acid (vol/vol). The LC gradient was 2–20% solvent B for 95 min followed by 20–80% solvent B for 10 min and finally 80% solvent B for 15 min. Tryptic peptides from purified E-cad were separated by using a 60-min LC gradient method; the LC gradient was 2–25% solvent B for 35 min followed by 25–80% solvent B for 10 min and finally 80% solvent B for 15 min.
Precursor MS1 scan (m/z 355–1,700) was acquired in the Orbitrap mass spectrometer at a resolution setting of 120,000 (AGC, 4.0e5; injection time, 100 ms) followed by Orbitrap HCD-MS/MS (resolution, 50,000; AGC, 5.0e4; injection time, 75 ms) and ETD-MS/MS (resolution, 60,000; AGC, 1.0e5; injection time, 150 ms) of multiply charged precursors (z = 2–7) in the MS1 spectrum; a minimum MS1 signal threshold of 50,000 ions was used for triggering data-dependent fragmentation events. To improve fragmentation, ETD supplemental activation (ETcid = 25%) was used in all analyses.
Data Analyses.
Data processing used Proteome Discoverer 1.4 software (Thermo Fisher Scientific) with minor modifications as outlined as follows (13). Raw data files (.raw) were processed by using SequestHT or the MS Amanda (42) node and searched against the human proteome (January 2013) downloaded from the UniProtKB database (www.uniprot.org/). In all cases, the precursor mass tolerance was set to 5 ppm and fragment ion mass tolerance to 0.02 Da. Carbamidomethylation on Cys was used as a fixed modification, and oxidation of Met and hexose modification of Ser and Thr residues were used as variable modifications. Enzyme specificity (full or semi-) was trypsin or chymotrypsin; a maximum of two missed cleavage sites were tolerated. Final results were filtered for high-confidence (P < 0.01) identifications only. All O-Man glycopeptides identified by the aforementioned stringent constrains were further manually validated to confirm the accuracy of ion assignments as previously described (6, 13, 43). For dimethyl-labeled samples, glycopeptide medium/light ratios were determined by using the Event Detector Node and the Precursor Ion Node of the Proteome Discoverer workflow as previously described (13). Relative quantification on the total proteolytic digests from HEK293SC/HEK293SC/TMTC1,2,3,4 and HEK293SC/HEK293SC/TMTC1,3 cells was performed to determine the technical/biological variability of the assay. A ±10-fold change was estimated for the assay because >97% of the quantified peptides showed <10-fold relative change in abundance (Fig. S2). We therefore chose to use >100-fold change as a stringent cutoff value when categorizing differentially regulated glycopeptides in this assay.
Supplementary Material
Acknowledgments
This work was supported by A. P. Møller og Hustru Chastine Mc-Kinney Møllers Fond til almene Formaal, Kirsten og Freddy Johansen Fonden, Læge Sofus Carl Emil Friis og hustru Olga Doris Friis’ Legat, The Lundbeck Foundation, The Carlsberg Foundation, The Novo Nordisk Foundation, The European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie Grant 704228, The Danish Research Councils, National Science Foundation Grant MCB-1412472 (to B.H.), National Institutes of Health Grants R01-GM107571 and R01GM118584 (to L. Shapiro), the University of Copenhagen Program of Excellence, and Danish National Research Foundation Grant DNRF107.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708319114/-/DCSupplemental.
References
- 1.Lommel M, Strahl S. Protein O-mannosylation: Conserved from bacteria to humans. Glycobiology. 2009;19:816–828. doi: 10.1093/glycob/cwp066. [DOI] [PubMed] [Google Scholar]
- 2.Loibl M, Strahl S. Protein O-mannosylation: What we have learned from baker’s yeast. Biochim Biophys Acta. 2013;1833:2438–2446. doi: 10.1016/j.bbamcr.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Halim A, Anonsen JH. Microbial glycoproteomics. Curr Opin Struct Biol. 2017;44:143–150. doi: 10.1016/j.sbi.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Neubert P, et al. Mapping the O-mannose glycoproteome in Saccharomyces cerevisiae. Mol Cell Proteomics. 2016;15:1323–1337. doi: 10.1074/mcp.M115.057505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barresi R, Campbell KP. Dystroglycan: From biosynthesis to pathogenesis of human disease. J Cell Sci. 2006;119:199–207. doi: 10.1242/jcs.02814. [DOI] [PubMed] [Google Scholar]
- 6.Vester-Christensen MB, et al. Mining the O-mannose glycoproteome reveals cadherins as major O-mannosylated glycoproteins. Proc Natl Acad Sci USA. 2013;110:21018–21023. doi: 10.1073/pnas.1313446110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Praissman JL, et al. The functional O-mannose glycan on α-dystroglycan contains a phospho-ribitol primed for matriglycan addition. Elife. 2016;5:e14473. doi: 10.7554/eLife.14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Live D, Wells L, Boons GJ. Dissecting the molecular basis of the role of the O-mannosylation pathway in disease: α-dystroglycan and forms of muscular dystrophy. ChemBioChem. 2013;14:2392–2402. doi: 10.1002/cbic.201300417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wells L. The o-mannosylation pathway: Glycosyltransferases and proteins implicated in congenital muscular dystrophy. J Biol Chem. 2013;288:6930–6935. doi: 10.1074/jbc.R112.438978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lommel M, et al. Protein O-mannosylation is crucial for E-cadherin-mediated cell adhesion. Proc Natl Acad Sci USA. 2013;110:21024–21029. doi: 10.1073/pnas.1316753110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartels MF, et al. Protein O-mannosylation in the murine brain: Occurrence of mono-O-mannosyl glycans and identification of new substrates. PLoS One. 2016;11:e0166119. doi: 10.1371/journal.pone.0166119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carvalho S, et al. O-mannosylation and N-glycosylation: Two coordinated mechanisms regulating the tumour suppressor functions of E-cadherin in cancer. Oncotarget. 2016;7:65231–65246. doi: 10.18632/oncotarget.11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsen ISB, et al. Mammalian O-mannosylation of cadherins and plexins is independent of protein O-mannosyltransferases 1 and 2. J Biol Chem. 2017;292:11586–11598. doi: 10.1074/jbc.M117.794487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Racapé M, et al. The involvement of SMILE/TMTC3 in endoplasmic reticulum stress response. PLoS One. 2011;6:e19321. doi: 10.1371/journal.pone.0019321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sunryd JC, et al. TMTC1 and TMTC2 are novel endoplasmic reticulum tetratricopeptide repeat-containing adapter proteins involved in calcium homeostasis. J Biol Chem. 2014;289:16085–16099. doi: 10.1074/jbc.M114.554071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jerber J, et al. Biallelic mutations in TMTC3, encoding a transmembrane and TPR-containing protein, lead to cobblestone lissencephaly. Am J Hum Genet. 2016;99:1181–1189. doi: 10.1016/j.ajhg.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farhan SMK, et al. Identification of a novel synaptic protein, TMTC3, involved in periventricular nodular heterotopia with intellectual disability and epilepsy. Hum Mol Genet. 2017 doi: 10.1093/hmg/ddx316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Runge CL, et al. Association of TMTC2 with human nonsyndromic sensorineural hearing loss. JAMA Otolaryngol Head Neck Surg. 2016;142:866–872. doi: 10.1001/jamaoto.2016.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yun EJ, Vu TH. mSmile is necessary for bronchial smooth muscle and alveolar myofibroblast development. Anat Rec (Hoboken) 2012;295:167–176. doi: 10.1002/ar.21475. [DOI] [PubMed] [Google Scholar]
- 20.Geer LY, Domrachev M, Lipman DJ, Bryant SH. CDART: Protein homology by domain architecture. Genome Res. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeytuni N, Zarivach R. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure. 2012;20:397–405. doi: 10.1016/j.str.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Steentoft C, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32:1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrou VI, et al. Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation. Science. 2016;351:608–612. doi: 10.1126/science.aad1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tavares-Carreón F, Fathy Mohamed Y, Andrade A, Valvano MA. ArnT proteins that catalyze the glycosylation of lipopolysaccharide share common features with bacterial N-oligosaccharyltransferases. Glycobiology. 2016;26:286–300. doi: 10.1093/glycob/cwv095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, et al. The I-TASSER suite: Protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steentoft C, et al. Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat Methods. 2011;8:977–982. doi: 10.1038/nmeth.1731. [DOI] [PubMed] [Google Scholar]
- 27.Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- 28.Schjoldager KT, et al. Deconstruction of O-glycosylation–GalNAc-T isoforms direct distinct subsets of the O-glycoproteome. EMBO Rep. 2015;16:1713–1722. doi: 10.15252/embr.201540796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buettner FF, Ashikov A, Tiemann B, Lehle L, Bakker H. C. elegans DPY-19 is a C-mannosyltransferase glycosylating thrombospondin repeats. Mol Cell. 2013;50:295–302. doi: 10.1016/j.molcel.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Jurado LA, Coloma A, Cruces J. Identification of a human homolog of the Drosophila rotated abdomen gene (POMT1) encoding a putative protein O-mannosyl-transferase, and assignment to human chromosome 9q34.1. Genomics. 1999;58:171–180. doi: 10.1006/geno.1999.5819. [DOI] [PubMed] [Google Scholar]
- 31.Willer T, Amselgruber W, Deutzmann R, Strahl S. Characterization of POMT2, a novel member of the PMT protein O-mannosyltransferase family specifically localized to the acrosome of mammalian spermatids. Glycobiology. 2002;12:771–783. doi: 10.1093/glycob/cwf086. [DOI] [PubMed] [Google Scholar]
- 32.Lommel M, Schott A, Jank T, Hofmann V, Strahl S. A conserved acidic motif is crucial for enzymatic activity of protein O-mannosyltransferases. J Biol Chem. 2011;286:39768–39775. doi: 10.1074/jbc.M111.281196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bausewein D, Engel J, Jank T, Schoedl M, Strahl S. Functional similarities between the protein O-mannosyltransferases Pmt4 from bakers’ yeast and human POMT1. J Biol Chem. 2016;291:18006–18015. doi: 10.1074/jbc.M116.739128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hutzler J, Schmid M, Bernard T, Henrissat B, Strahl S. Membrane association is a determinant for substrate recognition by PMT4 protein O-mannosyltransferases. Proc Natl Acad Sci USA. 2007;104:7827–7832. doi: 10.1073/pnas.0700374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lubas WA, Hanover JA. Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J Biol Chem. 2000;275:10983–10988. doi: 10.1074/jbc.275.15.10983. [DOI] [PubMed] [Google Scholar]
- 37.van der Knaap MS, et al. Magnetic resonance imaging in classification of congenital muscular dystrophies with brain abnormalities. Ann Neurol. 1997;42:50–59. doi: 10.1002/ana.410420110. [DOI] [PubMed] [Google Scholar]
- 38.Vuillaumier-Barrot S, et al. Identification of mutations in TMEM5 and ISPD as a cause of severe cobblestone lissencephaly. Am J Hum Genet. 2012;91:1135–1143. doi: 10.1016/j.ajhg.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radmanesh F, et al. Mutations in LAMB1 cause cobblestone brain malformation without muscular or ocular abnormalities. Am J Hum Genet. 2013;92:468–474. doi: 10.1016/j.ajhg.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lonowski LA, et al. Genome editing using FACS enrichment of nuclease-expressing cells and indel detection by amplicon analysis. Nat Protoc. 2017;12:581–603. doi: 10.1038/nprot.2016.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Z, et al. Fast and sensitive detection of indels induced by precise gene targeting. Nucleic Acids Res. 2015;43:e59. doi: 10.1093/nar/gkv126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dorfer V, et al. MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. J Proteome Res. 2014;13:3679–3684. doi: 10.1021/pr500202e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Halim A, et al. Discovery of a nucleocytoplasmic O-mannose glycoproteome in yeast. Proc Natl Acad Sci USA. 2015;112:15648–15653. doi: 10.1073/pnas.1511743112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akasaka-Manya K, Manya H, Hayashi M, Endo T. Different roles of the two components of human protein O-mannosyltransferase, POMT1 and POMT2. Biochem Biophys Res Commun. 2011;411:721–725. doi: 10.1016/j.bbrc.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 45.Praissman JL, Wells L. Mammalian O-mannosylation pathway: Glycan structures, enzymes, and protein substrates. Biochemistry. 2014;53:3066–3078. doi: 10.1021/bi500153y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.