Significance
The pandemic potential of coronaviruses was recently demonstrated twice by global outbreaks of deadly pneumonia. The spike (S) glycoprotein initiates infection through conformational changes that remain largely uncharacterized. Here we report the cryoEM structure of an S glycoprotein in the postfusion state, showing large-scale rearrangements compared with the prefusion trimer. We further characterized the refolding of the metastable prefusion conformation using limited proteolysis, mass spectrometry, and single-particle EM. The observed similarity to paramyxovirus F structures demonstrates a conserved refolding trajectory and supports the evolutionary relatedness of their fusion subunits. Finally, our data provide a structural framework for understanding antibody neutralization and for engineering vaccines against this medically important virus family.
Keywords: coronavirus, fusion proteins, proteolytic activation, cryoEM, membrane fusion
Abstract
The tremendous pandemic potential of coronaviruses was demonstrated twice in the past few decades by two global outbreaks of deadly pneumonia. The coronavirus spike (S) glycoprotein initiates infection by promoting fusion of the viral and cellular membranes through conformational changes that remain largely uncharacterized. Here we report the cryoEM structure of a coronavirus S glycoprotein in the postfusion state, showing large-scale secondary, tertiary, and quaternary rearrangements compared with the prefusion trimer and rationalizing the free-energy landscape of this conformational machine. We also biochemically characterized the molecular events associated with refolding of the metastable prefusion S glycoprotein to the postfusion conformation using limited proteolysis, mass spectrometry, and single-particle EM. The observed similarity between postfusion coronavirus S and paramyxovirus F structures demonstrates that a conserved refolding trajectory mediates entry of these viruses and supports the evolutionary relatedness of their fusion subunits. Finally, our data provide a structural framework for understanding the mode of neutralization of antibodies targeting the fusion machinery and for engineering next-generation subunit vaccines or inhibitors against this medically important virus family.
Coronaviruses are large enveloped viruses associated with up to 30% of respiratory tract infections in humans. Coronaviruses have also emerged as a global pandemic threat due the outbreaks of severe acute respiratory syndrome (SARS) and of Middle-East respiratory syndrome (MERS). SARS coronavirus (SARS-CoV) and MERS coronavirus (MERS-CoV) are the causative agents of these deadly pneumonias that demonstrated that coronaviruses could cross the species barrier from bats, camels, raccoons, or palm civets to humans (1–4). These observations, along with surveillance studies, suggest that additional emergence events could occur.
Coronavirus entry is mediated by the trimeric transmembrane spike (S) glycoprotein, which is responsible for receptor binding and fusion of the viral and host membranes. S is a class I viral fusion protein that is synthesized as a single-chain precursor of ∼1,300 amino acids and trimerizes upon folding. It forms an extensive crown decorating the virus surface and is the main target of neutralizing antibodies upon infection.
Coronavirus S proteins are comprised of two functional subunits, termed “S1” and “S2” (5). S1 mediates binding to the host receptor and exhibits the most diversity among coronaviruses, partially accounting for the wide host range of this virus family. S2 induces fusion of the viral envelope with cellular membranes and is conserved among coronaviruses. The S glycoprotein exists as a metastable prefusion trimer at the viral surface, and its structure has recently been characterized (6–11). Receptor binding and proteolytic processing promote large-scale conformational changes allowing initiation of the fusion reaction by insertion of the hydrophobic fusion peptide into the host membrane (12, 13). The subsequent irreversible refolding of the fusion machinery provides the energy required to juxtapose the viral and host membranes, promoting fusion and delivery of the viral genome into the cytoplasm.
The only available structural information about the conformational changes undergone by coronavirus fusion machinery comes from X-ray crystallography studies of short polypeptide fragments spanning the heptad-repeat motifs (14–16). The data are limited to a small portion of the fusion machinery and do not reveal how most of the S2 subunit refolds. A detailed knowledge of the conformational changes driving fusion is important to define the accessibility of epitopes targeted by neutralizing antibodies and to engineer improved subunit vaccine candidates, as was reported for the Respiratory syncytial virus (RSV) fusion (F) protein (17–19). Alternatively, heptad-repeat–mimicking peptides have been successfully used to inhibit type I fusion machineries, including coronavirus S glycoproteins (5). Furthering our understanding of the structural rearrangements underlying fusion bears the promise of developing next-generation inhibitors targeting this viral family.
We report here the characterization of the molecular determinants associated with the triggering of several β-coronavirus S glycoproteins using a combination of limited proteolysis, mass spectrometry, and single-particle EM. We describe a near-atomic-resolution cryoEM reconstruction of a coronavirus fusion machinery ectodomain in the postfusion conformation. Our data reveal that the postfusion S trimer adopts a 180-Å-long cone-shaped architecture arranged around a prominent central triple-helical bundle and is the longest structure observed for any class I fusion protein. Despite weak sequence conservation, the structure demonstrates structural similarity to paramyxovirus F proteins, thereby reinforcing the relatedness of their fusion mechanisms and their evolutionary connection. Finally, the results provide a structural framework to rationalize the mode of neutralization of antibodies targeting the conserved fusion machinery.
Results
Protease-Mediated Fusion Activation of Coronavirus S Proteins.
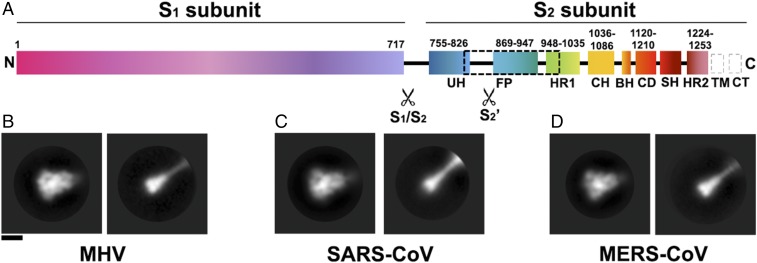
Coronavirus S proteins harbor up to two protease cleavage sites located at the boundary between the S1 and S2 subunits (S1/S2 site) and upstream from the fusion peptide (S2′ site) (Fig. 1A) (12, 13). Cleavage at S1/S2 occurs upon biogenesis and viral egress for some coronaviruses, such as Mouse hepatitis virus (MHV) or MERS-CoV (20). This cleavage event, along with subsequent binding to the host receptor, is essential to promote cleavage at the S2′ site and fusion activation in the case of MERS-CoV (12). The critical importance of cleavage at the S1/S2 site is also exemplified by the Bat coronavirus (Bat-CoV) HKU4. Bat-CoV HKU4 shares a high degree of sequence similarity with MERS-CoV and can bind to the same human receptor (DPP4), although it is unable to infect human cells (3). Engineering two point mutations in the Bat-CoV HKU4 S1/S2 region, which introduces two protease cleavage sites similar to the ones found in the MERS-CoV S sequence, is sufficient to allow efficient entry into human cells (4). These results demonstrate that both receptor and protease specificity are important determinants of host range. Proteolytic fusion activation at the S2′ site, which occurs for all coronaviruses, can take place in several cellular compartments (20). For instance, transmembrane protease/serine protease (TMPRSS) processing of SARS-CoV and MERS-CoV S at the cell membrane, furin-mediated processing of human coronavirus (HCoV)-NL63 and MERS-CoV S in the early endosomes, or lysosomal protease-mediated triggering of SARS-CoV S (cathepsin L) or MHV S are key events that enable fusion activation and coronavirus entry into host cells (12, 20–22). Several coronaviruses have redundancy built into their S protein sequence, enabling activation via multiple proteases and increasing the spectrum of cell types that can be infected (20).
Fig. 1.
Proteolytic activation of coronavirus S proteins. (A) Schematic of the MHV S glycoprotein organization with emphasis on the S2 subunit. The dashed black box shows the region of the S2 polypeptide chain that is unresolved in the map. Gray dashed boxes show regions that were not part of the construct. BH, beta hairpin; CD, connector domain; CH, central helix; CT, cytoplasmic tail; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; SH, stem helix; TM, transmembrane domain; UH, upstream helix. (B–D) 2D class averages of negatively stained MHV S (B), SARS-CoV S (C), and MERS-CoV S (D) trimers in the prefusion state (Left) and in the trypsin-cleaved postfusion state (Right). (Scale bar, 10 nm.)
We incubated the prefusion uncleaved MHV, SARS-CoV, and MERS-CoV S glycoprotein ectodomains with trypsin, under limited-proteolysis conditions, before analysis by mass spectrometry and negative-staining EM. We identified that cleavage occurred at or near the S1/S2 and S2′ sites, which suggested that we recapitulated the proteolytic activation mechanism taking place in vivo (Fig. S1 A–E). We observed particles exhibiting the globular prefusion architecture and particles featuring an elongated cone-like structure reminiscent of the postfusion conformation described for paramyxovirus F proteins (Fig. 1 B–D) (19, 23–25). These observations suggest that the three S ectodomains could refold to the postfusion conformation upon proteolytic activation and release of the S1 subunit. We also detected the formation of rosettes, previously described for the SARS-CoV S protein (26), which are presumed to result from interactions of multiple postfusion trimers via their hydrophobic fusion peptides (Fig. S1F). In agreement with previous reports, our data show that β-coronavirus S proteins can undergo pH-independent conformational changes upon proteolytic cleavage to promote fusion of the viral and host membranes.
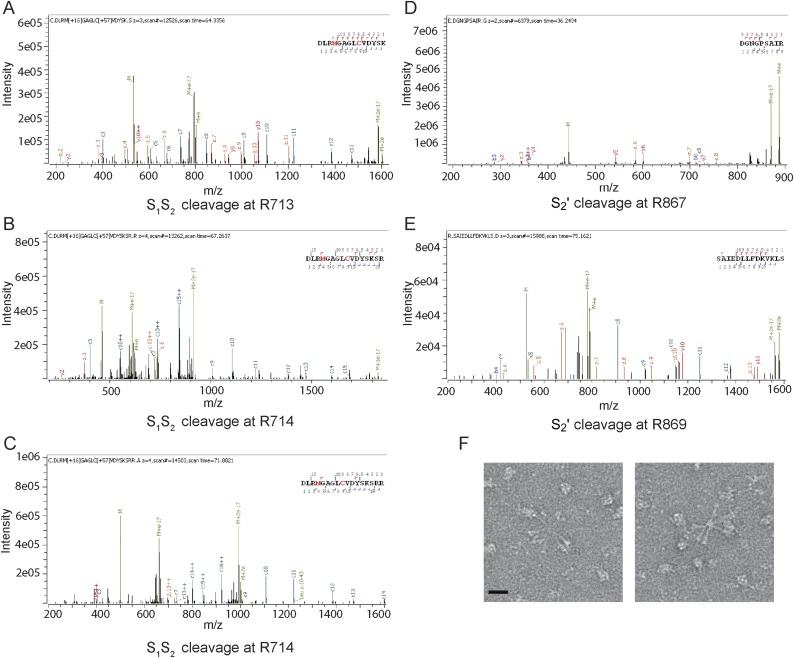
Fig. S1.
Characterization of the MHV S pre- to postfusion transition. (A–E) Annotated mass spectra showing that trypsin cleavage can occur at or near the predicted S1/S2 (A–C) and S2′ (D and E) sites. (F) Section of a micrograph with negatively stained MHV S2 rosettes formed upon incubation of prefusion S with 1:100 trypsin. (Scale bar, 10 nm.)
Structure Determination and Validation.
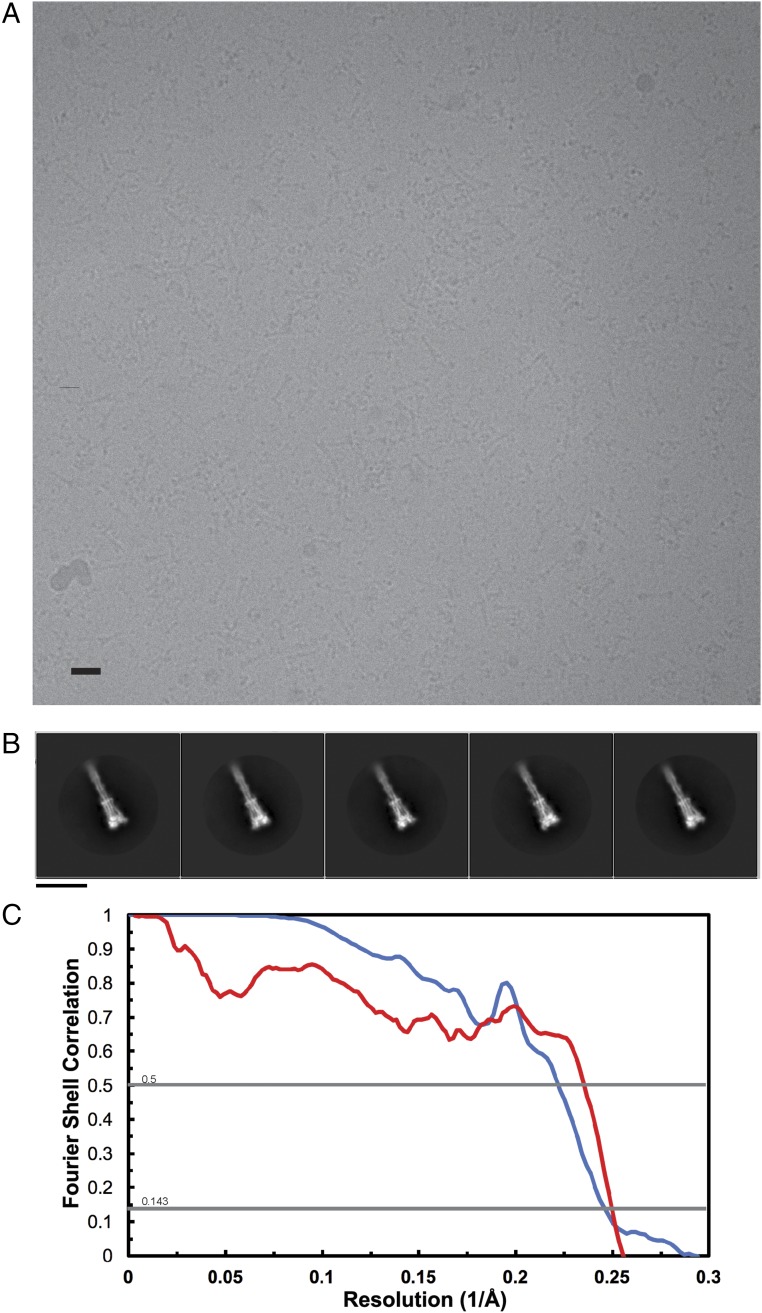
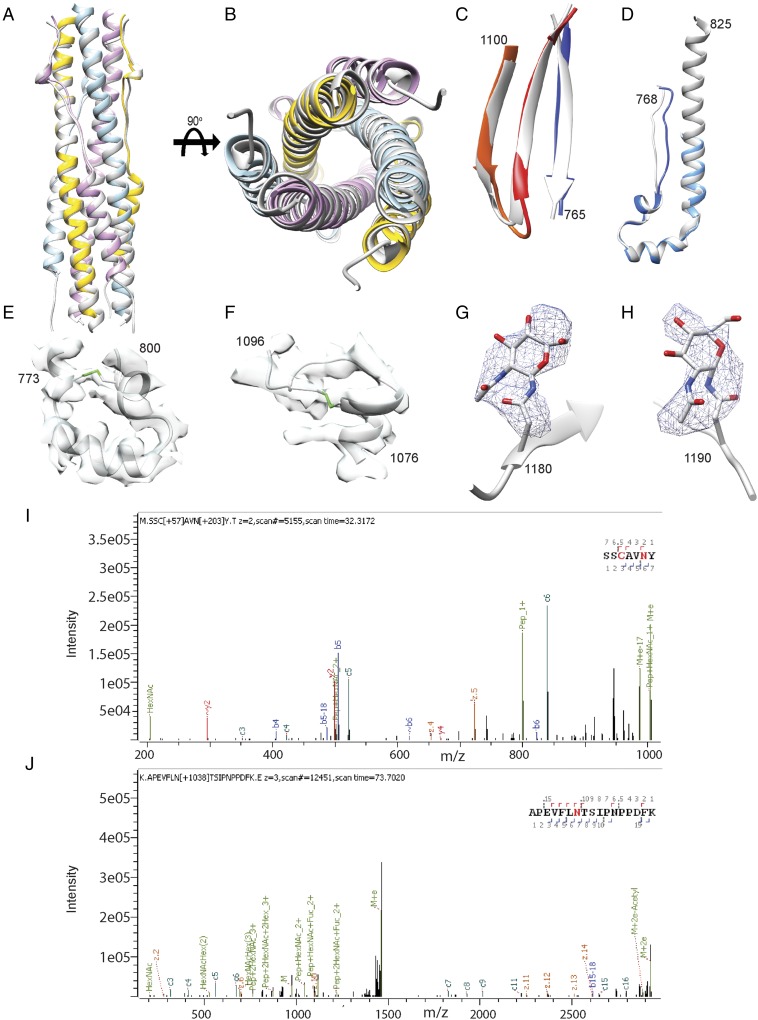
To understand the molecular basis of this transition, we used Drosophila S2 cells to produce only the MHV S2 subunit ectodomain (residues 718–1,252) and to recapitulate the expected large-scale conformational rearrangements of the fusion machinery (lacking the receptor-binding subunit) driving membrane fusion. This construct was fused to a GCN4 trimerization motif at the C-terminal end (27). Frozen-hydrated MHV S2 trimers are 190-kDa cone-shaped particles resembling those shown in Fig. 1B, which suggested that we captured a biologically relevant conformation and that the presence of the GCN4 motif was not sufficient to preserve the prefusion state (Fig. S2 A and B). We determined a 3D reconstruction at 4.1-Å resolution and built and refined a model including residues 741–1,248, with an internal break between residues 808-971, using Coot (28), Rosetta de novo (29), RosettaES (30), and Rosetta density-guided iterative refinement (Fig. 2 and Fig. S2C and Table S1) (31).
Fig. S2.
CryoEM analysis of MHV S2. (A) Representative micrograph. (Scale bar, 10 nm.) (B) 2D class averages of frozen-hydrated MHV S2 particles. (Scale bar, 10 nm.) (C) Gold-standard (blue) and model/map (red) Fourier shell correlation curves. The 0.143 and 0.5 cutoff values are indicated by horizontal gray lines. The resolution was determined to be 4.1 Å.
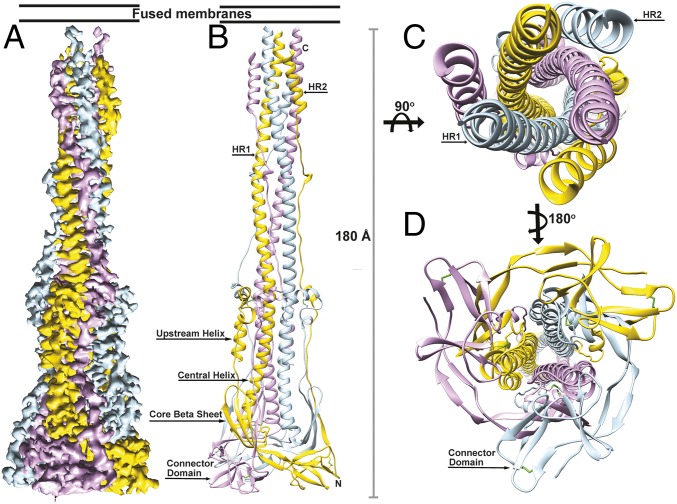
Fig. 2.
CryoEM structure of the MHV S2 postfusion machinery. (A) 3D map colored by protomer and viewed from the side with the fused membranes located at the top. (B) Ribbon diagram of the MHV S2 atomic model oriented as in A. (C and D) Ribbon diagram showing the atomic model from the extremity proximal (C) or distal (D) to the fused membranes.
Table S1.
Data collection, refinement, and model-validation statistics
| Parameter | Value |
| Data collection | |
| No. of particles | 106,272 |
| Pixel size, Å | 1.36 |
| Defocus range, µm | 1.5–3.0 |
| Voltage, kV | 300 |
| Electron dose, e−/Å2 | 48 |
| Refinement | |
| Resolution, Å | 4.1 |
| Map-sharpening B factor, Å2 | −280 |
| Model validation | |
| Favored rotamers, % | 99.32 |
| Poor rotamers, % | 0.34 |
| Ramachandran favored, % | 97.65 |
| Ramachandran outliers, % | 0.29 |
| Clash score | 1.77 |
| MolProbity score | 1.01 |
The atomic model was validated using a multipronged approach including comparison with known fragment structures, analysis of disulfide bond positions, and mass spectrometry. First, available crystal structures of coronavirus S six-helix bundle fragments (14–16), spanning part of the heptad-repeat 1 (HR1) and 2 (HR2) motifs, could be superimposed with good agreement onto the refined structure (Fig. S3 A and B). Also, the tertiary structure of the polypeptide chain corresponding to the upstream helix and the core β-sheet remain virtually identical to the prefusion structure, although the rest of the S2 subunit undergoes refolding (Fig. S3 C and D) (6). Second, the reconstruction resolves several intrachain disulfide bonds (Cys775–Cys797, Cys780–Cys786, Cys1082–Cys1093, and Cys1132–Cys1177) between pairs of cysteine residues that are conserved in the MHV, HCoV-NL63, SARS-CoV, and HCoV-HKU1 S glycoproteins (Fig. S3 E and F) (6–9). As expected, these covalent links are maintained during the transition from the prefusion to the postfusion conformation. Finally, we identified several N-linked glycosylation sites using on-line reversed-phase liquid chromatography with electron transfer/high-energy collision-dissociation tandem mass-spectrometry (32). Some of these sites are resolved in the cryoEM map (Asn754, Asn1180, and Asn1190) with the correct sequence context, further validating the assigned register (Fig. S3 G–J and Table S2).
Fig. S3.
Validation of the atomic model. (A and B) Two orthogonal views of the superimposition of the HR1/HR2 six-helix bundle structure derived from the MHV S2 cryoEM map (colored by protomer) and from X-ray crystallography (Protein Data Bank ID code 1WDF, gray). (C) Comparison of the core β-sheet structure in the prefusion (gray) and postfusion (colored as in Fig. 2) MHV S spike glycoprotein. (D) Comparison of the upstream helix structure in the prefusion (gray) and postfusion (colored as in Fig. 2) MHV S spike glycoprotein. (E and F) The MHV S2 model is shown in gray ribbon with the cryoEM density shown in transparent light gray. Disulfide bonds are shown in green for residues 775–797 (E) and 1,082–1,093 (F). (G and H) Glycans linked to Asn-1180 (G) and Asn-1190 (H) are rendered as ball and sticks with the corresponding region of the cryoEM map shown as a blue mesh. Carbon, nitrogen, and oxygen atoms are colored gray, blue, and red, respectively. (I and J) Mass spectra for Asn1180 (I) and Asn1190 (J) showing the presence of glycans at these glycosylation sequons.
Table S2.
Identification of MHV S2 N-linked glycosylation sites using glycoproteomics and cryoEM
| Site | Sequon | Combined | Intact | EndoH- and EndoF-treated | Unglycosylated | CryoEM map |
| 737 | NDSV | 737 | 737 | 737 | Not detected | Not detected |
| 754 | NFTI | 754 | 754 | 754 | Not detected | 754 |
| 844 | NFSP | Not detected | Not detected | Not detected | 844 | Not detected |
| 893 | NCTG | 893 | 893 | 893 | Not detected | Not detected |
| 1,126 | NVSP | Not detected | Not detected | Not detected | 1,126 | Not detected |
| 1,180 | NYTK | 1,180 | 1,180 | 1,180 | 1,180 | 1,180 |
| 1,190 | NTSI | 1,190 | Not detected | 1,190 | 1,190 | 1,190 |
| 1,209 | NQTS | Not detected | Not detected | Not detected | Not detected | Not detected |
| 1,225 | NVTL | Not detected | Not detected | Not detected | Not detected | Not detected |
| Total no. of sites | 8 | 5 | 4 | 5 | 4 | 3 |
The Coronavirus S Protein Forms a Conical Structure in the Postfusion Conformation.
Previous studies have shown that the HR1 and HR2 motifs of coronavirus S proteins assemble as a stable, six-helix bundle when expressed in isolation (14–16), a canonical feature of class I viral fusion proteins. Our structure of the MHV S2 fusion machinery ectodomain resolves a long central triple helical bundle, assembled from the central helices and HR1 motifs, surrounded by three antiparallel HR2 helices forming a six-helix bundle at one end (Fig. 2 B and C). At the opposite end exists a triangular pyramidal base comprised of the N terminus of the S2 subunit and the connector domain (Fig. 2 B and D). The HR1 motif of each protomer interacts exclusively with the other two protomers, forming an intertwined network (Fig. 2 B and C). In line with studies on structural rearrangements of other class I fusion proteins, the antiparallel orientation of the HR2 helices relative to the central coiled coil is indicative of the postfusion conformation, as the fusion peptide and transmembrane regions are located at the same end of the cone-like structure, unlike in the prefusion structure (Fig. 2 B and C). These conformational changes are believed to be crucial to mediate membrane fusion. Neither the fusion peptide region nor the adjacent N terminus of HR1 is resolved, suggesting a high degree of flexibility of this polypeptide segment in the absence of a membrane. The conformation of the MHV S2 trimer observed in this structure, along with the coronavirus S limited-proteolysis experiments, provides structural and biochemical evidence that the fusion machinery is maintained in its metastable prefusion conformation by the S1 subunits, as removal of the latter results in spontaneous refolding to the more stable postfusion state (even in the presence of a GCN4 motif). Additionally, our data show that refolding can occur in the absence of proteolytic processing in the context of an isolated S2 trimer ectodomain.
Comparison with Prefusion CoV S Structures.
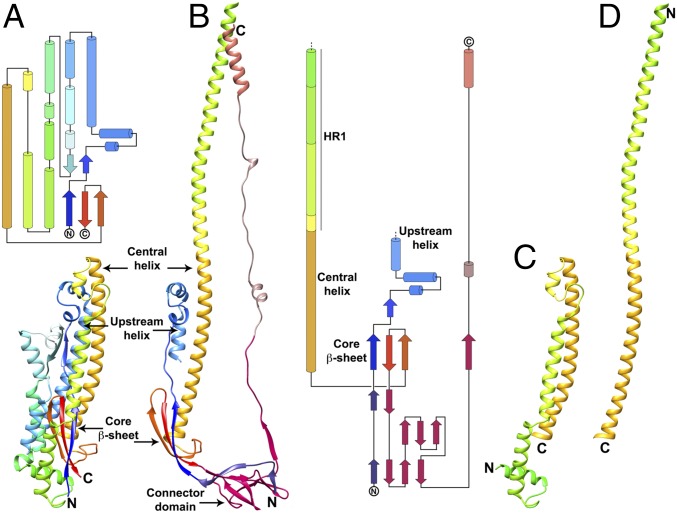
Transition of the MHV S2 fusion machinery from the pre- to the postfusion state is accompanied by conformational changes of spectacular magnitude (Fig. 3). The prefusion S2 trimer is short and wide (88 Å long and ∼40 Å wide) (Fig. 3A) (6), whereas the postfusion structure adopts an elongated cone-shaped conformation (185 Å long and 15–50 Å wide) (Fig. 3B). The tertiary structure of the core β-sheet and of the upstream helix remains mostly unchanged in the two conformations, although their relative orientation is modified (Fig. S3 C and D). Multiple disulfide bonds constrain S2 around the core β-sheet and the upstream helix, which appear to act as scaffolds around which the conformational changes in the rest of the polypeptide chain take place (Fig. S3 E and F). Based on the relatively high sequence conservation of the S2 subunit among coronaviruses and the conservation of these disulfide bonds, we postulate that similar conformational changes take place in all coronaviruses during fusion.
Fig. 3.
Conformational changes associated with the fusion reaction. (A) Ribbon and topology diagrams of the MHV S2 subunit in the prefusion conformation (6). (B) Ribbon and topology diagrams of the MHV S2 subunit in the postfusion conformation. (C and D) Ribbon rendering of the MHV S central helix and HR1 in the prefusion (C) and postfusion (D) states highlighting the jack-knife refolding of the four HR1 helices and intervening regions into a single continuous helix.
One of the most noticeable rearrangements between the pre- and postfusion structures occurs at the level of the HR1 motif (residues 948–1,035). In the prefusion S trimer, HR1 consists of four α-helices connected by extended loops and runs in an antiparallel orientation relative to the central helix (Fig. 3C) (6). In the postfusion structure, the four HR1 α-helices and connecting loops refold and reorient to append to the N terminus of the central helix, leading to the formation of a single, continuous 130-residue-long α-helix (for which 113 residues are resolved in our reconstruction) (Fig. 3D). This extremely long helix assembles as a homotrimeric helical bundle, stabilized partly via HR1-mediated coiled-coil interactions, and forms the core of the S2 postfusion trimer (Fig. 2 A–C). These conformational changes are accompanied by an alteration of the quaternary organization of the central helix. Although the fusion peptide region (22, 33) and the HR1 N-terminal moiety were resolved in the MHV S prefusion conformation (except for the S2′ loop), these residues are not visible in the postfusion map, suggesting that they are highly dynamic in the absence of the target membrane and transmembrane domain. Since this region is directly adjacent to HR1, which is completely refolded, it is expected that repositioning and potentially reorganization of the polypeptide chain occur for the fusion peptide region as well. As a result, we propose that targeting this polypeptide segment with antibodies or other types of inhibitors could prevent the insertion of the fusion peptide into the host membrane and putatively block refolding. The suitability of this approach is supported by in vivo neutralization data for MHV and SARS-CoV that identified this region as a major antigenic determinant, containing neutralization epitopes, upon infection by these viruses (34, 35).
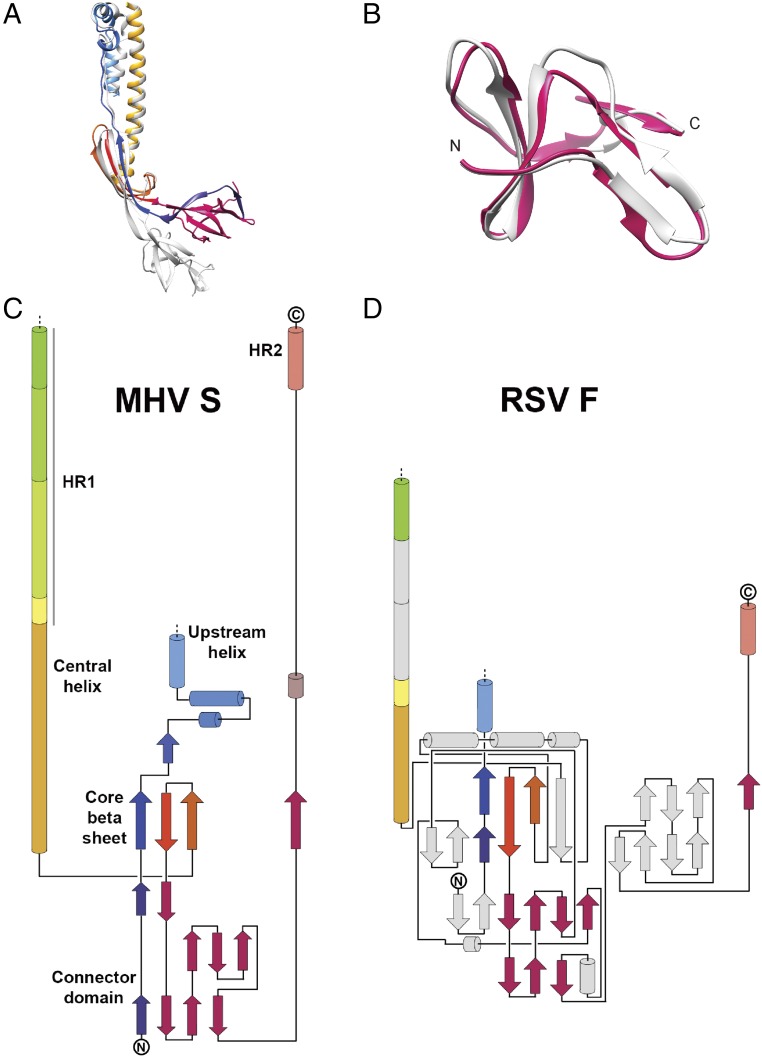
The C-terminal connector domain and stem helices are not resolved in the MHV S prefusion structure (6). These domains, however, are visible in the recently determined HCoV-NL63, SARS-CoV, and MERS-CoV S structures and can be used for comparison with the MHV S2 postfusion structure (7, 10, 11). MHV S residues 1,120–1,210, which span the connector domain and stem helix, share 22%, 33%, and 36% sequence identity with the equivalent residues of the HCoV-NL63, SARS-CoV, and MERS-CoV S glycoproteins, respectively. Although the tertiary structures of the prefusion S and postfusion MHV S connector domains are similar, the latter is repositioned underneath the core β-sheet of a neighboring subunit due to the refolding of HR1 and the fusion peptide region (Fig. S4 A and B). The most C-terminal part of the postfusion connector domain undergoes extensive refolding to adopt a domain-swapped organization with interactions formed exclusively with neighboring protomers. It contributes a fourth β-strand (residues 1,178–1,184) to the three-stranded core β-sheet of a different protomer and folds as an extended segment interacting with the central coiled coil in an antiparallel orientation (Figs. 3B and 4A). The N terminus of the stem helix is refolded into an extended loop, whereas the previously unresolved C-terminally adjacent region folds as a short helix docked near the upstream helix, perpendicular to the central coiled coil (Figs. 3B and 4A). In the context of the trimer, these two motifs form a pseudotriangular scaffold wrapping around the central triple helical bundle, potentially stabilizing it in the postfusion conformation. Since the region corresponding to HR2 has not been observed in any prefusion S structure, the structural rearrangements taking place in this segment are unknown. However, it is likely that the C terminus of the polypeptide chain is oriented in the opposite direction to connect to the viral membrane before fusion occurs.
Fig. S4.
Comparison of the MHV S postfusion S2 structure with other fusion proteins. (A) Superimposition of the HCoV-NL63 S2 (gray) and MHV postfusion S2 (colored as in Fig. 2) subunits. Only the upstream and central helices, core β-sheet, and connector domain are shown. (B) Superimposition of the HCoV-NL63 S2 (gray) and MHV postfusion S2 (pink) connector domains emphasizing their structural conservation. (C and D) Topology diagrams comparing postfusion MHV S2 and RSV F glycoproteins. Conserved elements in the RSV F subunit are shown using the same colors; nonconserved elements are shown in gray.
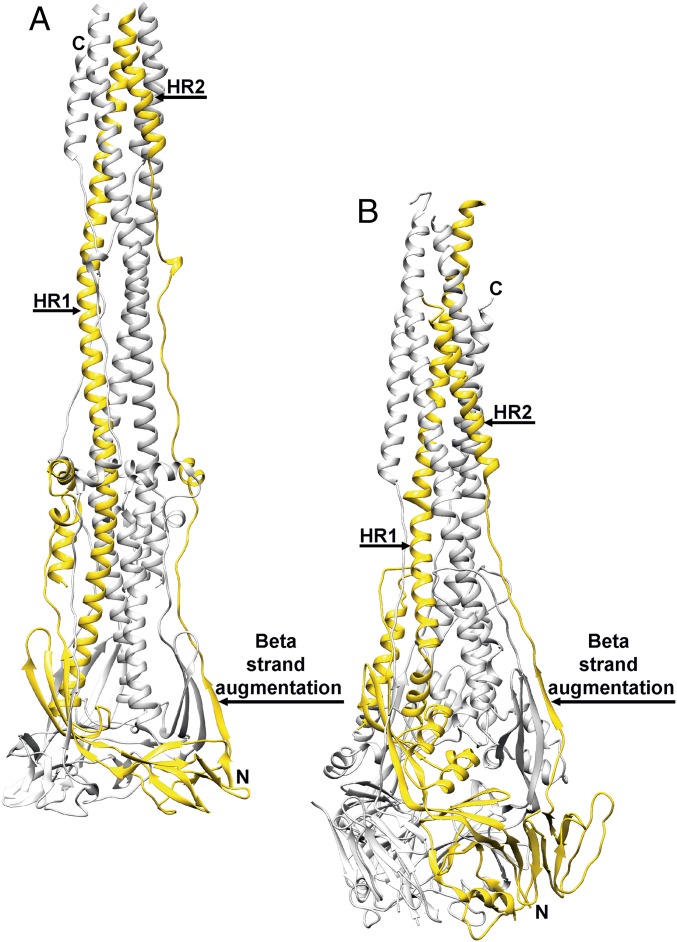
Fig. 4.
Comparison of the coronavirus and paramyxovirus fusion machineries. (A and B) Postfusion structures of MHV S (A) and RSV F (B) with one protomer of each trimer colored yellow and the other two colored gray.
As for all other known class I viral fusion proteins, the postfusion state of coronavirus S glycoproteins represents the lowest-energy point of the conformational landscape (36). This is supported by the spontaneous assembly of the isolated heptad repeats of all characterized coronavirus S proteins as six-helix bundles recapitulating the postfusion state (14–16). The free energy released upon refolding is believed to promote merging of the viral and host membranes and subsequent fusion. Strikingly, the postfusion MHV fusion machinery buries 6,150 Å2 of surface area per protomer at the interface with other subunits of the trimer through a large contribution from domain swapping of the connector domain C-terminal part and HR2. This observation provides a molecular basis explaining the unusually high stability of the postfusion S2 conformation and its representing the ground-state of the fusion reaction (36).
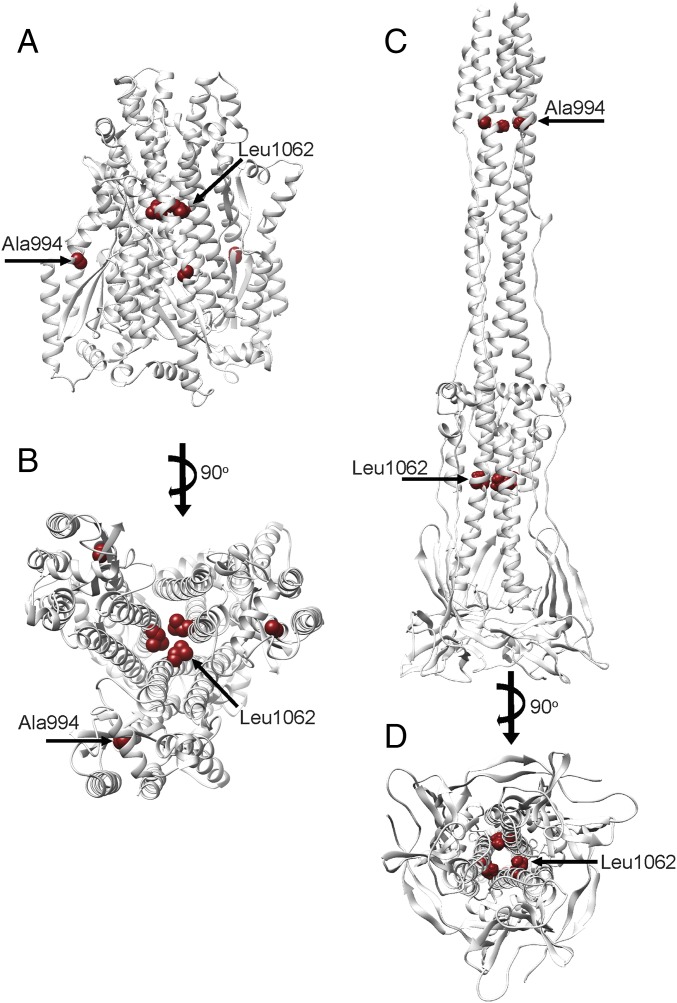
Mapping of Mutations That Affect Membrane Fusion.
Previous genetic and biochemical studies have identified amino acid substitutions in the MHV S2 subunit affecting fusogenicity, which can be analyzed based on the structural data reported here (37, 38). The L1062F mutation introduces a bulkier amino acid side chain that would negatively affect packing of the central helix trimer and/or the interactions formed by the central helix with the adjacent upstream helix. Although modeling this mutation in the MHV S pre- and postfusion structures suggests comparable outcomes, the fact the upstream helix region appears to act as a pseudorigid scaffold around which refolding of the fusion machinery takes place may make this mutation more destabilizing for the postfusion conformation (Fig. S5). The A994V substitution in the HR1 region introduces a bulkier amino acid side chain that could alter the packing of the postfusion S2 central coiled coil as well as potentially disturb the formation of the six-helix bundle with the HR2 C-terminal helices, the latter being a major contributor of the extensive surface area buried between protomers, and might explain its destabilizing effect on the postfusion conformation (Fig. S5). In summary, it appears that substitutions at these two strictly conserved positions would be more destabilizing for the postfusion conformation, which is the ground-state for all known class I fusion proteins, than for the prefusion state, thereby explaining the reported reduction of S-mediated membrane fusion.
Fig. S5.
Location of fusion-inhibiting mutations in MHV S. (A and B) Prefusion MHV S2 subunit shown in gray ribbon from the side (A) and from the viral membrane (B). (C and D) Postfusion MHV S2 shown in gray ribbon from the side (C) and looking toward the long axis (D). Two residues known to attenuate fusogenicity upon substitution are shown as red spheres.
Comparison with Other Postfusion Class I Viral Fusion Proteins.
We previously reported that the prefusion architectures of coronavirus S and paramyxovirus F proteins share a similar topology, which suggests they use conserved mechanisms for mediating fusion of the viral and host membranes and that they may have arisen from a common ancestral gene (6, 7, 18, 39). Comparison of the F protein structures of RSV (8, 19), Parainfluenza virus 3 (PIV3) (23), or Newcastle disease virus (25) with MHV S2 also shows that their tertiary structures are related in the postfusion conformation (Fig. 4 and Fig. S4 C and D). Although large-scale conformational changes occur during fusion, the core β-sheet and upstream helix remain mostly unchanged in the prefusion and postfusion S and F states (Fig. 4). CoV S2 features a 30-turn helix, resulting from refolding of the four α-helices and connecting loops spanning the HR1 motif in the prefusion S state and appending to the central helix. Refolding of the multiple secondary structural motifs spanning the HR1 sequence into a single helix is the signature of class I fusion proteins and is reminiscent of what has been reported for Influenza virus hemagglutinin (40), HIV envelope (41), and Ebola virus GP (42) in addition to paramyxovirus F proteins (18, 19, 23, 24, 39).
The quaternary reorganization of the postfusion coronavirus S2 and paramyxovirus F glycoproteins results in the formation of similar cone-shaped rods tapering toward the end proximal to the merged membranes (Fig. 4). The coronavirus fusion machinery, however, is at least 10 Å longer than paramyxovirus F glycoproteins (without considering the unresolved S HR1 N-terminal region) and is the longest structure observed for any class I fusion protein. Both fusion subunits are characterized by the formation of a triple helical-bundle core, resulting from the association of the extended central/HR1 helices, which participates in the formation of a six-helix bundle with the HR2 C-terminal helices. The polypeptide segment corresponding to the C-terminal region of the connector domain and HR2 is also largely refolded in coronavirus S2 and paramyxovirus F glycoproteins in which they are observed extending along the periphery of each neighboring protomer (8, 19, 23, 25). Of note, the core β-sheet of each fusion subunit is supplemented by a β-strand contributed in an identical manner by the connector domain of a neighboring subunit (Fig. 4).
These observations reinforce the similarity between coronavirus S and other characterized class I viral fusion proteins, especially paramyxovirus F proteins, as they share a conserved refolding trajectory to promote membrane fusion. Refolding of coronavirus S and paramyxovirus F likely proceeds through a similar zippering reaction during which the C-terminal moiety enhances its interactions with the central triple-helical bundle to bring the transmembrane region of the fusion machinery in proximity to the fusion peptide (Fig. 5).
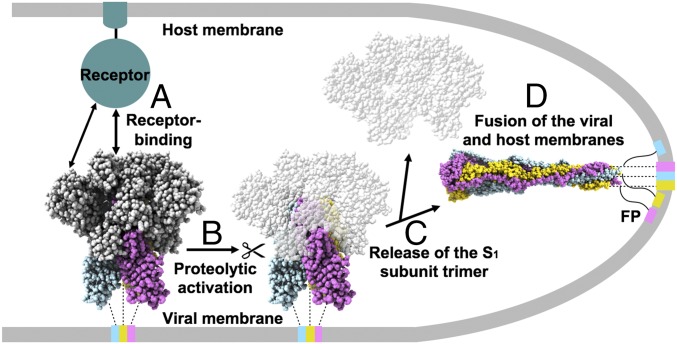
Fig. 5.
Proposed model of coronavirus entry. (A) The S glycoprotein promotes virus attachment to a host cell via binding to a transmembrane receptor using either domain A (e.g., MHV S) or domain B (e.g., SARS-CoV or MERS-CoV S). The prefusion MHV S trimer is shown with the S1 subunit depicted in gray and the S2 subunits colored by protomer. (B) Upon receptor binding, activation of the S trimer occurs via protease cleavage at the S2′ site. (C) Shedding of the S1 subunit trimer frees the fusion machinery, as reported for MERS-CoV (10). (D) Subsequent conformational changes of the S glycoprotein result in fusion of the viral and host membranes. The postfusion MHV S2 trimer is depicted with each protomer in a different color. The transmembrane helices and the fusion peptides (FP) are connected to the MHV S trimer with dotted and solid lines, respectively.
Discussion
We previously postulated that the S1 subunits make interactions with the S2 subunits that are key to stabilizing the prefusion conformation of coronavirus S glycoproteins (6). Similar observations were also made based on the HCoV–HKU1 prefusion S structure (9). The results of our limited-proteolysis experiments and the near-atomic-resolution reconstruction of the MHV S2 trimer further corroborate this hypothesis. Proteolytic processing at the S1/S2 and S2′ cleavage sites removes the covalent linkage between the two functional subunits and frees the fusion peptide, which allows shedding of the S1 crown and subsequent refolding of the fusion machinery (Fig. 5). Inspection of coronavirus prefusion structures show that the HR1 region of each protomer contacts neighboring protomers via the B and C domains (6–9). These interactions act as a molecular clamp, stabilizing the metastable prefusion trimer until receptor binding and cleavage triggering at the S2′ site. Similar mechanisms of stabilization have been described for influenza hemagglutinin in which the HA1 globular head contacts the HR1 polypeptide segment to prevent early refolding. Subsequent exposure to the acidic environment of the endosomes promotes refolding of HA2 and membrane fusion (40). Structurally related paramyxovirus F proteins exist in a metastable prefusion state in the viral membrane. Upon receptor engagement, conformational changes in the receptor-binding protein H, HN, or G on the surface of measles virus, PIV5, or Nipah virus, respectively, promote F-triggering and subsequent membrane fusion (43). These similarities between influenza HA, paramyxovirus F, coronavirus S, and other class I viral-fusion proteins suggest that comparable mechanisms have evolved to ensure proper spatial and temporal coordination of their fusion proteins and productive infection.
The tertiary structure of the fusion machinery is highly conserved among coronaviruses across multiple genera, in agreement with the relatively high sequence conservation of the S2 subunit (6–9). Based on these observations, along with the presence of several conserved disulfide bonds constraining the fusion machinery, we propose that the tectonic conformational changes reported here for MHV S2 are representative of the rearrangements taking place in all coronavirus S proteins during fusion. Most vaccine-design initiatives aim at targeting the prefusion state of viral fusion proteins, which correspond to the conformation that could be detected by the immune system before infection. Comparison of the prefusion S trimer structures with the MHV postfusion structure reported here provides a blueprint to analyze the accessibility of neutralization epitopes in each conformation. Although most known neutralization epitopes characterized to date are present in the S1 subunit, due to its higher immunogenicity than the S2 subunit, the marked sequence and structural diversity of S1 has so far led to the elicitation of species-specific antibodies (44, 45). In contrast, the higher conservation of the S2 fusion machinery bears the promise that epitopes could potentially be targeted by broadly neutralizing antibodies cross-reacting with multiple coronaviruses (6). For instance, the fusion peptide region is highly conserved among coronavirus S glycoproteins and has been identified as a major antigenic determinant upon infection by MHV and SARS-CoV (34, 35). The postfusion MHV S2 structure supports our previous hypothesis that antibodies binding to this site could hinder insertion of the fusion peptide into the target membrane and putatively prevent fusogenic conformational changes. The stem helix and part of the HR2 motif are also conserved among S glycoproteins and therefore represent attractive targets to elicit broadly neutralizing antibodies. In the prefusion state, the yet unresolved HR2 C terminus of the fusion machinery would likely point in the opposite direction, compared with the postfusion conformation, corresponding to a 180° reorientation of the polypeptide chain with potential changes in secondary, tertiary, and quaternary structures. The large-scale nature of the predicted conformational changes in HR2 reinforces the idea that antibodies targeting this region will be neutralizing, in agreement with previous reports (46, 47). For example, the 10G monoclonal antibody is known to inhibit MHV S-mediated cell-to-cell fusion and to block virus infectivity (47) upon binding to an epitope comprised within residues 1,212–1,226 that fold as an extended loop upstream of the HR2 C-terminal helix in the postfusion structure. Finally, the data reported here provide a structural framework for designing coronavirus S-based subunit vaccine immunogens. Knowledge of the precise structural rearrangements taking place during the fusion reaction paves the way for rationally engineering modifications enhancing the stability of the prefusion trimer. Designed disulfide bonds cross-linking residues that are close to each other in the prefusion state but far apart in the postfusion conformation and/or cavity-filling mutations are strategies that have been successfully used to enhance the stability of the prefusion RSV F trimer and could also be implemented for S glycoproteins (17). Finally, the recently stabilization of the MERS-CoV prefusion S via introduction of proline residues at the junction between the central helix and the HR1 motif can also be rationalized with our data (11).
Methods
CryoEM data were collected on an FEI Titan Krios microscope operated at 300 kV and equipped with a Gatan Quantum GIF energy filter and a Gatan K2 Summit direct electron detector. Frame alignment was carried out with dose weighting using MotionCor2 (48), and Relion 2.0 was used for downstream processing and 3D reconstruction (49).
Plasmids
A human codon-optimized gene encoding for the MHV S glycoprotein (UniProt accession no. P11224) was synthesized, and the fragments encoding the MHV S ectodomain (residues 15–1,252) or the MHV S2 ectodomain (residues 718–1,252) were PCR amplified and ligated to a gene fragment encoding a GCN4 trimerization motif (27, 50), a thrombin cleavage site, an eight-residue-long Strep-tag (IBA), and a stop codon. A human-codon–optimized gene encoding for the MERS-CoV S glycoprotein (UniProt accession no. W6A028) with the R748K substitution was synthesized, and the fragment encoding the S ectodomain (residues 19–1,262) was ligated to a gene fragment encoding a foldon trimerization motif, a thrombin cleavage site, an eight-residue-long Strep-tag, and a stop codon. These genes were cloned into the pMT\BiP\V5\His expression vector (Invitrogen) in frame with the Drosophila BiP secretion signal downstream of the metallothionein promoter.
A human codon-optimized gene encoding for the ectodomain (residues 14–1,180) of the SARS-CoV S protein (UniProt accession no. P59594) was cloned into a modified pOPING vector (Addgene) (51) introducing an N-terminal Mu-phosphatase signal peptide and a C-terminal Tobacco etch virus (TEV) protease cleavage site, a foldon, and a hexa-histidine tag at the C terminus of the construct.
Production of Recombinant MHV S, MHV S2, and MERS S Ectodomain in Drosophila S2 Cells
To generate a stable Drosophila S2 cell line expressing the recombinant MHV-S2 or MERS S ectodomain, we used Effectene (Qiagen) and 2 μg of plasmid. Puromycin N-acetyltransferase was cotransfected and used as a dominant selectable marker. Stable MHV-S2– or MERS S-expressing cell lines were selected by the addition of 7 μg/mL puromycin (Invivogen) to the culture medium 48 h after transfection. For large-scale production, the cells were cultured in spinner flasks and induced by 5 μM of CdCl2 at a density of ∼107 cells/mL. After 1 wk at 28 °C, clarified cell supernatants were concentrated 40-fold with Vivaflow tangential filtration cassettes (10-kDa cutoff) (Sartorius) and adjusted to pH 8.0 before affinity purification with a Strep-Tactin Superflow column (IBA) followed by gel-filtration chromatography with a Superose 6 10/300 GL column (GE Life Sciences) equilibrated in 20 mM Tris⋅HCl (pH 7.5) and 100 mM NaCl or, in the case of MERS S, 20 mM sodium phosphate (pH 7.0) and 250 mM NaCl. The purified protein was concentrated to ∼3 mg/mL.
Production of Recombinant SARS S Ectodomain in HEK293F Cells
Transient transfection of 250 mL of HEK293F cells at a density of 106 cells/mL was performed using 293fectin (Thermo Fisher) and Opti-MEM (Thermo Fisher). After 3 d the cells were harvested before affinity purification with a Talon 5-mL cobalt column equilibrated in 25 mM sodium phosphate (pH 8.0), 300 mM NaCl, 5 mM imidazole, and 0.5 mM PMSF. The purified protein was buffer exchanged into 20 mM Tris (pH 8.0), 150 mM NaCl and was concentrated to 1.0 mg/mL.
Limited Proteolysis of MHV S, SARS S, and MERS S
Five microliters of 1 mg/mL protein were incubated with either 1:100 or 1:10 trypsin (Sigma Aldrich) at room temperature for 1 h. This reaction was then used for negative-staining EM (3 μL of ∼0.001 mg/mL).
Negative-Stain EM
Three microliters of trypsin-treated CoV S proteins were incubated for 30 s on carbon-coated copper mesh grids before staining with 2% uranyl formate. Micrographs were acquired using the Leginon software (52) on a 120-kV FEI Tecnai G2 Spirit transmission electron microscope with a Gatan UltraScan 4000 4k × 4k CCD camera at 52,000 or 67,000 nominal magnification (pixel size of 2.07 Å or 1.62 Å at the specimen level). The defocus ranged from 1.0 to 2.0 μm. Particles were picked automatically in a reference-free manner using DoGpicker (53) and were extracted using a box size of 320 Å. Contrast transfer function (CTF) estimation was performed using GCTF (54). 2D classifications were performed using RELION 2.0 (49) and included correction for the effect of the microscope CTF. Each displayed class contains ∼1,000–5,000 particles.
CryoEM Specimen Preparation and Data Collection
Two microliters of purified MHV S2 spike at 0.6 mg/mL in 0.025% amphipole (Anatrace) were double blotted (55) onto a 1.2/1.3 C-flat grid (Protochips), which had been glow-discharged for 30 s at 20 mA. Grids were then plunge-frozen in liquid ethane with an FEI Mark I Vitrobot with a 13-s blot time and an offset of −1 mm at 100% humidity and 25 °C. Data were collected with Leginon automatic data-collection software on an FEI Titan Krios electron microscope operated at 300 kV and equipped with a Gatan Quantum GIF energy filter, operated in zero-loss mode with a slit width of 20 eV, and a Gatan K2 Summit direct electron detector camera (56). The dose rate was adjusted to eight counts per pixel per second, and each movie was acquired in counting mode fractionated in 50 frames of 200 ms. In a single session, 2,200 micrographs were collected with a defocus range between 1.5 and 3.0 μm.
CryoEM Data Processing
Movie frame alignment was carried out with dose weighting using MotionCorr2 (48). The parameters of the microscope CTF were estimated with GCTF (54). Particles were manually selected to generate a template. The template was low-pass filtered to 25 Å and used to pick particles on all micrographs with FindEM (57) integrated in Appion (58). Particle images were extracted and processed with Relion 2.0 (49) with a box size of 320 × 320 pixels and a pixel size of 1.36 Å. Following two rounds of 2D classification, 150,000 of 900,000 particles remained to run 3D classification with C3 symmetry. Then 106,000 particles were used to run gold-standard 3D refinement with Relion using the solvent_correct_fsc_flag and a solvent mask with a soft edge of five pixels and a cosine-edge falloff of five pixels. Postprocessing was performed with Relion to apply a B-factor of −280 Å2. Reported resolutions were based on the gold-standard criterion [Fourier shell correlation (FSC) = 0.143], and FSC curves were corrected for the effects of soft masking by high-resolution noise substitution. The soft mask used for FSC calculation had a 10-pixel cosine edge falloff.
Model Building and Analysis
University of California, San Francisco (UCSF) Chimera (59) and Coot (60) were used to fit and rebuild atomic models into the cryoEM map. The central helix, core β-sheet, and upstream helix of the prefusion MHV S structure were fit into the S2 map and were both manually adjusted and extended with Rosetta ES (30). The connector domain was built using the HCoV-NL63 (7) and SARS-CoV (10) prefusion connector domains. The final model was refined by application of strict noncrystallographic symmetry constraints with Rosetta (31, 61), with a training map corresponding to one of the two maps generated by the gold-standard refinement procedure in Relion. The second map (the testing map) was used only for calculation of the FSC compared with the atomic model and preventing overfitting (62). The quality of the final model was analyzed with MolProbity (63). All figures were generated with UCSF Chimera (59) or UCSF ChimeraX (www.rbvi.ucsf.edu/chimerax).
Mass Spectrometry
MHV-S after limited proteolysis with trypsin was separated by SDS/PAGE and cut from gel for digestion with AspN. The gel bands were cut to small pieces, washed in Milli-Q water, and treated with acetonitrile to shrink the gel pieces. The sample was then incubated in 1,4-DTT (1 g/L) for 60 min at 60 °C, treated with acetonitrile, alkylated with iodoacetamide (10 g/L) for 30 min at room temperature in the dark, and subsequently washed with ammonium bicarbonate and treated with acetonitrile, twice. The gel pieces were then incubated with 30 mg/L AspN for 90 min on ice. Excess AspN was removed, the gel pieces were covered in ammonium bicarbonate, and the samples were subsequently incubated overnight at 37 °C. The digested samples were collected, and remaining sample was extracted from the gel pieces by treatment with acetonitrile. The solution with the peptides was subsequently dried in a SpeedVac, and the peptides were resuspended in 10% formic acid, 5% dimethyl sulfoxide in water.
For analysis of N-linked glycosylation, an estimated 250 pmol of MHV S2 was denatured, reduced, and alkylated by dilution to 5 µM in 50 µL of buffer containing 100 mM Tris (pH 8.5), 10 mM Tris(2-carboxyethyl)phosphine (TCEP), 40 mM iodoacetamide, and 2% (wt/vol) sodium deoxycholate. Samples were first heated to 95 °C for 10 min and then incubated for an additional 20–30 min at room temperature in the dark. The samples were split in two for digestion with trypsin and chymotrypsin (Sigma Aldrich) in parallel, by diluting 20 µL of sample for each protease in a total volume of 100 µL 50 mM ammonium bicarbonate (pH 8.5). Protease was added to the samples in a ratio of 1:75 by weight and left to incubate at 37 °C overnight. After digestion, 2 µL of formic acid was added to the samples to precipitate the sodium deoxycholate from solution. After centrifugation at 17,000 × g for 20 min, 80 μL of the supernatant was collected.
For each sample 8 µL was injected on a Thermo Scientific Orbitrap Fusion Tribrid mass spectrometer. A 35-cm analytical column and a 3-cm trap column filled with ReproSil-Pur C18AQ 5 μm (Dr. Maisch) beads were used. Nanospray LC-MS/MS was used to separate peptides over a 110-min gradient from 5 to 30% acetonitrile with 0.1% formic acid. A positive spray voltage of 2,100 was used with an ion transfertube temperature of 350 °C. An electron-transfer/higher-energy collision dissociation ion-fragmentation scheme (32) was used with calibrated charge-dependent entity-type definition (ETD) parameters and supplemental higher-energy collision dissociation energy of 0.15. A resolution setting of 120,000 with an AGC target of 2 × 105 was used for MS1, and a resolution setting of 30,000 with an AGC target of 1 × 105 was used for MS2. Data were searched with Protein Metrics Byonic software (64), using a small custom database of recombinant protein sequences including the proteases used to prepare the glycopeptides. Reverse decoy sequences were also included in the search. Specificity of the search was set to C-terminal cleavage at R/K (trypsin) or F/W/Y/M/L (chymotrypsin), allowing up to two missed cleavages, with EThcD fragmentation (b/y- and c/z-type ions). We used a precursor mass and product mass tolerance of 12 ppm and 24 ppm, respectively. Carbamidomethylation of cysteines was set as fixed modification, methionine oxidation as variable modification, and all four software-provided N-linked glycan databases were used to identify glycopeptides. All glycopeptide hits were manually inspected, and only those with quality peptide sequence information are reported here.
Acknowledgments
This work was supported by the National Institute of General Medical Sciences Grants 1R01GM120553 (to D.V.) and T32GM008268 (to A.C.W.); a Pew Scholars Award (to D.V.); Netherlands Organization for Scientific Research Award Rubicon 019.2015.2.310.006 (to J.S.); European Molecular Biology Organization Grant ALTF 933-2015 (to J.S.); the Pasteur Institute (F.A.R.); National Institute of Health Grants 1S10RR23057, 1S10OD018111, and 1U24GM116792, as well as National Science Foundation Grant DBI-1338135 (to the Electron Imaging Center of the California NanoSystems Institute at the University of California, Los Angeles); and University of Washington’s Proteomics Resource Grant UWPR95794.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The cryoEM map and atomic model have been deposited to the Electron Microscopy Data Bank and Protein Data Bank, respectively (EMDB accession code EMD-7040 and PDB ID code 6B3O).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708727114/-/DCSupplemental.
References
- 1.Ge XY, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sabir JS, et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016;351:81–84. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- 3.Wang Q, et al. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y, et al. Two mutations were critical for bat-to-human transmission of Middle East respiratory syndrome coronavirus. J Virol. 2015;89:9119–9123. doi: 10.1128/JVI.01279-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walls AC, et al. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walls AC, et al. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat Struct Mol Biol. 2016;23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gui M, et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirchdoerfer RN, et al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan Y, et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pallesen J, et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Millet JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci USA. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci USA. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duquerroy S, Vigouroux A, Rottier PJ, Rey FA, Bosch BJ. Central ions and lateral asparagine/glutamine zippers stabilize the post-fusion hairpin conformation of the SARS coronavirus spike glycoprotein. Virology. 2005;335:276–285. doi: 10.1016/j.virol.2005.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Supekar VM, et al. Structure of a proteolytically resistant core from the severe acute respiratory syndrome coronavirus S2 fusion protein. Proc Natl Acad Sci USA. 2004;101:17958–17963. doi: 10.1073/pnas.0406128102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y, et al. Structural basis for coronavirus-mediated membrane fusion. Crystal structure of mouse hepatitis virus spike protein fusion core. J Biol Chem. 2004;279:30514–30522. doi: 10.1074/jbc.M403760200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLellan JS, et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLellan JS, et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLellan JS, Yang Y, Graham BS, Kwong PD. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol. 2011;85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Millet JK, Whittaker GR. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bosch BJ, Bartelink W, Rottier PJ. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J Virol. 2008;82:8887–8890. doi: 10.1128/JVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burkard C, et al. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10:e1004502. doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci USA. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swanson KA, et al. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc Natl Acad Sci USA. 2011;108:9619–9624. doi: 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L, et al. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure. 2001;9:255–266. doi: 10.1016/s0969-2126(01)00581-0. [DOI] [PubMed] [Google Scholar]
- 26.Li F, et al. Conformational states of the severe acute respiratory syndrome coronavirus spike protein ectodomain. J Virol. 2006;80:6794–6800. doi: 10.1128/JVI.02744-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walls A, et al. Crucial steps in the structure determination of a coronavirus spike glycoprotein using cryo-electron microscopy. Protein Sci. 2017;26:113–121. doi: 10.1002/pro.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71:136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang RY, et al. De novo protein structure determination from near-atomic-resolution cryo-EM maps. Nat Methods. 2015;12:335–338. doi: 10.1038/nmeth.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frenz B, Walls AC, Egelman EH, Veesler D, DiMaio F. RosettaES: A sampling strategy enabling automated interpretation of difficult cryo-EM maps. Nat Methods. 2017;14:797–800. doi: 10.1038/nmeth.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DiMaio F, et al. Atomic-accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement. Nat Methods. 2015;12:361–365. doi: 10.1038/nmeth.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frese CK, et al. Unambiguous phosphosite localization using electron-transfer/higher-energy collision dissociation (EThcD) J Proteome Res. 2013;12:1520–1525. doi: 10.1021/pr301130k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madu IG, Roth SL, Belouzard S, Whittaker GR. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J Virol. 2009;83:7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daniel C, et al. Identification of an immunodominant linear neutralization domain on the S2 portion of the murine coronavirus spike glycoprotein and evidence that it forms part of complex tridimensional structure. J Virol. 1993;67:1185–1194. doi: 10.1128/jvi.67.3.1185-1194.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, et al. Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies. J Virol. 2004;78:6938–6945. doi: 10.1128/JVI.78.13.6938-6945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingallinella P, et al. Structural characterization of the fusion-active complex of severe acute respiratory syndrome (SARS) coronavirus. Proc Natl Acad Sci USA. 2004;101:8709–8714. doi: 10.1073/pnas.0402753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krueger DK, Kelly SM, Lewicki DN, Ruffolo R, Gallagher TM. Variations in disparate regions of the murine coronavirus spike protein impact the initiation of membrane fusion. J Virol. 2001;75:2792–2802. doi: 10.1128/JVI.75.6.2792-2802.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taguchi F, Matsuyama S. Soluble receptor potentiates receptor-independent infection by murine coronavirus. J Virol. 2002;76:950–958. doi: 10.1128/JVI.76.3.950-958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 41.Buzon V, et al. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog. 2010;6:e1000880. doi: 10.1371/journal.ppat.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weissenhorn W, Carfí A, Lee KH, Skehel JJ, Wiley DC. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 43.Liu Q, et al. Unraveling a three-step spatiotemporal mechanism of triggering of receptor-induced Nipah virus fusion and cell entry. PLoS Pathog. 2013;9:e1003770. doi: 10.1371/journal.ppat.1003770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rockx B, et al. Structural basis for potent cross-neutralizing human monoclonal antibody protection against lethal human and zoonotic severe acute respiratory syndrome coronavirus challenge. J Virol. 2008;82:3220–3235. doi: 10.1128/JVI.02377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pascal KE, et al. Pre- and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc Natl Acad Sci USA. 2015;112:8738–8743. doi: 10.1073/pnas.1510830112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elshabrawy HA, Coughlin MM, Baker SC, Prabhakar BS. Human monoclonal antibodies against highly conserved HR1 and HR2 domains of the SARS-CoV spike protein are more broadly neutralizing. PLoS One. 2012;7:e50366. doi: 10.1371/journal.pone.0050366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Routledge E, Stauber R, Pfleiderer M, Siddell SG. Analysis of murine coronavirus surface glycoprotein functions by using monoclonal antibodies. J Virol. 1991;65:254–262. doi: 10.1128/jvi.65.1.254-262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng SQ, et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimanius D, Forsberg BO, Scheres SH, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife. 2016;5:e18722. doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eckert DM, Malashkevich VN, Kim PS. Crystal structure of GCN4-pIQI, a trimeric coiled coil with buried polar residues. J Mol Biol. 1998;284:859–865. doi: 10.1006/jmbi.1998.2214. [DOI] [PubMed] [Google Scholar]
- 51.Berrow NS, et al. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 2007;35:e45. doi: 10.1093/nar/gkm047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suloway C, et al. Automated molecular microscopy: The new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 53.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snijder J, et al. Vitrification after multiple rounds of sample application and blotting improves particle density on cryo-electron microscopy grids. J Struct Biol. 2017;198:38–42. doi: 10.1016/j.jsb.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roseman AM. FindEM–A fast, efficient program for automatic selection of particles from electron micrographs. J Struct Biol. 2004;145:91–99. doi: 10.1016/j.jsb.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 58.Lander GC, et al. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 60.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DiMaio F, Leaver-Fay A, Bradley P, Baker D, André I. Modeling symmetric macromolecular structures in Rosetta3. PLoS One. 2011;6:e20450. doi: 10.1371/journal.pone.0020450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DiMaio F, Zhang J, Chiu W, Baker D. Cryo-EM model validation using independent map reconstructions. Protein Sci. 2013;22:865–868. doi: 10.1002/pro.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bern M, Kil YJ, Becker C. Byonic: Advanced peptide and protein identification software. Curr Protoc Bioinformatics. 2012;Chapter 13:Unit13 20. doi: 10.1002/0471250953.bi1320s40. [DOI] [PMC free article] [PubMed] [Google Scholar]